Abstract
Combinatorial libraries of de novo amino acid sequences can provide a rich source of diversity for the discovery of novel proteins. Randomly generated sequences, however, rarely fold into well ordered protein-like structures. To enhance the quality of a library, diversity must be focused into those regions of sequence space most likely to yield well folded structures. We have constructed focused libraries of de novo sequences by designing the binary pattern of polar and nonpolar amino acids to favor structures that contain abundant secondary structure, while simultaneously burying hydrophobic side chains in the protein interior and exposing hydrophilic side chains to solvent. Because binary patterning specifies only the polar/nonpolar periodicity, but not the identities of the side chains, detailed structural features, including packing interactions, cannot be designed a priori. Can binary patterned libraries nonetheless encode well folded proteins? An unambiguous answer to this question requires determination of a 3D structure. We used NMR spectroscopy to determine the structure of S-824, a novel protein from a recently constructed library of 102-residue sequences. This library is “naïve” in that it has not been subjected to high-throughput screens or directed evolution. The experimentally determined structure of S-824 is a four-helix bundle, as specified by the design. As dictated by the binary-code strategy, nonpolar side chains are buried in the protein interior, and polar side chains are exposed to solvent. The polypeptide backbone and buried side chains are well ordered, demonstrating that S-824 is not a molten globule and forms a unique structure. These results show that amino acid sequences that have neither been selected by evolution, nor designed by computer, nor isolated by high-throughput screening, can form native-like structures. These findings validate the binary-code strategy as an effective method for producing vast collections of well folded de novo proteins.
Attempts to devise proteins de novo are motivated by two considerations: (i) Recapitulation of natural systems is the ultimate test of our understanding of such systems, and (ii) construction of novel proteins is an essential step toward future biotechnologies using “tailor-made” proteins with desired properties. Progress toward the construction of novel proteins emanates from two approaches, rational design and combinatorial methods. Rational design typically uses computational techniques to design individual amino acid sequences by considering the atom-by-atom interactions of an entire macromolecule (1, 2). In contrast, combinatorial methods typically make no attempt to design particular structural features, but instead rely on the numerical power of large libraries to (hopefully) generate desired structures and functions (3). We have developed an alternative approach that incorporates both rational design and combinatorial methods to produce focused libraries of novel proteins (4, 5). In these libraries, the binary patterning of polar and nonpolar residues is designed rationally, but the exact identities of the individual polar and nonpolar residues are not defined and are varied combinatorially. Here, we present the structure of a protein from a binary-patterned library and thereby demonstrate that designed combinatorial libraries can encode well folded de novo proteins.
The main premise of the binary-code approach is that combinatorial libraries must be designed such that all sequences in a library are consistent with the formation of protein structures that (i) contain abundant secondary structure (α-helices or β-strands), while (ii) simultaneously burying hydrophobic and exposing hydrophilic side chains. To satisfy these requirements, libraries of sequences are designed such that the periodicity of polar and nonpolar residues in the linear sequence matches the structural periodicity of the desired secondary structure. For α-helices, the structural periodicity is 3.6 residues per repeat, and so de novo sequences targeted to form amphiphilic α-helices are designed with a binary pattern that places a nonpolar residue (•) every three or four positions (e.g., ○•○○••○○•○○••○). Conversely, β-strands have a structural periodicity of 2 residues per repeat, so sequences targeted to form amphiphilic β-strands are designed with an alternating pattern (e.g., ○•○•○•○). Because a binary pattern specifies only the type (polar or nonpolar) of side chain and not its identity, these patterns are compatible with enormous combinatorial diversity. Incorporation of this diversity into libraries of sequences expressed in vivo is made possible by the organization of the genetic code: Five nonpolar amino acids (Met, Leu, Ile, Val, and Phe) can be encoded by the degenerate DNA codon NTN, whereas six polar amino acids (Lys, His, Glu, Gln, Asp, and Asn) can be encoded by the degenerate codon VAN. (N represents the DNA bases A, G, C, or T, and V represents A, G, or C.)
