Abstract
Guanylyl cyclase activating protein 1 (GCAP1), a member of the neuronal calcium sensor (NCS) subclass of the calmodulin superfamily, confers Ca2+-dependent activation of retinal guanylyl cylcase (RetGC) during phototransduction in vision. Here we analyze the energetics of Ca2+ and Mg2+ binding to the individual EF-hands, characterize metal-induced conformational changes, and evaluate structural effects of myristoylation as studied by isothermal titration calorimetry (ITC), differential scanning calorimetry (DSC) and NMR. GCAP1 binds cooperatively to Ca2+ at EF3 and EF4 (ΔHEF3 = −3.5 kcal/mol and ΔHEF4 = −0.9 kcal/mol) with nanomolar affinity (KEF3 = 80 nM and KEF4 = 200 nM), and a third Ca2+ binds entropically at EF2 (ΔHEF2 = +3.1 kcal/mol and KEF2 = 0.9 μM). GCAP1 binds functionally to Mg2+ at EF2 (ΔHEF2 = +4.3 kcal/mol and KEF2 = 0.7 mM) required for RetGC activation. Ca2+ and/or Mg2+ binding to GCAP1 dramatically alter DSC and NMR spectra, indicating metal-induced protein conformational changes in EF2, EF3 and EF4. Myristoylation of GCAP1 does not significantly alter its metal binding energetics or NMR spectra, suggesting that myristoylation does not influence the structure of the metal-binding EF-hands. Myristoylation also has almost no effect on protein folding stability measured by DSC. NMR resonances of myristate attached to GCAP1 are exchange broadened, upfield shifted and insensitive to Ca2+, consistent with the myristoyl group being sequestered inside the protein as seen in the crystal structure. We conclude that the protein environment near the myristate is not influenced by Mg2+ or Ca2+ binding, but instead is constitutively dynamic and may play a role in promoting GCAP1 interactions with the cyclase.
Keywords: GCAP1, calcium, NMR, EF-hand, RetGC, phototransduction, retina, neuronal calcium sensor
Guanylyl cyclase activating proteins (GCAPs) belong to the neuronal calcium sensor (NCS) branch of the calmodulin superfamily (1-3) and regulate Ca2+-sensitive activity of retinal guanylyl cyclase (RetGC) in rod and cone cells (4, 5). Phototransduction in retinal rods and cones is modulated by intracellular Ca2+ sensed by GCAPs (6, 7) and defects in Ca2+ signaling by GCAPs are linked to retinal diseases (8). Light excitation of photoreceptor cells triggers a phototransduction cascade, leading to the closure of cGMP-gated channels, which hyperpolarizes the plasma membrane and generates a neural signal (9). The recovery of the dark state requires activation of RetGC to restore the cytosolic cGMP level. The activity of RetGC is Ca2+-sensitive (10) and is mediated by GCAPs (4, 5, 7). Channel closure during phototransduction blocks the entry of Ca2+ and lowers the cytosolic Ca2+ concentration from ∼250 nM in the dark down to ∼25 nM in the light (11). This drop in Ca2+ causes the formation of Ca2+-free/Mg2+-bound GCAPs that activate RetGC (12), whereas Ca2+-bound GCAPs inhibit RetGC at high Ca2+ levels generated in the dark (7).
The GCAPs (GCAP1 (5), GCAP2 (13), GCAP3 (14) and GCAP4−8 (15)) are all ∼200-residue proteins containing a covalently attached N-terminal myristoyl group and four EF-hand motifs (Fig. 1). Mg2+ binds to three EF-hands (EF2, EF3 and EF4) when cytosolic Ca2+ levels are low and Mg2+-bound GCAP1 activates RetGC (12, 16). Ca2+ also binds functionally to these three EF-hands (12, 17) when Ca2+ levels are high in the dark. The first EF-hand (EF1) is unable to bind Ca2+ or Mg2+ due to conserved subsitutions in the binding loop (Cys29 and Pro30 in GCAP1). The x-ray crystal structure of Ca2+-bound GCAP1 (18) and NMR structure of GCAP2 (19) clearly showed that Ca2+ is bound at EF2, EF3 and EF4. In addition, the N-terminal myristoyl group in GCAP1 is buried inside the Ca2+-bound protein flanked by hydrophobic residues at the N- and C-termini (see italicized residues in Fig. 1). The structure of the physiologically active form of GCAPs (Mg2+-bound/Ca2+-free state) is currently unknown.
Figure 1.
Amino acid sequence alignment of GCAP1 with various NCS proteins. Secondary structural are elements indicated schematically. The four EF-hands (EF1, EF2, EF3 and EF4) are underlined. Mutated residues in the EF-hand loops are indicated in bold. N- and C-terminal residues that interact with myristate are shown in italics.
What are the Ca2+- and Mg2+-dependent conformational changes in GCAPs that promote activation of RetGC? Recoverin is the only NCS protein whose structure is known in both the Ca2+-free and Ca2+-bound states (Fig. 1) (20, 21). Ca2+-free recoverin contains a highly sequested myristoyl group buried inside the protein that interacts intimately with residues from EF1, EF2 and EF3 (22, 23). Ca2+ binding at EF2 and EF3 leads to large overall conformational changes in recoverin that promotes the extrusion of the fatty acyl group outward, enabling it to interact with membrane targets (20, 24). Since the target for GCAPs (RetGC) is a membrane protein, we wondered if GCAPs might undergo a similar Ca2+-induced conformational change and if Ca2+ and/or Mg2+ binding to GCAPs alter the protein environment around the myristoyl group. Here, we perform ITC, DSC and NMR studies on GCAP1 to analyze the thermodynamics of Ca2+ and Mg2+ binding to the individual EF-hands, characterize metal-induced protein conformational changes, and evaluate the Ca2+- and Mg2+-dependent structural environment around the myristoyl group.
