Abstract
A new approach, the immuno-spin trapping assay, used a novel rabbit polyclonal anti-DMPO (5,5-dimethyl-1-pyrroline N-oxide) antiserum to detect protein radical-derived DMPO nitrone adducts in the hemoprotein Mycobacterium tuberculosis catalase-peroxidase (KatG). This work demonstrates that the formation of protein nitrone adducts is dependent on the concentrations of tert-BuOOH and DMPO as shown by Western blotting and an enzyme-linked immunosorbent assay (ELISA). We have also detected protein-protein cross-links formed during turnover of Mtb KatG driven by tert-butyl peroxide (tert-BuOOH) or enzymatic generation of hydrogen peroxide. DMPO inhibits this dimerization due to its ability to trap the amino acid radicals responsible for the cross-linkage. Chemical modifications by tyrosine and tryptophan blockage suggest that tyrosine residues are one site of formation of the dimers. The presence of the tuberculosis drug isoniazid (INH) also prevented cross-linking as a result of its competition for the protein radical. Protein-DMPO nitrone adducts were also generated by a continuous flux of hydrogen peroxide. These findings demonstrated that the protein-based radicals were formed not only during Mtb KatG turnover with alkyl peroxides but also in the presence of hydrogen peroxide. Furthermore, the formation of protein-DMPO nitrone adducts was accelerated in the presence of isoniazid.
Tuberculosis, a disease caused by the bacillus Mycobacterium tuberculosis, is responsible for more than 2 million deaths each year (1). Isoniazid (INH)1 is one of the oldest (2, 3) but still extensively used tuberculosis drugs, and is activated by the bifunctional heme protein catalase-peroxidase Mtb KatG (4, 5). It has been proposed that the activated form of the drug is an isonicotinoyl radical, which is able to react with β-nicotinamide adenine dinucleotide (NADH) to form an IN-NAD adduct molecule (6-8). The main role of this adduct is to inhibit the protein InhA (9), a reductase involved in biosynthesis of mycolic acids, one of the components of the mycobacterial cell wall (10). Despite extensive efforts aimed at elucidating the mechanism, the activated form of the drug and the mechanistic details of the reaction are still debated.
The activating protein M. tuberculosis KatG is a dimeric heme protein with a molecular mass of ∼160 kDa and belongs to the family of class I peroxidases (11) that have both catalase (H2O2 → H2O + ½O2) and peroxidase (2AH + H2O2 → 2A· + 2H2O) activities. It has also been proposed that Mtb KatG activates INH in a two-step peroxidatic cycle characteristic of class III peroxidases such as horseradish peroxidase (HRP) (12), and, very recently, a mechanism of INH binding and oxidation has been proposed (13).
The ESR spin trapping technique has been successfully applied to detect various types of radicals from small molecules (14-16) to protein radicals (17-19). For the reaction of isoniazid with peroxidases HRP and KatG, a number of radical species have been reported in the literature as intermediates (20-22). Additionally, from direct electron paramagnetic resonance spectroscopy, two types of protein radicals (tyrosyl and tryptophanyl) have been detected during turnover of Mtb KatG with organic peroxides (23, 24). Lately, two Compound I intermediates have been reported, the classical porphyrin π-cation radical (FeIV=O Por·+) and an exchange-coupled oxoferryl-Trp radical intermediate (FeIV=O Trp·+) (25). It has also been shown that the intermediates formed in KatG have different reactivities with isoniazid, but it is still unclear what role these radicals play and how they are related to the enzyme’s function. In general, besides participating in catalytic processes, protein radicals can lead to the formation of cross-links between two identical or nonidentical amino acid residues (26), and such covalent bond formation can be produced within the same or between different protein molecules (27). One of the most common kinds of cross-links that has been observed in heme proteins is Tyr-Tyr dimerization (28) and, in some cases, oligomerization (29).
In the past few years, we have used a new approach (30), immuno-spin trapping, to investigate formation of protein-centered free radicals. This technique uses a novel polyclonal antiserum against the nitrone spin trap 5,5-dimethyl-1-pyroline N-oxide (DMPO) (31). The immunoassay has many advantages compared to spin trapping ESR spectroscopy; among them, it requires much less material (micrograms of protein). Also, the stability of the final oxidation product, DMPO nitrone adduct (a nonradical species), is much higher than that of the paramagnetic DMPO-radical adducts (t1/2 of seconds to minutes) required for ESR, resulting in outstanding sensitivity. The antibody specifically and effectively detects protein radicals in their DMPO nitrone adduct form and has been successfully used in our laboratory to detect the presence of DMPO nitrone adducts covalently attached to protein molecules (16, 18, 19, 28, 32, 33).
In this study, we have combined the immuno-spin trapping technique with the complementary ESR spectroscopy to further investigate the radical formation picture. Western blot and ELISA techniques have shown generation of Mtb KatG radical-nitrone adducts formed in the reaction of the protein with tert-BuOOH or by a constant flux of H2O2. Chemical modifications of tyrosine and tryptophan residues provide evidence that the radicals located on the surface and involved in the dimerization process are most likely tyrosyl. For the first time, both immuno-spin trapping and ESR have been used to study the formation of protein-based radicals during activation of isoniazid by H2O2. Finally, we present evidence of the participation of the isoniazid-derived radical in the generation of more KatG-DMPO nitrone adducts than are formed by hydrogen peroxide turnover alone.
MATERIALS AND METHODS
Reagents
Potassium iodide, N-bromosuccinimide (NBS), 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB), isoniazid (INH), o-dianisidine, tert-butyl peroxide (tert-BuOOH), potassium cyanide (KCN), and glucose oxidase (from Aspergillus niger, low in catalase) were from Sigma. Glucose was from ICN Biomedicals, Inc. Solutions of isoniazid (recrystallized from methanol and stored at 4 °C) were freshly prepared for all experiments due to its relatively rapid decomposition in aqueous solution or buffer. DMPO (high purity, ≥99%) from Alexis Biochemicals (San Diego, CA) was purified twice under vacuum at room temperature and stored under an argon atmosphere at -80 °C before being used. DBNBS was synthesized according to ref 34. M. tuberculosis KatG was prepared from an overexpression system in Escherichia coli strain UM262 containing the M. tuberculosis katG gene in plasmid vector pKatII (a gift from S. Cole, Institut Pasteur, Paris, France). A purification procedure was performed in 20 mM potassium phosphate buffer (pH 7.2) as described previously (35, 36). The protein concentration was determined from the Soret maximum at 407 nm (ε = 100 mM-1 cm-1).
Enzyme-Linked Immunosorbent Assay (ELISA)
The rabbit anti-DMPO nitrone adduct polyclonal antiserum was obtained in our laboratory as previously described (30, 32). Mtb KatG radical-derived nitrone adducts were determined using standard ELISA plates (96-well, Greiner Labortechnik). Purified Mtb KatG [final concentration of 10 μM in Chelex-100-treated 100 mM phosphate buffer (pH 7.4) containing 100 μM DTPA] was reacted with tert-BuOOH in the presence of 100 mM DMPO. After incubation for 1 h at 37 °C, reactions were stopped by the addition of 2 mM potassium cyanide. One microliter of the reaction mixture (1.6 μg of protein) was added to each well containing 300 μL of coating buffer [100 mM sodium bicarbonate (pH 9.6)] and incubated for 90 min at 37 °C with mixing at 300 rpm. ELISA was then performed as previously described (16, 37). Data are means ± the standard deviation (SD) from three independent examinations using freshly prepared solutions of all reactants.
Protein Staining and Western Blots
After Mtb KatG, tert-BuOOH, and DMPO were reacted, samples (1.6 μg of protein per lane) were electrophoresed under reducing conditions through duplicate 4 to 12% BisTris NuPAGE acylamide gels (Invitrogen). The gels were used for Coomassie blue staining (Bio-Rad Laboratories, Hercules, CA) to visualize protein loading and for electrophoretic transfer onto a nitrocellulose membrane for Western blots. The membrane was blocked with a 4% cold-water fish gelatin solution in 100 mM bicarbonate buffer (pH 9.6) overnight at 4 °C. After being washed with wash buffer [Tris-buffered saline (pH 7.4) containing 0.2% cold-water fish gelatin and 0.05% Tween 20], the membrane was incubated for 60 min at room temperature with 1:5000 anti-DMPO rabbit serum dissolved in wash buffer, followed by four washes. The secondary antiserum, anti-rabbit IgG, conjugated to alkaline phosphatase (Pierce), was dissolved in wash buffer (1:5000), added to the membrane, and incubated for 60 min at room temperature. After another four washings to eliminate any excess of secondary antiserum, the membrane was treated with 250 μM CDP-Star (Roche) and 0.1 mg/mL Nitro-Block-II Enhancer (Tropix) in Tris-buffered saline (pH 9.6) for 5 min. The positive immunocomplexes were detected on CL-Xposure X-ray film (Pierce) and digitized using Adobe PhotoShop (Adobe Systems). Densitometric analysis was performed by using Kodak (Rochester, NY) 1D, version 4.0.
