Abstract
Calmodulin (CaM) is a second messenger protein that has evolved to bind tightly to a variety of targets and, as such, exhibits low binding specificity. We redesigned CaM by using a computational protein design algorithm to improve its binding specificity for one of its targets, smooth muscle myosin light chain kinase (smMLCK). Residues in or near the CaM/smMLCK binding interface were optimized; CaM interactions with alternative targets were not directly considered in the optimization. The predicted CaM sequences were constructed and tested for binding to a set of eight targets including smMLCK. The best CaM variant, obtained from a calculation that emphasized intermolecular interactions, showed up to a 155-fold increase in binding specificity. The increase in binding specificity was not due to improved binding to smMLCK, but due to decreased binding to the alternative targets. This finding is consistent with the fact that the sequence of wild-type CaM is nearly optimal for interactions with numerous targets.
Identifying the interactions responsible for conveying binding specificity in proteins is critical to understanding protein function and is a prerequisite for the design of novel protein receptors and ligands. Here, we explore the basis of binding specificity in calmodulin (CaM). CaM is a ubiquitous Ca2+-binding protein that binds to and regulates a variety of proteins (1–5). Numerous biochemical studies on CaM (6–9), as well as several x-ray and NMR structures of CaM-target complexes (10–17), have revealed many aspects of the target recognition mechanism in this molecule. CaM usually interacts with proteins by binding to stretches of 14- to 30-aa residues capable of forming amphipathic α-helices. Although specific interactions between CaM and its targets differ depending on the target identity, the general topology of CaM-target complexes remains the same (Fig. 1). On binding, CaM embraces its targets with its C- and N-terminal globular domains, inducing a coil–to–helix transition in the target (18).
Fig. 1.
X-ray structure of CaM in complex with smMLCK (PDB code 1CDM) generated with molmol (40). Ca2+ atoms are shown as yellow spheres. The CaM surface and boundary residues selected for optimization are shown in red. The core residues in the CaM-binding interface, shown in cyan, and the smMLCK residues, shown in light yellow, were allowed to change conformation during the optimization procedure. The identities of the CaM core residues were fixed to those of CaMcore, a core-optimized CaM variant.
CaM has evolved to bind to multiple targets with equally high affinity and, as such, exhibits low binding specificity. To explore the nature of the interactions conferring (the lack of) binding specificity in CaM, we redesigned CaM so it would favor binding to one of its natural targets, smooth muscle myosin light chain kinase (smMLCK). Starting from the crystal structure of the CaM–smMLCK complex (11), we optimized the CaM-binding interface by using ORBIT (optimization of rotamers by iterative techniques), a program that has been successfully applied to protein design and stabilization (19–23). In previous work, we demonstrated that the binding specificity of CaM could be improved by redesigning a number of core positions in the CaM-binding interface (24). The designed 8-fold CaM variant, CaMcore (designated CaM_8 in previous work), exhibited binding affinity for the desired target that was similar to wild type and showed an increase in binding specificity of up to 120-fold.
In this study, we sought to increase CaM-binding specificity further by extending the optimization to boundary and surface positions in the CaM–smMLCK-binding interface. These positions, occupied mostly by glutamates, are likely to play a major role in the CaM target recognition mechanism because of the positively charged nature of the target sequences. Initially, we used computational protein design methods that were developed for stabilization of single proteins (25). However, when these methods yielded poor results, we refined the computational procedure for optimization of protein–target complexes by exploring the use of a modified energy function as well as different optimization parameters. Finally, we discuss the importance of including negative design when optimizing protein interfaces for improved binding specificity.
Materials and Methods
Computational Methods. All residues in the CaM–smMLCK crystal structure (11) were divided into core, boundary, and surface classes as described (19). Eight CaM surface and boundary positions (positions 14, 83, 87, 114, 116, 120, 123, and 127) that are within 4 Å of smMLCK were selected for optimization. The following amino acids were allowed at each of the optimized positions: Ala, Val, Leu, Ile, Met, Phe, Tyr, Trp, Ser, Thr, Asp, Asn, His, Glu, Gln, Lys, and Arg. The identities of the core residues in the CaM–smMLCK interface were set to those of a previously designed CaM variant, CaMcore (designated CaM_8 in previous work; ref. 24). The side chain conformations of the core residues, as well as those of the smMLCK residues, were allowed to change during the optimization. The designed sequences were obtained by using either previously described procedures (24) (CaMcore and CaMboundary) or by using the variations described in the main text ( and
and  ). Rotamer libraries used for the CaM optimizations were based on the backbone dependent library of Dunbrack and Karplus (26). A potential energy function that included terms for van der Waals, electrostatic, and hydrogen bonding interactions, and surface area-based solvation was used to calculate side chain/side chain and side chain/backbone pairwise interactions as described (19, 25, 27). The calculated energies served as input to a side-chain selection procedure that used either the Dead-End Elimination theorem (28, 29) (CaMcore and CaMboundary) or a Monte Carlo algorithm (30, 31) (
). Rotamer libraries used for the CaM optimizations were based on the backbone dependent library of Dunbrack and Karplus (26). A potential energy function that included terms for van der Waals, electrostatic, and hydrogen bonding interactions, and surface area-based solvation was used to calculate side chain/side chain and side chain/backbone pairwise interactions as described (19, 25, 27). The calculated energies served as input to a side-chain selection procedure that used either the Dead-End Elimination theorem (28, 29) (CaMcore and CaMboundary) or a Monte Carlo algorithm (30, 31) ( and
and 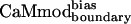 ). All CaM optimizations were performed by using SGI R10000 processors (Silicon Graphics, Mountain View, CA) running at 195 MHz.
