Abstract
Molecular mechanisms that initiate meiosis have been studied in fungi and mammals, but little is known about the mechanisms directing the meiosis transition in other organisms. To elucidate meiosis initiation in plants, we characterized and cloned the ameiotic1 (am1) gene, which affects the transition to meiosis and progression through the early stages of meiotic prophase in maize. We demonstrate that all meiotic processes require am1, including expression of meiosis-specific genes, establishment of the meiotic chromosome structure, meiosis-specific telomere behavior, meiotic recombination, pairing, synapsis, and installation of the meiosis-specific cytoskeleton. As a result, in most am1 mutants premeiotic cells enter mitosis instead of meiosis. Unlike the genes involved in initiating meiosis in yeast and mouse, am1 also has a second downstream function, whereby it regulates the transition through a novel leptotene–zygotene checkpoint, a key step in early meiotic prophase. The am1 gene encodes a plant-specific protein with an unknown biochemical function. The AM1 protein is diffuse in the nucleus during the initiation of meiosis and then binds to chromatin in early meiotic prophase I when it regulates the leptotene–zygotene progression.
Keywords: chromosomes, plant development, genetics, recombination
Anumber of processes taking place in early stages of meiotic prophase I, such as chromosome pairing and recombination, are known as critical for the chromosome number reduction in meiosis (1). These processes are specific to meiosis and must be regulated by mechanisms that control the transition from the somatic (mitotic) cell cycle to meiosis. However, in contrast to the overall evolutionary conservation of the basic meiotic processes, the mechanisms controlling the transition to meiosis show substantial diversity among species (2). Initiation of meiosis has so far been studied in budding and fission yeasts and more recently also in mouse. In budding yeast, meiosis is initiated by expression of the IME1 gene encoding a meiosis-specific transcription factor, which upregulates expression of the IME2 gene (3). IME2 encodes a protein kinase, directing the activation of a cyclin-dependent kinase that promotes entry into meiosis (4). In fission yeast, the master switch for meiosis initiation is the Mei2 gene encoding an RNA-binding protein (5). The Mei2 protein acts as a regulator of a complex system that sequesters meiotic transcript during vegetative growth (6). To initiate meiosis, Mei2 turns off this regulatory system so that meiosis-specific RNAs are no longer intercepted. In the mouse, Stra8, a vertebrate-only gene, is required for the transition to meiosis (7, 8). Stra8 expression is specifically induced in the germ cells by retinoic acid, a derivative of vitamin A, which is produced in the cells of the excretory organ of the embryo, and from there enters the gonads (8, 9).
Because the meiosis-initiating mechanisms in budding yeast, fission yeast, and mouse show little in common, it is likely that the transition to meiosis in other taxa is also regulated by species-dependent mechanisms. Although mammals and plants are both multicellular eukaryotes with complex developmental patterns, the transition from the mitotic cell cycle to meiosis in plants takes place in a very different developmental context than in mammals. To elucidate the mechanisms regulating the transition to meiosis in maize we examined the ameitic1 (am1) mutants that disrupt the entry into meiosis (10, 11). We cloned the am1 gene and determined that it encodes a plant-specific protein, indicating that meiosis in plants is initiated by a novel, plant-specific mechanism.
Results
Chromosome Behavior in am1 Mutants.
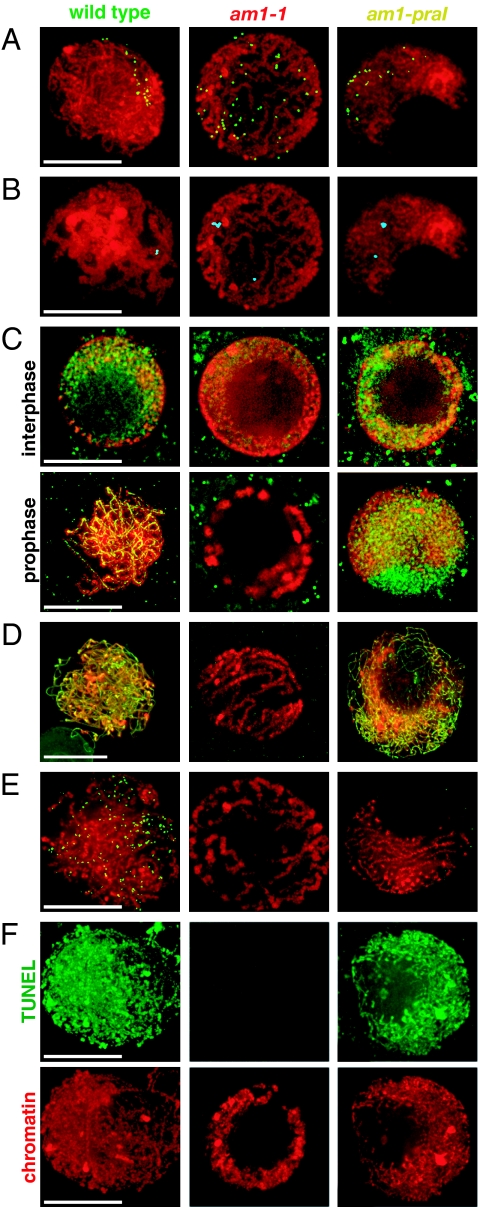
The 5 known mutants at the maize am1 locus can be grouped into 2 classes (Table 1). In the first class, comprising am1-1, am1-2, am1-485, and am1-489, male meiocytes, and in most cases also female meiocytes, do not enter a normal meiosis but instead undergo 2 or 3 rounds of equational division (11). The am1-praI mutant allele confers a dramatically different phenotype, in which male and female meiocytes enter meiotic prophase and arrest during its early stages (10). To understand the nature of the nuclear divisions in am1 mutants and discern whether the equational division is a mitosis or a defective meiosis, we used 3-dimensional deconvolution microscopy to examine chromosome behavior in wild-type meiocytes, as well as meiocytes from the am1-1 and am1-praI mutants. In wild-type meiocytes, prophase chromosomes are first visible in early leptotene as thin, elongated chromatin threads that surround a centrally located nucleolus. In late leptotene, the nucleolus moves to the side of the nucleus, and the bulk of the chromatin is located off-center during zygotene and most of pachytene (see chromosomes in Fig. 1A). In the am1-1 mutant, chromosomes also surround a centrally located nucleolus, but they do not move to the side of the nucleus at any time during prophase (Fig. 1A). Overall, chromosomes in am1-1 display behavior resembling the prophase of a mitotic division rather than meiosis. Male meiocytes in the am1-2, am1-485, and am1-489 mutants also behave in this way (supporting information Fig. S1). In contrast to the other am1 mutants, chromosomes in am1-praI meiocytes exhibit behavior similar to that of chromosomes in early stages of a normal meiosis (Fig. 1A).