EXPERIMENTAL PROCEDURES
GCAP1 Protein Expression and Purification
Plasmids (pET11d) encoding wild-type GCAP1 and its mutants for disabling both Ca2+ and Mg2+ binding (D68G/E75Q (EF2−), D100N/D102G (EF3−), D144N/D148G (EF4−), and D100N/D102G/D144N/D148G (EF34−)) were prepared as described previously (12). All plasmids contain the D6S mutation for the recognition of yeast N-myristoyl-transferase (NMT) needed for expressing recombinant myristoylated GCAP1 in E. coli. Plasmid vector (pET11d) harboring GCAP1 or its mutants were co-transformed into BL21(DE3) cells with or without pBB131 vector encoding NMT. The bacterial cells were typically pre-cultured until optical density at 600 nm (A600) reached 1.0 in 20 ml Luria-Bertani (LB) medium at 37 °C with antibiotics (100 μg/ml ampicillin or along with 50 μg/ml kanamycin). They were then inoculated into 1 L LB medium with the antibiotics and grown until A600 = 1.3. GCAP1 and NMT proteins were co-expressed upon adding isopropyl-β-D-1-thiogalactopyranoside (IPTG) into the cell culture to a final concentration of 0.5 mM at 25 °C or at 37 °C for 4 ∼ 16 hours. GCAP1 was expressed in either the soluble fraction or as inclusion bodies by adjusting the induction temperature: Soluble GCAP1 expression occurs at 25 °C, whereas GCAP1 expression in inclusion bodies occured at 37 °C. For the expression of myristoylated proteins, 5 mg/L (final concentration) myristic acid (from 5mg/ml ethanol stock) was added to the culture 20 minutes prior to the induction. The purification of myristoylated GCAP1 and mutants (EF2−, EF3−, EF4− and EF34−) expressed in inclusion bodies has been described previously (12).
The purification of recombinant unmyristoylated GCAP1 (expressed in soluble lysate) was as follows. Cells were harvested by centrifugation and the cell pellet was resuspended and sonicated in the lysis buffer containing 20 mM Tris-HCl (pH 7.5), 0.1 M KCl, 2 mM EGTA, 1 mM DTT, 10% glycerol and 0.1 mM PMSF. Cell lysate was centrifuged down at 100,000 g by ultra centrifugation. The supernatant was mixed with CaCl2 to a final concentration of 3 mM and then applied onto butyl-Sepharose column (HiPrep 16/10 Butyl FF, Amersham) which was pre-equilibrated with buffer A containing 20 mM Tris (pH 7.5), 0.3 M KCl, 2 mM CaCl2, 1 mM MgCl2 and 1 mM DTT. The elution of the protein was done by removing Ca2+ ions by adding the buffer B containing 20 mM Tris (pH 7.5), 0.1 M KCl, 2 mM EGTA and 1 mM DTT. The fractions containing target proteins were pooled and diluted 3-fold with ddH2O lowering ionic strength for anion exchange column application. For myristoylated GCAP1, a similar purification procedure was followed except that the lysis buffer contained 0.5 M ammonium sulfate instead of 0.1 M KCl. This modification was necessary to cause more efficient binding and elution of myristoylated GCAP1 using Butyl-Sepharose chromatography. GCAP1 protein samples were then further purified by DEAE anion exchange chromatography (HiTrap 5ml DEAE FF, Amersham) pre-equilibrated with 20 mM Tris (pH 7.5) buffer containing 1 mM EGTA and 1 mM DTT. GCAP1 was eluted using a linear NaCl gradient (0 to 0.5 M over 20 column volumes). Lastly, proteins were purified by a size exclusion chromatography (HiLoad 26/60 Superdex 200, Amersham) pre-equilibrated with 20 mM Tris (pH 7.5) buffer containing 1 mM EGTA and 1 mM DTT.. The final purity (>95%) of the GCAP1 sample was verified by SDS-PAGE and mass spectrometry.
Metal-free GCAP1 was prepared as described previously (16). Briefly, the final purified GCAP1 samples in the presence of 5 mM EGTA were concentrated 10-fold (Amicon-10, 10 kDa cut-off), diluted 10-fold with de-calcified buffer (prepared as described previously (25, 26)) and then concentrated again. This procedure was repeated four times to completely exchange GCAP1 into de-calcified and EGTA-free buffer used in the metal-binding studies. The free Ca2+ concentration in the apo-GCAP1 sample was verified to be less than 10 nM using fluo-3 flourescent indicator dye analysis (27) and atomic absorption measurements. The lack of Ca2+ in apo GCAP1 was also verified by monitoring the absence of downfield-shifted amide proton NMR resonances of Gly 69, Gly 105 and Gly 149 that report on Ca2+ binding to EF2, EF3 and EF4, respectively. The apo-GCAP1 samples were also verified by NMR to not contain any lingering EGTA.
Isothermal titration calorimetry (ITC)
All ITC experiments were performed using a VP-ITC calorimeter (MicroCal) and the data were processed with the Origin 7 software package from MicroCal as described previously (28). GCAP1 samples (myristoylated, unmyristoylated, wildtype and mutants) for ITC studies were prepared in 20 mM Tris buffer (pH 7.5), 100 mM NaCl and 1 mM β-mercaptoethanol. The protein concentration was either 25 μM or 50 μM determined by measuring optical density at 280 nm. A series of 5 μL aliquots of 2 mM CaCl2 were injected into the protein sample (1.6 mL) in the presence and absence of 2 mM Mg2+ ions and corresponding heat signals were monitored calorimetrically. For the Mg2+ titration, 10 μL aliquots of 40 mM MgCl2 were titrated into the protein sample. All titrations were typically performed at 30 °C. Additional titrations were also performed at 23, 25 and 27 °C for measuring ΔCp (29, 30). The baseline from a buffer blank titration was subtracted from the raw data. A sequential binding sites model was used to fit ITC data using a non-linear least squares minimization method (31) and calculate the dissociation constant (Kd) and enthalpy change (ΔH) for each site.
Fluorescence
Tryptophan fluorescence emission spectra of myrGCAP1 were observed with a Varian Cary Eclipse spectrometer. Protein samples were prepared in 20 mM Tris-HCl (pH 7.5) buffer containing 5 mM DTT in a 1 cm path-length cell with increasing amounts of guanidine HCl (0 − 6 M) in the presence and absence of 5 mM Ca2+ or Mg2+ ions. Protein concentration was 10 μM. The emission spectra were recorded in range of 300 − 500 nm with the excitation wavelength at 295 nm at room temperature. The protein samples were incubated at room temperature for 30 min before each measurement.