Binary patterning has been used as the key feature for designing focused libraries of both α-helical and β-sheet proteins (4–6). These libraries have successfully produced cofactor-binding proteins (7), catalytically active enzymes (8), amyloid-like fibrils (5), and protein-based biomaterials (9). However, the high-resolution structure of a protein from these libraries had not been determined. Recently, we constructed a second generation binary-patterned library (10), designed to form four-helix bundles with α-helices significantly longer than those in earlier libraries. Proteins from this new library were shown to be α-helical and quite stable (10). The ultimate test of whether the designed binary pattern actually encodes four-helix bundles requires determination of a 3D structure. This structure is presented here.
Materials and Methods
Sample Preparation. Protein S-824 was expressed in Escherichia coli strain BL21(DE3). Uniformly 15N-labeled sample was prepared by growing cells in M9 minimal media supplemented with 1 g/liter [15N]NH4Cl and 10 g/liter glucose as the sole nitrogen and carbon sources. 15N and 13C doubly labeled sample was prepared similarly, except that 2 g/liter [13C]glucose was used instead of unlabeled glucose. Protein was purified as described (4, 10). The extent of isotope labeling was checked for the 15N-labeled sample by comparing the observed mass (by mass spectroscopy) with that expected from the sequence. The protein was determined to be >98% labeled. Sample concentration was ≈1.5 mM as determined by absorbance at 280 nm. NMR samples were in 50 mM HAc-NaAc buffer (pH 4.0) and 92% H2O/8% D2O.
NMR Spectroscopy. Spectra were collected at 25°C by using a Varian INOVA 600 MHz spectrometer equipped with a triple-resonance probe and pulse-field gradients. Assignments were reported (11). Mixing time for the NOESY experiments was 80 ms. The 1H chemical shift was referenced to the DOH line, and 13C and 15N dimensions were referenced indirectly by using standard parameters from the BMRB (BioMagResBank) (12). Spectra were processed and analyzed with FELIX 97 software (Molecular Simulations, Waltham, MA).
Interproton distance restraints were derived from nuclear Overhauser effect (NOE) cross peaks. Based on peak volumes, the peaks were divided into three classes: strong (1.8–2.9 Å), medium (1.8–3.3 Å), and weak (1.8–5.0 Å). The three classes were calibrated by using NOEs corresponding to the known distances of local intrahelical interactions. The upper limits of the distance restraints were then modified to account for the absence of stereo-specific assignments (13).
φ angle restraints were derived from HNHA experiments (14). A range of ±25° was allowed for the φ restraints for the helical regions, and a more generous range of ±60° was allowed for other interhelical regions.
Structure Calculation. The structure was calculated by using the dynamic annealing protocol of CNS, Version 1.1 (15). The calculation started with a fully extended structure with ideal geometry. Restraints were then incorporated as input files. The annealing has four stages: (i) high (constant)-temperature torsion-angle annealing, (ii) slow-cooling torsion-angle dynamic annealing, (iii) Cartesian dynamics annealing, and (iv) energy minimization (15). The parameters suggested by the program were used throughout the process. The protocol started with an initial search of the torsion-angle space at 50,000 K for 1,000 steps at 0.015 ps per step. The van der Waals (VDW) scale factor was reduced to 0.1 to enable crossings of rotational barriers. Next, the temperature was reduced from 50,000 K to 0 K in 1,000 steps at 0.015 ps per step. Then the system was reheated to 2,000 K and gradually cooled to 0 K in 3,000 steps at 0.003 ps per step. The VDW scale factor was increased gradually in these two cooling stages, from 0.1 to 1.0 in the first cooling stage, and then from 1.0 to 4.0 in the second cooling stage. The cooling stages were followed by 2,000 steps of energy minimization with a VDW scale factor of 1.0. In this final step, bond lengths and angles were allowed to relax to minimize the overall energy.
The structure calculation was done iteratively. Initially, only the unambiguously assigned NOE peaks were converted into distance restraints and used for the calculation. The resulting conformations were then used to resolve the ambiguities of more NOE peaks and thereby generate more distance restraints. These newly generated restraints were incorporated into another round of calculation, and improved structures were generated. This process was repeated until 95% of the NOE peaks were incorporated into the calculation. Meaningful assignments could not be made for the remaining 5% of the NOE peaks. These peaks might result from spin diffusion or local flexibility. In the final iteration, 100 structures were calculated by using 1,644 NOE distance restraints and 91 φ angle restraints. The 10 lowest energy structures were used for the analysis shown in Table 1.