Table 1.
The series of am1 mutant alleles
Arrow indicates the dominance relationship of the phenotype in female meiocytes (11). am1-489 is the most recessive; am1-praI is the most dominant of all am1 mutant alleles. am1-2 and am1-485 are codominant.
Fig. 1.
Chromosome behavior in male meiocytes in wild-type maize and in the am1-1 and am1-praI mutants. (A) Telomere bouquet analysis using FISH with the telomere probe (green). (B) Chromosome pairing analyzed using FISH with the 5S rRNA locus probe (cyan). (C) Installation of meiotic cohesin AFD1 (green), the maize Rec8 homologue, on chromosomes. The pictured wild-type meiocyte is in zygotene. (D) Installation of ASY1 (green), a meiosis-specific protein associated with chromosome axis. (E) Installation of the RAD51 recombination protein (green) on meiotic chromosomes. (F) Cytologic detection of meiotic DSB formation using a modified TUNEL assay. Green: TUNEL assay staining meiotic DSBs. (A–F) Red: DAPI-stained chromatin. (Scale bars, 10 μm.)
am1 Mutants Show Defective Telomere Bouquet Formation.
Clustering of telomeres on a single site on the nuclear envelope, known as telomere bouquet formation, is a central event of a general repositioning of chromosomes during zygotene (12). Bouquet formation is thought to facilitate homologous chromosome pairing by bringing the chromosome ends together and confining chromosomes to a limited region within the nucleus. To examine bouquet formation, we used FISH with a telomere-specific probe (Fig. 1A). In wild-type maize meiocytes, telomeres attached to the nuclear envelope during leptotene, clustered in early zygotene, and remained clustered until late zygotene. In contrast, telomeres in am1-1 mutant meiocytes did not attach to the nuclear envelope or cluster but were scattered throughout the nuclear volume (Fig. 1A). In am1-praI some bouquet formation was observed, although it did not progress to completion. A tight bouquet was present in approximately 8% of am1-praI meiocytes (n = 24), whereas another 37.5% of meiocytes displayed a “loose” bouquet or multiple bouquet clusters (Fig. 1A). The remaining am1-praI meiocytes showed no bouquet.
Homologous Chromosomes Do Not Pair in am1 Mutants.
To determine whether homologous chromosomes pair in am1 mutant meiocytes, we monitored chromosome behavior using FISH with the 5S rRNA locus probe. The maize genome contains a single 5S rRNA locus (13). In wild-type maize meiocytes, the 2 unpaired 5S foci are visible at the beginning of zygotene. At the beginning of pachytene, a single focus representing the homologously paired 5S loci is observed in each cell (Fig. 1B). In contrast, 2 separate foci were always visible in am1-1 meiocytes (Fig. 1B), indicating that homologous chromosomes did not pair. In am1-praI meiocytes, the 5S rRNA locus probe also showed only unpaired 5S loci (Fig. 1B).
Sister-Chromatid Cohesion and Meiotic Chromosome Axis Formation Are Defective in am1 Mutants.
Correct chromosome segregation in meiosis requires meiosis-specific sister-chromatid cohesion. A meiosis-specific α-kleisin Rec8 is a key component of the meiotic cohesin complex (14). Rec8 is recruited to chromosomes during the premeiotic S phase and is essential for both arm and centromere cohesion (15). To discern whether the meiotic cohesion complex is installed on chromosomes in am1 mutants, we used an antibody against the AFD1 protein, the maize homologue of Rec8 (16). In wild-type meiocytes, the antibody exhibits weak diffuse staining in premeiotic interphase (Fig. S2A) (16). In leptotene, it shows strong staining mostly localized to chromatin (Fig. S2B). In zygotene and pachytene, AFD1 is seen as mostly continuous stretches along chromosome cores (Figs. 1C and S2 C and D). We observed no AFD1 staining in mitotically dividing anther tapetal cells (Fig. S2 F and G). Although AFD1 staining was present in nuclei of dividing root meristem cells (Fig. S2 H and J), it was diffuse and did not localize to chromatin, indicating that just the presence of AFD1 in the nucleoplasm is not sufficient to ensure meiosis-specific cohesion.