Enzyme Activity Assays
M. tuberculosis KatG peroxidase activity was determined by monitoring the color development of o-dianisidine (38, 39). The reaction mixture containing 0.1 mM o-dianisidine and 23 mM tert-BuOOH in 50 mM sodium acetate buffer (pH 5.5) was added to a quartz cuvette, and after the absorbance at 460 nm was measured (ε460 = 11.33 mM-1 cm-1), the Mtb KatG protein (final concentration of 0.01 μM) was added and the absorbance increase recorded.
Enzymatic Generation of H2O2
Hydrogen peroxide was generated enzymatically with a glucose (10 mM)/glucose oxidase (0.4-2 units/mL) system. The amount of generated H2O2 was determined using an optically isolated, multichannel, electrode-based free radical analyzer, APOLLO 4000 (World Precision Instruments, Inc., Sarasota, FL), with constant stirring of the solution in the chamber. A calibration curve was created by plotting changes in current (picoamperes) generated by addition of different amounts of hydrogen peroxide to the calibration solution. The calibration was carried out at 37 °C. Biphasic kinetic behavior was observed for all used concentrations of glucose oxidase. In the presence of 0.4, 0.8, and 2 units/mL glucose oxidase, the initial rates of H2O2 formation were calculated to be 20, 40, and 100 μM/min, respectively. The slow phase of peroxide release by the G/GOx system (8 μM/min of H2O2 production in the presence of 2 units/mL GOx) is due to the relatively high rate of turnover of the oxidase with respect to the oxygen solubility (∼250 μM in air-saturated solutions) and its diffusion, which nearly depletes oxygen even from a stirred solution.
Chemical Modification of Tyrosine, Tryptophan, and Cysteine Residues on Mtb KatG
Tyrosine residues were blocked by using Iodo-Beads Iodination Reagent (Pierce). The protein (1 mg/mL) was incubated with two N-chlorobenzenesulfonamide-immobilized beads in sodium iodide (200 mM final concentration, pH 7.4), for 15 min at room temperature in the dark. Iodination was stopped by removal of the beads and dialysis at 4 °C against a 1000-fold excess of PBS buffer with one change of buffer. Blocking of tryptophan residues was performed by mixing N-bromosuccinimide (NBS) in 50 mM sodium acetate buffer (pH 4.5) with KatG at an NBS:tryptophan molar ratio of 2:1 for 30 min at room temperature in the dark (40). The reaction was stopped by adding free tryptophan (0.8 mM final concentration), and then the mixture was dialyzed. Cysteine residues were modified by alkylation with 5,5′-dithiobis(2-nitrobenzoic acid). The mixture was incubated at a DTNB:cysteine molar ratio of 20:1 for 30 min, and the excess reagent was removed by dialysis. The concentration of recovered protein was determined using an extinction coefficient at 407 nm of 100 mM-1 cm-1.
Electron Spin Resonance (ESR) Spectroscopy
ESR spectra were recorded using a Bruker E500 ESR spectrometer equipped with an ER4122SHQ microwave cavity and operating at 9.78 GHz and a modulation frequency of 100 kHz. The ESR spin trapping experiments were performed by placing the samples in a 10 mm flat cell (250 μL final volume) immediately after peroxide addition, and the spectra were recorded within 1 min of the start of the reaction. The instrumental settings were as follows: field sweep, 80 G; microwave frequency, 9.78 GHz; microwave power, 20 mW; modulation amplitude, 0.4 G; conversion time, 164 ms; time constant, 164 ms; receiver gain, 2 × 104; and number of scans, 3. Computer simulation was performed using WinSim that is available to the public (http://epr.niehs.nih.gov) (41).
RESULTS
KatG Dimer Formation and Its Analysis by SDS-PAGE, ELISA, and Western Blot
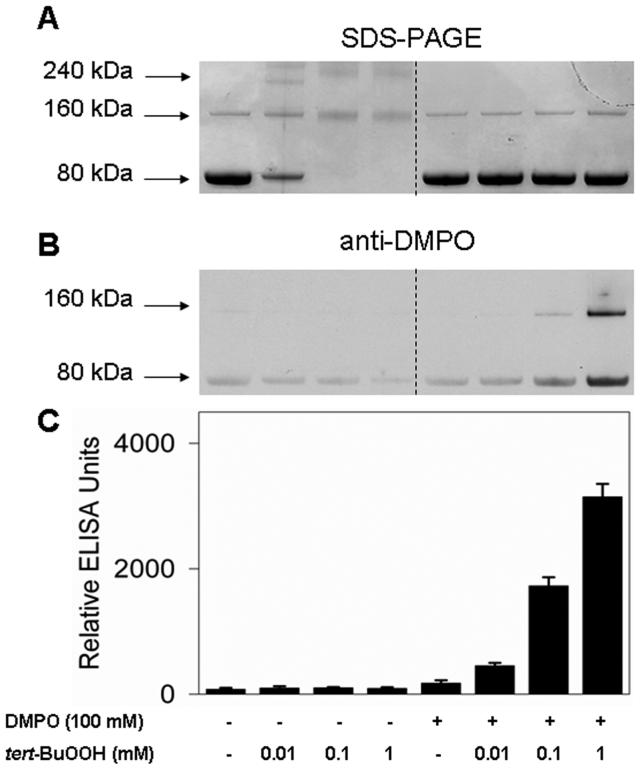
To demonstrate the formation of DMPO-KatG nitrone adducts by immuno-spin trapping, 10 μM Mtb KatG was incubated with tert-BuOOH in the presence or absence of DMPO. SDS-PAGE was used to visualize the resulting protein products. For an untreated KatG sample, Coomassie blue staining of the gel confirmed the presence of essentially a single band at approximately 80 kDa together with a small amount of a 160 kDa dimer band (<10% of the total amount of protein) (Figure 1A). When KatG was allowed to react with an equimolar amount of tert-BuOOH, SDS-PAGE analysis showed some dimerization of the protein (Figure 1A, left), with a concomitant decrease in the intensity of the monomer band. Increasing the amount of oxidant led to a conversion of KatG to dimers and trimers and a loss of 80 kDa monomer. The protein also appears to have suffered oxidative fragmentation, resulting in smearing of bands and a reduction in the amount of detectable stained protein. Another possibility for the loss of the protein is formation of very high-molecular mass aggregates which are not resolved under the electrophoretic conditions that are used. However, when DMPO was present, the loss of the monomer band was totally prevented (Figure 1A, right). This result demonstrates that DMPO prevents dimerization.
Figure 1.
Dimerization of Mtb KatG by reaction with tert-BuOOH in the absence of DMPO and immunochemical detection of protein-derived DMPO nitrone adducts. (A) SDS-PAGE and protein staining. (B) Anti-DMPO immunostain as shown with Western blot. (C) Chemiluminescent detection by ELISA of KatG radical-derived DMPO nitrone adducts. Wild-type KatG (10 μM) exposed to various concentrations of tert-BuOOH (0.01-1 mM) was incubated for 1 h at 37 °C in the presence or absence of 100 mM DMPO in 0.1 M phosphate buffer (pH 7.4). Reactions were stopped by the addition of 20 mM potassium cyanide. Each lane contained 1.6 μg of Mtb KatG. After electrophoresis, the proteins were visualized by being stained with Coomassie blue protein gel stain or transferred to nitrocellulose for Western blot analysis.
