Abstract
We report here our systematic studies of excited-state dynamics of two common flavin molecules, FMN and FAD, in five redox states of oxidized form, neutral and anionic semiquinones, and neutral and anionic fully-reduced hydroquinones in solution and in inert protein environments with femtosecond resolution. Using protein environments, we are able to stabilize two semiquinone radicals and thus observed their weak emission spectra. Significantly, we observed a strong correlation between their excited-state dynamics and the planarity of their flavin isoalloxazine ring. For a bent ring structure, we all observed ultrafast dynamics from a few to hundreds of picoseconds and strong excitation-wavelength dependence of emission spectra, indicating deactivation during relaxation. A butterfly bending motion is invoked to get access to conical intersection(s) to facilitate deactivation. These states include the anionic semiquinone radical and fully-reduced neutral and anionic hydroquinones in solution. In a planar configuration, flavins have a long lifetime in nanoseconds except for the stacked conformation of FAD, where the intramolecular electron transfer between the ring and the adenine moiety in 5-9 ps as well as the subsequent charge recombination in 30-40 ps were observed. These observed distinct dynamics, controlled by the flavin ring flexibility, are fundamental to flavoenzyme’s functions as observed in photolyase with a planar structure to lengthen the lifetime to maximize DNA repair efficiency and in insect Type 1 cryptochrome with a flexible structure to vary the excited-state deactivation to modulate the functional channel.
Introduction
Flavin molecule is one of the most important cofactors in enzymatic funtions.1-8 Flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD) are the most commonly occurring flavins in flavoproteins, which consist of a heterocyclic isoalloxazine ring with ribityl phosphate and ribityl adenine diphosphate, respectively (scheme 1). Flavin can have three different redox states: oxidized form, one-electron reduced radical semiquinone and two-electron fully-reduced hydroquinone. Semiquinone and hydroquinone have a pKa of 8.3 and 6.7, respectively,3 and can be present in their neutral or anionic forms under physiological conditions (scheme 2). Among fives redox states, two redox pairs, oxidized flavin/anionic semiquinone (FAD/FAD•-) and neutral semiquinone/anionic hydroquinone (FADH•/FADH-), are often involved in electron-transfer reactions (scheme 2). Because of flavin’s chemical versatility, flavoproteins are ubiquitous and participate in a broad spectrum of biological activities, such as cell apoptosis,9,10 detoxification,11,12 dehydrogenation of metabolites,13 oxygen activation,14 redox reactions,4,6 halogenation of aromatic substrates,15,16 light-driven DNA repair,17,18 and blue-light photoreceptors.19-23
Scheme1.

Structure formulae of flavin in oxidized state.
Scheme 2.

Different redox states of flavin (FAD) under physiological conditions
The steady-state spectroscopic properties of these redox flavins have been extensively studied, especially their absorption spectra in different proteins and solution.4,6,24-28 Among the emission spectra of flavoproteins, there was no report about the anionic and neutral radical semiquinones, while the emission spectra of both oxidized and fully-reduced states are well characterized.25,29 Only recently, we reported the emission spectrum of the flavin anionic radical (FAD•-) in blue-light photoreceptors.30 In the case of free flavins in solution, besides the well-known oxidized state emission, only a few spectra of fully-reduced flavin derivatives in a rigid glassy solvent at 77K were reported25 and the radical semiquinones, both neutral and anionic, are not stable. Surprisingly, the emission spectra of fully-reduced flavins in solution, such as FADH- and FADH2, have not been reported.
Due to the essential role of flavoproteins in light-driven biological activities, such as photoinduced DNA repair and signal transduction, photochemistry and photophysics of flavins have drawn considerable attention.17-23 Also, flavoproteins are ideal systems for studies of intraprotein electron transfer (ET) at short distances.30-38 In flavoproteins, oxidized flavin, FAD or FMN, can be photoreduced to anionic radical flavin, FAD•- or FMN•-, through intraprotein ET from neighboring aromatic residues (Tryptophan or Tyrosine) in hundreds of femtoseconds to a few picoseconds.30-32 Without aromatic residues in proximity, excited FAD or FMN has a lifetime in several nanoseconds.39-41 In solution, excited FAD exhibits an ultrafast intramolecular ET in 9.0 ps42 with a stacked conformation between the isoalloxazine ring and the adenine moiety in close proximity,42-47 while excited FMN shows a predominant lifetime of 4.7 ns.46,48 In contrast to the rapid dismutation of flavin radicals in solution, flavin semiquinones steadily maintain their radical forms in protein environments. The flavin neutral radical FADH• can also be photoreduced to FADH- through intraprotein ET with neighboring aromatic residues in tens of picoseconds.34-38 For fully-reduced flavins, the excited states in solution show the dynamics on the picosecond time scales,49-53 while in flavoproteins they exhibit a predominant lifetime in a few nanoseconds.54,55
In this work, we systematically examine the excited-state dynamics of five redox states of flavins from oxidized FAD and FMN, to anionic radical FAD•-, to neutral radical FMNH•, to fully-reduced anionic FADH- and FMNH-, and finally to fully-reduced neutral FADH2 with femtosecond (fs) resolution. Here, we mainly focus on the photophysical properties of flavin molecules in solution. Because the flavin radicals are unstable in solution, we study their intrinsic photophysics in protein environments, such as FAD•- in cryptochrome and FMNH• in flavodoxin. To avoid the photochemical reaction through intraprotein ET, we mutate the neighboring electron donors in flavodoxin to make an inert protein environment around FMNH•. In the absence of electron acceptors, such as enzyme’s substrates, both anionic radical and fully-reduced flavins (FAD•- and FADH-) are inert to photochemical ET reactions in flavoproteins. Therefore, we further compare photophysics of flavin cofactors in solution and in inert protein environments for fully-reduced as well as oxidized states. By determining the time scales of these dynamics in five redox states, we are able to understand the excited-state behavior of flavins, thus facilitating our understanding of the functionality of flavins in flavoproteins for their biological activities.