In am1-1 mutant meiocytes, the AFD1 antibody showed weak diffuse staining that did not localize to chromatin (Fig. 1C) and was comparable, although less intense, to the staining in wild-type root meristem cells. In am1-praI meiocytes, the antibody showed strong staining that was partly localized to chromatin and similar to the staining in wild-type leptotene cells (Fig. 1C). These results indicate that AM1 is required for proper recruitment of AFD1 to meiotic chromosomes.
As meiocytes enter prophase I, the establishment of the leptotene chromosome structure relies at least partially on the installation of axial elements onto the chromosomes (17). In budding yeast, the HOP1 protein is an essential component of the axial element (18). ASY1, a meiosis-specific protein that associates with chromosome axes, has been proposed as the HOP1 homologue in Arabidopsis (19). We used an antibody against the Arabidopsis ASY1 protein to investigate the axial element assembly in am1-1, am1-praI, and wild-type meiocytes. In wild-type meiosis, the ASY1 antibody showed punctate staining along chromosomes when the axial elements form in leptotene (16, 19). In early zygotene, the signal became associated with the entire length of the axis (Fig. 1D). In am1-1 meiocytes, we detected no ASY1 staining (Fig. 1D) suggesting that a proper chromosome axis does not form. In contrast, am1-praI meiocytes showed punctuate ASY1 signals and some continuous stretches as in wild-type leptotene meiocytes (Fig. 1D), suggesting partial formation of the chromosome axis.
am1 Is Required for the Initiation and Progression of Meiotic Recombination.
Meiotic recombination, which includes the formation and repair of meiotic DSBs, is required for segregation of chromosomes in the first meiotic division and is one of the hallmarks of meiotic prophase I. To examine whether the meiotic recombination machinery is installed on chromosomes in am1 mutant meiocytes, we analyzed nuclear distribution of the recombination protein RAD51, which is involved in repair of meiotic DSBs and forms distinct foci on chromosomes on the sites of DSBs (20). In somatic plant cells, the foci are absent (20). In wild-type maize meiocytes, RAD51 foci are first visible in early zygotene and reach their peak number of 500 ± 47 per nucleus (n = 9) at mid-zygotene (Fig. 1E). In contrast, we did not find any RAD51 foci in male meiocytes in plants homozygous for the am1-1 and am1-praI mutations (Fig. 1E) or any of the other am1 mutant alleles (data not shown). These data indicate that the am1 gene is required for the installation of the recombination machinery on meiotic chromosomes.
To establish whether the absence of RAD51 foci was caused by the absence of meiotic DSBs, we used a modification of the TUNEL assay (21), which allows cytologic detection of the presence of meiotic DNA breaks. In wild-type maize meiocytes, the TUNEL assay produces a strong signal on chromosomes from leptotene through early pachytene (Fig. 1F). In contrast, we did not observe TUNEL staining in am1-1 meiocytes, implying the absence of DSBs (Fig. 1F). In am1-praI meiocytes, the staining pattern resembled that of the wild-type meiocytes, indicating the presence of meiotic DSBs (Fig. 1F).
am1 Regulates Meiocyte Cytoskeleton Organization.
During the G2 phase of the mitotic cell cycle, microtubules in plant cells are organized into a dense preprophase band that marks the future cell division plane. The preprophase band is, however, absent from meiotic cells (22). It has been reported previously that the preprophase band is present in am1-1 mutant meiocytes before they enter division (22). We have now determined that the am1-2, am1-485, and am1-489 meiocytes also show the mitotic pattern of microtubule organization (Fig. S3). In contrast, in am1-praI mutant meiocytes, the microtubules form an extensive network filling the cytoplasm, similar to what is observed in wild-type meiocytes (Fig. S3). Thus, different alleles of am1 regulate meiocyte microtubule behavior in a cell cycle stage-specific manner.
am1 Affects Meiotic Expression of dmc1.
In addition to examining the cellular processes that take place during meiotic prophase I, we investigated whether the am1 mutation affects expression of meiotic genes. To do this, we analyzed expression of dmc1, hop2, rad51, and spo11–1 in wild-type male flowers undergoing meiosis and in male flowers of the am1-1 and am1-praI mutants. Products of these 4 genes encode homologues of key meiotic recombination proteins (1). SPO11–1 is responsible for creating DSBs in chromosomal DNA that initiate meiotic recombination (23). DMC1, HOP2, and RAD51 are involved in repair of meiotic DSBs (24–27). DMC1, HOP2, and SPO11–1 act exclusively during meiosis (23–25). RAD51 is, in addition, thought to act in somatic DNA repair, although rad51 mutants in maize and Arabidopsis show only defects in meiosis (20, 26, 27). In contrast to their strictly meiotic roles, the HOP2 and SPO11–1 genes are expressed in a range of somatic tissues, at least in Arabidopsis (23, 25), suggesting that some meiotic genes in plants are not regulated at the transcriptional level but perhaps at the protein level. RAD51 is also expressed in somatic tissues in maize and Arabidopsis, but this expression pattern is consistent with its role in somatic DNA repair. In contrast to HOP2, SPO11–1, and RAD51, the DMC1 gene in Arabidopsis shows exclusively meiotic expression (28).
To compare expression of dmc1, hop2, rad51, and spo11–1 in wild-type and mutant flowers, we used a custom maize oligonucleotide GeneChip array containing 82,000 probe sets to maize genes and EST clusters. Among the tested genes, only dmc1 showed a significantly reduced (by approximately 60%) level of expression in am1-1 mutant flowers, although its expression in the am1-praI mutant was similar to that in wild-type flowers (Table 2). In contrast, hop2, rad51, and spo11–1 did not exhibit significant changes in their expression levels in either am1-1 or am1-praI as compared with the wild type. The most plausible explanation for these data is that genes showing meiosis-specific expression are under the control of am1, whereas am1 has no effect on genes whose expression is not under meiosis-specific control.
