Abstract
Background and Aims
Epidemiologic studies have linked nutritional folate deficiency to an increased risk of cancer, but recent trials suggest that folate supplementation does not protect against tumor formation. Our aim was to analyze the genetic and epigenetic consequences of folate deficiency and to investigate whether impairment of the uracil base excision repair pathway can enhance its effects.
Methods
Wild-type mice and those deficient in uracil DNA glycosylase (Ung−/−) were placed on a folate-deficient diet for 8 months. We measured tumor incidence in major organs, DNA mutation rates, DNA mutation spectra, local DNA methylation, and global DNA methylation in colon epithelial cells.
Results
The experimental diet increased plasma homocysteine (60%, P < .001) and DNA uracil content (24%, P < .05) but not tumor formation. Global DNA methyllation was slightly decreased in splenocytes (9.1%) and small intestinal epithelial cells (4.2%), and significantly reduced in colon epithelial cells (7.2%, P < .04). No gene-specific changes in methylation were detected at the mouse B1 element, the H19 DMR, or the Oct4 gene. By λ CII assay and sequencing analysis of 730 mutants, we found that Ung−/− mice had a higher frequency of point mutations and increased C:G to T:A transitions at non-CpG sites. However, folate deficiency had no additional effect on the DNA mutation frequency or spectrum in Ung−/− or wild-type mice.
Conclusions
Contradicting current concepts, these findings indicate that the effects of a low-folate diet on DNA methylation and point mutations are insufficient to promote tumor development, even in the presence of Ung deficiency.
Several epidemiologic studies have shown a correlation between deficiency in the B-vitamin folate and an increased risk of cancer, in particular colorectal cancer1,2 and breast cancer.3 Also, cancer of the oropharynx,4 esophagus, stomach, and pancreas has been linked to folate deficiency.5 Other evidence pointing towards folate’s role in tumor development consists of epidemiologic studies linking variants of the enzyme methylene tetrahydrofolate reductase, which is involved in cellular folate metabolism, to a reduced risk of colon cancer6 and leukemia.7 However, recent human trials reported that folate supplementation either had no effect8 or increased the risk of intestinal tumors,9 and one study reported that deficiency of folate protects against intestinal tumor formation.10
The contradictory results of these human studies are further confounded by uncertainties about the molecular pathways that link folate deficiency to tumor formation. Three main molecular mechanisms have been proposed:(1) a global decrease in DNA methylation, (2) increased uracil misincorporation during DNA replication, and (3) increased cytosine deamination at sites of DNA methylation. Folate metabolites are required for the conversion of homocysteine to methionine, which in the activated form of S-adenosyl-methionine (SAM) is required for DNA methylation. Folate deficiency can therefore decrease global DNA methylation, which is associated with genetic instability and tumor formation.11,12 Indeed, a methyl donor–deficient diet combining vitamin B12, choline, methionine, and folate deficiencies has been shown to diminish methylation of transposable elements in DNA with functional consequences such as a change in coat color,13 but these experiments do not address the consequences of folate deficiency alone. Studies on isolated folate deficiency have shown conflicting results14 ranging from a 20% decrease in DNA methylation,15 through no change,16 to a 56% increase in global DNA methylation.17 The interpretation of these data is difficult because in some cases, tissues with low cellular turnover such as liver were analyzed,17 or the periods of dietary intervention were short (eg, 3 months),16 even though a decrease in global DNA methylation resulting from nutrient deficiency is considered a passive process requiring multiple cell divisions. Also, when studying tumor formation, it is important to distinguish between effects on regional and global methylation.
The second potential tumorigenic pathway of folate deficiency is increased uracil misincorporation during DNA replication. Folic acid and its metabolites are involved in nucleotide synthesis, and it has been shown that folate deficiency impairs thymidine synthesis with secondary increases in dUTP/dTTP ratios and increased uracil misincorporation into DNA.18 The resulting U:A base pairs are potentially mutagenic because of secondary repair activity that can generate abasic sites,19 causing point mutations or chromosomal breaks at T:A base pairs. Indeed, increases in DNA strand breaks as a consequence of folate deficiency have been reported,18,20 but direct proof of mutations by sequence analysis is lacking. The relevance of uracil misincorporation into DNA as a mutagenic mechanism is therefore still unclear.21 It is possible that uracil-related DNA strand breaks inhibit transcription of tumor-relevant genes without causing DNA mutations.20
A third potentially mutagenic consequence of folate deficiency is enzymatic cytosine deamination at sites of DNA methylation. In bacteria, cytosine methyltrans-ferases deaminate cytosine when SAM concentrations are low, resulting in C→U transitions at methylation sites.22 Because lack of folate decreases SAM levels,14 it is possible that this will cause cytosine deamination and increased C:G to T:A transitions at CpG sites. However, this mechanism has not yet been explored in the context of mammalian folate deficiency.
An open question is whether the effect of folate deficiency is enhanced by mutations in DNA base excision repair enzymes, such as uracil DNA glycosylase (Ung). Ung removes misincorporated uracil during DNA replication23 and repairs U:G mispairs that occur as a consequence of cytosine deamination.24 Based on these functions, it is possible that Ung deficiency might amplify mutagenic effects of low folate.
To address these questions and to determine the genetic and epigenetic effects of folate deficiency, we subjected wild-type and Ung knockout mice (Ung−/−) to a folate-deficient diet. After 8 months of treatment, we analyzed tumor incidence in major organs, DNA mutation rates, DNA mutation spectra, local DNA methylation, and global DNA methylation in colon epithelial tissue. This target tissue was selected for molecular analysis because of its high cell turnover rate and because of the current debate questioning the link between folate deficiency and colorectal cancer in humans.