Table 1. Structural statistics for the 10 lowest-energy structures.
| NOE distance restraints | |
| Intraresidue (|i — j| = 0) | 594 |
| Sequential (|i — j| = 1) | 405 |
| Medium-range (1 < |i — j| < 5) | 447 |
| Long-range (|i — j| ≥ 5) | 198 |
| Φ angle | 91 |
| Total | 1,735 |
| Mean rms deviations from experimental restraints | |
| Distance restraints, Å | 0.016 ± 0.001 |
| Dihedral angle, ° | 0.207 ± 0.029 |
| Mean rms deviations from ideal geometry | |
| Bonds, Å | 0.0027 ± 0.0001 |
| Angles, ° | 0.461 ± 0.007 |
| Impropers, ° | 0.336 ± 0.014 |
| rms deviations to the mean structure, Å | |
| All backbone | 0.52 ± 0.06 |
| All heavy atoms | 1.06 ± 0.10 |
| Backbone in helical region* | 0.34 ± 0.05 |
| Heavy atoms in helical region* | 0.97 ± 0.11 |
| Heavy atoms in hydrophobic core† | 0.51 ± 0.09 |
| Ramachandran statistics from PROCHECK‡ | |
| Residues in most favored regions, % | 88.9 ± 1.3 |
| Residues in additional allowed regions, % | 10.0 ± 1.3 |
| Residues in generously allowed regions, % | 1.1 ± 0 |
| Residues in disallowed regions, % | 0 |
| Ramachandran statistics from MOLPROBITY§ | |
| Residues in favored region, % | 85.0 ± 2.1 |
| Residues in allowed regions (including favored regions), % | 97.1 ± 0.9 |
Ten lowest energy structures of 100 calculated structures. None of the structures contain distance violations of >0.2 Å or dihedral-angle violations of >5°. Resonance assignments were described (11).
Residues 5—20, 28—48, 57—72, and 80—99
Backbone and side chains of 29 nonpolar residues
Ref. 30
http://kinemage.biochem.duke.edu and ref. 31
Results and Discussion
The first binary-patterned library was designed to encode a collection of 74-residue four-helix bundles (4). The resulting proteins were invariably α-helical, and several displayed features consistent with well ordered structures (17–19). However, most proteins in the original library formed fluctuating structures that did not maintain sufficient order for structure determination by x-ray crystallography or NMR. Because the 74-residue sequences in this original library were considerably shorter than those of natural four-helix bundles (which are almost always >100 residues), we surmised that the dynamic structures of the first-generation proteins might be caused by short helices, rather than by shortcomings in the binary-code strategy. Indeed, previous work with bundles of α-helices had shown a dramatic relationship between chain length and protein stability (20, 21). Therefore, we hypothesized that a true test of the potential of the binary-code strategy to encode well folded proteins required construction of a library of proteins with chain lengths similar to those found in natural four-helix bundles.
Recently, we constructed a second-generation binary-patterned library in which the α-helices were designed to be ≈50% longer (10). Each helix was extended from 14 residues to 21 residues to more closely match the lengths of helices in natural four-helix bundles. The overall length of the protein sequences was increased from 74 to 102 residues. The interhelical turns in the second generation sequences were designed to be relatively flexible by incorporating multiple glycines in each turn. Flexibility in the turns was desired to accommodate variations in how the four helices pack together in the different proteins in the library. The design of the second generation library was described in more detail by Wei et al. (10). Five proteins from the second-generation library were purified and characterized (10). These five proteins were chosen arbitrarily from the naïve library: They were not selected by high-throughput screening or directed evolution. All five proteins were α-helical and quite stable. Four of the five displayed thermodynamic properties and NMR spectra consistent with well ordered and/or native-like structures (10). One of these four proteins, S-824, was chosen for structure determination by NMR.