Table 2.
Expression analysis of known meiotic genes in am1 mutants in maize
| Gene | Wild-type meiosis | am1-1 | am1-praI |
|---|---|---|---|
| dmc1 | 1,230 ± 88 | 485 ± 86 | 1,346 ± 150 |
| spo11-1 | 217 ± 10 | 227 ± 32 | 185 ± 47 |
| hop2 | 297 ± 81 | 297 ± 67 | 204 ± 90 |
| rad51A1 | 517 ± 40 | 530 ± 129 | 341 ± 226 |
| rad51A2 | 262 ± 55 | 175 ± 10 | 200 ± 14 |
Expression levels were measured using a custom GeneChip array and are in arbitrary units (mean ± SE). Number of biologic replicates: wild-type meiosis n = 3, am1-1 n = 4, and am1-praI n = 3.
am1 Is a Plant-Specific Gene.
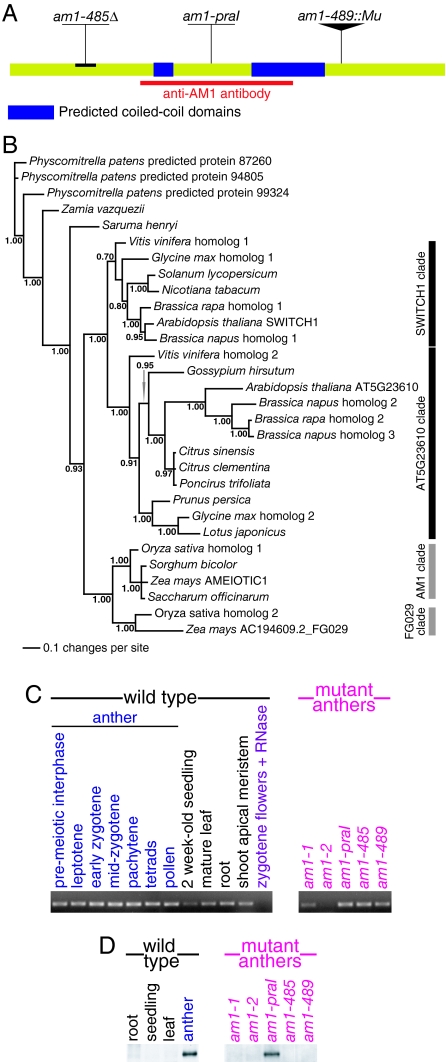
To further understand the regulation of meiotic prophase initiation and progression in maize, we cloned the am1 gene. The am1-485 and am1-489 alleles were recovered in a screen of maize populations with a high activity of the Mutator (Mu) transposon. We discovered that a DNA fragment containing a terminal inverted repeat of the transposon cosegregated with the mutant phenotype in a population of plants segregating for the am1-489 allele. Analysis of the adjacent genomic region revealed a Mu copy inserted into the ninth exon of a 10-exon-long ORF. The putative am1 gene encodes a 780-aa protein with a predicted molecular mass of 85.6 kDa. BLAST searches showed that the AM1 protein does not exhibit significant similarity to any protein with a known biochemical function. However, the AM1 sequence contains 2 putative coiled-coil domains (AA 260–287 and AA 441–547; Fig. 2A), which may be involved in protein–protein interactions.
Fig. 2.
(A) Diagram of the AM1 protein with marked positions of lesions in the am1-praI, am1-485, and am1-489 mutant alleles. Also noted are positions of 2 predicted coiled-coil domains and the protein region used to raise the anti-AM1 antibody. (B) Bayesian reconstruction of phylogeny of AM1 homologues in plants. Numbers next to branches are clade credibility values. (C) Expression of the am1 gene detected by RT-PCR in various wild-type maize tissues and in the 5 am1 mutants. (D) Western blot analysis of AM1 protein accumulation in wild-type maize tissues and in the 5 am1 mutants.
We found sequence homologues of AM1 in several plant species but not outside of plants. The genomes of a number of monocot and dicot species, including maize, rice, and Arabidopsis, harbor 2 copies of am1-like genes. This is a result of a gene duplication event that, interestingly, took place independently in both the monocot and dicot lineages after the 2 groups diverged (Fig. 2B). One of the 2 AM1 homologues in Arabidopsis, SWITCH1 (SWI1)/DYAD, has been previously characterized and proposed to regulate meiotic chromosome structure and cohesion (29–32). AM1 and SWI1/DYAD share approximately 30% identity, mostly in the central portion of the protein. This region also shows strong similarity to several other plant proteins (Fig. S4), including MS1 and MMD1 in Arabidopsis, both involved in regulating meiotic gene expression (33, 34), and 2 predicted proteins in Arabidopsis and rice.
We identified the lesions in the am1-praI, am1-485, and am1-489 alleles of am1. The am1-praI allele carries a point mutation that results in an arginine-to-tryptophan substitution at AA position 358 (Fig. 2A). This position is conserved in both monocots and dicots (Fig. S4). The am1-485 allele carries a deletion in the 5′ region of the gene introducing a premature stop codon, which would result in a severely truncated protein (Fig. 2A). The am1-489 allele contains a Mu transposon insertion in the 3′ end of the am1 coding region (Fig. 2A), which would produce an abnormal protein consisting of truncated AM1 fused to a short protein resulting from translation of a portion of the Mu sequence. The am1-485 and am1-489 transcripts are stable, at least in anthers of the mutant plants (Fig. 2C). The am1-1 and am1-2 mutations have no lesions in the am1 ORF, but anthers from the 2 mutants showed significantly reduced levels of the am1 transcript (Fig. 2C), suggesting that the mutant lesions may be in the am1 regulatory regions.