Materials and Methods
Chemical Flavins
Oxidized FAD and FMN were purchased from Sigma-Aldrich without further purification and used in 50 mM phosphate buffer at pH=7.4 with 100 mM NaCl. Anionic hydroquinones (FADH- and FMNH-) and a neutral hydroquinone (FADH2) were prepared at 350 μM in 12.5 mM phosphate buffers at pH 8.5 and 5.0, respectively,3 and special care was taken to maintain samples in anaerobic condition. These fully-reduced flavins were generated from 350-μM oxidized flavins either by chemical reduction with 2-4 mM sodium dithionite56 or 250 mM sodium borohydride,57 or by photochemical reduction with 10-20 mM sodium oxalate58 under 360-nm irradiation from a UVB lamp (8W) with more than 10 hours. Complete reduction was confirmed by both the UV/visible absorption25 and, more importantly, the fluorescence spectra as judged by a single emission peak, indicating a single species without a mixture of different redox flavins.
Flavoproteins
The flavodoxin mutant, W60F/Y98F, from Desulfovibrio vulgaris contains oxidized FMN after purification and then was converted to the neutral radical FMNH• or fully-reduced FMNH- as described previously.25,59-61 A flavodoxin concentration at 300 μM was used in 50 mM MOPS buffer at pH 7.0 for neutral radical form and in 50 mM phosphate buffer at pH 8.6 for anionic fully-reduced form. The insect Type 1 cryptochrome from Anopheles gambiae (AgCRY1) or Antheraea pernyi (ApCRY1) has FAD in oxidized state after purification and then was photoreduced to anionic radical FAD•- under anaerobic condition as detailed elsewhere.62,63 The final protein concentrations of 20∼50 μM in 50 mM Tris-HCl buffer at pH 7.5 with 100 mM NaCl, 1mM EDTA, 5 mM dithiothreitol, and 50% (v/v) glycerol were used. Similarly, the folate (MTHF)-depleted photolyase from Escherichia coli was purified with neutral radical FADH• and then converted to fully-reduced FADH- under anaerobic condition.64,65 A photolyase concentration at 400 μM was prepared in 50 mM Tris-HCl buffer at pH 7.4 with 50 mM NaCl, 1 mM EDTA, 10 mM dithiothreitol, and a percentage of glycerol of 10%, 30% or 50% (v/v) for different studies.
Femtosecond Spectroscopy
All the femtosecond-resolved measurements were carried out using fluorescence up-conversion and transient absorption methods. The experimental layout has been detailed elsewhere.37 Briefly, the femtosecond pulse after the two-stage amplifier (Spitfire, Spectra-Physics) has a temporal width of 110 fs with energy of more than 2 mJ and a repetition rate of 1 kHz. The laser beam is then split into two equal parts to pump two optical parametric amplifiers (OPA-800C, Spectra-Physics). The pump wavelengths of 400, 480, and 580 nm were then generated by doubling of 800 nm or mixing of 800 nm with either the signal beam (1200 nm) or the idler beam (2109 nm) from the first optical parametric amplifier in a 0.2-mm thick β-barium borate crystal (BBO, type I). To generate a pump wavelength at 325 nm, a signal beam (1300 nm) from the first optical parametric amplifier was frequency-doubled twice by two 0.2-mm thick BBO crystals. The pump pulse energy typically was attenuated to 100–140 nJ before being focused into the sample cell. For fluorescence up-conversion experiments, the fluorescence emission was gated by another 800-nm beam in a 0.2 mm BBO crystal to obtain fluorescence transients at desired wavelengths. For transient-absorption measurements, the various probe wavelengths from 400 to 800 nm were generated by the second optical parametric amplifier. The sensitivity of the transient absorption method can reach 10-4 to 10-5 of the absorbance change. The instrument response time is between 400 and 500 fs for fluorescence detection and less than 300 fs for transient-absorption measurements. All experiments were done at the magic angle (54.7°). Samples were kept in stirring quartz cells during irradiation to avoid heating and photobleaching. Experiments were carried out under anaerobic conditions for radical and fully-reduced flavins and under aerobic condition for oxidized flavins.
Results and Discussion
Steady-State Spectroscopic Properties
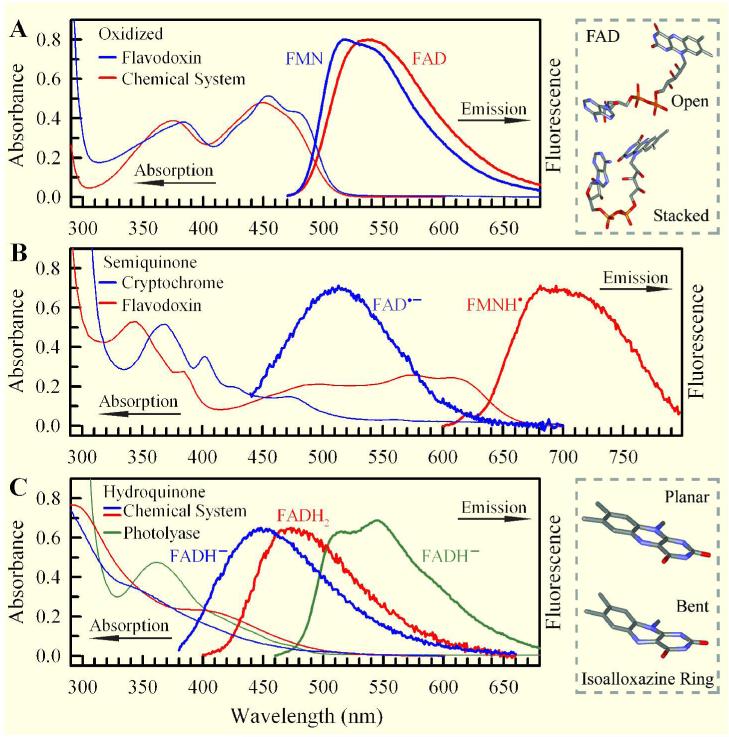
Figure 1 shows the steady-state absorption and emission spectra of flavin cofactors in various redox states and environments. Oxidized FAD in solution exhibits two broad absorption bands, with the peaks at 450 nm for S0→S1 and at 375 nm for S0→S2,43 while the FMN enclosed in the flavodoxin mutant shows some peak features at 476, 454, and 432 nm in S0→S1 and a broad band at 384 nm for S0→S2 absorption. The emission peak of oxidized flavin is around 530 nm in solution and shifts to the blue side in a hydrophobic protein environment in flavodoxin (Figure 1A).66,67 It has been reported that the fluorescence intensity of FAD* in solution is about 10-fold weaker than that of excited riboflavin and FMN,44-46 due to intramolecular ET quenching with a stacked conformation between the isoalloxazine ring and the adenine moiety. Numerous experimental and theoretical studies44-46,48,68-73 suggested that two FAD conformations, open and stacked, exist in solution (Figure 1A).