Immunochemical detection of protein-DMPO nitrone adducts was performed using Western blotting in parallel with ELISA analysis. As shown in the left panel of Figure 1B, omission of DMPO resulted in little immunostaining at the 80 kDa band (indicating comparatively little anti-DMPO cross-reacting material), but in the presence of the spin trap, the samples treated with 0.1 and 1 mM peroxide were positive, confirming the production of Mtb KatG protein radical-derived nitrones. The dependence of protein-derived nitrone adduct formation on tert-BuOOH concentration was demonstrated by ELISA analysis, which showed a significant increase in the level of nitrone adducts above background at a concentration of tert-BuOOH as low as 0.01 mM (Figure 1C). In addition, formation of a nitrone adduct was noted in the dimers (Figure 1B, right).
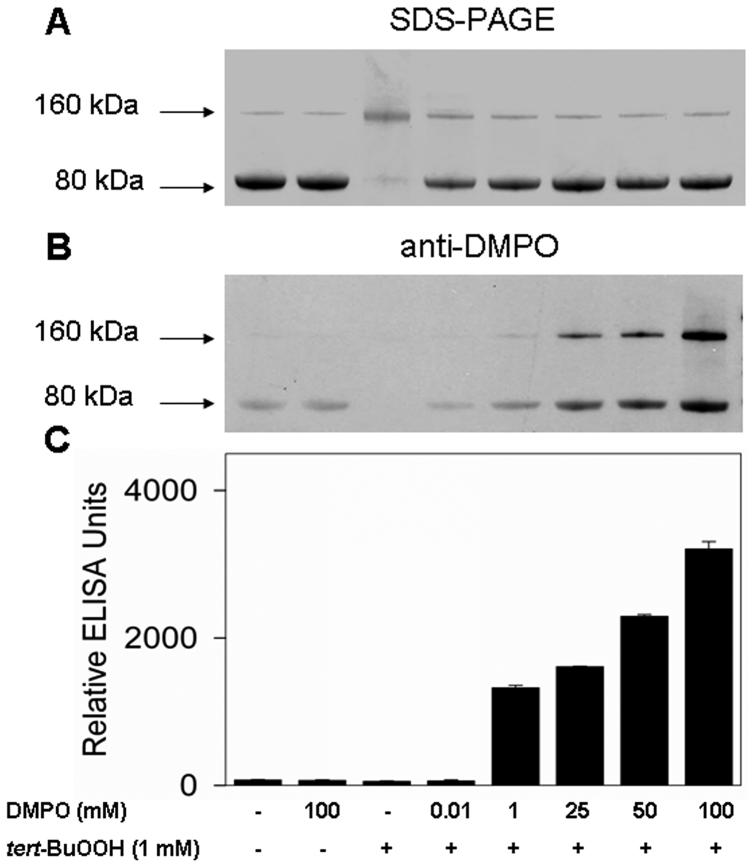
Next we evaluated the DMPO concentration dependence of nitrone adduct formation (Figure 2). When 10 μM DMPO and 10 μM KatG were reacted in the presence of 1 mM tert-BuOOH, no nitrone adducts were detected (Figure 2C). Incubation of Mtb KatG with DMPO (1-100 mM) showed that protein-derived nitrone adduct formation was positively correlated with the spin trap concentration. Interestingly, when 100 mM DMPO was applied, immunostained samples showed a significant amount of nitrone adducts on the dimerized KatG-DMPO complex. Clearly, SDS-PAGE demonstrated that DMPO protected the protein against dimerization (Figure 2A), showing that nitrone adduct formation (Figure 2B,C) and dimerization are competitive.
Figure 2.
Concentration-dependent effects of DMPO on the dimerization and formation of the protein radical-derived nitrone adducts. (A) SDS-PAGE. (B) Anti-DMPO immunostain. (C) ELISA analysis. Reactions including Mtb KatG (10 μM) and DMPO as indicated were initiated with 1 mM tert-BuOOH, and the mixtures were incubated for 1 h at 37 °C in 0.1 M phosphate buffer (pH 7.4). Reactions were stopped by the addition of 20 mM potassium cyanide. Each lane contained 1.6 μg of Mtb KatG.
Effect of Tyrosine, Tryptophan, and Cysteine Modification on KatG Radical Formation
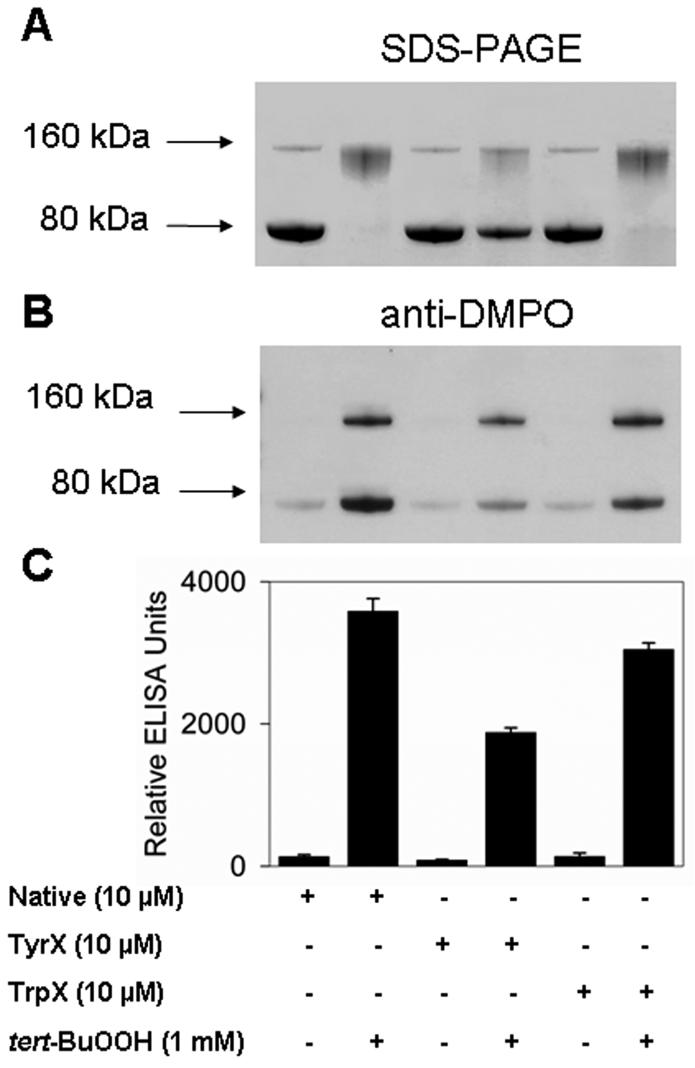
The experiments described above indicate that Mtb KatG forms radicals in the presence of tert-BuOOH that can either form cross-links leading to dimerization of the protein or be trapped by DMPO. Rapid freeze-quench and high-field ESR experiments had previously demonstrated the formation of tyrosyl and tryptophanyl radicals in Mtb KatG (23-25, 39). Since tyrosine, tryptophan, and cysteine are the most commonly oxidized amino acids that produce free radicals (28, 33, 42, 43), the studies reported above led us to suspect these types of residues to be involved in the dimerization process. Next, we reacted the native and chemically modified (iodination reaction for tyrosine blocking, NBS treatment for tryptophan blocking, and DTNB treatment for cysteine blocking) KatG samples with tert-BuOOH. SDS-PAGE analysis indicated that iodination of the tyrosines of Mtb KatG to form iodotyrosine partially prevents nitrone adduct and cross-link formation (Figure 3A). Production of protein-DMPO nitrone adducts was also assessed by ELISA and Western blotting. As shown in Figure 3C, modification of tryptophans caused an only 15% decrease in the amount of nitrone adducts. The same effect was observed for cysteine-blocked KatG (data not shown). However, when tyrosine residues were iodinated, the ELISA results showed a decrease to approximately half in the radical-derived nitrone adducts versus in the native KatG. Western blot data paralleled those from ELISA. These observations together with the protein stain analysis suggested that tyrosine residues are significant sites of dimerization and formation of radical-DMPO nitrone adducts.
Figure 3.
Effect of chemical modifications of tyrosines and tryptophans on dimerization and Mtb KatG-derived radical nitrone adducts and peroxidase activity. (A) SDS-PAGE. Unmodified KatG (native), iodo-KatG (TyrX), and NBS-KatG (TrpX) were incubated with or without 1 mM tert-BuOOH in the absence of DMPO. (B) Western blotting. (C) ELISA. Basically, the experiment shown in panel A was repeated in the presence of DMPO (100 mM), and the immuno-spin trapping analysis was performed. Incubations were performed for 1 h at 37 °C in 0.1 M phosphate buffer (pH 7.4), and the reactions were stopped by the addition of 20 mM KCN.