NMR spectroscopy
For 1H-15N-HSQC experiments, samples of myristoylated and unmyristoylated GCAP1 were prepared in 5 mM Tris-d11-HCl (pH7.4) buffer containing 10 % D2O, 1 mM Dithiothreitol-d10 (DTT) and either 5 mM CaCl2 or 5 mM MgCl2. For 1H-13C HMQC experiments, NMR sample were prepared in the similar condition as above except the sample was prepared in 100 % D2O. The reading of the pH meter was not corrected for the deuterium isotope effect. The sample concentration was typically around 0.3 mM.
All NMR spectra were acquired using a Bruker Advance 600 MHz spectrometer equipped with a triple resonance cryo-probe and z gradient. Typically, spectra were obtained at 300 K, 310 K and 320 K with 8 ∼ 32 scans per increment. For 15N-labeled myristoylated GCAP1 (apo-, Mg2+-bound or Ca2+-bound form), two dimensional 1H-15N-HSQC spectra with total number of 256 increments were acquired. For samples of GCAP1 with 13C-labeled myristic group covalently attached to N-terminus, 2D 1H-13C-HMQC spectra were recorded with 1024×256 points. Three dimensional (13C/F1)-edited 13C/F3-filtered HMQC-NOESY spectra (120 ms mixing time) were obtained with 1024×64×180 points (23, 32).
Differential scanning calorimetry
A VP-DSC calorimeter from MicroCal was used for all DSC measurements and the data were processed with the Origin 7 software package from MicroCal as described previously (28). Scanning was done in the temperature range of 10 − 120 °C at a scan rate of 60 °C/h. A buffer baseline was subtracted from each scan. Myristoylated and unmyristoylated GCAP1 with concentrations of 50 ΔM were prepared in 20 mM Tris buffer (pH 7.5) containing 100 mM NaCl and 1 mM β-mercaptoethanol with/without 2 mM CaCl2 or MgCl2. Samples were degassed before each scan.
RESULTS
Thermodynamics of Ca2+-binding to GCAP1
Isothermal titration calorimetry (ITC) was used in this study to analyze the energetics of Ca2+ and Mg2+ binding to the individual EF-hands of GCAP1 (Fig. 1). Previous equilibrium binding studies revealed that GCAP1 binds Ca2+ at EF2, EF3 and EF4 in the nanomolar range (16), but precise values of dissociation constants for the individual EF-hands were difficult to resolve because their binding appeared somewhat overlapped. ITC can resolve dissociation constants (Kd) of multiple sites on the basis of differences in their binding enthalpy (ΔH). Hence, two or more sites with similar dissociation constants can be resolved if their binding enthalpies are sufficiently different or vice-versa.
The Ca2+ binding properties of both myristoylated and unmyristoylated GCAP1 were monitored by ITC (Fig. 2). N-terminal myristoylation of GCAP1 had a very small influence on Ca2+ binding (see circles in Figs. 2A-B) unlike that of recoverin (26, 33), and all subsequent analyses below were performed on myristoylated GCAP1. Titration of CaCl2 into myristoylated GCAP1 resulted in a binding isotherm that is multiphasic and best fit by a 3-site sequential binding model (solid lines in Figs. 2A-B and Table 1), consistent with Ca2+ binding at EF2, EF3 and EF4 as suggested by previous fluorescence equilibrium binding studies (12, 16). The ITC Ca2+-binding isotherm of apo-GCAP1 (absence of Mg2+, see Fig. 2A) exhibited exothermic binding of two Ca2+ (ΔH1 = −3.5 kcal/mol and ΔH2 = −0.9 kcal/mol) in the nanomolar range followed by endothermic binding of one Ca2+ (ΔH = +3.1 kcal/mol) with much lower affinity (Kd = 0.9 μM). The high resolution ITC data clearly indicated three binding sites having distinct values of ΔH and Kd. To assign each ITC binding phase as binding by a particular EF-hand, we have constructed various mutants that disable functional Ca2+ binding to the individual EF-hands (Fig. 1): D68G/E75Q (EF2−), D100N/D102G (EF3−), D144N/D148G (EF4−), and D100N/D102G/D144N/D148G (EF34−). In each mutant, a negatively charged Glu or Asp at the beginning and end of the EF-hand loop has been substituted with a neutral residue (Gln or Asn). Also the second negatively charged residue in the loop (Asp) was replaced by Gly. These substitutions dramatically lower the Ca2+ and/or Mg2+ binding affinity of the respective EF-hand outside of the physiological range as previously described in detail (16, 34).
Figure 2.
ITC Ca2+ titration of apo GCAP1WT (A), Mg2+-bound GCAP1WT (B), apo EF2−(C), apo EF4− (D) and apo EF34− (E). The upper panels show representative scans of ITC data from the Ca2+ titrations. The lower panels show plots of the corresponding integrated binding isotherms fit to a sequential binding model with 3-sites (wildtype), 2-sites (EF2− and EF4−), or 1-site (EF34−) as described in Methods (solid lines). Optimal fitting parameters are displayed in Table 1. Ca2+ binding isotherms of unmyristoylated GCAP1 are overlayed as circles in A and B.
Table 1.
Thermodynamic Parameters of Ca2+ Binding to GCAP1 and Mutants at 30 °C.
| KEF2 | KEF3 | KEF4 | ΔHEF2 | ΔHEF3 | ΔHEF4 | |
|---|---|---|---|---|---|---|
| 0 Mg2+ | ||||||
| Wildtype | 0.9 ±0.2 | 0.08 ±0.05 | 0.2 ±0.05 | +3.1 ±0.1 | −3.5 ±0.1 | −0.9 ±0.2 |
| EF2− | - | 0.2 ±0.1 | 0.7 ±0.5 | - | −3.0 ±0.1 | −0.05±0.1 |
| EF4− | 69 ±20 | 4.9 ±0.9 | - | +0.8 ±0.1 | −3.4 ±0.1 | - |
| EF34− | 11 ±1 | - | - | +2.1 ±0.2 | - | - |
| 2 mM Mg2+ | ||||||
| Wildtype | 1.6 ±0.3 | 0.1 ±0.05 | 0.2 ±0.05 | +1.4 ±0.1 | −3.9 ±0.1 | −0.6 ±0.2 |
| EF2− | - | 0.2 ±0.1 | 2.0 ±0.4 | - | −4.2 ±0.1 | −0.5±0.2 |
| EF4− | 1.1 ±0.2 | 0.4 ±0.1 | - | +0.4 ±0.1 | −4.0 ±0.1 | - |
| EF34− | 50 ±20 | - | - | +1.6 ±0.1 | - | - |
Dissociation constants (KEF2, KEF3, KEF4) are expressed in micromolar. The enthalphy differences (ΔHEF2, ΔHEF3, ΔHEF4) are in units of kcal/mol.