). All CaM optimizations were performed by using SGI R10000 processors (Silicon Graphics, Mountain View, CA) running at 195 MHz.
Protein Expression. CaM variant genes were constructed by using a recursive PCR procedure (32) with six primers. Wild-type and mutant CaM genes were cloned into pET-11 (Novagen) and expressed by isopropyl β-d-thiogalactoside (IPTG) induction in BL21(DE3) hosts (Invitrogen). Proteins were purified by reverse-phase HPLC with a water/acetonitrile gradient containing 0.1% trifluoroacetic acid, lyophilized, and stored at -20°C. Protein masses were verified by electrospray mass spectroscopy.
Peptide Synthesis. Seven peptides, corresponding to CaM-binding domains in target proteins, were selected to assess binding to the CaM variants: smMLCK (-ARRKWQKTGHAVRAIGRLSS-); skMLCK (-KRRWKKNFIAVSAANRFKKISSSGA-); melittin (-QQRKRKIWSILAPLGTTLVKLVAGIG-); spectrin (-KTASPWKSARLMVHTVATFNSIKE-); CaMKI (-AKSKWKQAFNATAVVRHMRKLQ-); CaMKII (-LKKFNARRKLKGAILTTMLATRNFS-); and CaMKK (-RFPNGFRKRHGMAKVLILTDLRPIRRV-). The eighth target, peptide 1 (-LKWKKLLKLLKKLLKLG-), was designed to bind CaM (33). The peptide targets were synthesized and purified as described (24).
Binding Experiments. Binding of the Trp-containing CaM targets (smMLCK, skMLCK, melittin, peptide 1, CaMKI, and spectrin) was measured by fluorescence spectroscopy, and binding of fluorescently silent targets (CaMKII and CaMKK) to CaM was assessed by circular dichroism spectroscopy as described (24).
Results and Discussion
Boundary and Surface Optimization with Standard Methods. Eight boundary and surface positions within 4 Å of the smMLCK peptide in the CaM–smMLCK structure (11) were selected for the optimization (Fig. 1). In addition, the 24 CaM core residues in the CaM-binding interface were allowed to change conformation, but their amino acid identities were fixed to those of CaMcore, a previously optimized CaM core variant that showed improved binding specificity for smMLCK (24). All of the smMLCK residues were allowed to change conformation. For the first CaM boundary and surface optimization, previously published computational protein design methods were used. The optimization produced CaMboundary, a protein with 6 boundary and surface substitutions in addition to the 8 mutations of CaMcore, for a total of 14 mutations with respect to wild-type CaM (CaMwt) (Table 1). CaMboundary was expressed, purified, and tested for binding to smMLCK by using fluorescence spectroscopy as described (24). The binding affinity (Kd) was determined to be 114 ± 80 nM. This value is substantially worse than the binding affinities observed for CaMwt and CaMcore (1.8 ± 1.3 and 1.3 ± 0.9 nM, respectively).
Table 1. Binding of boundary- and surface-optimized CaM variants to smMLCK.
| Designed positions*
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11 | 12 | 14 | 55 | 76 | 83 | 84 | 87 | 91 | 109 | 114 | 116 | 120 | 123 | 127 | 145 | Kd, nM | |
| CaMwt | E | F | E | V | M | E | E | E | V | M | E | L | E | E | E | M | 1.8 ± 1.3 |
| CaMcore | L | Y | — | I | E | — | Y | — | I | L | — | — | — | — | — | I | 1.3 ± 0.9 |
| CaMboundary | L | Y | D | I | E | R | Y | L | I | L | I | E | — | — | Q | I | 114 ± 83 |
| CaMmodboundaryno-bias | L | Y | — | I | E | R | Y | K | I | L | I | E | — | K | — | I | 6.1 ± 0.8 |
| CaMmodboundarybias | L | Y | — | I | E | R | Y | K | I | L | I | E | — | Q | — | I | 2.3 ± 0.7 |
Eight boundary and surface positions in the CaM—smMLCK binding interface were optimized. Dashes indicate sequence identity to CaMwt. Eight core mutations, shown in bold, were carried over from CaMcore
To determine the source of CaMboundary's decreased affinity for smMLCK, six additional CaM variants were constructed, each with a single boundary or surface substitution from the CaMboundary design in addition to the eight core mutations of CaMcore. Titrations of these variants with smMLCK revealed that the decrease in CaMboundary's affinity to smMLCK was largely due to a single mutation, E87L. This mutation produced a CaMcore variant, CaMcoreE87L, with a binding affinity of 50 ± 8 nM. CaMcoreE127Q had a significant but less deleterious effect on binding, giving a binding affinity of 8.2 ± 2.8 nM. The remaining four CaMcore mutants (CaMcoreE14D, CaMcoreE83R, CaMcoreE114I, and CaMcoreL116E) showed no decrease in binding to smMLCK compared with CaMwt and CaMcore.