Methods
Experimental Mice and Diet Schedule
Big Blue Mice with the CII transgene (C57BL/6J background) were purchased from Stratagene (La Jolla, CA) and crossed with Ung−/− mice (129 background) that were previously generated in our lab.25 Ung+/− intercrosses with one parent Big Blue+/− generated the 2 experimental genotypes Big Blue+/−, Ung+/+ and Big Blue+/−, Ung−/−. The genetic background of all mice was 50% C57BL/6J and 50% 129. To control for genetic background effects, we evenly distributed littermates of the same genotype between the 2 diet groups. Starting at 3 months of age, mice received either a control diet (purified synthetic diet with 4 ppm folic acid, diet #32271) or a matched folic acid– deficient diet with less than 0.05 ppm folic acid (diet #29691) for a total period of 8 months (32 weeks). Diets were supplied by Test Diet (Richmond, IN). Both control and deficient diets contained 1% succinyl sulfathiazole and were fully matched with the exception of folic acid content. Both diets contained 20 mg/kg vitamin B12, 1400 ppm choline chloride, and 0.69% methionine. Food was exchanged weekly.
Genotyping
Primers and polymerase chain reaction (PCR) conditions for genotyping are detailed in the Supplementary Material (see Supplementary Methods online at www.gastrojournal.org).
Tissue Collection Immunostaining
After 8 months of dietary intervention, mice were killed at 11 months of age. All organs were macroscopically inspected for tumor development. Colon and small intestinal epithelial cells were harvested as described previously. 26 For lymphoma diagnosis, paraffin sections were stained with anti-CD45R (abcam ab64100), 1:1000 dilution (details provided in Supplementary Methods online at www.gastrojournal.org).
Plasma Folate and Homocysteine
Mice were fasted overnight and killed by exsanguination under isoflurane anesthesia. Blood was collected by heart puncture into heparinized tubes on ice. Plasma was separated within an hour of collection and frozen at −80°C for further analysis. Plasma folate was measured using the Quantaphase II radioassay kit (Bio-Rad Laboratories, Hercules, CA). Plasma total homocysteine was determined by high-performance liquid chromatography according to Araki and Sako.27
Lambda CII Mutagenesis Assay and CII Mutant Sequencing
DNA was isolated with minimal fragmentation and dialyzed. The LIZ shuttle vector was rescued from genomic DNA using Transpack packaging extract (Strat-agene), and mutant selection was conducted following instructions of the manual. Only confirmed mutants were used to calculate mutant frequencies and used for mutant sequencing. Each mutant was sequenced forward and reverse, with repeats if necessary, to ensure double sequencing coverage of the entire CII region for every sample (for details, see Supplementary Methods online at www.gastrojournal.org).
Uracil Measurement in DNA
All DNA samples were Rnase treated and dialyzed for 48 hours before being used for uracil analysis. Uracil in DNA was measured by a previously described gas chromatography–mass spectrometry method18,28 (see Supplementary Methods online at www.gastrojournal.org).
Global DNA Methylcytosine Measurement
Global DNA cytosine methylation of colon epithelial cells and spleen was determined by liquid chromatography–mass spectrometry according to Friso et al,29 and methylation analysis of small intestinal epithelial cells was measured by liquid chromatography–mass spectrometry as described previously30 (see Supplementary Methods online at www.gastrojournal.org).
Mouse B1 Element Pyrosequencing
Bisulfite mutagenized DNA was used to amplify a 145-bp fragment of the mouse B1 element containing 4 CpG sites, and PCR products were analyzed by pyrosequencing (EpigenDx, Inc, Worcester, MA). Samples were sequenced in triplicates, and relative methylation rates at all 4 CpG sites were combined to determine the average methylation rate of each sample. Procedural details and primers are listed in the Supplementary Material (see Supplementary Methods online at www.gastrojournal.org).
Oct4 and H19 DMR Bisulfite Sequencing Analysis
Bisulfite sequencing analysis of the H19 DMR and the Oct4 promoter were conducted as described previously. 30 Only sequences with more than 95% non–CpG C conversion were used for final sequence analysis. For primers and PCR conditions, see Supplementary Methods online at www.gastrojournal.org.
Statistical Analysis
The Mann–Whitney U test was used for analysis of folate, homocysteine, DNA methylation, and uracil measurements. Student t test was used for body weight analysis. To test for independence of nutrient/genotype group and mutation type, we used Fisher’s exact test and Bonferroni correction. For full statistical analysis of CII mutation spectra, see Supplementary Methods online at www.gastrojournal.org.
Results
Folate Deficiency Causes Increased Plasma Homocysteine But Does Not Increase Tumor Numbers
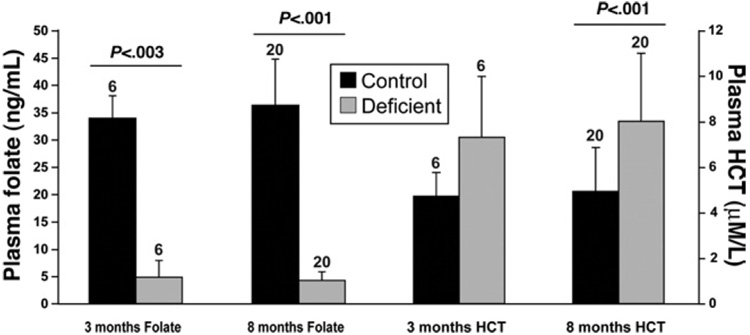
To verify that the experimental diet was successful in achieving folate deficiency, we analyzed plasma folate levels at 3 months and 8 months of dietary intervention (Figure 1). Severe reduction of folate levels was seen after 3 months and persisted until the end of the experiment (88% decrease, P < .001). Parallel to the observed decrease in plasma folate, we detected an increase in plasma homocysteine (60% increase at 8 months, P < .001). Folate deficiency caused a moderate but not significant growth retardation of 14% in wild-type mice (P =.17) and 5.3% in Ung−/− mice (P = .50), which correlates with previous studies (see Supplementary Figure 1 and Table 1 online at at www.gastrojournal.org)31.
Figure 1.
Folate-deficient diet causes increase in plasma homocysteine (HCT) and significant decrease in plasma folate levels. Plasma HCT and folate after 3 and 8 months of dietary intervention. Severe folate deficiency was detectable after 3 and 8 months, with parallel increases in plasma HCT. Average ± SD, n above bars, Mann–Whitney U test.