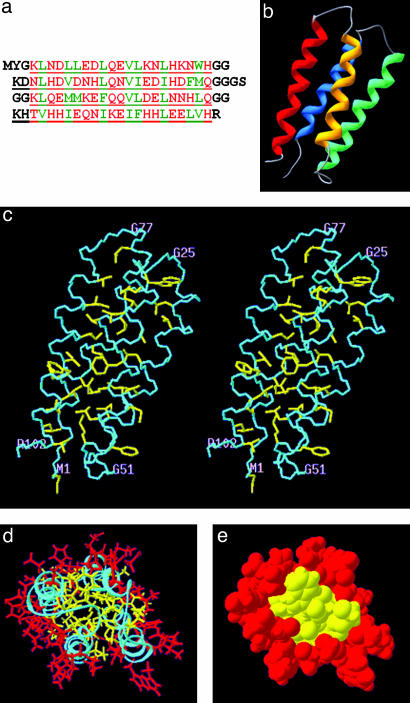
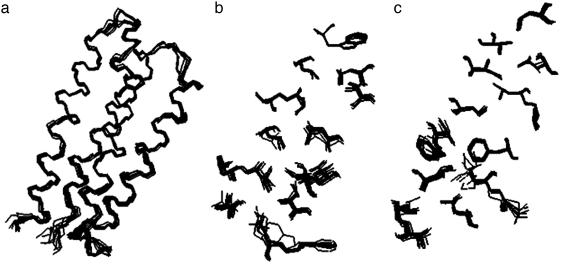
The overall structure of protein S-824 is an up-down-up-down four-helix bundle (Fig. 1). An overlay of the backbone and buried side chains shows that the 10 lowest energy structures are very similar to one another (Fig. 2). The rms deviation from the mean structure for all heavy atoms is 1.06 ± 0.10 Å. For backbone atoms in the α-helices, the rms deviation is 0.34 ± 0.05 Å. For the backbone and side-chain heavy atoms of the 29 nonpolar amino acids constituting the hydrophobic core, the rms deviation is 0.51 ± 0.09Å. These small deviations indicate that S-824 forms a very well defined structure similar to those of natural proteins (22).
Fig. 1.
(a) The amino acid sequence of protein S-824 shown in single-letter code. Polar and nonpolar residues of the binary-patterned α-helices are shown in red and green type, respectively. Turn residues (and the N and C termini) are shown in black. Residues that were encoded by degenerate DNA codons specifying combinatorial mixtures of amino acids are underlined. Of the 102 residues in this sequence, 88 were derived from combinatorially encoded mixtures of amino acids. (b) Solution structure of protein S-824 is an up-down-up-down four-helix bundle. Helix 1 is blue, and helix 4 is red. (c) Stereo view of the structure. Backbone and all nonpolar side chains are shown. (d) Head-on view with the polar and nonpolar side chains of the α-helices shown in red and yellow, respectively. (e) Same as d, but in space-filling representation.
Fig. 2.
Overlay of 10 lowest energy structures calculated from the NMR data. (a) Backbone. (b) Nonpolar side chains of α-helices 1 and 2. (c) Nonpolar side chains of α-helices 3 and 4.
Because the goal of the binary-code strategy is to produce proteins that fold into a chosen target structure, it is important to compare the experimentally determined structure with the structure expected from the design. Because binary patterning encodes enormous combinatorial diversity, individual side chains are not specified and detailed interactions are not designed a priori. Therefore, this comparison focuses on global structural features. The key predictions of the design were that the binary-patterned 102-residue sequences would (i) form four amphiphilic α-helices, (ii) fold into an antiparallel four-helix bundle, and (iii) bury nonpolar side chains in the interior and expose polar side chains on the surface (4, 10).
As shown in Fig. 1, the experimentally determined structure confirms these predictions. Four α-helices are formed. They span residues 5–20 (helix 1), 28–48 (helix 2), 56–72 (helix 3), and 80–99 (helix 4). These helices occur at sequence positions at or near their designed locations (10). The interhelical turns occur at the designed locations, although, in some cases, the turns are slightly longer than expected. Moreover, as specified by the binary-code strategy, the side chains in the experimentally determined structure are partitioned with nonpolar residues in the interior and polar residues on the surface (Fig. 1e).