Analysis of the Standard Computational Methods. The failure of CaMboundary to retain tight binding to smMLCK highlighted the inability of the basic design methods to predict favorable intermolecular interactions. This result was not surprising because the methods were developed for stabilization of isolated proteins rather than protein–protein interfaces. A similar observation was recently made for the use of protein folding-optimized potential functions for describing protein–protein interfaces. These potentials work well when applied to hydrophobic binding interfaces (equivalent to the CaMcore design) but fail for hydrophilic interfaces (equivalent to the CaMboundary design) (34). Examination of the interactions predicted in the CaMboundary–smMLCK complex revealed several possible problems with the computational methods.
In the CaM optimization, all of the molecular interactions were treated equally whether they were within CaM or between CaM and smMLCK. Such an approach is likely to produce CaM sequences with improved intramolecular interactions at the expense of eliminating favorable CaM–smMLCK intermolecular interactions, which are clearly important for binding. To address this problem, we modified the existing energy function, biasing it toward selection of more favorable intermolecular interactions. A parameter β was introduced into the pairwise portion of the energy function to enhance the intermolecular side chain/side chain interactions and to attenuate the intramolecular side chain/side chain interactions:
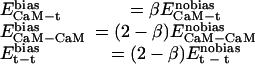 |
[1] |
Here, 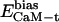 ,
, 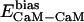 , and
, and 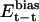 denote the modified energy functions for the interactions between CaM and the target, within CaM, and within the target, respectively.
denote the modified energy functions for the interactions between CaM and the target, within CaM, and within the target, respectively. 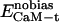 describes the unbiased intermolecular interactions between CaM and the target;
describes the unbiased intermolecular interactions between CaM and the target; 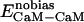 and
and 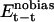 describe the unbiased intramolecular interactions within CaM and the target, respectively. The optimal value of the parameter β was estimated to be in the 1.2–1.6 range, which was determined by performing a number of CaM–smMLCK optimizations and by visually inspecting the computational results.
describe the unbiased intramolecular interactions within CaM and the target, respectively. The optimal value of the parameter β was estimated to be in the 1.2–1.6 range, which was determined by performing a number of CaM–smMLCK optimizations and by visually inspecting the computational results.
An additional concern was the large distance-dependent dielectric constant (ε = 40r) used in the calculation. Although historically satisfactory, the use of a large dielectric constant effectively underemphasized the long-range electrostatics term in the energy function relative to more local terms such as van der Waals and hydrogen bonding interactions. Electrostatic interactions, however, are likely to play a key role in the CaM target recognition mechanism. Their importance is supported by the presence of many negatively charged residues in the CaM-binding interface and the positively charged nature of all known CaM target sequences. Hence, lowering the dielectric constant in the CaM boundary- and surface-optimization might help to restore high binding affinity to smMLCK for the variants generated. A dielectric constant of 4r was selected because this value was found to be optimal for the design of β-sheet surface residues (35) and it is in the range of values generally used for calculations of protein binding free energies (36–38).
The small size of the rotamer library used in the calculation is also a concern. To reduce computational time, we selected a rotamer library similar to the one used in previous protein design calculations. In this library, no expansion about the χ1 and χ2 side chain dihedral angles was performed for polar residues, resulting in a relatively small number of rotamers for the polar amino acids. However, polar residues are thought to be especially important for CaM target binding. The absence of appropriate side chain rotamers could have resulted in selection of CaM sequences with suboptimal interactions with smMLCK. To reduce this concern, a rotamer library that contained rotamers representing expansion about both χ1 and χ2 for all amino acids was used as an option in subsequent calculations.
Redesign of the CaM–smMLCK-Binding Interface Using Modified Methods. Six additional CaM optimizations were performed with and without biasing of the energy function, by using dielectric constants of 4r or 40r, and the standard or large rotamer libraries (see Table 4, which is published as supporting information on the PNAS web site). Starting from the minimum-energy solution obtained for the CaM core optimization, CaMcore, a Monte Carlo search algorithm (30, 31) was used to optimize the eight boundary and surface residues in the CaM–smMLCK-binding interface. A Monte Carlo search algorithm was used (instead of the dead-end elimination method) to rapidly explore different computational protocols. Computationally predicted CaM variants were then constructed, and their binding affinities to smMLCK were determined.