To evaluate whether the dietary intervention had any effect on tumor formation, we analyzed all major organs and subjected suspicious lesions to histologic analysis. In addition, each enlarged spleen (>150 mg) was saved for further histologic analysis (details in Supplementary Methods online at www.gastrojournal.org). Almost all tumors were diagnosed as follicular lymphomas, with one exception (Table 1). As shown in Table 1, folate deficiency had no significant effect on tumor formation. Ung−/− mice showed a trend towards increased formation of follicular lymphomas, similar to a study that reported increased lymphoma formation in aging Ung−/− mice.32
Table 1.
Tumor Incidencea
| Ung+/+ | Ung−/− | Ung+/+ | Ung−/− | |
|---|---|---|---|---|
| (lymphoma) | (lymphoma) | (other) | (other) | |
| Control diet | — | 3 | — | — |
| Folate-deficient diet | 2 | 2 | — | 1b |
| Tumors control/deficient | P = .69 | |||
| Tumors Ung+/+/Ung−/− | P = .23 |
Incidence of lymphomas and nonlymphoma tumors (other) (n = 10 for all experimental categories). For stringent lymphoma diagnosis, we only included samples with histologic criteria of intermediate- or high-grade lymphoma, or additional evidence of systemic spreading to extrasplenic organs.
The single nonlymphoma tumor was diagnosed as bronchoalveolar adenoma/carcinoma. Fisher’s exact test was used to calculate P values.
Folate Deficiency Causes Global DNA Hypomethylation
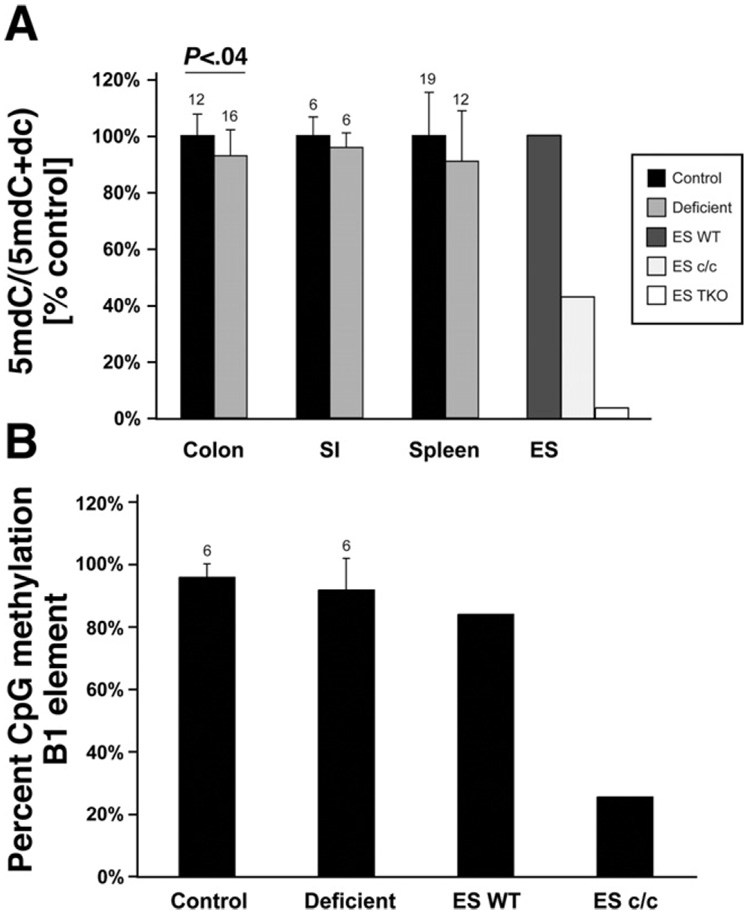
To evaluate whether folate deficiency affected global DNA methylation, we measured the DNA methylcytosine content by mass spectrometry in colon epithelial cells, small intestinal epithelial cells, and spleen. As controls we used wild-type ES cells, ES cells deficient for Dnmt1 (ES c/c, knockout of Dnmt1), and ES cells deficient for Dnmt1, 3a, and 3b (ES TKO, knockout of Dnmt3a and Dnmt3b and small interfering RNA [siRNA]-mediated knock-down of Dnmt1).33 As shown in Figure 2A, the DNA methylcytosine content showed a decrease in tissues of folate-deficient mice that was statistically significant for colon epithelial cells (P < .04, Mann–Whitney U test). No differences were detectable between wild-type and Ung −/− mice (data not shown). Confirming previous results, the ES c/c cells and ES TKO cells both showed very strong reductions in methylcytosine content.33 We also analyzed CpG methylation of the mouse B1 element in colon epithelial cells by using pyrosequencing. As shown in Figure 2B, a slight but not significant difference was detectable between B1 methylation of control samples and folate-deficient samples, whereas as expected, ES c/c cells showed a strong reduction in B1 element methylation.
Figure 2.
Folate deficiency induces mild genomic hypomethylation of intestinal epithelial cells, but methylation levels of the B1 repetitive element are not significantly affected. (A) Genomic deoxymethylcytosine (5mdC) relative to total deoxycytosine (5mdC + dC) measured by mass spectrometry. To demonstrate relative changes, the average of the control group in each tissue was set as 100%. DNA from colon epithelial cells, small intestinal epithelial cells (SI), and spleen was analyzed. As controls we used wild-type ES cells (ES WT), ES cells knocked out for Dnmt1 (ES c/c), and ES cells deficient for Dnmt1, Dnmt3a, and Dnmt 3b (ES TKO). All folate-deficient tissues showed hypomethylation, and for colon epithelial cells this was statistically significant (Mann–Whitney U test), n above bars. (B) Pyrosequencing analysis of 4 CpG sites within the B1 repetitive element of colon DNA derived from control mice and folate-deficient mice (n = 6 each). DNA from ES WT and ES c/c were used as controls. Both groups contain 3 samples each from wild-type mice and Ung knockout mice. Shown is the average methylation level of all CpG sites combined; values are given as average ± SD. The folate-deficient samples show a small decrease in B1 methylation, which is not statistically significant (U test), n above bar.