Additionally, enzyme peroxidase activity was determined for the native and chemically modified protein (not shown). The assays showed a 30% loss of enzymatic activity for tryptophan-modified KatG (TrpX) but an only 7% decrease for the iodinated protein (TyrX). These findings indicated that the decrease in the level of protein-derived nitrone adducts in Trp-modified KatG (Figure 3B,C) is probably due to its reduced peroxidase activity. However, the relatively undiminished specific activity together with the 50% decrease in yield of nitrone adducts for Tyr-modified protein indicated that formation of these protein-DMPO nitrone adducts is dependent on unmodified tyrosine and that tyrosyl radicals are required for most attachment of DMPO to Mtb KatG.
Electron Spin Resonance (ESR) Spectroscopy
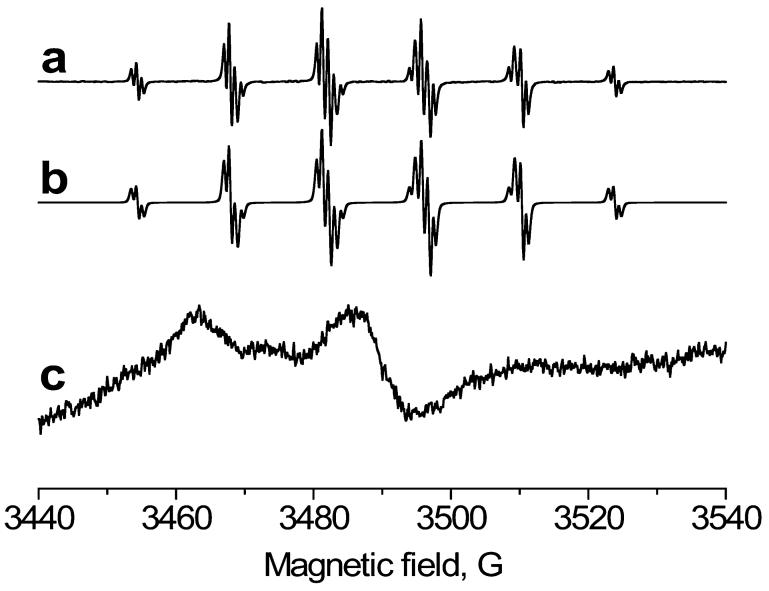
To provide further insight into dimerization of KatG and immunochemical detection of protein-DMPO nitrone adducts, parallel ESR spin trapping experiments were performed. Reactions to produce amino acid-based radicals were initiated by adding different amounts of tert-BuOOH to 50 μM Mtb KatG and 100 mM DMPO. Unfortunately, no Mtb KatG-trapped radicals were detected, possibly due to nitroxide spin adduct instability and/or further oxidation to nitrone products by KatG. However, with the same system in the presence of the spin trap DBNBS (where the nitroxide spin adducts cannot be oxidized to nitrones) (Figure 4, spectra a and b), we could observe and simulate a species previously assigned on the basis of its hyperfine couplings (aN = 14.47 G; 3 methyl H, aH = 13.48 G; 2 meta H, aH = 0.70 G) as the methyl radical adduct (44), thus unambiguously confirming its presence. The trapped methyl radical was previously assigned as a secondary radical product of tert-butyl hydroperoxide decomposition in reaction with chloroperoxidase (14, 45), which, after β-scission, produces acetone and methyl radical. When Mtb KatG was allowed to react under similar conditions in the presence of DBNBS, and the sample was immediately passed over a G-25 column to remove small molecules, an anisotropic spectrum characteristic of an immobilized protein-derived nitroxide adduct was detected (Figure 4, spectrum c). When the adduct was subjected to digestion (using Pronase), the ESR signal decayed rapidly.
Figure 4.
ESR spectra obtained from the reaction of Mtb KatG with tert-BuOOH in the presence of spin trap DBNBS. (a) The reaction mixture containing KatG (50 μM), DBNBS (0.6 mM), and tert-BuOOH (0.5 mM) was placed immediately into a 10 mm flat cell. (b) Computer simulation of the species in spectrum a. (c) Same as spectrum a, except the reaction mixture was passed over a Sephadex G-25 size exclusion column. The instrument parameters were as follows: field sweep, 80 G; microwave frequency, 9.78 GHz; microwave power, 20 mW; modulation amplitude, 0.4 G (4 G for spectrum c); conversion time, 164 ms; time constant, 164 ms; receiver gain, 2 × 104; and number of scans, 3.
Formation of Radicals from Mtb KatG by Enzymatic Generation of H2O2 As Determined by Immuno-Spin Trapping and ESR Spectroscopy
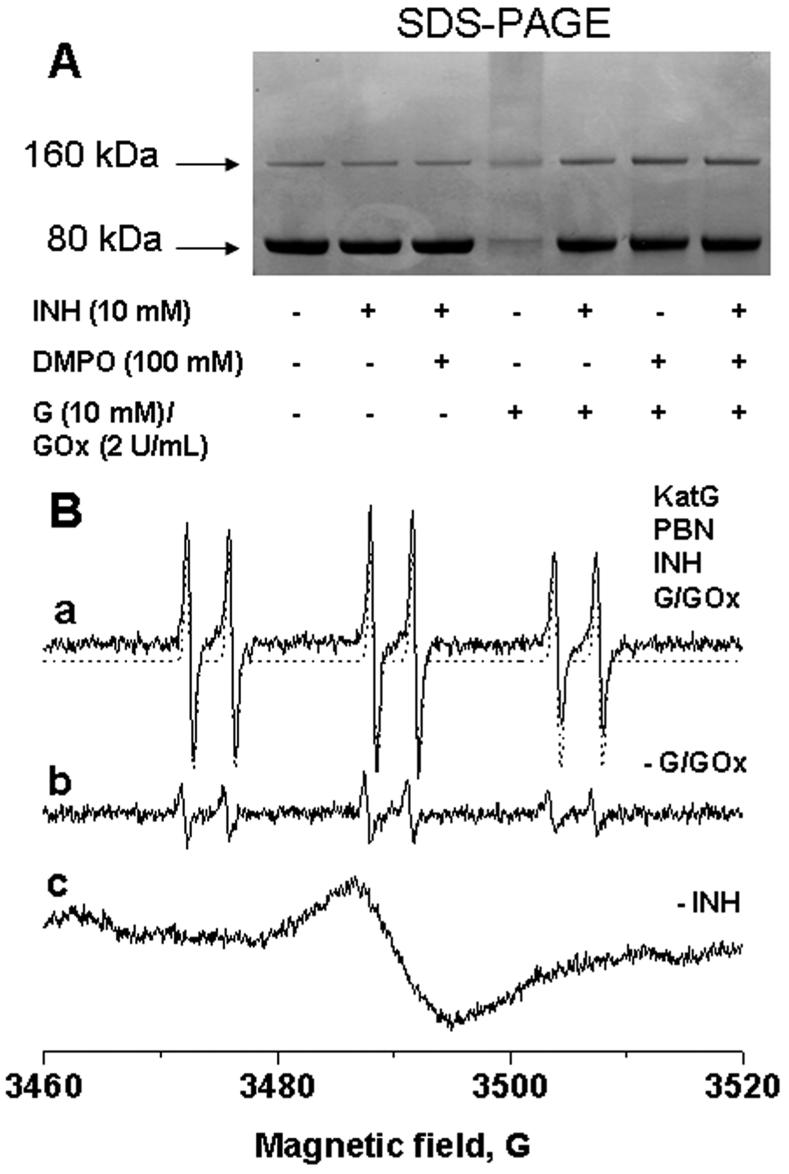
To further explore dimerization and protein radical formation under more physiologically relevant conditions, we investigated the effect of hydrogen peroxide as an oxidant in the system. Hydrogen peroxide (10 μM to 1 mM) did not result in ESR-detectable radical adduct formation or protein-derived nitrone adducts (by ELISA and Western blot). This could be explained by the high catalase activity of the enzyme and extremely fast cycling of Compound I intermediates back to the resting state (35). Therefore, we decided to use exogenous H2O2 generated by G/GOx and, additionally, to study the effect of the presence of isoniazid. Protein staining (Figure 5A) showed that Mtb KatG formed dimer and protein, smearing when a continuous flux of hydrogen peroxide with an initial rate of 100 μM/min was applied. When the protein was reacted with H2O2 in the presence of either DMPO, isoniazid, or both, the dimerization process was prevented. The fact that INH was previously shown to react with tyrosyl and/or tryptophanyl radicals (23, 25) strongly suggests that there is a competition between the drug and the spin trap for reaction with protein radicals.