Two of six tested variants showed good binding to smMLCK. Both variants were predicted by using a dielectric constant of 4r and the large rotamer library. The first variant, 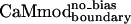 , was obtained without introducing biasing into the energy function; the second variant,
, was obtained without introducing biasing into the energy function; the second variant,  , was obtained by using the biased energy function (Table 1).
, was obtained by using the biased energy function (Table 1). 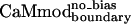 bound to smMLCK almost as well as CaMwt and CaMcore, showing a Kd value of 6.1 ± 0.8 nM (Table 1).
bound to smMLCK almost as well as CaMwt and CaMcore, showing a Kd value of 6.1 ± 0.8 nM (Table 1). 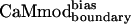 performed even better, exhibiting a binding affinity of 2.3 ± 0.7 nM, similar to that of CaMwt. Analysis of the computational methods and binding data for all of the variants (see Table 4) suggests that reducing the dielectric constant to 4r and increasing the rotamer library size have a roughly equal effect on improving binding affinity, with each modification improving binding by about a factor of 4.5. Biasing the energy function to emphasize intermolecular interactions provides an additional factor of 2.7 in binding affinity. Combining the three modifications gives the maximal improvement.
performed even better, exhibiting a binding affinity of 2.3 ± 0.7 nM, similar to that of CaMwt. Analysis of the computational methods and binding data for all of the variants (see Table 4) suggests that reducing the dielectric constant to 4r and increasing the rotamer library size have a roughly equal effect on improving binding affinity, with each modification improving binding by about a factor of 4.5. Biasing the energy function to emphasize intermolecular interactions provides an additional factor of 2.7 in binding affinity. Combining the three modifications gives the maximal improvement. 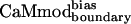 was selected for further analysis.
was selected for further analysis.
Binding Specificity of 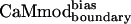 . To determine the binding specificity of
. To determine the binding specificity of 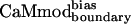 , we assessed its binding to a set of seven alternative targets; these targets were not included in the model used for the CaM optimization calculations. Binding of
, we assessed its binding to a set of seven alternative targets; these targets were not included in the model used for the CaM optimization calculations. Binding of 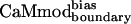 to the Trp-containing targets (skMLCK, spectrin, melittin, peptide 1, and CaMKI) was assessed by fluorescence spectroscopy; binding of
to the Trp-containing targets (skMLCK, spectrin, melittin, peptide 1, and CaMKI) was assessed by fluorescence spectroscopy; binding of 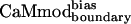 to the fluorescently silent targets (CaMKII and CaMKK) was monitored by circular dichroism spectroscopy. Fig. 2 A and B shows the titration curves for
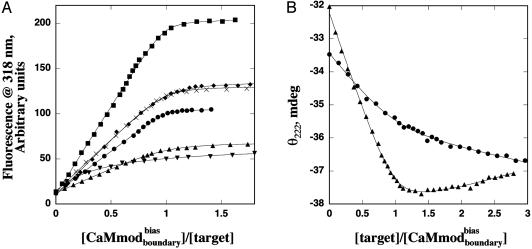
to the fluorescently silent targets (CaMKII and CaMKK) was monitored by circular dichroism spectroscopy. Fig. 2 A and B shows the titration curves for  binding to the selected targets. CaMwt binds to seven of eight targets equally well, with dissociation constants in the low nM range; weaker binding (Kd of 28 ± 5 nM) is observed for melittin (Table 2). Binding of
binding to the selected targets. CaMwt binds to seven of eight targets equally well, with dissociation constants in the low nM range; weaker binding (Kd of 28 ± 5 nM) is observed for melittin (Table 2). Binding of 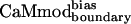 to all of the alternative targets except melittin was reduced with respect to CaMwt. Melittin exhibited similar binding affinities for
to all of the alternative targets except melittin was reduced with respect to CaMwt. Melittin exhibited similar binding affinities for 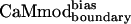 and CaMwt. Furthermore, binding of
and CaMwt. Furthermore, binding of 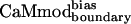 to four of seven of the alternative targets was lowered in comparison to CaMcore. When compared with CaMcore,
to four of seven of the alternative targets was lowered in comparison to CaMcore. When compared with CaMcore, 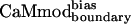 showed similar binding to spectrin, melittin, and CaMKII, and reduced binding to skMLCK, peptide 1, CaMKI, and CaMKK. The greatest loss (200-fold) in binding affinity to
showed similar binding to spectrin, melittin, and CaMKII, and reduced binding to skMLCK, peptide 1, CaMKI, and CaMKK. The greatest loss (200-fold) in binding affinity to 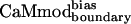 is observed for peptide 1, which contains a nonnatural target sequence that was engineered to bind CaM without considering specific CaM-target interactions (33). Particularly noteworthy is the ability of
is observed for peptide 1, which contains a nonnatural target sequence that was engineered to bind CaM without considering specific CaM-target interactions (33). Particularly noteworthy is the ability of 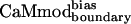 to discriminate against the skMLCK peptide. Unlike CaMcore, which showed little decrease in binding to skMLCK,
to discriminate against the skMLCK peptide. Unlike CaMcore, which showed little decrease in binding to skMLCK,  exhibits a 6-fold decrease in binding to this target when compared with CaMwt. The sequence of skMLCK is very similar to that of smMLCK, and, hence, these peptides would be expected to present very similar binding surfaces, making it difficult to discriminate between them. Overall,
exhibits a 6-fold decrease in binding to this target when compared with CaMwt. The sequence of skMLCK is very similar to that of smMLCK, and, hence, these peptides would be expected to present very similar binding surfaces, making it difficult to discriminate between them. Overall, 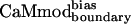 showed increased binding specificity compared with CaMwt and CaMcore.