Figure 1.
Steady-state absorption and emission spectra of flavins in five redox states. (A) oxidized state, (B) radical state, and (C) fully-reduced state. The excitation wavelengths are 400 nm for oxidized FAD and FMN, 420 nm for anionic radical FAD•-, 580 nm for neutral radical FMNH•, and 360 nm for fully-reduced FADH- and FADH2. The fluorescence emissions of FAD•- in the cryptochrome and FADH- and FADH2 in solution are very weak. Upper right frame: Schematic representation of FAD in open and stacked conformations. The stacked conformation was adopted from U—shaped FAD in photolyase. Lower right frame: Schematic representation of the isoalloxazine ring in planar and “butterfly” bent conformations.
Figure 1B shows the absorption and emission spectra of stable radical semiquinones in two flavoproteins. Although the absorption of radical flavins in solution was reported by pulse radiolysis,26,27 the emission spectra simply lack. The emission spectrum of the neutral radical semiquinones is reported here for the first time. The absorption of the neutral radical FMNH• in flavodoxin mutant extends until 700 nm and a weak broad emission spectrum peaked at 700 nm was observed (Figure 1B). We also observed a similar fluorescence profile from FADH• in photolyase with a peak at 715 nm. The anionic radical FAD•- in insect Type 1 cryptochrome also gives a weak emission spectrum with a peak at 513 nm upon 420-nm excitation (Figure 1B) and it was shown that the emission peak depends on excitation wavelength30 (also see below).
Figure 1C shows the absorption and emission spectra of fully-reduced anionic and neutral flavins (FADH- and FADH2) in solution and in photolyase. The absorption spectra in solution are consistent with those previously reported for reduced FMNH- and FMNH2,25 but the emission spectra are first reported here. Upon 360-nm excitation, we observed weak fluorescence emission peaked at 455 nm for anionic hydroquinone (FADH-) and at 480 nm for neutral hydroquinone (FADH2). However, in photolyase, the anionic hydroquinone (FADH-) clearly shows an absorption band with a peak at 360 nm and its emission spectrum shows a structured fluorescence profile peaked at 515 and 545 nm (Figure 1C). The fluorescence intensity of fully-reduced flavins in solution is much weaker than that obtained in the protein, suggesting very different dynamic behaviors (see below), which may be involved with a bending motion of the isoalloxazine ring (Figure 1C).
Oxidized Flavins: Intramolecular Charge Separation and Recombination
As mentioned earlier, excited FMN and FAD in inert protein environments, as well as FMN* in solution, all have a lifetime in several nanoseconds.39-41,46,48 For example, in the flavodoxin mutant we observed a 6.0-ns decay of FMN* (Figure 2A inset). For FAD* in solution, at 530-nm emission we observed a biexponential decay in 9.2 ps (50%) and 2.5 ns (50%), due to the heterogeneous conformations, stacked and open,44 respectively (Figure 2A inset). The observed 2.5 ns reflects the lifetime of FAD* in the open conformation without significant intramolecular ET reaction. Here, we systematically study the excited-state dynamics of FAD in the stacked conformation with focus on intramolecular charge separation and recombination. The observed decay in 9.2 ps represents an ET process from the adenine moiety to the excited isoalloxazine ring in the stacked conformation, consistent with the recent observation of 9.0 ps.42 Our recent 10-ns molecular dynamics (MD) simulations as well as by others in 4-ns simulations46 showed the fluctuation of conformations with a center-to-center distance of the two stacked rings at 5-6 Å, consistent with prediction of the ET dynamics in 1-10 ps at such a distance.74 The oxidation potential of adenine is 1.96V vs. NHE75 and the reduction potential of FAD is about -0.22 V vs. NHE.76 Considering the S0→S1 (0-0) transition at 480 nm (2.59 eV), we obtained a net ΔG of -0.41 eV. Thus, the intramolecular ET between the two rings is energetically favorable. Similar to our recent studies of protein hydration,77,78 when we gated the blue side of the emission peak, we observed faster dynamics, a mixture of solvation dynamics and the ET reaction; for example, at 490-nm emission, the transient dynamics shows a fast component of 1.4 ps (64%) besides ∼7.9-ps (29%) ET process and ∼1.8-ns (7%) lifetime emission of the open conformation (Figure 2A).
Figure 2.

Femtosecond-resolved oxidized flavin dynamics in solution and in flavodoxin. (A) Normalized fluorescence transients of excited FAD gated at 490 and 530 nm with removal of the long lifetime components in nanosecond. The complete signal is shown in inset with comparison to the transient of FMN* in the flavodoxin mutant with a predominant lifetime in nanosecond. (B) Transient-absorption detection of oxidized FAD in solution for various pump/probe schemes. Note that the nanosecond decay from non-ET dynamics has been subtracted for clarity. The coupled solvation and ET dynamics is in 4-5 ps and the charge recombination takes 30-40 ps.