The ITC isotherms for wildtype GCAP1 and each of the EF-hand mutants were fit to a sequential binding model (solid lines in Fig. 2) and optimal Ca2+-binding parameters (ΔH and Kd) are shown in Table 1. All ITC Ca2+ binding measurements were performed both in the presence (Fig. 2B) or absence (Fig. 2A) of physiological Mg2+ levels (2 mM Mg2+) at 30 °C. We show that Mg2+ has relatively small effects on Ca2+ binding, indicating that GCAP1 binds selectively to Ca2+ with at least 500-fold preference over Mg2+ (Table 1). The ITC Ca2+-binding isotherm of the EF2− mutant (Fig. 2C) exhibited exothermic binding of 2 Ca2+ ions but lacked the endothermic phase seen in wildtype, indicating that EF2 must be the low affinity endothermic site. Correspondingly, the ITC isotherm of EF34− (Fig. 2E) exhibited only endothermic binding (ΔH = +2.1 kcal/mol), further confirming that EF2 is the endothermic site. The ITC Ca2+-binding isotherm of EF4− (Fig. 2D) exhibited an exothermic phase (ΔH = −3.4 kcal/mol) assigned as Ca2+ binding at EF3 followed by a lower affinity, endothermic phase (Kd = 69 μM and ΔH = +0.8 kcal/mol) assigned above to EF2. By the process of elimination, the lower affinity exothermic site in the wildtype isotherm (ΔH = −0.9 kcal/mol and Kd = 200 nM), therefore, must be assigned to EF4.
Our ITC Ca2+-binding analysis summarized in Table 1 reveals that EF2 is an endothermic site (KEF2 = 0.9 μM and ΔHEF2 = +3.1 kcal/mol); EF3 and EF4 exhibit exothermic binding in the nanomolar range (KEF3 = 80 nM and ΔHEF3 = −3.5 kcal/mol; KEF4 = 200 nM and ΔHEF4 = −0.9 kcal/mol). The Ca2+-binding dissociation constants of EF3 and EF4 are in the physiological range of free Ca2+ concentration measured in rod outer segments (11) and are similar to binding constants measured previously by direct equilibrium Ca2+ binding assays (12, 16). The relatively low apparent Ca2+-binding affinity of EF2 measured by ITC (KEF2 = 0.9 μM) is ∼10-fold weaker than the intrinsic binding affinity determined by fluorescent Ca2+ binding assays (16). The discrepancy is explained in part by a protein conformational change in GCAP1 that is thermodynamically coupled to Ca2+ binding at EF2. The intrinsic Ca2+ binding at EF2 measured by the fluorescence binding assay (Ka = 107 M−1) coupled to an unfavorable conformational change (Keq ∼ 10−1) probed by ITC yields an overall equilibrium constant of Ktot = Ka*Keq ∼ 106 M−1, consistent with the overall Kd measured by ITC. Hence, the overall low apparent binding affinity of EF2 suggests that Ca2+ binding at this site drives an unfavorable protein conformational change. Consistent with this conformational change are large Ca2+-induced NMR chemical shift changes for residues in EF2 (see below) and the relatively steep temperature dependence of ΔHEF2 (ΔCp = −139 cal mol−1 K−1), suggesting a change in the solvent accessible hydrophobic surface area caused by Ca2+ binding at EF2 (29, 30).
Ca2+-induced conformational changes in EF2 are further supported by its Mg2+-dependent Ca2+ binding properties. Physiological Mg2+ levels (∼1 mM in rod outer segments (35)) are required for GCAP1 to activate RetGC at low Ca2+, implying that Ca2+-free GCAP1 must bind Mg2+ (see below) (16). The presence of physiological Mg2+ levels caused a ∼1.8-fold increase in KEF2 and two-fold decrease in ΔHEF2 with smaller effects on binding to EF3 and EF4 (Table 1). This suggests that Mg2+ competes with Ca2+ for binding at EF2 even at high Ca2+ concentrations (∼1 μM in dark-adaped rods), but Mg2+ does not effectively compete for binding at EF3 and EF4 under these conditions. We conclude that both Mg2+ and Ca2+-induced conformational changes in EF2 are consistent with recent studies showing that binding of GCAP1 to RetGC requires Mg2+ binding to EF2 under conditions that mimick light adaptation of rods (12, 17).
We do not see any evidence of Ca2+ binding at EF1. The primary structure of EF1 contains unfavorable substitutions (e.g. Cys29 and Pro30 in Fig. 1) known to abolish physiological Ca2+ binding at this site in other NCS proteins (3, 36, 37). Accordingly, our ITC Ca2+ binding measurements on GCAP1 do not detect any heat signal from Ca2+ binding at EF1, suggesting that either EF1 does not bind Ca2+ under physiological conditions or the enthalpy of binding is zero.
Mg2+ Binding to GCAP1
The presence of physiological levels of Mg2+ (2 mM Mg2+) is required for activation of RetGC by GCAP1 at low Ca2+ levels (16), suggesting that Mg2+ binds to Ca2+-free GCAP1. Indeed, we showed the Ca2+-binding affinity of EF2 is Mg2+-dependent (Table 1). These Mg2+-dependent effects prompted us to quantitate the direct Mg2+ binding to GCAP1 using ITC. Titration of MgCl2 into apo GCAP1 resulted in a biphasic binding isotherm best fit by a sequential 2-site model (solid lines in Fig. 3A). The binding isotherm shows endothermic Mg2+ binding with high micromolar affinity (ΔH = +4.3 kcal/mol and Kd = 700 μM) followed by weaker binding in the millimolar range (ΔHlow = +0.9 kcal/mol and Klow = 2 mM). Mg2+ binding experiments were also performed on the EF-hand mutants to assign each Mg2+ binding phase.