showed increased binding specificity compared with CaMwt and CaMcore. 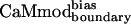 retained the tight binding affinity of CaMwt for the desired target and exhibited a up to 200-fold decrease in binding to the alternative targets (Table 2).
retained the tight binding affinity of CaMwt for the desired target and exhibited a up to 200-fold decrease in binding to the alternative targets (Table 2).
Fig. 2.
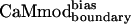 binding to target peptides. (A) Titration curves corresponding to binding of the Trp containing targets to
binding to target peptides. (A) Titration curves corresponding to binding of the Trp containing targets to 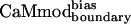 monitored by fluorescence at 318 nm: smMLCK (•), skMLCK (♦), spectrin (×), peptide 1 (▾), melittin (▴), and CaMKI (▾). (B) Titration curves corresponding to binding of
monitored by fluorescence at 318 nm: smMLCK (•), skMLCK (♦), spectrin (×), peptide 1 (▾), melittin (▴), and CaMKI (▾). (B) Titration curves corresponding to binding of 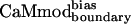 to the fluorescently silent targets monitored by circular dichroism: CaMKK (•) and CaMKII (▴).
to the fluorescently silent targets monitored by circular dichroism: CaMKK (•) and CaMKII (▴).
Table 2. Binding affinities (Kd, nM) of WT and redesigned CaM to selected targets.
| smMLCK | skMLCK | Spectrin | Melittin | Peptide1 | CaMKI | CaMKII | CaMKK | |
|---|---|---|---|---|---|---|---|---|
| CaMwt | 1.8 ± 1.3 | 3.3 ± 0.8 | 3.3 ± 1.5 | 28 ± 5 | 1.7 ± 0.8 | 1.7 ± 0.7 | 5.1 ± 1.5 | 1.0 ± 3.0 |
| CaMcore | 1.3 ± 0.9 | 4.9 ± 1.2 | 16 ± 6.0 | 54 ± 18 | 147 ± 48 | 1.3 ± 0.8 | 54 ± 20 | 32 ± 13 |
| Decrease in binding* | 0.72 | 1.5 | 4.8 | 1.9 | 86 | 0.76 | 11 | 32 |
| Specificity increase† | 1.0 | 2.1 | 6.7 | 2.6 | 120 | 1.1 | 15 | 44 |
| CaMmodboundarybias | 2.3 ± 0.7 | 21 ± 3 | 11 ± 2 | 33 ± 6 | 343 ± 66 | 7 ± 2 | 27 ± 6 | 67 ± 28 |
| Decrease in binding* | 1.3 | 6.4 | 3.3 | 1.2 | 202 | 4.1 | 5.3 | 67 |
| Specificity increase† | 1.0 | 4.9 | 2.6 | 0.9 | 155 | 3.2 | 4.1 | 51 |
Decrease in binding was calculated by taking the ratio of binding affinities of a CaM variant and CaMwt for a particular target
Specificity increase was calculated by taking the ratio of binding affinity of a CaM variant and CaMwt for a particular target and dividing it by the same ratio for smMLCK
Analysis of the Interactions Predicted in the Redesigned CaM–smMLCK Complexes. Six mutations that distinguish CaMboundary from CaMcore produced almost a 100-fold decrease in binding to the desired target, smMLCK (Table 1). Among these mutations, only two had a substantial effect on binding. The E87L mutation was identified as causing the majority of the decrease in binding affinity. This result was unexpected because Glu-87 does not interact with the target in the CaMwt–smMLCK complex. Rather, it forms an intramolecular salt bridge to Arg-90, the disruption of which in 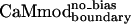 and
and 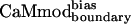 does not diminish binding. It is possible that the E87L mutation reduced binding by decreasing the overall stability of CaMboundary. The 4-fold drop in binding to smMLCK caused by E127Q in the CaMboundary background was consistent with the disruption of an intermolecular salt bridge to an Arg on smMLCK.
does not diminish binding. It is possible that the E87L mutation reduced binding by decreasing the overall stability of CaMboundary. The 4-fold drop in binding to smMLCK caused by E127Q in the CaMboundary background was consistent with the disruption of an intermolecular salt bridge to an Arg on smMLCK.