To test whether folate deficiency affects gene-specific DNA methylation and also to analyze whether some cells are more affected by demethylation than others, we analyzed the H19 DMR and the Oct4 gene of colon epithelial cells by using bisulfite sequencing. Typically, the Oct4 promoter is fully methylated in differentiated tissues,34 and the H19 DMR shows a monoallelic methylation pattern characteristic of imprinted regions.35 As shown in Figure 3, folate deficiency did not cause a significant change in the methylation pattern of these two loci.
Figure 3.
Methylation of the Oct4 promoter and the H19 DMR in colon epithelial cells was not affected by folate-deficient diet. Methylation status of the Oct4 promoter (A) and the H19 DMR (B) in colon epithelial cells analyzed using bisulfite sequencing, covering 12 and 15 CpG sites, respectively. Samples derived from control diet (control, n = 5) or folate-deficient diet (n = 5); in each case, 3 samples were taken from wild-type (WT) mice and 2 samples from Ung−/− mice. Black boxesindicate methylated CpGs, white boxes unmethylated CpGs. KO, knockout.
Folate Deficiency Increases Uracil Misincorporation in DNA
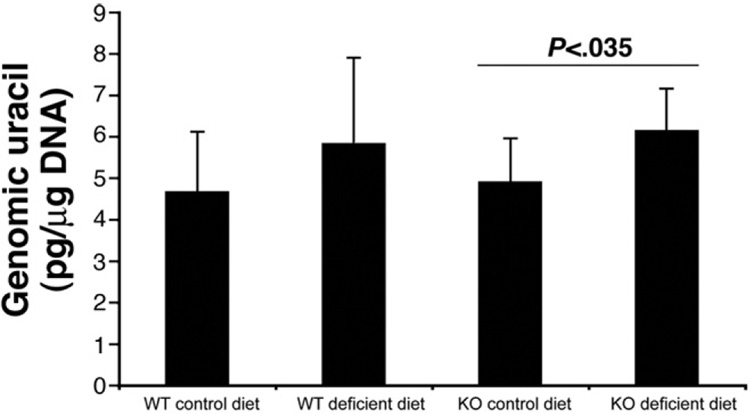
The folate metabolite methylene tetrahydrofolate is required for thymidine synthesis, and folate deficiency has been shown to increase dUTP/dTTP ratios and uracil misincorporation into DNA.18 We therefore tested whether folate deficiency increases the uracil content of colon epithelial cells. Also, since the Ung enzyme is involved in removal of uracil during DNA replication,23 we analyzed whether Ung deficiency can amplify this effect. Using mass spectrometry, we found that DNA uracil content was indeed increased in folate-deficient samples, and this effect was significant in Ung−/−mice (24% increase, P < .05) (Figure 4). However, the average DNA uracil content of cells from Ung−/− mice did not differ from wild-type mice, and the relative change of DNA uracil content secondary to folate deficiency was not amplified in Ung −/− mice, indicating possible adaptation and partial compensation of Ung deficiency by other DNA repair enzymes such as Smug1.36
Figure 4.
Folate-deficient diet increases genomic uracil content of colon epithelial cells. Genomic uracil content of colon epithelial cells measured by mass spectrometry after 8 months of dietary intervention. Uracil was increased in folate-deficient samples, and this was significant for Ung−/− mice (n = 9 each, P= .03, Mann–Whitney U test). Average genomic uracil levels of Ung−/− mice were not significantly higher than in wild-type (WT) mice. KO, knockout.
Ung Deficiency But Not Folate Deficiency Affects Point Mutation Frequencies
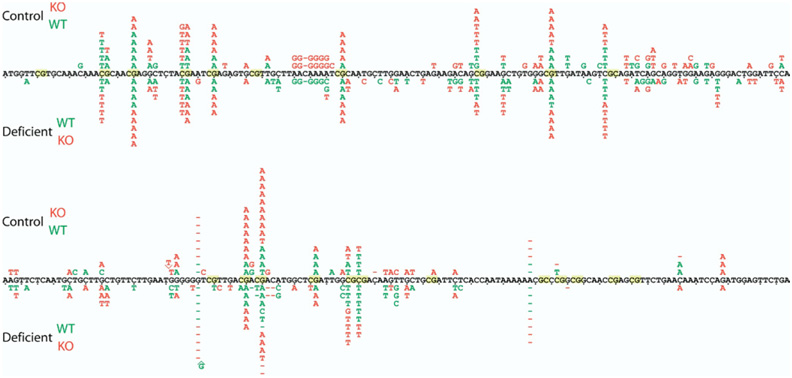
To allow in vivo mutation analysis, all mice in this study carried the Big Blue mutational reporter.37 For measurement of point mutation frequencies (Table 2), DNA was isolated from colon epithelial cells and used in the λ CII assay as previously described.38 To analyze whether Ung or folate deficiency caused a different spectrum of mutations, we sequenced the DNA isolated from a total of 730 phages that were previously identified as mutants in the λ CII assay. The spatial distribution of all mutations is summarized in Figure 5, and the categorized frequency of point mutations is shown in Table 3. Details of the statistical analysis are shown in the Supplementary Material (see Supplementary Methods online at www.gastrojournal.org).
Table 2.
λ CII Assay, Mutant Frequencya
| WT control | WT deficient | Ung−/− control | Ung−/− deficient | |
|---|---|---|---|---|
| No. of mice | 7 | 7 | 8 | 7 |
| Total plaques screened | 1,648,500 | 1,097,250 | 1,708,050 | 1,429,083 |
| Total mutants | 64 | 86 | 210 | 138 |
| Mutant frequency × 10−5, SD | 4.4 ± 2.7 | 7.2 ± 4.2 | 11.9 ± 6.1 | 10 ± 3.6 |
WT, wild-type.