Figure 3.
ITC Mg2+ titration of myristoylated GCAP1WT (A), EF2− (B) and EF34− (C). The upper panels show representative scans of ITC data from the Mg2+ titrations. The lower panels show integrated binding isotherms fit to 2-site (WT) or 1-site (EF2− and EF34−) model as described in Methods (solid lines). Optimal fitting parameters are displayed in Table 2.
The ITC isotherms for wildtype and the EF-hand mutants were fit to a sequential binding model (solid lines in Fig. 3) and optimal Mg2+-binding parameters (ΔH and Kd) are shown in Table 2. The ITC Mg2+-binding isotherm of EF34− (Fig. 3D) exhibits a single endothermic phase representing binding of Mg2+ with high micromolar affinity, suggesting that EF2 must be the highest affinity endothermic site (ΔHEF2 = +4.3 kcal/mol and KEF2 = 0.7 mM). Correspondingly, the isotherm of EF2− (Fig. 3B) exhibits millimolar binding of 1 or more Mg2+, but lacks the micromolar phase seen in the wildtype isotherm, confirming that EF2 is the highest affinity site. The lower affinity endothermic phase in the wildtype isotherm likely represents Mg2+ binding to EF3 and/or EF4. However, individual values of ΔH and Kd for Mg2+ binding at EF3 and EF4 could not be accurately resolved due to their low affinity and non-stoichiometric binding (Kd >> protein concentration).
Table 2.
Thermodynamic Parameters of Mg2+ Binding to GCAP1 and Mutants at 30 °C.
| KEF2 | Klow | ΔHEF2 | ΔHlow | |
|---|---|---|---|---|
| Wildtype | 0.7 ±0.2 | 2 ±0.8 | +4.3 ±0.1 | +0.9 ±0.5 |
| EF2− | - | 2 ±0.8 | - | +2.9 ±0.2 |
| EF4− | 0.7 ±0.2 | 2 ±0.8 | +0.5 ±0.3 | +3.9 ±0.3 |
| EF34− | 0.7 ±0.2 | - | +4.2 ±0.1 | - |
Dissociation constants (KEF2 and Klow - low-affinity phase) are expressed in millimolar. The enthalphy differences (ΔHEF2 and ΔHlow) are in units of kcal/mol.
Our ITC Mg2+-binding analysis reveals that EF2 is an endothermic site with sub-millimolar affinity (KEF2 = 0.7 mM and ΔHEF2 = +4.3 kcal/mol); EF3 and/or EF4 bind Mg2+ in the millimolar range. Mg2+ competes somewhat with Ca2+ for binding at EF2, explaining its Mg2+-dependent Ca2+ binding affinity and enthalpy (Table 1). By contrast, EF3 and EF4 each bind Ca2+ with at least 10000-fold higher affinity than Mg2+, explaining why these sites are more selective for Ca2+ and less sensitive to Mg2+. Mg2+ binding at EF2 may be functionally important and explain why the activation of RetGC by Ca2+-free GCAP1 requires physiological levels of Mg2+. Indeed recent experiments have shown that apo GCAP1 does not regulate RetGC, and Mg2+ binding at EF2 is required for light-dependent activation of the cyclase (17).
Folding Stability vs. Ca2+, Mg2+ and Myristoylation
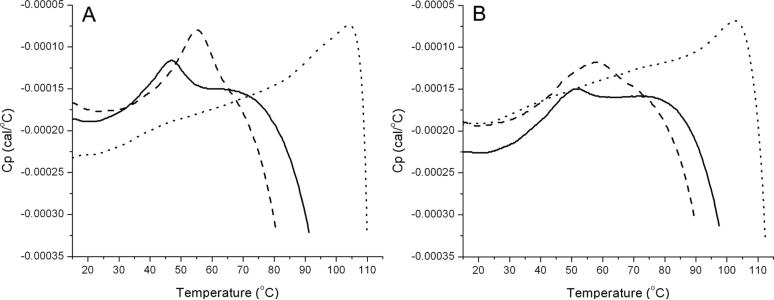
Differential scanning calorimetry (DSC) experiments were performed on GCAP1 to quantitatively assess the effect of Ca2+, Mg2+ and myristoylation on protein folding stability. Representative DSC scans of GCAP1 are shown in Fig. 4. The peak maximum of unmyristoylated metal-free (apo) GCAP1 (transitition temperature, Tm = 47 °C) is lower than the peak maximum of Mg2+-bound (Tm = 55 °C) and Ca2+-bound (Tm = 104 °C) unmyristoylated GCAP1, indicating that both Mg2+ and particularly Ca2+ increase the folding stability quite substantially. N-terminal myristoylation of GCAP1 has a much smaller effect on the folding stability. The DSC peak maxima of myristoylated GCAP1 in the apo (transitition temperature, Tm = 52 °C), Mg2+-bound (Tm = 58 °C) and Ca2+-bound (Tm = 103 °C) forms are slightly higher than the corresponding melting temperatures of unmyristoylated GCAP1, indicating that myristoylated and unmyristoylated forms of GCAP1 have similar folding stabilities (Fig. 4). The transition peaks in all the thermograms did not fully reappear upon re-scanning each of the samples, suggesting irreversible unfolding due to aggregation and/or denaturation. In addition, the DSC thermograms of apo, Mg2+-bound, and Ca2+-bound GCAP1 were all quite broad with steeply sloped post-transitional baselines, making it impossible to accurately fit these thermograms by any quantitative models. The cause of the sloping and highly curved baselines is not fully understood, but may be explained in part by visible protein aggregation that occurred during unfolding at the very high melting temperatures.
Figure 4.