After charge separation, the redox pair of adenine+ and isoalloxazine- (Aden+-Iso-) must proceed to charge recombination to restore the original form of FAD. In Figure 2B, we used the transient-absorption method to detect all putative intermediates and products. Note that the nanosecond dynamics from open conformations, non-ET processes, has been removed for clarity. At 480-nm excitation, we observed positive signals in two probing ranges of 800-600 nm and 550-400 nm, resulting from the excited-state FAD and intermediates of Aden+ and Iso-.79,80 Specifically, at 800-nm probing, we observed a decay component of 4.5 ps (100%) for FAD* and a formation (4.5 ps) and decay (33 ps) component (16%) for Aden+.80 The observed 4∼5 ps decay of FAD* here, consistent with the previous study,43 is faster than that obtained from the fluorescence detection (9.2 ps), indicating that the transient absorption may detect coupled solvation and ET dynamics. When we tuned the probe to 680 nm, we observed a similar dynamics with a larger amplitude of Aden+ (23%), resulting from the relatively stronger absorption of Aden+.80 The observed ∼ 33 ps is important and represents the dynamics of charge recombination between Aden+ and Iso-. When we tuned the probe to 540 nm and 510 nm, we are also able to detect the intermediate Iso- and observed a 4.9-5.9 ps decay of FAD* (100%) and a formation (4.9-5.9 ps) and decay (37-38 ps) component for Aden+ and/or Iso- (15%). However, from 600 to 550 nm probing, we observed negative signals due to the simulated emission from FAD*.43 For example, at 560-nm probing, the transient shows an emission dynamics in 1.2 ps (68%) and 8.6 ps (32%), consistent with our fs-resolved fluorescence results (Figure 2A). More importantly, at 400-nm excitation and 480-nm probing, we observed the ground-state FAD recovery in ∼30 ps, resulting from charge recombination, as well as the formation and decay dynamics of the intermediates Aden+ and Iso-. The successful detection of the ground-state recovery is critical to further confirmation of the intramolecular charge separation in 5-9 ps and charge recombination in 30-40 ps for stacked FAD* in solution.
Anionic and Neutral Radical Flavins: Fluorescence Emission and Intrinsic Lifetime
The observation of the flavin radical emission is significant (Figure 1B) to studies of the spectroscopic property of flavoproteins and, more importantly, provides a way to preciously measure their excited-state dynamics using the fs-resolved fluorescence method.81 In Figure 3A, we show the fluorescence transient of anionic radical FAD•-* gated at 550-nm emission. The transient exhibits three exponential decay components: 2.2 ps (41%), 30 ps (35%) and 530 ps (24%). In the absence of electron acceptors, the observed multiple decays reflect the deactivation dynamics of excited FAD•- in the cryptochrome.30 Also, we observed the excitation-wavelength dependence of emission spectra, as shown in Figure 3A inset, indicating that the excited FAD•- does not completely relax to the lowest excited state and deactivates during relaxation. Our previous studies30 proposed that the isoalloxazine ring undergoes a “butterfly” bending motion in the ground or excited state that leads to multiple deactivation processes through conical intersection(s) in which the rapid decay pathways predominate. The proposed bending motion here is attributed to the higher plasticity of the cryptochrome and the less rigidity of its active site.82 Such conical intersections are often observed in large organic molecules, such as in DNA bases.83,84
Figure 3.
Femtosecond-resolved fluorescence transients of anionic radical FAD•- in the insect cryptochrome (ApCRY1) gated at 550 nm (A) and of neutral radical FMNH• in the flavodoxin mutant gated at 680 nm (B). Insets show the excitation-wavelength dependence of weak FAD•- emission spectra, indicating deactivation during relaxation, and the excitation-wavelength independence of weak FMNH• emission spectra, resulting from the lowest excited-state emission. For all emission spectra, the Raman scattering signals at the blue side of emission peaks were all removed for clarity.
Figure 3B shows the fluorescence transient of the neutral radical FMNH•* in the flavodoxin mutant gated at the 680-nm emission. The transient follows a single-exponential decay in 230 ps. Comparing the free energy changes between the oxidized FMN and neutral radical FMNH• reductions, the excited FMN favors photoreduction.60 However, we observed a 6.0-ns lifetime of FMN* in the protein (Figure 2A inset), indicating that no ET reaction occurs between oxidized FMN with F60 or F98. Thus, the observed 230-ps dynamics represents the intrinsic lifetime of excited FMNH• in the flavodoxin mutant. The weak fluorescence profile of FMNH•*in the mutant is independent of excitation wavelengths (Figure 3B inset) and must result from the lowest excited-state emission, consistent with the observed single lifetime of 230 ps and the fact that the isoalloxazine ring is rigidly stacked between F60 and F98 residues and has a nearly planar structure.85
Anionic and Neutral Fully-Reduced Flavins: Dynamic Deactivation and “Butterfly” Bending Motion
In Figure 4, we show the fluorescence transients of excited fully-reduced hydroquinones in solution and in inert protein environments. Figure 4A shows the dynamics of anionic hydroquinone FADH-* in photolyase and FMNH-* in flavodoxin with dominant long lifetimes of 1.3 ns and 0.8 ns, respectively. However, in solution we observed very rapid deactivation of FADH-* and FMNH-* (Figure 4B). Upon 325-nm excitation, the fluorescence transient at 450 nm emission exhibits the multiple decay dynamics in 5.0 ps (84%), 31 ps (13%) and 2.0 ns (3%) for FADH- and in 5.8 ps (80%), 35 ps (19%) and 1.5 ns (< 1%) for FMNH-. Similarly, we also examined the transient at 470 nm emission for neutral hydroquinone FADH2* (Figure 4C) and the dynamics show a similar ultrafast decay behavior in 2.9 ps (50%) and 40 ps (24%) with a significant nanosecond component (1.0 ns and 26%). Previous studies also observed the multiple decay dynamics with limited time resolution.49-51 For example, the fluorescence decay of FMNH-* in aqueous solution at 4°C was reported with a predominant fast component of 115 ps (80%) and slower nanosecond components (20%).50
Figure 4.

Femtosecond-resolved fluorescence transients of fully-reduced flavins. Fully-reduced anionic FADH- and FMNH- in the proteins (A), photolyase and the flavodoxin mutant, and in solution (B). (C) Fully-reduced neutral FADH2 in solution. Insets show strong excitation-wavelength dependence of weak FADH- and FADH2 emission spectra, indicating deactivation during relaxation. The Raman scattering signals at the blue side of emission peaks were all removed for clarity.
We also examine their dynamics using transient absorption detection with probing wavelengths from 585 nm to 690 nm. All these fully-reduced flavins in solution show similar multiple decay dynamics on similar time scales for all pump/probe schemes (Figure 5). These transients can be globally fitted with three exponential decays: 8∼18 ps (50∼60%), 40∼70 ps (20∼30%) and 2∼3 ns (10∼15%), slightly slower than those obtained from fluorescence measurements. Previous studies mainly showed the dynamics with a biexponential decay in 4∼60 ps and 1.5 ns,53 but with high excitation energy, low time resolution, and different probing wavelengths.
Figure 5.