The difference between WT control and Ung−/− control is significant (P = .017), whereas differences between WT control and WT deficient are not significant (P = .28) (Mann–Whitney U test). Average mutant frequencies and SD were calculated by averaging frequencies of individual mice. For calculation of mutant frequencies mice with failed, not scorable, titering plates were disregarded (details see supplementary methods).
Figure 5.
Spatial distribution of point mutations in the λ CII gene. Compiled sequence data of the CII gene generated from 730 phage mutants from all 4 experimental groups illustrating the spatial distribution of point mutations. All mutations in control diet samples are shown above the reference sequence, and all mutations from folate-deficient samples are shown below the CII sequence. Letters (individual base substitutions), lines (deletions), ^ (insertions), green (wild-type [WT]), and red (Ung−/−) mice. The spatial distribution of point mutations in the control diet group is almost identical to the folate-deficient group, with the majority of mutational events occurring at CpG sites. The asymmetric mutation frequencies within some CpG dinucleotides are caused by underrepresented mutations that don’t affect the CII amino acid sequence and therefore are not detected by the λ CII assay. KO, knockout.
Table 3.
λ CII Mutant Analysis
| Ung−/− | Ung−/− | |||
|---|---|---|---|---|
| WT control | WT deficient | Control | deficient | |
| No. of mice | 8 | 10 | 8 | 9 |
| Mutants sequenced | 108 | 194 | 215 | 213 |
| Sequences with CII mutation | 82 | 158 | 154 | 161 |
| Nonredundant eventsa | 70 | 120 | 133 | 119 |
| Transition | 46 (66%) | 63 (53%) | 89 (67%) | 70 (59%) |
| C/G → T/A | 40 (57%) | 49 (41%) | 67 (50%) | 66 (55%) |
| non CpG | 2 (3%) | 8 (7%) | 21 (16%) | 20 (17%) |
| at CpG | 38 (54%) | 41 (34%) | 46 (35%) | 46 (39%) |
| T/A → C/G | 6 (9%) | 14 (12%) | 22 (17%) | 4 (3%) |
| Transversion | 20 (29%) | 43 (36%) | 29 (22%) | 33 (28%) |
| C/G → A/T | 12 (17%) | 24 (20%) | 22 (17%) | 28 (24%) |
| C/G → G/C | 4 (6%) | 6 (5%) | 2 (2%) | 2 (2%) |
| T/A → A/T | 3 (4%) | 8 (7%) | 3 (2%) | 3 (3%) |
| T/A → G/C | 1 (1%) | 5 (4%) | 2 (2%) | 0 |
| Events at T/A | 10 (14%) | 27 (23%) | 27 (20%) | 7 (6%) |
| Frameshift | 4 (6%) | 14 (12%) | 15 (11%) | 16 (13%) |
| +1 | 0 | 1 (1%) | 1 (1%) | 0 |
| − 1 | 4 (6%) | 13 (11%) | 14 (11%) | 16 (13%) |
WT, wild-type.
Identical mutations occurring at the same site in more than one phage from the same mouse (jackpot mutants) were tabulated as a single mutational event. The difference between C:G transition frequency at non–CpG sites of Ung−/− mice and WT mice is significant (P = .0003, Fisher’s exact test). For sequencing and analysis of mutation patterns, all available mutant plaques were used.
In all experimental groups, the dominant mutation was C:G to T:A transitions that occurred mostly at CpG sites (Table 3). As demonstrated in Table 2, Ung deficiency alone increased point mutation frequencies in the λ CII assay, and the spectra of mutations showed significantly increased C:G to T:A transitions at non–CpG sites (P = .0003, Fisher’s exact test). Folate deficiency, on the other hand, did not cause a significant increase in mutation frequencies or a significant change of the mutation spectrum in wild-type or Ung −/− mice (Table 2 and Table 3). In particular, we did not observe an increase in point mutation frequencies at T:A base pairs as might be expected from increased uracil misincorporation, and there was no increase of transitions at CpG sites that would indicate increased cytosine deamination (Table 3).
Discussion
The primary goal of our study was to evaluate the genetic and epigenetic consequences of folate deficiency to help clarify the role of this nutrient in tumor development. Also, we wanted to investigate whether lack of the DNA repair enzyme Ung can increase point mutations during folate deficiency. We improved on previous studies by using a long-term dietary intervention (8 months) and by focusing on tissues with a high cell turnover rate, which is important for detecting passive loss of DNA methylation. Furthermore, all animals carried the Big Blue mutational reporter gene, allowing us to directly analyze in vivo point mutation frequencies and spectra.38
Our experimental diet achieved significantly reduced plasma folate levels even during the initial period of the experiment. We also observed that plasma homocysteine was increased, consistent with previous reports.20
A central aim of our study was to directly test whether folate deficiency increases DNA mutation rates and whether this is amplified by additional lack of the Ung enzyme. Folate deficiency is known to increase uracil misincorporation during DNA replication,18 and faulty repair at resulting U:A mispairs has been proposed to cause secondary DNA mutations.19 In addition, we wanted to investigate whether conditions of folate deficiency would induce cytosine deamination by DNA methyltransferases by analogy to bacteria,22 which generates mutagenic U:G mispairs.
The Ung enzyme is involved in the repair of U:A base pairs that occur as a consequence of uracil misincorporation, and of U:G base pairs that occur as a consequence of cytosine deamination.23,36 We therefore wanted to test whether Ung deficiency can amplify possible mutagenic effects of low folate.