DSC scans showing thermal denaturation of GCAP1. Data were obtained for unmyristoylated GCAP1 (A) and myristoylated GCAP1 (B). Thermograms of apo, Mg2+-bound and Ca2+-bound GCAP1 are indicated by solid, dashed and dotted lines, respectively.
In summary, the DSC analysis on GCAP1 reveals that Mg2+ and Ca2+ binding both increase the protein folding stability. N-terminal myristoylation of GCAP1 also stabilizes the protein fold slightly as expected based on the sequestered myristoyl group observed in the x-ray crystal structure (18). However, the increased folding stability conferred by myristoylation of GCAP1 is much smaller than that observed by myristoylated recoverin (Supplemental Fig. 1). We suggest that the vast majority of the folding stability in GCAP1 is derived by structural interactions among the EF-hands, which would explain the strong dependence of Tm on Mg2+ and Ca2+ binding. Therefore, the myristoyl group interaction with the N- and C-terminal helices observed in the crystal structure appears to contribute very little to the overall protein stability. By contrast, the myristoyl group in Ca2+-free recoverin interacts intimately with hydrophobic residues in EF1, EF2 and EF3 (22), which explains how Ca2+-binding is coupled to extrusion of the myristoyl group and why myristoylation of recoverin has a much more dramatic effect on the overall protein folding stability.
Mg2+ and Ca2+ Alter the Tertiary Structure of GCAP1
NMR spectroscopy was used to probe protein conformational changes in GCAP1 induced by Mg2+ and/or Ca2+ binding (Fig. 5). The peaks in the 1H-15N HSQC NMR spectra of GCAP1 represent main chain and side-chain amide groups and provide a residue-specific fingerprint of the overall protein conformation. HSQC spectra of myristoylated (red) and unmyristoylated (black) forms of GCAP1 look very similar, indicating once again that myristoylation has very little effect on the overall main chain structure of GCAP1. The structural similarity between myristoylated and unmyristoylated GCAP1 is also supported by tryptophan fluorescence emission spectra that have nearly the same emission wavelength maxima (Supplemental Fig. 2). This is in stark contrast to the spectral properties of Ca2+-free myristoylated recoverin, in which myristoylation has a profound influence on both NMR and fluorescence spectra (32, 33). The myristoyl group of recoverin interacts intimately with the various EF-hands and induces large overall structural changes in the protein (20). By contrast, the myristoyl group of GCAP1 appears to have very little effect on the overall protein structure.
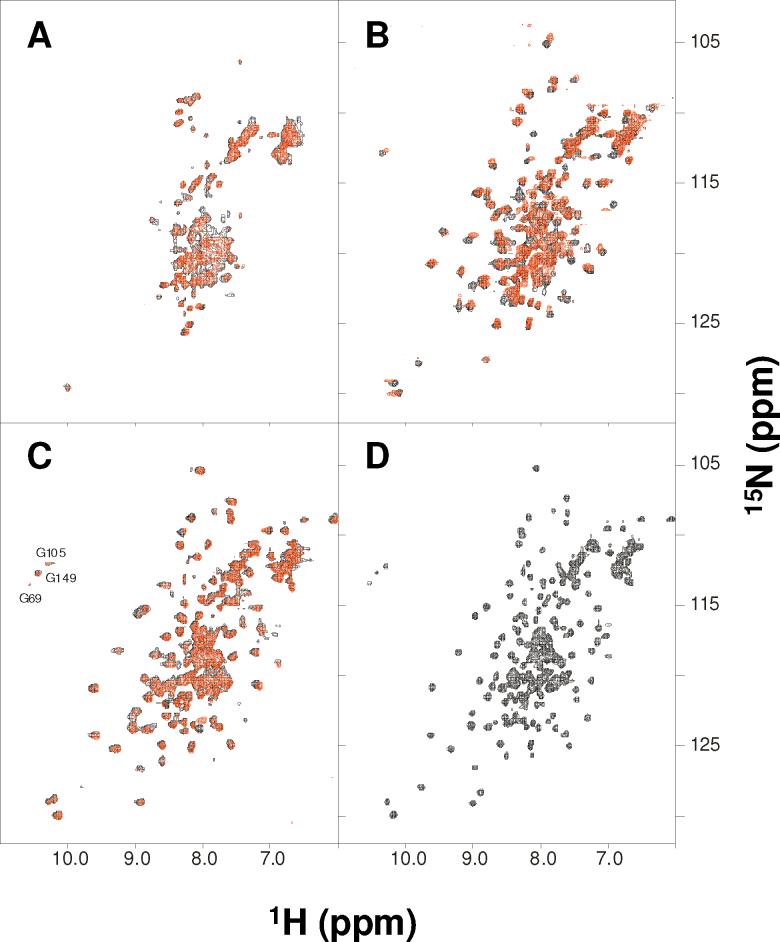
Figure 5.
NMR spectra of myristoylated and unmyristoylated GCAP1. Two-dimensional (1H-15N HSQC) NMR spectra of 15N-labeled unmyristoylated (black) and myristoylated GCAP1 (red) in the apo (A), Mg2+-bound (B), Ca2+-bound (C), and detergent solubilized, Ca2+-bound (D) states. Spectra in A-C were obtained at 37 °C and the spectrum in D was obtained in the presence of 25 mM octylglucoside at 47 °C.
The two-dimensional 1H-15N HSQC spectrum of apo GCAP1 (both myristoylated and unmyristoylated) exhibited poorly resolved and overlapping peaks with narrow chemical shift dispersion in the amide proton dimension (Fig. 5A). The number of observed peaks was far less than the expected number of amide groups and the intensities of many peaks were quite weak due to broadening caused by dimerization or conformational heterogeneity. The poor chemical shift dispersion suggests that apo GCAP1 may form an unstructured molten-globule state similar to that described for the apo states of many other Ca2+-binding proteins such as GCAP-2 (19), Frq1 (38), CIB (39), calexcitin (40), protein S (41), and DREAM (25). However, circular dichroism analysis (data not shown) suggests that apo GCAP1 adopts a high degree of helical content, consistent with the formation of the four EF-hands, and that the helical content does not change much upon binding Mg2+ and/or Ca2+. Together, our structural studies suggest that apo GCAP1 forms native secondary structure but lacks stable tertiary structure.