Unlike CaMboundary, both molecules predicted by the modified methods, 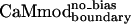 and
and 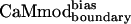 , retained tight binding to smMCLK. These results support the use of a stronger electrostatic potential and a larger rotamer library in the CaM–smMLCK optimization. Introducing bias into the energy function to obtain
, retained tight binding to smMCLK. These results support the use of a stronger electrostatic potential and a larger rotamer library in the CaM–smMLCK optimization. Introducing bias into the energy function to obtain 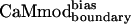 resulted in a 3-fold improvement in Kd relative to
resulted in a 3-fold improvement in Kd relative to 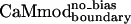 (Table 1). At the sequence level,
(Table 1). At the sequence level,  and
and 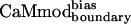 differ only at position 123, where a wild-type glutamate is substituted by a lysine in
differ only at position 123, where a wild-type glutamate is substituted by a lysine in 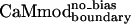 and a glutamine in
and a glutamine in 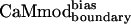 . Lys-123 in
. Lys-123 in 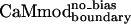 could reduce binding to smMLCK by forming favorable intramolecular electrostatic interactions with neighboring glutamates (Glu-120 or Glu-127), at the expense of forming unfavorable intermolecular electrostatic interactions with the positively charged target peptide.
could reduce binding to smMLCK by forming favorable intramolecular electrostatic interactions with neighboring glutamates (Glu-120 or Glu-127), at the expense of forming unfavorable intermolecular electrostatic interactions with the positively charged target peptide.
These results support previous findings on the importance of glutamates for the target recognition mechanism of CaM (11, 12, 17). CaMwt is able to bind tightly to a variety of targets by using a subset of available glutamates in salt-bridge interactions. Removal of a single intermolecular salt bridge between CaM and smMLCK produces a 3- to 4-fold reduction in CaM's binding to the target.
Positive vs. Negative Design in Improving CaM-Binding Specificity. In the present work, we were able to increase the binding specificity of CaM by optimizing its interactions with the desired target, smMLCK. However, the increase in binding specificity was not due to CaM's improved binding affinity to smMLCK but due to its decreased binding affinity to the alternative targets. Considering that the alternative targets were not included in the optimization procedure, the results seem counterintuitive.
CaM has evolved to bind tightly to a large number of targets. The fitness of CaMwt is nearly optimal with respect to its sequence for all CaM targets. Fig. 3 illustrates the change in fitness of CaM for interactions with its targets during the computational optimization procedure. The two surfaces represent the fitness of CaM interacting with the desired target, smMLCK, and with alternative targets. The computational optimization of the CaM–smMLCK complex results in sequence changes that improve or maintain CaM's ability to bind smMLCK (Fig. 3). Because no information from the alternative targets is included in the optimization procedure, the sequence changes are essentially random with respect to interactions with the latter targets. Given that the fitness of CaMwt is near optimal for each CaM-target complex, the probability of increasing fitness by a random sequence change is very low. Hence, without directly including negative design in the calculations, we expect to obtain CaM sequences that on average show decreased binding affinities to the alternative targets, and thus increased binding specificity for the desired target. Energies calculated for CaMwt, CaMcore, and 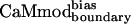 in the context of five available CaM-target complex structures (10–12, 15, 17) support this argument (Table 3). As expected, the energy of the CaM–smMLCK complex is decreased (improved) by changing the sequence from CaMwt to CaMcore to
in the context of five available CaM-target complex structures (10–12, 15, 17) support this argument (Table 3). As expected, the energy of the CaM–smMLCK complex is decreased (improved) by changing the sequence from CaMwt to CaMcore to 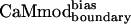 . For the alternative targets, the opposite trend is evident: the energies of the CaM–target complexes are most favorable for the CaMwt sequence, less favorable for the CaMcore sequence, and least favorable for the
. For the alternative targets, the opposite trend is evident: the energies of the CaM–target complexes are most favorable for the CaMwt sequence, less favorable for the CaMcore sequence, and least favorable for the 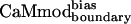 sequence.
sequence.
Fig. 3.
Fitness of CaM interacting with the desired target, smMLCK (red), and with alternative targets (blue). The CaMwt sequence lies near the respective maxima for both the desired target and the alternative targets as indicated by *. Arrows show the change in fitness due to mutations in the CaM sequence predicted by the optimization.
Table 3.
Energies of WT and redesigned CaM—target complexes*
| smMLCK | skMLCK | CaMKI | CaMKII† | CaMKK | |
|---|---|---|---|---|---|
| CaMwt | —579.4 | —510.5 | —704.8 | —612.0 | —748.4 |
| CaMcore | —621.3 | —494.3 | >1,000‡ | —478.2 | —731.7 |
| CaMmodboundarybias | —649.4 | —481.8 | >1,000‡ | —472.5 | —706.3 |
The energies of CaMwt and the redesigned CaM variants were calculated in context of the available structures for CaM—target complexes by using the set of positions shown in Table 1. All calculations were carried out by using no biasing of the energy function, the larger rotamer library, a dielectric constant of 4r, and the Dead-End Elimination theorem (28, 29)
CaM residues 74—83 are missing from the crystal structure of the CaM—CaMKII complex and, hence, were omitted from the calculations
The large energy is due to a van der Waals clash between Tyr-84 on CaM and Ala-154 on CaMKI, which results from the fixed backbone approximation used in the calculations
The further we move away from the CaMwt sequence, while improving or preserving the fitness of interaction with the desired target, the more specificity we achieve. This argument suggests that simultaneously optimizing a larger number of CaM positions would increase the chances of obtaining a protein with increased binding specificity. This finding is indeed confirmed by the experimental results: the binding specificity of 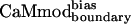 , which included additional boundary and surface positions, was greater than that of the core design, CaMcore. Expanding the number of optimization positions could increase CaM-binding specificity either through the cumulative effect of multiple, slightly improved interactions or through the effect of a small number of critical interactions. Although the proposed model (Fig. 3) accommodates both possibilities, the current data do not allow us to discriminate between these two extremes. It is likely, however, that both mechanisms operate to different extents for different targets.