Several features that could not have been predicted are also noteworthy. The overall topology of the bundle is left-turning (viewed from the outside, the chain turns left to traverse from α-helix 1 to α-helix 2). This topology was not consciously designed into the library, and it is notable that, although right-turning topologies occur somewhat more frequently in natural four-helix bundles (23), the left-turning topology is favored in this de novo protein. The angles between the helices were also not designed explicitly. In the experimentally determined structure, the angle between helices 1 and 4, and between helices 2 and 3 is ≈20°, similar to the angle in the “knobs-into-holes” packing of many natural α-helical proteins (24). In contrast, helices 1 and 2 and helices 3 and 4 are roughly antiparallel. Four-helix bundles in which some angles are 20° and others are more parallel (or antiparallel) also occur in nature (e.g., cytochrome b562) (25). Because the packing of side chains between neighboring helices influences interhelical angles, it will be interesting to see whether other structures from this combinatorial library pack with different angles. Finally, we note that the layering of buried side chains is not uniform throughout the structure. Near the top and bottom of the bundle, each layer is discreet, whereas the central layer appears as a large interdigitated “double layer.” This crowded central layer presumably is responsible for the kink near the center of helix 3. Because the compositions of the layers depend on the identities of the constituent side chains, we anticipate this level of structural detail will differ among the diverse sequences in the library.
As shown in Fig. 1 a, only 14 of the 102 positions in the sequence of S-824 were designed as specific amino acids. The other 88 positions (86% of the total) were encoded by degenerate DNA codons, allowing mixtures of amino acids. Consequently, the potential diversity encoded by this binary pattern is enormous. Among these diverse sequences, what fraction actually folds into α-helical structures? How many are well ordered and native-like? Finally, how many form four-helix bundles consistent with the design? Given the size of the potential library, we cannot fully answer these questions. Nonetheless, we can summarize what has been learned thus far from an initial sampling of sequences from this 102-residue library. Of the five proteins characterized to date, (i) all are α-helical and stable (10), (ii) only one resembles a molten globule and the other four appear well ordered (10), and, (iii) as shown in Figs. 1 and 2, the first structure determined from this library is indeed a four-helix bundle similar to that specified by the design. In considering whether these proteins are representative of the library as a whole, it is important to emphasize that these five proteins were chosen arbitrarily from a naïve library that had not been subjected (yet) to genetic selections or high-throughput screens. Therefore, we presume these stably folded protein structures are not “needles in a haystack.”
Creation of high-diversity libraries in fields ranging from pharmaceutical chemistry to materials science is increasingly being used as a first step toward the discovery of new functions. Latter steps usually involve screens or selections that facilitate isolation of rare “winners” from large collections of inactive candidates. Success in finding these winners depends on both the power of the screen (or selection) and the quality of the initial library. For proteins, several powerful methods have been developed to search for active molecules amid large collections of sequences (3, 26, 27). When these methods are applied to libraries of randomly generated sequences, functionally active proteins are found only very rarely (3). This scarcity is not surprising because randomly generated sequences will seldom yield well ordered structures (28, 29), and poorly folded de novo proteins are not likely to possess activities rivaling those of their natural counterparts. To enhance the likelihood of finding functional de novo proteins, it will be important to focus large libraries of novel sequences into the most productive regions of sequence space. As described here, the binary-code strategy can accomplish this goal by favoring the formation of well folded protein structures.
Acknowledgments
Y.W. and M.H.H. thank Istvan Pelczer for extensive advice and numerous helpful discussions on NMR. This work was supported by National Institutes of Health Grants R01-GM62869 (to M.H.H.) and R01-GM45302 (to J.B.).
Abbreviation: NOE, nuclear Overhauser effect.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1P68).