The AM1 Protein Is Localized to Chromosomes in Meiotic Prophase I.
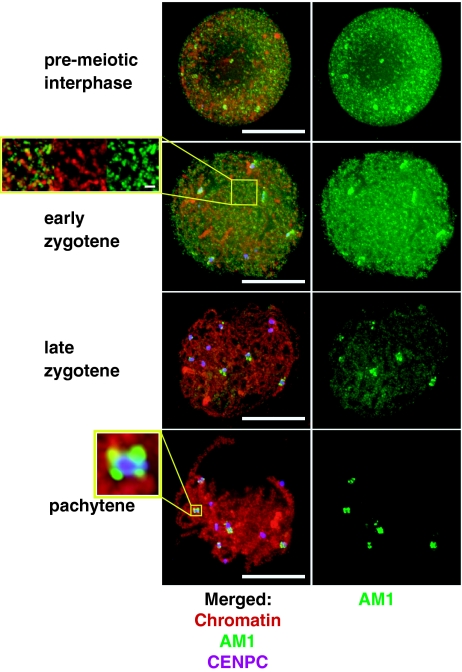
Although phenotypes of the am1 mutants are confined to meiosis, we found that the am1 gene is expressed throughout the plant and at most stages of development (Fig. 2C). To better understand the spatial and temporal pattern of am1 function, we generated an antibody against the central portion of the AM1 protein (AA 233–501). Western blot analysis showed that the protein was present in anthers but not in root, shoot, and leaf tissues (Fig. 2D), suggesting that translation and/or stability of the protein are developmentally regulated. To further understand the function of the protein, we conducted immunolocalization experiments to identify where it localizes during meiosis (Fig. 3). The protein was absent from the tapetal (somatic) cells in the anther and only present in the meiocytes, where it accumulated starting with the premeiotic interphase. During the premeiotic interphase, the anti-AM produced diffuse antibody staining inside the nucleus. During leptotene and early zygotene, AM1 localized to the chromatin, forming punctuated foci, with more elongated patches in some chromosome regions. The protein largely disappeared by late-zygotene, except for pericentromeric regions of 5 to 6 out of the 10 maize chromosomes, where it persisted until late pachytene.
Fig. 3.
Immunolocalization of the AM1 protein in male meiocytes in wild-type maize. Red: DAPI-stained chromatin; green: AM1; purple: CENPC, which is a constitutive component of the inner kinetochore and marks centromeres. (Inset) Magnification of a centromeric region of one of the pachytene chromosomes, with CENPC identifying the kinetochore and AM1 staining in the pericentromeric region. (Scale bars, 10 μm; small bar Inset, 1 μm.)
AM1 Does Not Localize to Chromosomes in am1-praI Meiocytes.
We examined accumulation of the AM1 protein in anthers of am1 mutants using Western blot analysis (Fig. 2D) and immunolocalization (Fig. S5). In the am1-praI mutant, we found that the size and amount of the AM1 protein in the anthers was similar to that seen in wild type (Fig. 2D). The protein showed diffuse staining in nuclei in premeiotic interphase, similar to that seen in wild type. However, during early leptotene, when chromosome threads become clearly visible, the defective AM1 protein remained diffuse in most am1-praI nuclei, and staining of AM1 in nucleoli was observed (Fig. S5). Only a fraction of cells (8 of 54) showed faint AM1 staining on chromosomes.
In the am1-1, am1-2, am1-485, and am1-489 mutants, we did not detect the AM1 protein in Western blot analyses of meiotic anthers (Fig. 2D) or immunolocalization experiments on male meiocytes (Fig. S5). These results correlate well with previous observations that am1-1, am1-2, am1-485, and am1-489 show similar mutant phenotypes in male meiosis (Table 1) (11). In female meiosis, am1 alleles form an allelic series (Table 1). We hypothesize that the differences between the male and female meiocyte defects are caused by a sex-specific regulation of am1 translation and/or by a difference in how the AM1 protein acts in male and female meiocytes. However, we could not examine am1 gene expression or AM1 protein accumulation in female meiocytes, because these cells in maize are not easily accessible to analysis.
Discussion
Our analyses of am1 mutants indicate that the am1 gene plays 2 distinct roles in regulating meiosis. First, it is required for the transition from mitotic cell cycle to meiosis, and in its absence meiocytes continue undergoing mitotic divisions. Later, am1 also regulates the transition from leptotene to zygotene.
am1 Controls the Transition from the Mitotic Cell Cycle to Meiosis in Maize.
Previous observations suggested that chromosomes in am1-1, am1-2, am1-485, and am1-489 meiocytes undergo an equational, “mitosis-like” division (10, 11, 35). However, they did not establish whether this equational division was mitosis or an aberrant meiosis. If the former is true, then the am1 gene can be regarded as a major regulator of meiotic transition in maize. However, if the latter were the case, it would mean that the am1 gene is involved in early meiotic processes but acts downstream from meiosis initiation. To discern between the 2 possibilities, we conducted an in-depth analysis of several key events of meiotic prophase I in am1-1 mutant meiocytes. These analyses established that chromosomes in am1-1 mutant meiocytes (i) exhibit arrangement in the nucleus that resembles the prophase of mitosis rather than meiosis, (ii) do not show the meiosis-specific telomere bouquet formation, (iii) do not exhibit homologous chromosome pairing, (iv) lack installation of AFD1, (v) lack installation of ASY1, (vi) do not show formation of meiotic DSBs in chromosomal DNA and lack chromosomal foci of RAD51, (vii) exhibit cytoskeleton features typical for mitosis rather than meiosis, and (viii) show reduced levels of expression of a meiosis marker dmc1. Overall, this evidence suggests that the cell division in am1-1 male meiocytes is a normal mitosis rather than an abnormal meiosis. Analyses of male meiocytes in the am1-2, am1-485, and am1-489 mutants show that they exhibit chromosome behavior similar to that of am1-1, suggesting that they too undergo normal mitotic divisions. All these data indicate that am1 is required in multiple early meiosis processes and is likely involved in the initiation of meiosis.