Femtosecond-resolved transient-absorption detection of fully-reduced flavins in solution upon 325-nm excitation with probing from 690 to 585 nm. (A) FMNH- (B) FADH- and (C) FADH2. The dynamics of each species at different probing wavelengths are all similar.
Due to the similar dynamics of FADH-* and FMNH-* and the lack of the adenine moiety in FMNH-, we exclude the possible intramolecular ET reaction in FADH-* in solution and the observed multiple-decay dynamics truly reflects the ultrafast deactivation of the excited state, as observed for FAD•-* in the cryptochrome. Thus, given by the similarity of all transients, the multiple decays of FMNH-*, FADH-* and FADH2* also reflect their ultrafast deactivation dynamics. Similar to FAD•-* in the cryptochrome, all weak fluorescence emission shows strong dependence of excitation wavelengths (Figure 4B and 4C inset), consistent with the deactivation mechanism by “butterfly” bending motions across conical intersection(s). Both 13C and 15N MNR studies86,87 and various theoretical calculations88-93 indicated that the isoalloxazine ring of fully-reduced flavins is in a bent conformation, whereas it is planar in oxidized flavins and close to planar in neutral radical form. The activation barrier of the “butterfly” bending motion in the isoalloxazine ring is small (<20 kJ/mole).87 The planarity of oxidized form originates from the electron delocalization among the three rings of isoalloxazine and the oxidized isoalloxazine appears to be stiffer than its reduced form.85 The MD studies72 showed clear “butterfly” characteristics of reduced flavins in water. In light of the striking excitation-wavelength dependence of emission spectra and all these relevant evidence,30,49,85-93 a flexible “butterfly” bending motion of the isoalloxazine ring is proposed to access CI(s) for the observed multiple deactivation processes, as shown in Figure 6. The observed striking different dynamics of FADH-* in solution and in photolyase indicate very different environment; in photolyase, the FADH- is highly restricted, geometrically and electrostatically, while in solution it is more flexible with a “butterfly” bending motion responsible for its deactivation (Figure 6).
Figure 6.
Schematic potential energy landscape of fully-reduced flavins in solution and in proteins. With a flexible bending motion to access CI(s), the flavin molecule in solution exhibits complex deactivation dynamics. In photolyase and the flavodoxin mutant, the bending motion is restricted at the local region and the flavin molecule could not get access to CI(s), leading to a long nanosecond lifetime. The bending coordinate ( θ) could be defined for the “butterfly” motion of the isoalloxazine ring along N5—N10 axis.
To ascertain the deactivation mechanism through CI(s), we further performed a series of dynamics studies by varying pump wavelengths, solvent viscosity, and temperature. Figure 7A and 7B show the fluorescence transients of FADH-* and the absorption transients of FMNH-*, respectively, at 400 and 325 nm excitation. Clearly, the dynamics slows down with longer excitation wavelength, consistent with the fact that the higher the excitation, the faster the deactivation (Figure 6). Because the deactivation is accomplished by the “butterfly” bending motion to cross conical intersection(s), the increase in solvent viscosity would slow down the bending motion and thus the molecules take longer time to deactivate. Figure 7C shows that the dynamics of FMNH-* in solution with 50% (v/v) glycerol is clearly slower than that without glycerol, consistent with the deactivation mechanism of the “butterfly” bending motion mainly responsible for accesses to the CI region. We also studied the dynamics of FADH-* in photolyase with changes in solvent viscosity and temperature. Figure 7D and 7E show the fluorescence dynamics under those conditions. Clearly, the more flexible the environment, the shorter the lifetime. Although the molecule FADH-* may not get access to the CI region by constraints in photolyase, the flexibility is clearly correlated to its lifetime and thus the bending motion also seems to be a critical motion coordination for facilitating the coupling in internal conversion to ground state.
Figure 7.

Comparative studies on fully-reduced flavins with various excitation wavelengths, solvent viscosity, and temperatures. (A) Normalized fluorescence transients of excited FADH- in solution with 400 and 325 nm excitation. (B) Normalized absorption transients of excited FMNH- in solution with 400 and 325 nm excitation and 585 nm probing. (C) Normalized absorption transients of excited FMNH- in 0% and 50% glycerol solutions. (D) Normalized fluorescence transients of excited FADH- in photolyase with 10%, 30% and 50% glycerol buffers. (E) Normalized fluorescence transients of FADH- in photolyase under 0°C, 12 °C and 25°C.
Conclusion
We reported here the systematic studies of excited-state dynamics of flavin molecules in five redox states from oxidized form, to neutral and anionic semiquinone radicals, and to neutral and anionic fully-reduced hydroquinones in solution and in inert protein environments. With femtosecond resolution, we characterized photophysics and photochemistry of each redox state with two common flavins of FMN and FAD. Specifically, for oxidized state, the FAD molecule can fold into a stacked conformation in solution and we observed ultrafast intramolecular charge separation in ∼5-9 ps from the adenine moiety to the excited isoalloxazine ring and subsequently charge recombination in ∼30-40 ps, while the FAD in an open configuration as well as the FMN have a lifetime in nanoseconds in solution and in inert proteins. For semiquinones, we observed weak fluorescence emission from both neutral and anionic states for the first time by utilizing protein’s environments to stabilize the radicals. For the neutral state, we obtained the intrinsic lifetime in 230 ps but for the anionic state, we observed multiple deactivation dynamics on the picosecond time scale (2-530 ps), which is related to the planarity of the isoalloxazine ring. A butterfly bending motion is believed to play a central role to get access to conical interaction(s) to facilitate deactivation. Such ultrafast deactivation dynamics are also observed for the fully-reduced states (3 ps-2 ns), both neutral and anionic, in solution without environmental restriction. In the protein with more constraints, the lifetime becomes longer in nanoseconds. Significantly, for all species with ultrafast deactivation, the weak emission spectra show significant dependence of excitation wavelengths and the molecules deactivate during relaxation.