References
- 1.DeGrado, W. F., Summa, C. M., Pavone, V., Nastri, F. & Lombardi, A. (1999) Annu. Rev. Biochem. 68, 779-819. [DOI] [PubMed] [Google Scholar]
- 2.Dahiyat, B. I. & Mayo, S. L. (1997) Science 278, 82-87. [DOI] [PubMed] [Google Scholar]
- 3.Keefe, A. D. & Szostak, J. W. (2001) Nature 410, 715-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamtekar, S., Schiffer, J. M., Xiong, H., Babik, J. M. & Hecht, M. H. (1993) Science 262, 1680-1685. [DOI] [PubMed] [Google Scholar]
- 5.West, M. W., Wang, W., Patterson, J., Mancias, J. D., Beasley, J. R. & Hecht, M. H. (1999) Proc. Natl. Acad. Sci. USA 96, 11211-11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang, W. & Hecht, M. H. (2002) Proc. Natl. Acad. Sci. USA 99, 2760-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rojas, N. R. L., Kamtekar, S., Simons, C. T., Mclean, J. E., Vogel, K. M., Spiro, T. G., Farid, R. S. & Hecht, M. H. (1997) Protein Sci. 6, 2512-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moffet, D. A., Certain, L. K., Smith, A. J., Kessel, A. J., Beckwith, K. A. & Hecht, M. H. (2000) J. Am. Chem. Soc. 122, 7612-7613. [Google Scholar]
- 9.Brown, C. L., Aksay, I. A., Saville, D. A. & Hecht, M. H. (2002) J. Am. Chem. Soc. 124, 6846-6848. [DOI] [PubMed] [Google Scholar]
- 10.Wei, Y., Liu, T., Sazinsky, S. L., Moffet, D. A., Pelczer, I. & Hecht, M. H. (2003) Protein Sci. 12, 92-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei, Y., Fela, D., Kim, S., Hecht, M. H. & Baum, J. (2003) J. Biomol. NMR 27, 395-396. [DOI] [PubMed] [Google Scholar]
- 12.Wishart, D. S., Bigam, C. G., Yao, J., Abildgaard, F., Dyson, H. J., Oldfield, E., Markley, J. L. & Sykes, B. D. (1995) J. Biomol. NMR 6, 135-140. [DOI] [PubMed] [Google Scholar]
- 13.Wuthrich, K. (1986) NMR of Proteins and Nucleic Acids (Wiley, New York).
- 14.Geerten, V. W. & Bax, A. (1993) J. Am. Chem. Soc. 115, 7772-7777. [Google Scholar]
- 15.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 16.Stein, E. G., Rice, L. M. & Brunger, A. T. (1997) J. Magn. Reson. 124, 154-164. [DOI] [PubMed] [Google Scholar]
- 17.Roy, S., Ratnaswamy, G., Boice, J. A., Fairman, R., McLendon, G. & Hecht, M. H. (1997) J. Am. Chem. Soc. 119, 5302-5306. [Google Scholar]
- 18.Rosenbaum, D. M., Roy, S. & Hecht, M. H. (1999) J. Am. Chem. Soc. 121, 9509-9513. [Google Scholar]
- 19.Roy, S. & Hecht, M. H. (2000) Biochemistry 39, 4603-4607. [DOI] [PubMed] [Google Scholar]
- 20.Fairman, R., Chao, H., Mueller, L., Lavoie, T. B., Shen, L., Novotny, J. & Matsueda, G. R. (1995) Protein Sci. 4, 1457-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Litowski, J. R. & Hodges, R. S. (2001) J. Peptide Res. 58, 477-492. [DOI] [PubMed] [Google Scholar]
- 22.Güntert, P. (1998) Q. Rev. Biophys. 31, 145-237. [DOI] [PubMed] [Google Scholar]
- 23.Richardson, J. S. & Richardson, D. C. (1989) in Prediction of Protein Structure and Principles of Protein Conformation, ed. Fasman, G. (Plenum, New York), pp. 1-98.
- 24.Crick, F. H. C. (1953) Acta Crystallogr. 6, 689-697. [Google Scholar]
- 25.Lederer, F., Glatigny, A., Bethge, P. H., Bellamy, H. D. & Mathews, F. S. (1981) J. Mol. Biol. 148, 427-448. [DOI] [PubMed] [Google Scholar]
- 26.Smith, G. P. (1985) Science 228, 1315-1317. [DOI] [PubMed] [Google Scholar]
- 27.Hanes, J. & Plückthun, A. (1997) Proc. Natl. Acad. Sci. USA 94, 4937-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davidson, A. R., Lumb, K. J. & Sauer, R. T. (1995) Nat. Struct. Biol. 2, 856-864. [DOI] [PubMed] [Google Scholar]
- 29.Mandecki, W. (1990) Protein Eng. 3, 221-226. [DOI] [PubMed] [Google Scholar]
- 30.Laskowski, R. A., Macarthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Chrystallogr. 26, 283-291. [Google Scholar]
- 31.Lovell, S. C., Davis, I. W., Arendall, W. B., III, de Bakker, P. I. W., Word, J. M., Prisant, M. G., Richardson, J. S. & Richardson, D. C. (2003) Proteins Struct. Funct. Genet. 50, 437-450. [DOI] [PubMed] [Google Scholar]