Figure 5.
(A) SDS-PAGE analysis of reaction mixtures containing KatG (10 μM), DMPO (100 mM), INH (10 mM), or subsets of these as indicated, incubated for 1 h at 37 °C in 0.1 M phosphate buffer (pH 7.4). Reactions were initiated with 10 mM glucose and 2 units/mL glucose oxidase and stopped by the addition of 20 mM potassium cyanide. Each lane contained 1.6 μg of Mtb KatG. (B) ESR spectra obtained from the reaction of Mtb KatG with INH and G/GOx in the presence of spin trap PBN. Basically, the experiment shown in panel A was repeated in the presence of PBN (10 mM), except the concentrations of the protein and the drug were 50 μM and 50 mM, respectively. Reaction mixtures were incubated for 30 min at 37 °C before the measurements. The instrument settings were the same as described in the legend of Figure 4 (4 G modulation amplitude for spectrum c).
To test the radical formation mediated by hydrogen peroxide, ESR and immuno-spin trapping were performed. In a system with hydrogen peroxide furnished from G/GOx, neither DMPO nor DBNBS yielded ESR-active radical adducts either from the enzyme or from the drug. With α-phenyl-N-tert-butylnitrone (PBN) (Figure 5B, spectrum a), a six-line spectrum was observed in the presence of INH, suggesting only one radical adduct. On the basis of the hyperfine couplings from computer simulation (aN = 15.75 G and aH = 3.60 G) (41), this ESR signal was identified as an oxygen-centered radical adduct, and in accordance with the results of Wengenack and Rusnak (21) using tert-BuOOH as a peroxide source, it was tentatively assigned as an acyl peroxo INH-derived radical. In the presence of INH and PBN, ESR spin trapping experiments were reported to produce three types of isoniazid-derived radicals generated via the KatG-mediated peroxidase Compound I/II cycle using tert-BuOOH as an oxidant (21).
A control reaction mixture in the absence of G/GOx contained a small amount of the radical adduct, probably due to partial decomposition of isoniazid after incubation for 30 min at 37 °C. This result confirms the dependence of radical formation on isoniazid (Figure 5B, spectrum b). The sample prepared without INH was devoid of any radical adducts using the same instrumental parameters (not shown), but increasing the modulation amplitude to 4 G allowed detection of a broad triplet characteristic of anisotropic protein-derived nitroxide adducts (Figure 5B, spectrum c). The signal of the immobilized protein-derived nitroxide adduct (as in Figure 4, spectrum c) was still detectable after the sample was passed over a G-25 column (not shown).
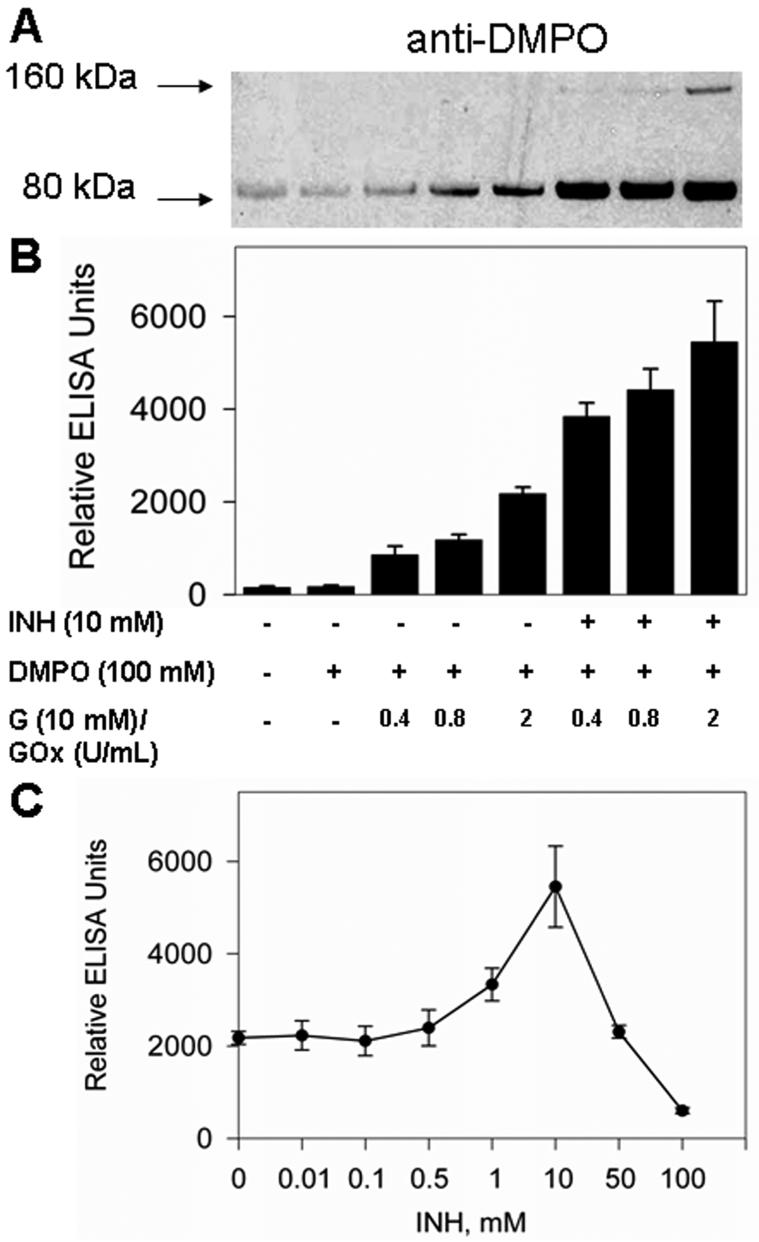
The studies presented above clearly demonstrate that isoniazid, like DMPO, protects Mtb KatG against cross-linking (Figure 5A). ESR experiments with exogenous hydrogen peroxide confirmed that protein-derived nitroxide adducts were generated. Thus, to investigate the role of isoniazid in protein radical production, we applied the immuno-spin trapping technology to detect protein-DMPO nitrone adducts in the system. According to the left panel of Figure 6A, treatments of KatG with fluxes of H2O2 with three different initial rates (20, 40, and 100 μM/min) resulted in nitrone adduct formation. When isoniazid was added to the system, there was a significant increase in the level of radical-derived nitrone adducts with Mtb KatG (Figure 6A, right panel) under the same conditions of H2O2 generation, indicating an essential role of isoniazid-derived radicals in further protein radical production. ELISA results paralleled those from the Western blot analysis (Figure 6B). The effect of variable concentrations of isoniazid was also determined with ELISA analysis (Figure 6C); formation of the KatG-DMPO nitrone adduct was positively correlated with drug concentration up to 10 mM INH. A further increase in isoniazid concentration (to 100 mM) decreased the level of adduct formation. This could be explained with the possibility of a bifunctional role of INH: a pro-oxidant at low concentrations and an antioxidant at high concentrations. Similar behavior was recently found in the myeloperoxidase-aminoglutethimide and myeloperoxidase-procainamide systems (16, 46).
Figure 6.
Western blot (A) and ELISA (B) analyses of isoniazid-dependent KatG radical-derived nitrone adducts in the presence of H2O2 generation. Reactions including Mtb KatG (10 μM) and DMPO (100 mM) were initiated with glucose (10 mM)/glucose oxidase (0.4-2 units/mL) in the presence or absence of isoniazid (10 mM); the mixtures were incubated for 1 h at 37 °C in 0.1 M phosphate buffer (pH 7.4), and the reactions were stopped by the addition of 20 mM potassium cyanide. Each lane contained 1.6 μg of Mtb KatG. (C) Concentration-dependent effects of isoniazid on the formation of KatG radical-derived nitrone adducts. The experimental conditions were the same as those described for panels A and B, with the concentration of glucose oxidase fixed (2 units/mL) and the isoniazid concentration varied.