It is striking that the excited-state dynamics are directly correlated with the flexibility of the isoalloxazine ring. For all observed ultrafast photophysics, flavins favor a bent ring structure while in a planar configuration, flavins have a nanosecond lifetime. The proposed butterfly bending is a critical motion coordinate and directly controls the excited-state dynamics of flavins. Thus, enzymes can utilize geometric restriction and electrostatic interactions to manipulate the flavin cofactor’s configuration, planar or bent, to achieve various biological functions. In photolyase, the fully-reduced flavin has a nearly planar structure to make excited flavin molecules stay long enough (1 ns) to react with substrates to repair damaged DNA and thus to reach the highest efficiency, while in insect Type 1 cryptochrome, the plasticity of the protein makes the anionic semiquinone more flexible, resulting in complex deactivation dynamics from a few to hundreds of picoseconds to modulate the functional channel. With the understanding of excited-state dynamics of five redox states now, more and more fundamental molecular mechanisms of various biological functions by flavoproteins will be elucidated.
Acknowledgements
We like to thank Chuang Tan for help with the cryptochrome experiments. This work was supported in part by the Packard Foundation Fellowship to DZ and the National Institute of Health (GM31082 and GM74813) to DZ and AS. We thank Prof. Richard Swenson (Ohio State Univ.) for the generous gift of flavodoxin plasmid and purification procedures.
References
- (1).Walsh C. Acc. Chem. Res. 1980;13:148–155. [Google Scholar]
- (2).Massey V, Hemmerich P. Biochem. Soc. Trans. 1980;8:246–257. doi: 10.1042/bst0080246. [DOI] [PubMed] [Google Scholar]
- (3).Müller F. The Flavin Redox-System and Its Biological Function. In: Boschke FL, editor. Topics in Current Chemistry. Vol. 108. Springer -Verlag; Berlin: 1983. pp. 71–107. [DOI] [PubMed] [Google Scholar]
- (4).Müller F, editor. Chemistry and Biochemistry of Flavoenzymes. I-III. CRC Press; Boca Raton, FL: 19901991. [Google Scholar]
- (5).Fraaije MW, Mattevi A. Trends Biochem. Sci. 2000;25:126–132. doi: 10.1016/s0968-0004(99)01533-9. [DOI] [PubMed] [Google Scholar]
- (6).Massey V. Biochem. Soc. Trans. 2000;28:283–296. [PubMed] [Google Scholar]
- (7).Bornemann S. Nat. Prod. Rep. 2002;19:761–772. doi: 10.1039/b108916c. [DOI] [PubMed] [Google Scholar]
- (8).Joosten V, van Berkel WJH. Curr. Opin. Chem. Biol. 2007;11:195–202. doi: 10.1016/j.cbpa.2007.01.010. [DOI] [PubMed] [Google Scholar]
- (9).Susin SA, Lorenzo HK, Zamzami N, Marzo I, Snow BE, Brothers GM, Mangion J, Jacotot E, Costantini P, Loeffler M, Larochette N, Goodlett DR, Aebersold R, Siderovski DP, Penninger JM, Kroemer G. Nature. 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- (10).Chen Y-C, Lin-Shiau S-Y, Lin J-K. J. Cell. Physiol. 1998;177:324–333. doi: 10.1002/(SICI)1097-4652(199811)177:2<324::AID-JCP14>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- (11).Dagley S. Annu. Rev. Microbiol. 1987;41:1–23. doi: 10.1146/annurev.mi.41.100187.000245. [DOI] [PubMed] [Google Scholar]
- (12).Chen HZ, Hopper SL, Cerniglia CE. Microbiology. 2005;151:1433–1441. doi: 10.1099/mic.0.27805-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Fitzpatrick PF. Acc. Chem. Res. 2001;34:299–307. doi: 10.1021/ar0000511. [DOI] [PubMed] [Google Scholar]
- (14).Massey V. J. Biol. Chem. 1994;269:22459–22462. [PubMed] [Google Scholar]
- (15).van Pée K-H, Patallo EP. Appl. Microbiol. Biotechnol. 2006;70:631–641. doi: 10.1007/s00253-005-0232-2. [DOI] [PubMed] [Google Scholar]
- (16).Dong C, Flecks S, Unversucht S, Haupt C, van Pée K-H, Naismith JH. Science. 2005;309:2216–2219. doi: 10.1126/science.1116510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Sancar A. Chem. Rev. 2003;103:2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- (18).Kao Y-T, Saxena C, Wang L, Sancar A, Zhong D. Proc. Natl. Acad. Sci. U.S.A. 2005;102:16128–16132. doi: 10.1073/pnas.0506586102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Partch CL, Sancar A. Photochem. and Photobiol. 2005;81:1291–1304. doi: 10.1562/2005-07-08-IR-607. [DOI] [PubMed] [Google Scholar]
- (20).Lin C, Shalitin D. Annu. Rev. Plant Biol. 2003;54:469–496. doi: 10.1146/annurev.arplant.54.110901.160901. [DOI] [PubMed] [Google Scholar]
- (21).Lin C, Todo T. Genome Biol. 2005;6:220.1–220.9. doi: 10.1186/gb-2005-6-5-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Kennis JTM, Groot M-L. Curr. Opin. Struct. Biol. 2007;17:623–630. doi: 10.1016/j.sbi.2007.09.006. [DOI] [PubMed] [Google Scholar]
- (23).Christie JM. Annu. Rev. Plant Biol. 2007;58:21–45. doi: 10.1146/annurev.arplant.58.032806.103951. [DOI] [PubMed] [Google Scholar]