Our in vivo data demonstrate that folate deficiency indeed increased the uracil content of colon epithelial cells.18 This likely reflects misincorporated uracil, which is the quantitatively dominant source of uracil in DNA. Surprisingly, however, the baseline uracil content was not higher in Ung−/− mice, nor was uracil accumulation amplified in these animals. This observation differs from previous reports showing increased baseline genomic uracil levels in Ung-deficient mouse embryonic fibroblasts.23 A possible explanation is the reduced half-life of intestinal epithelial cells relative to cultured mouse embryonic fibroblasts that results in less cumulative cell divisions per cell and consequently less uracil misincorporation per cell.39 Also it is possible that backup repair activity of Smug1 uracil DNA glycosylase could partially compensate for Ung deficiency in intestinal epithelial cells of Ung−/− mice.24,36 To analyze the effects of folate and Ung deficiency on DNA point mutations, we used the previously described λ CII assay.38 We found that folate deficiency had no significant effect on the frequency or spectrum of point mutations of wild-type or Ung−/− mice (full statistical analysis in Supplementary Methods online at www.gastrojournal.org). In particular, a relative increase in point mutations at T:A base pairs, resulting from repair of uracil misincorporation,19 was not detected. Our data suggest that although folate deficiency causes an increase in genomic uracil content through uracil misincorporation, this does not result in increased DNA point mutations; nor was there an increase in transition mutations at CpG sites, which suggests that folate deficiency does not induce enzymatic cytosine deamination. Our results are consistent with a previous study that found no correlation between folate status and adenomatosis polyposis coli or p53 mutations in dimethylhydrazine-induced tumors.40 However, we cannot exclude the possibility that DNA strand breaks and genetic rearrangements, both of which have been reported for folate deficiency,18,20 might have occurred in the DNA of these mice, since such alterations are not detected by the λ CII assay. It also has to be considered that diminished transcription of critical tumor suppressor genes caused by DNA strand breaks alone, even in the absence of mutations, could feasibly be a consequence of uracil misincorporation.20
In contrast to lack of folate, which had no effect on mutation rates, Ung deficiency alone did increase the frequency of DNA point mutations. This in vivo result is similar to previous in vitro studies reporting slightly increased mutation frequencies for Ung-deficient mouse embryonic fibroblasts.24,25 We also detected increased C:G to T:A transitions at non–CpG sites in Ung−/− mice, as had a study on the hprt locus of Ung-deficient mouse embryonic fibroblasts.24 Our in vivo data therefore support a role for the Ung enzyme in suppressing mutations at unmethylated C:G base pairs and suggest that impairment of this activity, not uracil misincorporation, is the underlying cause of increased mutation rates in Ung−/− mice. It is possible that uracil misincorporation is mutagenic in other circumstances, such as additional deletion of Smug1,36 which might have a stronger effect on uracil misincorporation than Ung deficiency alone.
In all experimental groups we found that the most frequent mutational event was a C:G to T:A transition at CpG sites. This fits well with published data and reflects the almost complete methylation of CpG sites in the CII transgene.41 In summary, our sequence analysis demonstrates that lack of folate has no effect on DNA point mutation frequencies and spectra, and that this is not changed by additional Ung deficiency.
In addition to the genetic analysis, we also tested the effects of low folate on DNA methylation. We found that folate deficiency caused a modest but statistically significant global DNA hypomethylation in colon epithelial cells. Hypomethylation was also observed in the small intestine and spleen, although this was not statistically significant. Sample numbers for the small intestine were small, and sample-to-sample variation in the spleen was high, which likely explains why the methylation differences did not reach statistical significance in these tissues. Similarly, using pyrosequencing analysis, we detected a trend towards less CpG methylation of the mouse B1 element, which is a highly abundant, repetitive element of the mouse genome; however, in analogy to the small intestine data, the difference was not significant, most likely due to the small sample number.
To determine whether the minimal loss of DNA methylation that was detected by mass spectrometry in the colon is stochastically distributed between cells and genomic regions, or whether individual cells or regions are more strongly affected than others, we analyzed the Oct4 promoter and the H19 DMR by using bisulfite sequencing analysis. This method yields allele-specific data; therefore, if individual cells are strongly affected by demethylation, one would expect to see demethylation of individual alleles, whereas a stochastic distribution should not show significant allele-specific changes. The Oct4 promoter was chosen because it is normally fully methylated in differentiated tissues, and aberrant expression of Oct4 has been shown to cause intestinal tumors in mice.42 Similarly, the H19 DMR was analyzed because it controls expression of the tumor-promoting growth factor Igf2. We did not detect any changes in genespecific methylation at the Oct4 promoter and the H19 DMR, suggesting that the minimal loss of DNA methylation is most likely randomly distributed between cells and genomic regions.
Taken together, our results suggest that chronic, isolated folate deficiency can induce a low degree of genomic hypomethylation with a distribution that is most likely stochastic. Previous studies on folate deficiency had shown contradictory results, possibly because of experimental design such as analyzed tissue or length of exposure to a folate-deficient diet.14
The relevance of such a low degree of genomic hypomethylation for the development of tumors is unclear. Severe global genomic hypomethylation can reactivate transposable elements and causes genetic instability,12 both of which can directly contribute to tumor formation. Gaudet et al11 found that reduction of Dnmt1 activity to 10% of the normal levels causes widespread T-cell lymphomas, whereas mice with 20% residual Dnmt1 activity (chip/chip) did not develop any tumors despite significant genomic hypomethylation. This suggests that only substantial hypomethylation causes detectable increases in tumor formation. It is therefore unlikely that an approximately 6% decrease in global methylation induces genetic instability and secondary tumor formation. Our observation that the tumor incidence in our study was not increased by folate deficiency further argues against a tumor-promoting effect, and it is very unlikely that the moderate and statistically not significant decrease in body weight of folate-deficient mice (14% in wild-type, P = .17; 5.3% in Ung−/− mice, P =.50) would have masked the outgrowth of early tumor lesions. It has to be kept in mind, however, that the mouse models used in our study are not specifically sensitized to the development of intestinal tumors. It is therefore possible that folate deficiency might promote intestinal tumors under conditions that more strongly predispose to the formation of such tumors.