, which included additional boundary and surface positions, was greater than that of the core design, CaMcore. Expanding the number of optimization positions could increase CaM-binding specificity either through the cumulative effect of multiple, slightly improved interactions or through the effect of a small number of critical interactions. Although the proposed model (Fig. 3) accommodates both possibilities, the current data do not allow us to discriminate between these two extremes. It is likely, however, that both mechanisms operate to different extents for different targets.
In recent work on binding specificity design in coiled coils, Havranek and Harbury (39) showed that both positive and negative design are necessary to achieve binding specificity for coiled-coil homodimers and heterodimers. The present work shows that, in the case of CaM, optimizing its interactions with only the desired target could be sufficient. Because CaM has a significantly more complex architecture compared with a coiled coil, it has a greater number of potential interactions to favor one target over another. Nevertheless, incorporating negative design in the optimization of the CaM–target interface is likely to improve the chances of obtaining increased binding specificity. Direct incorporation of negative design would ensure that the optimization procedure generates CaM variants with improved or maintained fitness for the interaction with the desired target, while simultaneously decreasing fitness for interactions with the alternative targets.
Supplementary Material
Acknowledgments
We thank K. Beckingham for providing a plasmid containing wild-type calmodulin, P. Huang for providing the program prpcr used for primer design, P. Shah for help with construction of some of the CaM mutants, and M. Ary for assistance with the manuscript. This work was supported by the Howard Hughes Medical Institute, the Ralph M. Parsons Foundation, an IBM Shared University research grant (to S.L.M.), a National Institutes of Health postdoctoral fellowship, and the Caltech Initiative in Computational Molecular Biology, awarded by the Burroughs Wellcome Fund (to J.M.S.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CaM, calmodulin; smMLCK, smooth muscle myosin light chain kinase.
References
- 1.Weinstein, H. & Mehler, E. L. (1994) Annu. Rev. Physiol. 56, 213-236. [DOI] [PubMed] [Google Scholar]
- 2.Crivici, A. & Ikura, M. (1995) Annu. Rev. Biophys. Biomol. Struct. 24, 85-116. [DOI] [PubMed] [Google Scholar]
- 3.Zielinski, R. E. (1998) Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 697-725. [DOI] [PubMed] [Google Scholar]
- 4.O'Neil, K. T. & DeGrado, W. F. (1990) Trends Biochem. Sci. 15, 59-64. [DOI] [PubMed] [Google Scholar]
- 5.Chin, D. & Means, A. R. (2000) Trends Cell Biol. 10, 322-328. [DOI] [PubMed] [Google Scholar]
- 6.Wintrode, P. L. & Privalov, P. L. (1997) J. Mol. Biol. 266, 1050-1062. [DOI] [PubMed] [Google Scholar]
- 7.Brokx, R. D., Lopez, M. M., Vogel, H. J. & Makhatadze, G. I. (2001) J. Biol. Chem. 276, 14083-14091. [DOI] [PubMed] [Google Scholar]
- 8.Gomes, A. V., Barnes, J. A. & Vogel, H. J. (2000) Arch. Biochem. Biophys. 379, 28-36. [DOI] [PubMed] [Google Scholar]
- 9.Moorthy, A. K., Gopal, B., Bhattacharya, S., Bhattacharya, A., Murthy, M. R. N. & Surolia, A. (1999) FEBS Lett. 461, 19-24. [DOI] [PubMed] [Google Scholar]
- 10.Ikura, M., Clore, G. M., Gronenborn, A. M., Guang, Z., Klee, C. B. & Bax, A. (1992) Science 256, 632-638. [DOI] [PubMed] [Google Scholar]
- 11.Meador, W. E., Means, A. R. & Quiocho, F. A. (1992) Science 257, 1251-1255. [DOI] [PubMed] [Google Scholar]