- (24).Heelis PF. Chem. Soc. Rev. 1982;11:15–39. [Google Scholar]
- (25).Ghisla S, Massey V, Lhoste J-M, Mayhew SG. Biochemistry. 1974;13:589–597. doi: 10.1021/bi00700a029. [DOI] [PubMed] [Google Scholar]
- (26).Land EJ, Swallow AJ. Biochemistry. 1969;8:2117–2125. doi: 10.1021/bi00833a050. [DOI] [PubMed] [Google Scholar]
- (27).Sakai M, Takahashi H. J. Mol. Struct. 1996;379:9–18. [Google Scholar]
- (28).Massey V, Palmer G. Biochemistry. 1966;5:3181–3189. doi: 10.1021/bi00874a016. [DOI] [PubMed] [Google Scholar]
- (29).Li YF, Sancar A. Biochemistry. 1990;29:5698–5706. doi: 10.1021/bi00476a009. [DOI] [PubMed] [Google Scholar]
- (30).Kao Y-T, Tan C, Song S-H, Öztürk N, Li J, Wang L, Sancar A, Zhong D. J. Am. Chem. Soc. 2008;130:7695–7701. doi: 10.1021/ja801152h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Zhong D, Zewail AH. Proc. Natl. Acad. Sci. U.S.A. 2001;98:11867–11872. doi: 10.1073/pnas.211440398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Mataga N, Chosrowjan H, Taniguchi S, Tanaka F, Kido N, Kitamura M. J. Phys. Chem. B. 2002;106:8917–8920. [Google Scholar]
- (33).Park H-W, Kim S-T, Sancar A, Deisenhofer J. Science. 1995;268:1866–1872. doi: 10.1126/science.7604260. [DOI] [PubMed] [Google Scholar]
- (34).Li YF, Heelis PF, Sancar A. Biochemistry. 1991;30:6322–6329. doi: 10.1021/bi00239a034. [DOI] [PubMed] [Google Scholar]
- (35).Aubert C, Vos MH, Mathis P, Eker APM, Brettel K. Nature. 2000;405:586–590. doi: 10.1038/35014644. [DOI] [PubMed] [Google Scholar]
- (36).Byrdin M, Eker APM, Vos MH, Brettel K. Proc. Natl. Acad. Sci. U.S.A. 2003;100:8676–8681. doi: 10.1073/pnas.1531645100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Saxena C, Sancar A, Zhong D. J. Phys. Chem. B. 2004;108:18026–18033. [Google Scholar]
- (38).Wang H, Saxena C, Quan D, Sancar A, Zhong D. J. Phys. Chem. B. 2005;109:1329–1333. doi: 10.1021/jp044652b. [DOI] [PubMed] [Google Scholar]
- (39).Gauden M, Yeremenko S, Laan W, van Stokkum IHM, Ihalainen JA, van Grondelle R, Hellingwerf KJ, Kennis JTM. Biochemistry. 2005;44:3653–3662. doi: 10.1021/bi047359a. [DOI] [PubMed] [Google Scholar]
- (40).Kennis JTM, Crosson S, Gauden M, van Stokkum IHM, Moffat K, van Grondelle R. Biochemistry. 2003;42:3385–3392. doi: 10.1021/bi034022k. [DOI] [PubMed] [Google Scholar]
- (41).Holzer W, Penzkofer A, Fuhrmann M, Hegemann P. Photochem. Photobiol. 2002;75:479–487. doi: 10.1562/0031-8655(2002)075<0479:scofmb>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- (42).Chosrowjan H, Taniguchi S, Mataga N, Tanaka F, Visser AJWG. Chem. Phys. Lett. 2003;378:354–358. [Google Scholar]
- (43).Stanley RJ, MacFarlane AW. J. Phys. Chem. A. 2000;104:6899–6906. [Google Scholar]
- (44).Weber G. Biochem. J. 1950;47:114–121. doi: 10.1042/bj0470114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Barrio JR, Tolman GL, Leonard NJ, Spencer RD, Weber G. Proc. Natl. Acad. Sci. U.S.A. 1973;70:941–943. doi: 10.1073/pnas.70.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).van den Berg PAW, Feenstra KA, Mark AE, Berendsen HJC, Visser AJWG. J. Phys. Chem. B. 2002;106:8858–8869. [Google Scholar]
- (47).Li G, Glusac KD. J. Phys. Chem. A. 2008;112:4573–4583. doi: 10.1021/jp7117218. [DOI] [PubMed] [Google Scholar]
- (48).Visser AJWG. Photochem. Photobiol. 1984;40:703–706. doi: 10.1111/j.1751-1097.1984.tb04640.x. [DOI] [PubMed] [Google Scholar]
- (49).Visser AJWG, Ghisla S, Lee J. Picosecond Fluorescence Dynamics of Reduced Flavins. In: Curti B, Ronchi S, Zanetti G, editors. Flavins and Flavoproteins 1990. Wlater de Gruyter & Co.; Berlin: 1991. pp. 49–54. [Google Scholar]
- (50).Leenders R, Kooijman M, van Hoek A, Veeger C, Visser AJWG. Eur. J. Biochem. 1993;211:37–45. doi: 10.1111/j.1432-1033.1993.tb19867.x. [DOI] [PubMed] [Google Scholar]
- (51).Bruggeman YE, Schoenmakers RG, Schots A, Pap EHW, van Hoek A, Visser AJWG, Hilhorst R. Eur. J. Biochem. 1995;234:245–250. doi: 10.1111/j.1432-1033.1995.245_c.x. [DOI] [PubMed] [Google Scholar]
- (52).Heelis PF, Hartman RF, Rose SD. Photochem. Photobiol. 1993;57:1053–1055. doi: 10.1111/j.1751-1097.1993.tb02970.x. [DOI] [PubMed] [Google Scholar]
- (53).Enescu M, Lindqvist L, Soep B. Photochem. Photobiol. 1998;68:150–156. doi: 10.1562/0031-8655(1998)068<0150:esdofr>2.3.co;2. [DOI] [PubMed] [Google Scholar]
- (54).Kim S-T, Heelis PF, Okamura T, Hirata Y, Mataga N, Sancar A. Biochemistry. 1991;30:11262–11270. doi: 10.1021/bi00111a011. [DOI] [PubMed] [Google Scholar]
- (55).Visser AJWG, Ghisla S, Massey V, Müller F, Veeger C. Eur. J. Biochem. 1979;101:13–21. doi: 10.1111/j.1432-1033.1979.tb04210.x. [DOI] [PubMed] [Google Scholar]
- (56).Fox JL. FEBS Lett. 1974;39:53–55. doi: 10.1016/0014-5793(74)80015-3. [DOI] [PubMed] [Google Scholar]