DISCUSSION
In this study, we report the formation of protein dimers in the reaction of Mtb KatG with tert-BuOOH. The anti-DMPO polyclonal antiserum that was used has the specificity to recognize protein-derived radical nitrone adducts formed from KatG during the turnover of the enzyme. Our SDS-PAGE experiments have shown that in the absence of DMPO, the protein is capable of forming cross-linked products which, according to their molecular masses, are dimers and trimers. Addition of the spin trap reduced the amount of cross-links to their background level, indicating that DMPO was able to trap the amino acid radical(s) responsible for the dimerization process by forming a radical adduct with it. SDS-PAGE analysis showed that an equimolar amount of the spin trap (10 μM) with respect to the protein significantly decreased the intensity of the dimer band probably due to the relatively low rate of the KatG dimerization process compared to the rate of spin trapping. Moreover, we observed DMPO concentration-dependent formation of KatG-DMPO nitrone adducts by Western blot and ELISA. Formation of nitrone adducts on the protein dimers could be the result of trapping of amino acid radicals either on dimers present in the untreated protein or possibly the dimers formed upon treatment in the presence of DMPO. Clearly, the immuno-spin trapping approach is quite sensitive since we could detect no protein radical adducts of DMPO using the traditional ESR spectroscopy technique.
In earlier reports, tyrosyl and tryptophanyl radicals have been reported in Mtb KatG using direct ESR spectroscopy combined with site-directed mutagenesis (24, 25, 39). Different residues have been proposed for the radical sites, from residues located very close to the heme to residues positioned on the surface, and several of the 21 tyrosines and 24 tryptophans are involved in the radical pathway. We performed chemical modifications on the tyrosine and tryptophan residues because both types of amino acids can be oxidized to free radicals in KatG during turnover with organic peroxides. The results indicate loss of peroxidase activity in the tryptophan-modified protein (by 30%), while there is almost no change in the activity of tyrosine-blocked KatG. A possible explanation for the reduced activity of the NBS-treated protein could be inactivation of Trp107, one of the components of the novel amino acid covalent adduct in Mtb KatG (Trp107-Tyr229-Met255). Analysis using sitedirected mutagenesis showed a moderately reduced peroxidase activity of KatG[W107F] compared to that of wildtype KatG (S. Yu and R. Magliozzo, unpublished data).
Immuno-spin trapping showed that the level of production of nitrone adducts decreased by ∼50% in the case of the iodination reaction, but there was almost no change in the NBS-treated (TrpX) or DTNB-treated enzyme. Fluorescence experiments using peroxyacetic acid as an oxidant confirmed the presence of a peak at ∼410 nm (see the Supporting Information); a similar peak in the case of sperm whale myoglobin was unambiguously assigned to dityrosine cross-links (29). Presumably, dimerization in Mtb KatG is due, at least in part, to dityrosine cross-links.
In the reaction of KatG with alkyl peroxides, similar to monofunctional peroxidases like thyroid peroxidase (33) and cytochrome c peroxidase (47), an amino acid radical can be formed by migration of one electron to the porphyrin π-cation radical (Compound I). In the case of bovine liver catalase, Ivancich et al. (48) have proposed that the protein radical has no function in the catalatic reaction. However, recent kinetic and spectral stopped-flow analysis of KatGs from three different bacteria (M. tuberculosis, Synechocystis PCC 6803, and Burkholderia pseudomallei) (49) demonstrated some differences between treatment with H2O2 and organic peroxides in the formation of radical intermediates (24, 25, 50). Our immuno-spin trapping experiments have shown that, in the presence of a continuous flux of hydrogen peroxide, Mtb KatG produced protein radicals trapped by DMPO that were further oxidized to protein-derived radical nitrone adducts (Figure 6).
When the protein is exposed to a high level of H2O2, the peroxidation reaction is inhibited due to the high rate of hydrogen peroxide turnover, and Mtb KatG functions instead as a catalase. In fact, KatG is known to be the only catalase in the pathogen M. tuberculosis and is the key enzyme essential for catabolizing exogenous peroxides via reactive oxygen intermediates. It might also be responsible for detoxification of endogenous peroxides generated by bacterial respiration (51). The activation of isoniazid by Mtb KatG in the presence of hydrogen peroxide is not well studied because its catalase activity tends to dominate its peroxidase activity, although the hypothesis that the peroxidase activity of the protein is sufficient for drug activation is well-accepted (13, 52).
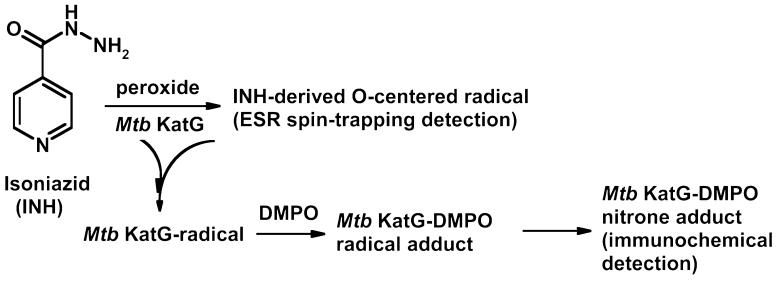
In previous studies, Rusnak and co-workers reported that the redox potential of Mtb KatG (E° ∼ -0.06 V) was approximately 200 mV higher than those of most monofunctional peroxidases such as HRP (53) and was insufficient for the oxidation of isoniazid (E° ∼ 0.5 V) without further activation by an exogenous oxidant. These authors suggested that much higher values for the redox potentials of Compounds I and II (E° ∼ 0.9 V) were enough to initiate drug activation. Furthermore, stopped-flow and double-mixing rapid freeze-quench ESR experiments confirmed not only the reduction of Compound I but also the quenching of tyrosyl radical by INH (23, 35). A recent work (25) showed detection of two types of Compound I (FeIV=O Por·+ and FeIV=O Trp·+), but the authors concluded that neither of these intermediates is involved in the reaction with the drug and, most probably, a neutral tryptophanyl radical is among the species oxidizing INH. Our Western blotting and ELISA analysis of tyrosine- and tryptophan-blocked KatG showed the same effect that was seen with unmodified KatG in increasing the yield of KatG-DMPO nitrone adducts compared to the samples lacking the drug (see Supporting Information). On the basis of this collection of data, the most likely candidates for the residue(s) involved in the accumulation of protein-derived nitrone adducts are tyrosines and/or tryptophans remaining unmodified upon chemical treatment or, alternatively, the INH-derived radical being able to oxidize different types of residue(s). Since ESR data showed only the presence of oxygen-centered INH-derived radicals using enzymatic generation of hydrogen peroxide, we propose that this species may be the reactive species that induced more protein radicals in Mtb KatG (Scheme 1).
Scheme 1.
In summary, our results from immuno-spin trapping experiments clearly demonstrate the formation of cross-links in Mtb KatG during turnover of peroxides, which was inhibited by addition of DMPO. Chemical modifications of tyrosine and tryptophan residues showed that tyrosyl radicals were involved in the dimer linkage. In addition, protein-derived nitrone adducts were detected in the presence of a continuous flux of hydrogen peroxide. This radical formation was accelerated by isoniazid oxidation and may play a role in the mode of action of the TB drug.
ACKNOWLEDGMENT
We are grateful to Mary Mason and Dr. Ann Motten for their editing of the manuscript. Also, we acknowledge Jean Corbett for the purification of DMPO and Dr. Wenjian Ma for performing densitometric analysis.
Footnotes
This work was supported by the Intramural Research Program of the National Institutes of Health and the National Institute of Environmental Health Sciences and NIH Grants AI-43582 and AI-060014 (National Institute of Allergy and Infections).