The HSQC spectrum of GCAP1 changed upon the addition of saturating Mg2+ (Fig. 5B). Mg2+ caused a greater number of peaks to appear and the NMR intensities were in general more uniform than those of apo GCAP1. Mg2+-binding to GCAP1 increased the NMR chemical shift dispersion and increased the number of observable long-range NOEs, demonstrating that Mg2+-bound GCAP1 adopts stable tertiary structure. Unfortunately, the number of observable HSQC peaks for Mg2+-bound GCAP1 was still only ∼50% of the expected number of amide groups, making it impossible to obtain enough sequence specific assignments necessary to solve the structure by NMR. Also, the average peak width in the spectrum appears much broader than expected for a monomeric protein, consistent with protein dimerization in the NMR samples as determined by size-exclusion chromatography studies (SEC). Pulsed-field gradient diffusion NMR studies (42) determined a hydrodynamic radius of 4.0 nm (corresponding to a spherical molecular mass of ∼60 kDa) for Mg2+-bound GCAP1, consistent with mostly dimer or higher order species present under conditions for NMR (500- μM protein concentration). However, this GCAP1 dimerization appears only at protein concentrations above 100 μM (as judged by SEC analysis) and therefore GCAP1 is monomeric at physiological concentrations in the rod cell (10−50 μM).
The HSQC spectrum of GCAP1 changed even further upon the addition of saturating Ca2+ (Fig. 5C). Spectral changes induced by the addition of saturating Ca2+ to the Mg2+-bound protein sample indicated that Ca2+ induced conformational changes are distinct and separate from the Mg2+-induced changes (Figs. 5B-C). Three downfield-shifted peaks near 10.5 ppm are characteristic of conserved glycine residues at the 6-position of Ca2+ occupied EF-hands (Gly69, Gly105 and Gly149), consistent with Ca2+ bound at EF2, EF3 and EF4. Additional unique peaks of Ca2+-bound GCAP1 (observed between 9 and 10 ppm) represent amino acid residues in EF3 and EF4 altered structurally by Ca2+-binding. The HSQC spectrum of Ca2+-saturated GCAP1 exhibited somewhat broadened peaks (like that of Mg2+-bound protein) with variable NMR intensities, consistent with protein dimerization as measured by SEC. Pulsed-field gradient diffusion NMR studies determined a hydrodynamic radius of 3.8 nm, corresponding to a Ca2+-bound protein dimer under NMR conditions.
The NMR peaks of Ca2+-bound GCAP1 sharpened quite substantially upon adding octylglucoside detergent and raising the sample temperature to 47 °C (Fig. 5D). Under these conditions the protein became monomeric, which greatly improved the overall NMR sensitivity. Also, the chemical shifts remained unperturbed by the detergent, consistent with a monomeric and properly folded protein. Initially, this improvement in spectral quality suggested that it might be feasible to determine the full three-dimensional structure by NMR. However upon closer inspection, we were only able to identify ∼80% of the main chain amide resonances in HSQC spectra and less than 50% of the resonances could be reliably assigned in triple resonance experiments. The resonances with strongest NMR intensities could be assigned mostly to residues in EF3 and EF4. However, many other resonances (associated with EF1 and EF2) had much weaker NMR intensities in triple resonance experiments and could not be accurately assigned. Furthermore, the first 16 residues from the N-terminus and a stretch of ∼20 residues following EF4 at the C-terminus have very weak NMR intensities due to exchange broadening as evidenced by their sharp temperature and magnetic field dependence. These N- and C-terminal residues (italicized in Fig. 1) are exchange broadened more so for myristoylated GCAP1 compared to that of the unmyristoylated protein. Interestingly, many of these same residues are found in helices that interact with the myristoyl group in the x-ray structure. The exchange broadening for the N- and C-terminal residues suggests that these residues undergo dynamical motions on the chemical shift time-scale to facilitate their interactions with the myristoyl group, and such dynamical behavior might be functionally important for recognition of RetGC.
Myristoyl Group Structural Environment vs. Ca2+ and Mg2+
The recent x-ray crystal structure of GCAP1 indicates that the myristoyl group is buried inside the Ca2+-bound protein (18). This is in contrast to a solvent exposed myristoyl group observed for GCAP2 (43), yeast frequenin (38), FCaBP (44), and recoverin (20). We performed NMR experiments on samples of apo, Mg2+-bound and Ca2+-bound forms of GCAP1 with a covalently attached 13C-labeled myristoyl group to probe its structural environment inside the protein and to look for any effects of Mg2+ or Ca2+-binding. Previously, two-dimensional (1H-13C HMQC) and three-dimensional (13C-filtered NOESY-HMQC) NMR experiments on samples of recoverin that contained a 13C-labeled myristoyl group were used to selectively probe Ca2+-induced changes to the chemical environment around the amino-terminal myristoyl group (23, 32). These studies revealed that the covalently-attached fatty acyl chain in recoverin is sequestered in a hydrophobic pocket in the Ca2+-free protein (22) and that binding of Ca2+ leads to conformational changes that extrude the myristoyl group into solvent.
Similar NMR experiments were performed on samples of GCAP1 that contained a covalently attached 13C-labeled myristoyl group (Fig. 6). Because the HMQC experiment selectively probes protons that are covalently attached to 13C, only the methylene and methyl resonances of the fatty acyl chain are expected to appear in these spectra. Extraneous peaks near 1.0, 1.4, 1.8 and 2.2 ppm (1H dimension) are due to background signals from the protein. The NMR spectrum of the myristoyl group of GCAP1 looks nearly identical for apo, Mg2+-bound and Ca2+-bound GCAP1 (Figs. 6A-C), indicating that the structural environment around the myristoyl group does NOT depend on Ca2+, in stark contrast to the large Ca2+-induced spectral changes for the myristoyl group in recoverin (20).
Figure 6.