In summary, this is the first in vivo study to evaluate whether chronic folate deficiency alone or with additional mutations in the DNA repair pathway affects DNA point mutation frequencies and spectra in normal tissue. Our data suggest that the effect of isolated folate deficiency on both DNA methylation and DNA point mutations is not sufficient to promote tumor formation, even in the presence of genetic susceptibility such as Ung deficiency. This may help to explain some inconsistencies in the epidemiologic and experimental data, and suggests that the hypothesis directly linking folate deficiency to cancer through changes in DNA has to be reevaluated.
Supplementary Material
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at doi: 10.1053/j.gastro.2008.10.016.
Acknowledgments
The authors thank Jessica Dausman, Ruth Flannery, and Dongdong Fu for help with maintenance of the mouse colony and histologic analysis; Tom DiCesare for .gure preparations; Seshamma Reddy for help with the pathologic assessment of histologic sections; Jacob Selhub for generously supporting this study; Bina M. Albuquerque for her technical assistance; Caroline Beard and Grant Welstead for critical reading of the manuscript; and the Fritz-Thyssen Stiftung for providing a fellowship to Heinz Linhart.
The authors disclose the following: H.G.L. is supported by a postdoctoral fellowship from the Fritz-Thyssen Stiftung. This work was supported by Philip Morris International and National Institutes of Health grant NIH RO1-CA087869 to R.J., and by the US Department of Agriculture, Agricultural Research Service (agreement No. 58-1950-7-707). Any opinions, .ndings, conclusion, or recommendations expressed in this publication are those of the authors and do not necessarily re.ect the view of the US Department of Agriculture.
Abbreviations used in this paper
- PCR
polymerase chain reaction
- SAM
S-adenosyl-L-methionine
- Ung
uracil DNA glycosylase.
References
- 1.Giovannucci E, Stampfer MJ, Colditz GA, et al. Multivitamin use, folate, and colon cancer in women in the Nurses’ Health Study. Ann Intern Med. 1998;129:517–524. doi: 10.7326/0003-4819-129-7-199810010-00002. [DOI] [PubMed] [Google Scholar]
- 2.Jacobs EJ, Connell CJ, Chao A, et al. Multivitamin use and colorectal cancer incidence in a US cohort: does timing matter? Am J Epidemiol. 2003;158:621–628. doi: 10.1093/aje/kwg190. [DOI] [PubMed] [Google Scholar]
- 3.Ericson U, Sonestedt E, Gullberg B, et al. High folate intake is associated with lower breast cancer incidence in postmenopausal women in the Malmo Diet and Cancer cohort. Am J Clin Nutr. 2007;86:434–443. doi: 10.1093/ajcn/86.2.434. [DOI] [PubMed] [Google Scholar]
- 4.Pelucchi C, Talamini R, Negri E, et al. Folate intake and risk of oral and pharyngeal cancer. Ann Oncol. 2003;14:1677–1681. doi: 10.1093/annonc/mdg448. [DOI] [PubMed] [Google Scholar]
- 5.Larsson SC, Giovannucci E, Wolk A. Folate intake, MTHFR polymorphisms, and risk of esophageal, gastric, and pancreatic cancer: a meta-analysis. Gastroenterology. 2006;131:1271–1283. doi: 10.1053/j.gastro.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Little J, Sharp L, Duthie S, et al. Colon cancer and genetic variation in folate metabolism: the clinical bottom line. J Nutr. 2003;133:3758S–3766S. doi: 10.1093/jn/133.11.3758S. [DOI] [PubMed] [Google Scholar]
- 7.Wiemels JL, Smith RN, Taylor GM, et al. Methylenetetrahydrofolate reductase (MTHFR) polymorphisms and risk of molecularly defined subtypes of childhood acute leukemia. Proc Natl Acad Sci U S A. 2001;98:4004–4009. doi: 10.1073/pnas.061408298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Logan RF, Grainge MJ, Shepherd VC, et al. Aspirin and folic acid for the prevention of recurrent colorectal adenomas. Gastroenterology. 2008;134:29–38. doi: 10.1053/j.gastro.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 9.Cole BF, Baron JA, Sandler RS, et al. Folic acid for the prevention of colorectal adenomas: a randomized clinical trial. JAMA. 2007;297:2351–2359. doi: 10.1001/jama.297.21.2351. [DOI] [PubMed] [Google Scholar]
- 10.Van Guelpen B, Hultdin J, Johansson I, et al. Low folate levels may protect against colorectal cancer. Gut. 2006;55:1461–1466. doi: 10.1136/gut.2005.085480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaudet F, Hodgson JG, Eden A, et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 12.Eden A, Gaudet F, Waghmare A, et al. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 13.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim YI. Nutritional epigenetics: impact of folate deficiency on DNA methylation and colon cancer susceptibility. J Nutr. 2005;135:2703–2709. doi: 10.1093/jn/135.11.2703. [DOI] [PubMed] [Google Scholar]
- 15.Balaghi M, Wagner C. DNA methylation in folate deficiency: use of CpG methylase. Biochem Biophys Res Commun. 1993;193:1184–1190. doi: 10.1006/bbrc.1993.1750. [DOI] [PubMed] [Google Scholar]
- 16.Le Leu RK, Young GP, McIntosh GH. Folate deficiency diminishes the occurrence of aberrant crypt foci in the rat colon but does not alter global DNA methylation status. J Gastroenterol Hepatol. 2000;15:1158–1164. doi: 10.1046/j.1440-1746.2000.02327.x. [DOI] [PubMed] [Google Scholar]
- 17.Song J, Sohn KJ, Medline A, et al. Chemopreventive effects of dietary folate on intestinal polyps in Apc+/−Msh2−/− mice. Cancer Res. 2000;60:3191–3199. [PubMed] [Google Scholar]