- (57).Müller F, Massey V, Heizmann C, Hemmerich P, Lhoste J-M, Gould DC. Eur. J. Biochem. 1969;9:392–401. doi: 10.1111/j.1432-1033.1969.tb00621.x. [DOI] [PubMed] [Google Scholar]
- (58).Ghisla S, Massey V. J. Biol. Chem. 1975;250:577–584. [PubMed] [Google Scholar]
- (59).Massey V, Stankovich M, Hemmerich P. Biochemistry. 1978;17:1–8. doi: 10.1021/bi00594a001. [DOI] [PubMed] [Google Scholar]
- (60).Swenson RP, Krey GD. Biochemistry. 1994;33:8505–8514. doi: 10.1021/bi00194a015. [DOI] [PubMed] [Google Scholar]
- (61).Traber R, Kramer HEA, Hemmerich P. Biochemistry. 1982;21:1687–1693. doi: 10.1021/bi00536a033. [DOI] [PubMed] [Google Scholar]
- (62).Song S-H, Öztürk N, Denaro TR, Arat NÖ, Kao Y-T, Zhu H, Zhong D, Reppert SM, Sancar A. J. Biol. Chem. 2007;282:17608–17612. doi: 10.1074/jbc.M702874200. [DOI] [PubMed] [Google Scholar]
- (63).Öztürk N, Song S-H, Selby CP, Sancar A. J. Biol. Chem. 2008;283:3256–3263. doi: 10.1074/jbc.M708612200. [DOI] [PubMed] [Google Scholar]
- (64).Sancar A, Smith FW, Sancar GB. J. Biol. Chem. 1984;259:6028–6032. [PubMed] [Google Scholar]
- (65).Heelis PF, Payne G, Sancar A. Biochemistry. 1987;26:4634–4640. doi: 10.1021/bi00389a007. [DOI] [PubMed] [Google Scholar]
- (66).Malicka J, Groth M, Karolczak J, Czaplewski C, Liwo A, Wiczk W. Biopolymers. 2001;58:447–457. doi: 10.1002/1097-0282(20010405)58:4<447::AID-BIP1020>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- (67).Reichardt C. Solvents and Solvent Effects in Organic Chemistry. 3rd Wiley-VCH; Weinheim: 2003. [Google Scholar]
- (68).Kotowycz G, Teng N, Klein MP, Calvin M. J. Biol. Chem. 1969;244:5656–5662. [PubMed] [Google Scholar]
- (69).Kainosho M, Kyogoku Y. Biochemistry. 1972;11:741–752. doi: 10.1021/bi00755a011. [DOI] [PubMed] [Google Scholar]
- (70).Copeland RA, Spiro TG. J. Phys. Chem. 1986;90:6648–6654. [Google Scholar]
- (71).Weber G, Tanaka F, Okamoto BY, Drickamer HG. Proc. Natl. Acad. Sci. U.S.A. 1974;71:1264–1266. doi: 10.1073/pnas.71.4.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Hahn J, Michel-Beyerle M-E, RÖsch N. J. Mol. Model. 1998;4:73–82. [Google Scholar]
- (73).Islam SDM, Susdorf T, Penzkofer A, Hegemann P. Chem. Phys. 2003;295:137–149. [Google Scholar]
- (74).Moser CC, Keske JM, Warncke K, Farid RS, Dutton PL. Nature. 1992;355:796–802. doi: 10.1038/355796a0. [DOI] [PubMed] [Google Scholar]
- (75).Seidel CAM, Schulz A, Sauer MHM. J. Phys. Chem. 1996;100:5541–5553. [Google Scholar]
- (76).Lowe HJ, Clark WM. J. Biol. Chem. 1956;221:983–992. [PubMed] [Google Scholar]
- (77).Qiu W, Kao Y-T, Zhang L, Yang Y, Wang L, Stites WE, Zhong D, Zewail AH. Proc. Natl. Acad. Sci. U.S.A. 2006;103:13979–13984. doi: 10.1073/pnas.0606235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Zhang L, Wang L, Kao Y-T, Qiu W, Yang Y, Okobiah O, Zhong D. Proc. Natl. Acad. Sci. U.S.A. 2007;104:18461–18466. doi: 10.1073/pnas.0707647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Jovanovic SV, Simic MG. J. Phys. Chem. 1986;90:974–978. [Google Scholar]
- (80).Candeias LP, Steenken S. J. Am. Chem. Soc. 1993;115:2437–2440. [Google Scholar]
- (81).Wan C, Xia T, Becker H-C, Zewail AH. Chem. Phys. Lett. 2005;412:158–163. [Google Scholar]
- (82).Partch CL, Clarkson MW, Özgür S, Lee AL, Sancar A. Biochemistry. 2005;44:3795–3805. doi: 10.1021/bi047545g. [DOI] [PubMed] [Google Scholar]
- (83).Sobolewski AL, Domcke W. Eur. Phys. J. D. 2002;20:369–374. [Google Scholar]
- (84).Levine BG, Martínez TJ. Ann. Rev. Phys. Chem. 2007;58:613–634. doi: 10.1146/annurev.physchem.57.032905.104612. [DOI] [PubMed] [Google Scholar]
- (85).Lennon BW, Williams CH, Jr., Ludwig ML. Protein Sci. 1999;8:2366–2379. doi: 10.1110/ps.8.11.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Moonen CTW, Vervoort J, Müller F. Biochemistry. 1984;23:4859–4867. doi: 10.1021/bi00316a007. [DOI] [PubMed] [Google Scholar]
- (87).Moonen CTW, Vervoort J, Müller F. Biochemistry. 1984;23:4868–4872. doi: 10.1021/bi00316a008. [DOI] [PubMed] [Google Scholar]
- (88).Dixon DA, Lindner DL, Branchaud B, Lipscomb WN. Biochemistry. 1979;18:5770–5775. doi: 10.1021/bi00593a004. [DOI] [PubMed] [Google Scholar]
- (89).Zheng Y-J, Ornstein RL. J. Am. Chem. Soc. 1996;118:9402–9408. [Google Scholar]
- (90).Hasford JJ, Kemnitzer W, Rizzo CJ. J. Org. Chem. 1997;62:5244–5245. [Google Scholar]
- (91).Rodríguez-Otero J, Martínez-Núñez E, Peña-Gallego A, Vázquez SA. J. Org. Chem. 2002;67:6347–6352. doi: 10.1021/jo011159c. [DOI] [PubMed] [Google Scholar]
- (92).Climent T, González-Luque R, Merchán M, Serrano-Andrés L. J. Phys. Chem. A. 2006;110:13584–13590. doi: 10.1021/jp065772h. [DOI] [PubMed] [Google Scholar]
- (93).Choe Y-K, Nagase S, Nishimoto K. J. Comput. Chem. 2007;28:727–739. doi: 10.1002/jcc.20533. [DOI] [PubMed] [Google Scholar]