It has been suggested that the decision to initiate meiosis in budding yeast, fission yeast, and mouse is made during, or before, the premeiotic S phase (5, 7, 36, 37). Our observations suggest that the timing of meiosis initiation in maize may be similar. This hypothesis is supported by (i) the nuclear localization of the AM1 protein during the premeiotic interphase, (ii) the role of AM1 in recruiting AFD1/Rec8, (iii) interphase arrest in female meiocytes in the am1-2, am1-485, and am1-489 mutants (11), and (iv) the presence of the mitosis-specific preprophase cortical microtubule band in am1-1, am1-2, am1-485, and am1-489 meiocytes.
am1 Controls Transition Through a Novel Leptotene–Zygotene Checkpoint.
In contrast to the other am1 mutants, am1-praI meiocytes enter meiosis and arrest during early meiotic prophase (10). Our analyses show now that the prophase arrest in am1-praI occurs specifically at the leptotene–zygotene transition [the prezygotene stage (38)]. This timing is indicated by (i) the incomplete formation of the telomere bouquet, which in wild-type meiocytes starts at the end of leptotene and is completed in early zygotene, (ii) the absence of chromosome pairing, which normally starts in early zygotene, (iii) the patterns of AFD1 and ASY1 localization, which resemble those of the wild-type leptotene cells, (iv) the presence of meiotic DSBs, which are formed in leptotene in wild-type meiocytes, and (v) the absence of RAD51 foci, which are normally installed on maize chromosomes in early zygotene. On the basis of these analyses, we propose that AM1 has a second function in meiosis in regulating the meiocyte progression through prezygotene. The leptotene–zygotene transition is a known major transition point in meiotic prophase, associated with a dramatic structural reorganization of chromosomes (38). The leptotene–zygotene transition arrest in am1-praI suggests the presence of a novel checkpoint. The existence of this checkpoint is intriguing because plants are thought to lack the typical meiotic checkpoints (39). It is likely that the conserved region in the AM1 protein sequence, which stretches from AA 324 to 436 and includes the arginine residue mutated in am1-praI, functions as a domain controlling meiotic prophase I progression. The arginine 358 residue is also required for the chromosomal localization of the AM1 protein in leptotene and zygotene.
Evolution of Mechanisms Regulating Meiosis Initiation.
The mechanisms that initiate meiosis have been studied extensively in fission and budding yeasts and in the mouse (5–9). The mechanisms of meiosis initiation in these species show little in common, in terms of the cues triggering meiosis, the signaling cascades that initiate the transition to meiosis, or the sequence conservation of the proteins involved in meiosis initiation. The fact that AM1 homologues are only found in plants suggests that the mechanisms of meiosis initiation in plants share little with analogous mechanisms in yeast or mammals. Also unique to the mechanism of meiosis initiation in maize is the fact that the AM1 protein required for the transition from the mitotic cell cycle to meiosis also plays a role in regulating the progression of meiotic prophase I once meiosis is initiated.
One of the 2 AM1 homologues in Arabidopsis, SWI1/DYAD, also affects the events of meiotic prophase I. However, its role in meiosis seems to be different from the role of AM1 (29–32). Chromosomes in swi1/dyad mutants in some cases undergo an equational-like segregation, but this division is an abnormal meiosis rather than a normal mitosis, based on chromosome behavior, installation of the meiosis-specific cohesin Rec8, and presence of the meiosis-specific expression of DMC1 (30, 31). This suggests that the SWI1/DYAD protein affects early stages of meiosis that are downstream from meiosis initiation but does not act directly to initiate meiosis in Arabidopsis. In addition, the SWI1/DYAD protein is present in the nucleus only during the premeiotic interphase, and it is not known to have a role in regulating progression of meiotic prophase I (30–32). Characterizing At5g23610, the second am1-like gene in Arabidopsis and the second am1 homologue in maize, should clarify the similarities and differences between the meiosis initiation mechanisms in the 2 species. Overall, our data imply that meiosis-initiating mechanisms show substantial variation not only between distant taxa but also among relatively more closely related groups like monocots and dicots.
Materials and Methods
am1 Mutants.
All am1 mutant alleles were reported previously (10, 11). The am1-485 and am1-489 mutants were isolated and initially characterized at the University of North Dakota.
Microscopy.
Immunolocalization using antibodies against RAD51 (40), AFD1 (16), ASY1 (19), α-tubulin, and AM1 was performed as previously described (16, 41). FISH and meiotic DSB detection were also performed as published (12, 21), except that the meiocyte-containing polyacrylamide pads were permeabilized in 0.1% sodium citrate + 0.1% Triton X-100 before the TUNEL reaction.
Analysis of Meiotic Gene Expression.