- 12.Meador, W. E., Means, A. R. & Quiocho, F. A. (1993) Science 262, 1718-1721. [DOI] [PubMed] [Google Scholar]
- 13.Elshorst, B., Hennig, M., Forsterling, H., Diener, A., Maurer, M., Schulte, P., Schwalbe, H., Griesinger, C., Krebs, J., Schmid, H., et al. (1999) Biochemistry 38, 12320-12332. [DOI] [PubMed] [Google Scholar]
- 14.Osawa, M., Swindells, M. B., Tanikawa, J., Tanaka, T., Mase, T., Furuya, T. & Ikura, M. (1998) J. Mol. Biol. 276, 165-176. [DOI] [PubMed] [Google Scholar]
- 15.Osawa, M., Tokumitsu, H., Swindells, M. B., Kurihara, H., Orita, M., Shibanuma, T., Furuya, T. & Ikura, M. (1999) Nat. Struct. Biol. 6, 819-824. [DOI] [PubMed] [Google Scholar]
- 16.Schumacher, M. A., Rivard, A. F., Bachinger, H. P. & Adelman, J. P. (2001) Nature 410, 1120-1124. [DOI] [PubMed] [Google Scholar]
- 17.Kurokawa, H., Osawa, M., Kurihara, H., Katayama, N., Tokumitsu, H., Swindells, M. B., Kainosho, M. & Ikura, M. (2001) J. Mol. Biol. 312, 59-68. [DOI] [PubMed] [Google Scholar]
- 18.Erickson-Viitanen, S. & DeGrado, W. F. (1987) Methods Enzymol. 139, 455-478. [DOI] [PubMed] [Google Scholar]
- 19.Dahiyat, B. I. & Mayo, S. L. (1997) Science 278, 82-87. [DOI] [PubMed] [Google Scholar]
- 20.Malakauskas, S. M. & Mayo, S. L. (1998) Nat. Struct. Biol. 5, 470-475. [DOI] [PubMed] [Google Scholar]
- 21.Shimaoka, M., Shifman, J. M., Jing, H., Takagi, J., Mayo, S. L. & Springer, T. A. (2000) Nat. Struct. Biol. 7, 674-678. [DOI] [PubMed] [Google Scholar]
- 22.Bolon, D. N. & Mayo, S. L. (2001) Proc. Natl. Acad. Sci. USA 98, 14274-14279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marshall, S. A. & Mayo, S. L. (2001) J. Mol. Biol. 305, 619-631. [DOI] [PubMed] [Google Scholar]
- 24.Shifman, J. M. & Mayo, S. L. (2002) J. Mol. Biol. 323, 417-423. [DOI] [PubMed] [Google Scholar]
- 25.Street, A. G. & Mayo, S. L. (1999) Struct. Fold. Des. 5, R105-R109. [DOI] [PubMed] [Google Scholar]
- 26.Dunbrack, R. L. & Karplus, M. (1993) J. Mol. Biol. 230, 543-574. [DOI] [PubMed] [Google Scholar]
- 27.Gordon, D. B., Marshall, S. A. & Mayo, S. L. (1999) Curr. Opin. Struct. Biol. 9, 509-513. [DOI] [PubMed] [Google Scholar]
- 28.Desmet, J., De Maeyer, M., Hazes, B. & Lasters, I. (1992) Nature 356, 539-542. [DOI] [PubMed] [Google Scholar]
- 29.Gordon, D. B., Hom, G. K., Mayo, S. L. & Pierce, N. A. (2003) J. Comp. Chem. 24, 232-243. [DOI] [PubMed] [Google Scholar]
- 30.Kirckpatrick, S., Gelatt, C. D. & Vecchi, M. P. (1983) Science 220, 671-680. [DOI] [PubMed] [Google Scholar]
- 31.Voight, C. A., Gordon, D. B. & Mayo, S. L. (2000) J. Mol. Biol. 299, 789-803. [DOI] [PubMed] [Google Scholar]
- 32.Prodromou, C. & Pearl, L. H. (1992) Protein Eng. 5, 827-829. [DOI] [PubMed] [Google Scholar]
- 33.O'Neil, K. T., Wolfe, H. R., Erickson-Viitanen, S. & DeGrado, W. F. (1987) Science 236, 1454-1456. [DOI] [PubMed] [Google Scholar]
- 34.Papoian, G. A., Ulander, J. & Wolynes, P. G. (2003) J. Am. Chem. Soc. 125, 9170-9178. [DOI] [PubMed] [Google Scholar]
- 35.Street, A. G., Datta, D., Gordon, D. B. & Mayo, S. L. (2000) Phys. Rev. Lett. 84, 5010-5013. [DOI] [PubMed] [Google Scholar]
- 36.Xu, D., Lin, S. L. & Nussinov, R. (1997) J. Mol. Biol. 265, 68-84. [DOI] [PubMed] [Google Scholar]
- 37.Hendsch, Z. S. & Tidor, B. (1999) Protein Sci. 8, 1381-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, L.-P. & Tidor, B. (2001) Protein Sci. 10, 362-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Havranek, J. J. & Harbury, P. B. (2003) Nat. Struct. Biol. 10, 45-52. [DOI] [PubMed] [Google Scholar]
- 40.Koradi, R., Billeter, M. & Wüthrich, K. (1996) J. Mol. Graphics 14, 51-55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.