- 18.Blount BC, Mack MM, Wehr CM, et al. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc Natl Acad Sci U S A. 1997;94:3290–3295. doi: 10.1073/pnas.94.7.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auerbach P, Bennett RA, Bailey EA, et al. Mutagenic specificity of endogenously generated abasic sites in Saccharomyces cerevisiae chromosomal DNA. Proc Natl Acad Sci U S A. 2005;102:17711–17716. doi: 10.1073/pnas.0504643102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim YI, Shirwadkar S, Choi SW, et al. Effects of dietary folate on DNA strand breaks within mutation-prone exons of the p53 gene in rat colon. Gastroenterology. 2000;119:151–161. doi: 10.1053/gast.2000.8518. [DOI] [PubMed] [Google Scholar]
- 21.Krokan HE, Drablos F, Slupphaug G. Uracil in DNA—occurrence, consequences and repair. Oncogene. 2002;21:8935–8948. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- 22.Shen JC, Rideout WM, III, Jones PA. High frequency mutagenesis by a DNA methyltransferase. Cell. 1992;71:1073–1080. doi: 10.1016/s0092-8674(05)80057-1. [DOI] [PubMed] [Google Scholar]
- 23.Nilsen H, Rosewell I, Robins P, et al. Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol Cell. 2000;5:1059–1065. doi: 10.1016/s1097-2765(00)80271-3. [DOI] [PubMed] [Google Scholar]
- 24.An Q, Robins P, Lindahl T, et al. C→T mutagenesis and gamma-radiation sensitivity due to deficiency in the Smug1 and Ung DNA glycosylases. EMBO J. 2005;24:2205–2213. doi: 10.1038/sj.emboj.7600689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Endres M, Biniszkiewicz D, Sobol RW, et al. Increased postischemic brain injury in mice deficient in uracil-DNA glycosylase. J Clin Invest. 2004;113:1711–1721. doi: 10.1172/JCI20926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujimoto K, Beauchamp RD, Whitehead RH. Identification and isolation of candidate human colonic clonogenic cells based on cell surface integrin expression. Gastroenterology. 2002;123:1941–1948. doi: 10.1053/gast.2002.37065. [DOI] [PubMed] [Google Scholar]
- 27.Araki A, Sako Y. Determination of free and total homocysteine in human plasma by high-performance liquid chromatography with fluorescence detection. J Chromatogr. 1987;422:43–52. doi: 10.1016/0378-4347(87)80438-3. [DOI] [PubMed] [Google Scholar]
- 28.Choi SW, Friso S, Ghandour H, et al. Vitamin B-12 deficiency induces anomalies of base substitution and methylation in the DNA of rat colonic epithelium. J Nutr. 2004;134:750–755. doi: 10.1093/jn/134.4.750. [DOI] [PubMed] [Google Scholar]
- 29.Friso S, Choi SW, Dolnikowski GG, et al. A method to assess genomic DNA methylation using high-performance liquid chromatography/electrospray ionization mass spectrometry. Anal Chem. 2002;74:4526–4531. doi: 10.1021/ac020050h. [DOI] [PubMed] [Google Scholar]
- 30.Linhart HG, Lin H, Yamada Y, et al. Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing. Genes Dev. 2007;21:3110–3122. doi: 10.1101/gad.1594007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clifford AJ, Bills ND, Peerson JM, et al. A depletion-repletion folate bioassay based on growth and tissue folate concentrations of rats. J Nutr. 1993;123:926–932. doi: 10.1093/jn/123.5.926. [DOI] [PubMed] [Google Scholar]
- 32.Nilsen H, Stamp G, Andersen S, et al. Gene-targeted mice lacking the Ung uracil-DNA glycosylase develop B-cell lymphomas. Oncogene. 2003;22:5381–5386. doi: 10.1038/sj.onc.1206860. [DOI] [PubMed] [Google Scholar]
- 33.Meissner A, Gnirke A, Bell GW, et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li JY, Pu MT, Hirasawa R, et al. Synergistic function of DNA methyltransferases Dnmt3a and Dnmt3b in the methylation of Oct4 and Nanog. Mol Cell Biol. 2007;27:8748–8759. doi: 10.1128/MCB.01380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waterland RA, Lin JR, Smith CA, et al. Post-weaning diet affects genomic imprinting at the insulin-like growth factor 2 (Igf2) locus. Hum Mol Genet. 2006;15:705–716. doi: 10.1093/hmg/ddi484. [DOI] [PubMed] [Google Scholar]
- 36.An Q, Robins P, Lindahl T, et al. 5-Fluorouracil incorporated into DNA is excised by the Smug1 DNA glycosylase to reduce drug cytotoxicity. Cancer Res. 2007;67:940–945. doi: 10.1158/0008-5472.CAN-06-2960. [DOI] [PubMed] [Google Scholar]
- 37.Hill KA, Buettner VL, Glickman BW, et al. Spontaneous mutations in the Big Blue transgenic system are primarily mouse derived. Mutat Res. 1999;436:11–19. doi: 10.1016/s1383-5742(98)00024-6. [DOI] [PubMed] [Google Scholar]
- 38.Jakubczak JL, Merlino G, French JE, et al. Analysis of genetic instability during mammary tumor progression using a novel selection- based assay for in vivo mutations in a bacteriophage lambda transgene target. Proc Natl Acad Sci U S A. 1996;93:9073–9078. doi: 10.1073/pnas.93.17.9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duthie SJ, Narayanan S, Brand GM, et al. DNA stability and genomic methylation status in colonocytes isolated from methyldonor- deficient rats. Eur J Nutr. 2000;39:106–111. doi: 10.1007/s003940070026. [DOI] [PubMed] [Google Scholar]
- 40.Sohn KJ, Puchyr M, Salomon RN, et al. The effect of dietary folate on Apc and p53 mutations in the dimethylhydrazine rat model of colorectal cancer. Carcinogenesis. 1999;20:2345–2350. doi: 10.1093/carcin/20.12.2345. [DOI] [PubMed] [Google Scholar]
- 41.Millar CB, Guy J, Sansom OJ, et al. Enhanced CpG mutability and tumorigenesis in MBD4-deficient mice. Science. 2002;297:403–405. doi: 10.1126/science.1073354. [DOI] [PubMed] [Google Scholar]
- 42.Hochedlinger K, Yamada Y, Beard C, et al. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at doi: 10.1053/j.gastro.2008.10.016.