Expression analysis was performed using a Syngenta-designed maize GeneChip genome array (SYNG007a520046). The array contains 82,661 probe sets, designed on the basis of 87,572 maize transcripts, of which 50,058 had protein similarity and 37,514 were unknown. Staged flowers of wild-type, am1-1, and am1-praI plants were collected and immediately frozen in liquid nitrogen to prevent expression of stress-response genes. Preparation of labeled cDNA, array hybridization, and data acquisition were carried out as previously described (42).
Cloning the am1 Gene.
Ligation-based expression analysis display (LEAD) (45) was used to PCR-amplify genomic DNA fragments adjacent to Mutator transposon insertions in the am1-489 mutant. Screening of a population of 82 plants from families segregating for the am1-489 mutation identified a fragment that cosegregated with the mutant gene. The sequence of the rest of the am1 ORF was obtained using 5′ and 3′ RACE. The GenBank accession number for the am1 gene is DQ663482.
RT-PCR.
RT-PCR was performed with the SuperScript One-Step RT-PCR kit (Invitrogen). To control for the amount of the RNA template, we ran 1.0 μg of each RNA sample in a 1% agarose gel and compared the intensities of the rRNA bands between samples.
AM1 Antibody Production.
A partial am1 cDNA was cloned into the pGEX-4T-3 plasmid. The fusion protein was expressed in Escherichia coli BL21 cells and purified with the GST purification kit (Amersham Biosciences). The tag was cleaved by thrombin digestion, and the purified antigen was used to raise an antibody in rabbits (Covance).
Supplementary Material
Acknowledgments.
We thank Tomoko Ogawa, Chris Franklin, and Kelly Dawe for antibodies, Charles Chilcott for technical assistance, and members of the Pawlowski and Cande laboratories for comments on the manuscript. This study was supported by National Institutes of Health, National Science Foundation, Syngenta, and Cornell University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810115106/DCSupplemental.
References
- 1.Ronceret A, Sheehan MJ, Pawlowski WP. Chromosome dynamics in meiosis. In: Verma DPS, Hong Z, editors. Cell Division Control in Plants. Heidelberg: Springer-Verlag; 2007. pp. 103–124. [Google Scholar]
- 2.Pawlowski WP, Sheehan MJ, Ronceret A. In the beginning: The initiation of meiosis. Bioessays. 2007;29:511–514. doi: 10.1002/bies.20578. [DOI] [PubMed] [Google Scholar]
- 3.Chu S, et al. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 4.Benjamin KR, Zhang C, Shokat KM, Herskowitz I. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. 2003;17:1524–1539. doi: 10.1101/gad.1101503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marston AL, Amon A. Meiosis: Cell-cycle controls shuffle and deal. Nat Rev Mol Cell Biol. 2004;5:983–997. doi: 10.1038/nrm1526. [DOI] [PubMed] [Google Scholar]
- 6.Harigaya Y, et al. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature. 2006;442:45–50. doi: 10.1038/nature04881. [DOI] [PubMed] [Google Scholar]
- 7.Baltus AE, et al. In germ cells of mouse embryonic ovaries, the decision to enter meiosis precedes premeiotic DNA replication. Nat Genet. 2006;38:1430–1434. doi: 10.1038/ng1919. [DOI] [PubMed] [Google Scholar]
- 8.Koubova J, et al. Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proc Natl Acad Sci USA. 2006;103:2474–2479. doi: 10.1073/pnas.0510813103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowles J, et al. Retinoid signaling determines germ cell fate in mice. Science. 2006;312:596–600. doi: 10.1126/science.1125691. [DOI] [PubMed] [Google Scholar]
- 10.Golubovskaya I, Grebennikova ZK, Avalkina NA, Sheridan WF. The role of the ameiotic1 gene in the initiation of meiosis and in subsequent meiotic events in maize. Genetics. 1993;135:1151–1166. doi: 10.1093/genetics/135.4.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golubovskaya I, Avalkina N, Sheridan WF. New insights into the role of the maize ameiotic1 locus. Genetics. 1997;147:1339–1350. doi: 10.1093/genetics/147.3.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golubovskaya IN, Harper LC, Pawlowski WP, Schichnes D, Cande WZ. The pam1 gene is required for meiotic bouquet formation and efficient homologous synapsis in maize (Zea mays, L. ) Genetics. 2002;162:1979–1993. doi: 10.1093/genetics/162.4.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mascia PN, Rubenstein I, Phillips RL, Wang AS, Xiang LZ. Localization of the 5S rRNA genes and evidence for diversity in the 5S rDNA region of maize. Gene. 1981;15:7–20. doi: 10.1016/0378-1119(81)90099-8. [DOI] [PubMed] [Google Scholar]
- 14.Watanabe Y. Sister chromatid cohesion along arms and at centromeres. Trends Genet. 2005;21:405–412. doi: 10.1016/j.tig.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- 16.Golubovskaya IN, et al. Alleles of afd1 dissect REC8 functions during meiotic prophase I. J Cell Sci. 2006;119:3306–3315. doi: 10.1242/jcs.03054. [DOI] [PubMed] [Google Scholar]
- 17.Zickler D, Kleckner N. Meiotic chromosomes: Integrating structure and function. Annu Rev Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- 18.Hollingsworth NM, Byers B. HOP1: A yeast meiotic pairing gene. Genetics. 1989;121:445–462. doi: 10.1093/genetics/121.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Armstrong SJ, Caryl AP, Jones GH, Franklin FC. Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J Cell Sci. 2002;115:3645–3655. doi: 10.1242/jcs.00048. [DOI] [PubMed] [Google Scholar]