- KatG
- catalase-peroxidase
- Mtb
- M. tuberculosis
- INH
- isonicotinic acid hydrazine (isoniazid)
- ESR
- electron spin resonance
- DMPO
- 5,5-dimethyl-1-pyrroline N-oxide
- PBN
- α-phenyl-N-tert-butylnitrone
- DBNBS
- 3,5-dibromo-4-nitrosobenzenesulfonate
- SDS-PAGE
- sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- G/GOx
- glucose/glucose oxidase
- NBS
- N-bromosuccinimide
- DTNB
- 5,5′-dithiobis(2-nitrobenzoic acid)
REFERENCES
- 1.World Health Organization . Fact Sheet 104. 2002. [Google Scholar]
- 2.Bernstein J, Lott WA, Steinberg BA, Yale HL. Chemotherapy of experimental tuberculosis. V. Isonicotinic acid hydrazide (nydrazid) and related compounds. Am. Rev. Tuberc. 1952;65:357–364. doi: 10.1164/art.1952.65.4.357. [DOI] [PubMed] [Google Scholar]
- 3.Fox HH. The chemical approach to the control of tuberculosis. Science. 1952;116:129–134. doi: 10.1126/science.116.3006.129. [DOI] [PubMed] [Google Scholar]
- 4.Winder F. Catalase and peroxidase in mycobacteria. Possible relationship to the mode of action of isoniazid. Am. ReV. Respir. Dis. 1960;81:68–78. doi: 10.1164/arrd.1960.81.1P1.68. [DOI] [PubMed] [Google Scholar]
- 5.Quémard A, Sacchettini JC, Dessen A, Vilcheze C, Bittman R, Jacobs WR, Jr., Blanchard JS. Enzymatic characterization of the target for isoniazid in Mycobacterium tuberculosis. Biochemistry. 1995;34:8235–8241. doi: 10.1021/bi00026a004. [DOI] [PubMed] [Google Scholar]
- 6.Rozwarski DA, Grant GA, Barton DHR, Jacobs WR, Jr., Sacchettini JC. Modification of the NADH of the isoniazid target (InhA) from Mycobacterium tuberculosis. Science. 1998;279:98–102. doi: 10.1126/science.279.5347.98. [DOI] [PubMed] [Google Scholar]
- 7.Miesel L, Rozwarski DA, Sacchettini JC, Jacobs WR. Mechanisms for isoniazid action and resistance. Novartis Found. Symp. 1998;217:209–221. doi: 10.1002/0470846526.ch15. [DOI] [PubMed] [Google Scholar]
- 8.Zhao X, Yu H, Yu S, Wang F, Sacchettini JC, Magliozzo RS. Hydrogen peroxide-mediated isoniazid activation catalyzed by Mycobacterium tuberculosis catalase-peroxidase (KatG) and its S315T mutant. Biochemistry. 2006;45:4131–4140. doi: 10.1021/bi051967o. [DOI] [PubMed] [Google Scholar]
- 9.Wilming M, Johnsson K. Spontaneous formation of the bioactive form of the tuberculosis drug isoniazid. Angew. Chem., Int. Ed. 1999;38:2588–2590. doi: 10.1002/(sici)1521-3773(19990903)38:17<2588::aid-anie2588>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 10.Dessen A, Quémard A, Blanchard JS, Jacobs WR, Jr., Sacchettini JC. Crystal structure and function of the isoniazid target of Mycobacterium tuberculosis. Science. 1995;267:1638–1641. doi: 10.1126/science.7886450. [DOI] [PubMed] [Google Scholar]
- 11.Klotz MG, Loewen PC. The molecular evolution of catalatic hydroperoxidases: Evidence for multiple lateral transfer of genes between prokaryota and from bacteria into eukaryota. Mol. Biol. Evol. 2003;20:1098–1112. doi: 10.1093/molbev/msg129. [DOI] [PubMed] [Google Scholar]
- 12.Pierattelli R, Banci L, Eady NAJ, Bodiguel J, Jones JN, Moody PCE, Raven EL, Jamart-Gregoire B, Brown KA. Enzyme-catalyzed mechanism of isoniazid activation in class I and class III peroxidases. J. Biol. Chem. 2004;279:39000–39009. doi: 10.1074/jbc.M402384200. [DOI] [PubMed] [Google Scholar]
- 13.Metcalfe C, Macdonald IK, Murphy EJ, Brown KA, Raven EL, Moody PCE. The tuberculosis prodrug isoniazid bound to activating peroxidases. J. Biol. Chem. 2008;283:6193–6200. doi: 10.1074/jbc.M707412200. [DOI] [PubMed] [Google Scholar]
- 14.Dikalov SI, Mason RP. Reassignment of organic peroxyl radical adducts. Free Radical Biol. Med. 1999;27:864–872. doi: 10.1016/s0891-5849(99)00134-3. [DOI] [PubMed] [Google Scholar]
- 15.Dikalov S, Jiang J, Mason RP. Characterization of the high-resolution ESR spectra of superoxide radical adducts of 5-(diethoxyphosphoryl)-5-methyl-1-pyrroline N-oxide (DEP-MPO) and 5,5-dimethyl-1-pyrroline N-oxide (DMPO). Analysis of conformational exchange. Free Radical Res. 2005;39:825–836. doi: 10.1080/10715760500155688. [DOI] [PubMed] [Google Scholar]
- 16.Siraki AG, Bonini MG, Jiang J, Ehrenshaft M, Mason RP. Aminoglutethimide-induced protein free radical formation on myeloperoxidase: A potential mechanism of agranulocytosis. Chem. Res. Toxicol. 2007;20:1038–1045. doi: 10.1021/tx6003562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barr DP, Gunther MR, Deterding LJ, Tomer KB, Mason RP. ESR spin-trapping of a protein-derived tyrosyl radical from the reaction of cytochrome c with hydrogen peroxide. J. Biol. Chem. 1996;271:15498–15503. doi: 10.1074/jbc.271.26.15498. [DOI] [PubMed] [Google Scholar]
- 18.Bonini MG, Siraki AG, Atanassov BS, Mason RP. Immunolocalization of hypochlorite-induced, catalase-bound free radical formation in mouse hepatocytes. Free Radical Biol. Med. 2007;42:530–540. doi: 10.1016/j.freeradbiomed.2006.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhattacharjee S, Deterding LJ, Jiang J, Bonini MG, Tomer KB, Ramirez DC, Mason RP. Electron transfer between a tyrosyl radical and a cysteine residue in hemoproteins: Spin trapping analysis. J. Am. Chem. Soc. 2007;129:13493–13501. doi: 10.1021/ja073349w. [DOI] [PubMed] [Google Scholar]
- 20.Sinha BK. Enzymatic activation of hydrazine derivatives. A spin-trapping study. J. Biol. Chem. 1983;258:796–801. [PubMed] [Google Scholar]
- 21.Wengenack NL, Rusnak F. Evidence for isoniazid-dependent free radical generation catalyzed by Mycobacterium tuberculosis KatG and the isoniazid-resistant mutant KatG(S315T) Biochemistry. 2001;40:8990–8996. doi: 10.1021/bi002614m. [DOI] [PubMed] [Google Scholar]
- 22.Timmins GS, Master S, Rusnak F, Deretic V. Requirements for nitric oxide generation from isoniazid activation in vitro and inhibition of mycobacterial respiration in vivo. J. Bacteriol. 2004;186:5427–5431. doi: 10.1128/JB.186.16.5427-5431.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chouchane S, Girotto S, Yu S, Magliozzo RS. Identification and characterization of tyrosyl radical formation in Mycobacterium tuberculosis catalase-peroxidase (KatG) J. Biol. Chem. 2002;277:42633–42638. doi: 10.1074/jbc.M207916200. [DOI] [PubMed] [Google Scholar]
- 24.Ranguelova K, Girotto S, Gerfen GJ, Yu S, Suarez J, Metlitsky L, Magliozzo RS. Radical sites in Mycobacterium tuberculosis KatG identified using electron paramagnetic resonance spectroscopy, the three-dimensional crystal structure, and electron transfer couplings. J. Biol. Chem. 2007;282:6255–6264. doi: 10.1074/jbc.M607309200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh R, Switala J, Loewen PC, Ivancich A. Two [Fe(IV)=O Trp·] intermediates in M. tuberculosis catalase-peroxidase discriminated by multifrequency (9-285 GHz) EPR spectroscopy: Reactivity toward isoniazid. J. Am. Chem. Soc. 2007;129:15954–15963. doi: 10.1021/ja075108u. [DOI] [PubMed] [Google Scholar]
- 26.Stadtman ER, Berlett BS. Reactive oxygen-mediated protein oxidation in aging and disease. Chem. Res. Toxicol. 1997;10:485–494. doi: 10.1021/tx960133r. [DOI] [PubMed] [Google Scholar]