- 20.Franklin AE, et al. Three-dimensional microscopy of the Rad51 recombination protein during meiotic prophase. Plant Cell. 1999;11:809–824. doi: 10.1105/tpc.11.5.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pawlowski WP, et al. Coordination of meiotic recombination, pairing, and synapsis by PHS1. Science. 2004;303:89–92. doi: 10.1126/science.1091110. [DOI] [PubMed] [Google Scholar]
- 22.Staiger CJ, Cande WZ. Ameiotic, a gene that controls meiotic chromosome and cytoskeletal behavior in maize. Dev Biol. 1992;154:226–230. doi: 10.1016/0012-1606(92)90063-m. [DOI] [PubMed] [Google Scholar]
- 23.Grelon M, Vezon D, Gendrot G, Pelletier G. AtSPO11–1 is necessary for efficient meiotic recombination in plants. EMBO J. 2001;20:589–600. doi: 10.1093/emboj/20.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Couteau F, et al. Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant Cell. 1999;11:1623–1634. doi: 10.1105/tpc.11.9.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schommer C, Beven A, Lawrenson T, Shaw P, Sablowski R. AHP2 is required for bivalent formation and for segregation of homologous chromosomes in Arabidopsis meiosis. Plant J. 2003;36:1–11. doi: 10.1046/j.1365-313x.2003.01850.x. [DOI] [PubMed] [Google Scholar]
- 26.Li W, et al. The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc Natl Acad Sci USA. 2004;101:10596–10601. doi: 10.1073/pnas.0404110101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, et al. Functional analysis of maize RAD51 in meiosis and double-strand break repair. Genetics. 2007;176:1469–1482. doi: 10.1534/genetics.106.062604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klimyuk VI, Jones JDG. AtDMC1, the Arabidopsis homologue of the yeast DMC1 gene: Characterization, transposon-induced allelic variation and meiosis-associated expression. Plant J. 1997;11:1–14. doi: 10.1046/j.1365-313x.1997.11010001.x. [DOI] [PubMed] [Google Scholar]
- 29.Mercier R, et al. SWITCH1 (SWI1): A novel protein required for the establishment of sister chromatid cohesion and for bivalent formation at meiosis. Genes Dev. 2001;15:1859–1871. doi: 10.1101/gad.203201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mercier R, et al. The meiotic protein SWI1 is required for axial element formation and recombination initiation in Arabidopsis. Development. 2003;130:3309–3318. doi: 10.1242/dev.00550. [DOI] [PubMed] [Google Scholar]
- 31.Agashe B, Prasad CK, Siddiqi I. Identification and analysis of DYAD: A gene required for meiotic chromosome organisation and female meiotic progression in Arabidopsis. Development. 2002;129:3935–3943. doi: 10.1242/dev.129.16.3935. [DOI] [PubMed] [Google Scholar]
- 32.Siddiqi I, Ganesh G, Grossniklaus U, Subbiah V. The dyad gene is required for progression through female meiosis in Arabidopsis. Development. 2000;127:197–207. doi: 10.1242/dev.127.1.197. [DOI] [PubMed] [Google Scholar]
- 33.Wilson ZA, Morroll SM, Dawson J, Swarup R, Tighe PJ. The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J. 2001;28:27–39. doi: 10.1046/j.1365-313x.2001.01125.x. [DOI] [PubMed] [Google Scholar]
- 34.Yang X, Makaroff CA, Ma H. The Arabidopsis MALE MEIOCYTE DEATH1 gene encodes a PHD-finger protein that is required for male meiosis. Plant Cell. 2003;15:1281–1295. [PMC free article] [PubMed] [Google Scholar]
- 35.Rhoades MM. Genic control of chromosomal behavior. Maize Genetics Cooperation Newsletter. 1956;30:38–42. [Google Scholar]
- 36.Watanabe Y, Yokobayashi S, Yamamoto M, Nurse P. Pre-meiotic S phase is linked to reductional chromosome segregation and recombination. Nature. 2001;409:359–363. doi: 10.1038/35053103. [DOI] [PubMed] [Google Scholar]
- 37.Borde V, Goldman AS, Lichten M. Direct coupling between meiotic DNA replication and recombination initiation. Science. 2000;290:806–809. doi: 10.1126/science.290.5492.806. [DOI] [PubMed] [Google Scholar]
- 38.Dawe RK, Sedat JW, Agard DA, Cande WZ. Meiotic chromosome pairing in maize is associated with a novel chromatin organization. Cell. 1994;76:901–912. doi: 10.1016/0092-8674(94)90364-6. [DOI] [PubMed] [Google Scholar]
- 39.Caryl AP, Jones GH, Franklin FC. Dissecting plant meiosis using Arabidopsis thaliana mutants. J Exp Bot. 2003;54:25–38. doi: 10.1093/jxb/erg041. [DOI] [PubMed] [Google Scholar]
- 40.Terasawa M, Shinohhara A, Hotta Y, Ogawa H, Ogawa T. Localization of RecA-like protein in chromosomes of the lily at various meiotic stages. Genes Dev. 1995;9:925–934. doi: 10.1101/gad.9.8.925. [DOI] [PubMed] [Google Scholar]
- 41.Pawlowski WP, Golubovskaya IN, Cande WZ. Altered nuclear distribution of recombination protein RAD51 in maize mutants suggests involvement of RAD51 in the meiotic homology recognition. Plant Cell. 2003;8:1807–1816. doi: 10.1105/tpc.012898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guimil S, et al. Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proc Natl Acad Sci USA. 2005;102:8066–8070. doi: 10.1073/pnas.0502999102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li BY, et al. Analysis of differential gene expression by ligation specificity-based transcript profiling. Omics. 2002;6:175–185. doi: 10.1089/153623102760092779. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.