- 27.Garrison WM. Reaction mechanisms in the radiolysis of peptides, polypeptides, and proteins. Chem. Rev. 1987;87:381–398. [Google Scholar]
- 28.Detweiler CD, Lardinois OM, Deterding LJ, Ortiz de Montellano PR, Tomer KB, Mason RP. Identification of the myoglobin tyrosyl radical by immuno-spin trapping and its dimerization. Free Radical Biol. Med. 2005;38:969–976. doi: 10.1016/j.freeradbiomed.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 29.Lardinois OM, Ortiz de Montellano PR. Intra- and intermolecular transfers of protein radicals in the reactions of sperm whale myoglobin with hydrogen peroxide. J. Biol. Chem. 2003;278:36214–36226. doi: 10.1074/jbc.M304726200. [DOI] [PubMed] [Google Scholar]
- 30.Detweiler CD, Deterding LJ, Tomer KB, Chignell CF, Germolec D, Mason RP. Immunological identification of the heart myoglobin radical formed by hydrogen peroxide. Free Radical Biol. Med. 2002;33:364–369. doi: 10.1016/s0891-5849(02)00895-x. [DOI] [PubMed] [Google Scholar]
- 31.Mason RP. Using anti-5,5-dimethyl-1-pyrroline N-oxide (anti-DMPO) to detect protein radicals in time and space with immuno-spin trapping. Free Radical Biol. Med. 2004;36:1214–1223. doi: 10.1016/j.freeradbiomed.2004.02.077. [DOI] [PubMed] [Google Scholar]
- 32.Ramirez DC, Chen Y-R, Mason RP. Immunochemical detection of hemoglobin-derived radicals formed by reaction with hydrogen peroxide: Involvement of a protein-tyrosyl radical. Free Radical Biol. Med. 2003;34:830–839. doi: 10.1016/s0891-5849(02)01437-5. [DOI] [PubMed] [Google Scholar]
- 33.Ehrenshaft M, Mason RP. Protein radical formation on thyroid peroxidase during turnover as detected by immuno-spin trapping. Free Radical Biol. Med. 2006;41:422–430. doi: 10.1016/j.freeradbiomed.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 34.Kaur H. A water soluble C-nitroso-aromatic spin-trap-3,5-dibromo-4-nitrosobenzenesulphonic acid. ‘The Perkins spin-trap’. Free Radical Res. 1996;24:409–420. doi: 10.3109/10715769609088040. [DOI] [PubMed] [Google Scholar]
- 35.Chouchane S, Lippai I, Magliozzo RS. Catalase-peroxidase (Mycobacterium tuberculosis KatG) catalysis and isoniazid activation. Biochemistry. 2000;39:9975–9983. doi: 10.1021/bi0005815. [DOI] [PubMed] [Google Scholar]
- 36.Yu S, Chouchane S, Magliozzo RS. Characterization of the W321F mutant of Mycobacterium tuberculosis catalase-peroxidase KatG. Protein Sci. 2002;11:58–64. doi: 10.1110/ps.09902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramirez DC, Gomez-Mejiba SE, Mason RP. Mechanism of hydrogen peroxide-induced Cu,Zn-superoxide dismutase-centered radical formation as explored by immuno-spin trapping: The role of copper- and carbonate radical anion-mediated oxidations. Free Radical Biol. Med. 2005;38:201–214. doi: 10.1016/j.freeradbiomed.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 38.Marcinkeviciene JA, Magliozzo RS, Blanchard JS. Purification and characterization of the Mycobacterium smegmatis catalase-peroxidase involved in isoniazid activation. J. Biol. Chem. 1995;270:22290–22295. doi: 10.1074/jbc.270.38.22290. [DOI] [PubMed] [Google Scholar]
- 39.Zhao X, Girotto S, Yu S, Magliozzo RS. Evidence for radical formation at Tyr-353 in Mycobacterium tuberculosis catalase-peroxidase (KatG) J. Biol. Chem. 2004;279:7606–7612. doi: 10.1074/jbc.M311884200. [DOI] [PubMed] [Google Scholar]
- 40.Lundblad RL. Techniques in Protein Modification. CRC Press; Boca Raton, FL: 1995. p. 75.p. 187.p. 211. [Google Scholar]
- 41.Duling DR. Simulation of multiple isotropic spin-trap EPR spectra. J. Magn. Reson., Ser. B. 1994;104:105–110. doi: 10.1006/jmrb.1994.1062. [DOI] [PubMed] [Google Scholar]
- 42.Chen Y-R, Gunther MR, Mason RP. An electron spin resonance spin-trapping investigation of the free radicals formed by the reaction of mitochondrial cytochrome c oxidase with H2O2. J. Biol. Chem. 1999;274:3308–3314. doi: 10.1074/jbc.274.6.3308. [DOI] [PubMed] [Google Scholar]
- 43.Gunther MR, Sturgeon BE, Mason RP. A long-lived tyrosyl radical from the reaction between horse metmyoglobin and hydrogen peroxide. Free Radical Biol. Med. 2000;28:709–719. doi: 10.1016/s0891-5849(00)00164-7. [DOI] [PubMed] [Google Scholar]
- 44.Athar M, Mukhtar H, Bickers DR, Khan IU, Kalyanaraman B. Evidence for the metabolism of tumor promoter organic hydroperoxides into free radicals by human carcinoma skin keratinocytes: An ESR-spin trapping study. Carcinogenesis. 1989;10:1499–1503. doi: 10.1093/carcin/10.8.1499. [DOI] [PubMed] [Google Scholar]
- 45.Chamulitrat W, Takahashi N, Mason RP. Peroxyl, alkoxyl, and carbon-centered radical formation from organic hydroperoxides by chloroperoxidase. J. Biol. Chem. 1989;264:7889–7899. [PubMed] [Google Scholar]
- 46.Siraki AG, Deterding LJ, Bonini MG, Jiang J, Ehrenshaft M, Tomer KB, Mason RP. Procainamide, but not N-acetylprocainamide, induces protein free radical formation on myeloperoxidase: A potential mechanism of agranulocytosis. Chem. Res. Toxicol. 2008;21:1143–1153. doi: 10.1021/tx700415b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoffman BM, Roberts JE, Kang CH, Margoliash E. Electron paramagnetic and electron nuclear double resonance of the hydrogen peroxide compound of cytochrome c peroxidase. J. Biol. Chem. 1981;256:6556–6564. [PubMed] [Google Scholar]
- 48.Ivancich A, Jouve HM, Sartor B, Gaillard J. EPR investigation of compound I in Proteus mirabilis and bovine liver catalases: Formation of porphyrin and tyrosyl radical intermediates. Biochemistry. 1997;36:9356–9364. doi: 10.1021/bi970886s. [DOI] [PubMed] [Google Scholar]
- 49.Jakopitsch C, Vlasits J, Wiseman B, Loewen PC, Obinger C. Redox intermediates in the catalase cycle of catalase-peroxidases from Synechocystis PCC 6803, Burkholderia pseudomallei, and Mycobacterium tuberculosis. Biochemistry. 2007;46:1183–1193. doi: 10.1021/bi062266+. [DOI] [PubMed] [Google Scholar]
- 50.Ivancich A, Jakopitsch C, Auer M, Un S, Obinger C. Protein-based radicals in the catalase-peroxidase of Synechocystis PCC6803: A multifrequency EPR investigation of wildtype and variants on the environment of the heme active site. J. Am. Chem. Soc. 2003;125:14093–14102. doi: 10.1021/ja035582+. [DOI] [PubMed] [Google Scholar]
- 51.Ng VH, Cox JS, Sousa AO, MacMicking JD, McKinney JD. Role of KatG catalase-peroxidase in mycobacterial pathogenesis: Countering the phagocyte oxidative burst. Mol. Microbiol. 2004;52:1291–1302. doi: 10.1111/j.1365-2958.2004.04078.x. [DOI] [PubMed] [Google Scholar]
- 52.Ghiladi RA, Medzihradszky KF, Rusnak FM, Ortiz de Montellano PR. Correlation between isoniazid resistance and superoxide reactivity in Mycobacterium tuberculosis KatG. J. Am. Chem. Soc. 2005;127:13428–13442. doi: 10.1021/ja054366t. [DOI] [PubMed] [Google Scholar]
- 53.Wengenack NL, Lopes H, Kennedy MJ, Tavares P, Pereira AS, Moura I, Moura JJG, Rusnak F. Redox potential measurements of the Mycobacterium tuberculosis heme protein KatG and the isoniazid-resistant enzyme KatG(S315T): Insights into isoniazid activation. Biochemistry. 2000;39:11508–11513. doi: 10.1021/bi001239v. [DOI] [PubMed] [Google Scholar]