Two-dimensional 1H-13C-HMQC NMR spectra of 13C-labeled myristoyl group attatched to unlabeled GCAP1 in the (A) Ca2+-bound state, (B) Mg2+-bound state, (C) apo state (1 mM EGTA), and (D) apo state in the presence of 8 M urea. The 13C2 resonance of myristate is folded and aliased shown in the upper left corner of the spectra. Extraneous peaks represent 13C natural abundance signals from the protein as determined by recording 1H-13C HMQC spectra on GCAP1 samples with an unlabeled myristoyl group.
The NMR chemical shifts of the myristoyl resonances of GCAP1 are consistent with the attached myristoyl group being sequestered inside a protein environment like was seen in the crystal structure (Table 3). Assignments of NMR resonances of the myristoyl group attached to GCAP1 were derived from assignments of free myristic acid in solution determined previously (32). The proton chemical shifts of the C14 methyl (0.62−0.73 ppm) and C4-C12 methylene resonances (0.9−1.1 ppm) in GCAP1 are all somewhat upfield shifted compared to the corresponding chemical shifts of free myristic acid in solution (32). These upfield chemical shifts in GCAP1 may be explained in part by aromatic groups (F39, F43, F63 and Y76) close the the myristoyl group in the crystal structure that might impose an aromatic ring-current effect on the nearby myristoyl group. Indeed, the myristoyl group methyl and methylene proton chemical shifts in GCAP1 became downfield shifted like those of myristate in solution upon unfolding GCAP1 by adding 8 M urea (Fig. 6D and Table 3).
Table 3.
1H and 13C (in parentheses) chemical shift assignments of myristoyl resonances.
| Position | Mg2+-GCAP1a | Ca2+-GCAP1b | denatured GCAP1c |
|---|---|---|---|
| C2 | 2.34 (38.5) | 2.34(38.4) | 2.35(38.2) |
| C3 | 1.5 (27.8) | 1.5 (27.5) | 1.55 (27.0) |
| C4 - C11 | 0.9−1.1 (31−33) | 0.9−1.1 (31−33) | 1.2−1.3 (32−33) |
| C12 | 0.88 (34.0) | 0.92 (33.8) | 1.27 (33.5) |
| C13 | 0.95 (24.4) | 0.98 (24.2) | 1.0 (25) |
| C14 | 0.62, 0.73 (16.5) | 0.62, 0.73 (16.8) | 0.92 (13) |
The NMR sample conditions were 0.3 mM GCAP1 in 5 mM Tris at pH 7.4 containing either 5 mM Mg2+ a, 5 mM Ca2+ b, or 8 M ureac.
The spectral widths of the myristoyl NMR resonances in GCAP1 are strikingly broad (50−80 Hz) compared to the widths of the natural abundance protein peaks as well as the widths of myristoyl resonances in recoverin (32) and related proteins (38, 44). The very broad NMR resonances of the myristoyl group in GCAP1 suggest that the attached myristoyl group may be located in a dynamic and/or heterogeneous protein environment. For free myristic acid in solution, a single proton resonance of the C14 methyl group is quite sharp because free rotation of the methyl group about the C13-C14 single bond causes the three methyl protons to experience a time-averaged, uniform environment. By contrast, the C14 methyl resonance (1H dimension) in GCAP1 is quite broad and is partially split into two resolved components at 0.73 and 0.62 ppm with approximately equal intensity (Fig. 6). This splitting (66 Hz) suggests that the C14 methyl group may occupy at least two different structural environments. These two chemical shift environments represent distinct conformational states of the protein (e.g. activator vs. inhibitor forms), but the relative intensities of the two components did not change much upon Ca2+ binding. The two methyl resonances might also represent the myristoyl group in folded and misfolded protein forms or in two subunits of an asymmetric dimer. However, the spectral heterogeneity persisted in the presence of 25 mM octylglucoside detergent (which causes GCAP1 to be monomeric), arguing against any artifacts caused by dimerization.
The apparent spectral heterogeneity and broadening implies that the myristoyl group must be in at least two different protein environments that are exchanging on the chemical shift time scale. The doublet methyl resonanace (66 Hz splitting) collapsed into a single broad peak (80 Hz linewidth) when the temperature was raised from 32 to 45 °C, consistent with an exchange rate on the millisecond time scale. The exchange behaviour of the myristoyl resonances could also relate and help explain why the 1H-15N HSQC peaks of the N- and C-terminal residues in GCAP1 (italicized in Fig. 1) are exchanged broadened as described above (Fig. 5). We propose that the N- and C-terminal residues, known to interact with the myristoyl group in the crystal structure (italicized in Fig. 1), can undergo conformational fluctuations on the chemical shift timescale and therefore give rise to the spectral heterogeneity observed around the myristoyl C14 methyl group.
To further probe the protein structural environment around the myristoyl group of GCAP1, three-dimensional (13C/F1)-edited and (13C/F3)-filtered NOESY experiments (32) were performed on unlabeled GCAP1 protein containing a 13C-labeled myristate. These spectra selectively probed atoms of residues in the protein that lie within 5 Å of the labeled CH3-group of the myristoyl chain. Nuclear Overhauser effect (NOE) dipolar interactions between the myristate methyl group and the protein could not be detected (data not shown), because the exchange broadening of the methyl resonances described above severely attenuates any NOE signals. However, the myristate methylene resonances at C3 and C4 did exhibit detectable NOEs to aromatic protons from the protein (Supplemental Fig. 3), providing further evidence that the myristoyl group is indeed surrounded by a hydrophobic and aromatic protein environment like what is observed in the crystal structure. The addition of Ca2+ had no effect on the filtered NOESY spectrum, further confirming that the myristate remains sequestered in a protein environment in both Ca2+-bound and Ca2+-free GCAP1. This Ca2+-independent sequestration of the myristate in GCAP1 is quite different from what is seen in recoverin that exhibits a sequestered myristoyl environment only in the Ca2+-free protein (20, 24).
DISCUSSION
In this study, we determined the energetics of Mg2+ and Ca2+ binding to GCAP1 as well as structural effects of myristoylation. Ca2+ binds enthalpically to both EF3 and EF4 in the nanomolar range with more than 1000-fold selectivity over Mg2+. EF2 binds entropically to both Ca2+ and Mg2+ and undergoes metal-induced conformational changes related to cyclase regulation (17). Myristoylation has almost no influence on the Ca2+/Mg2+ binding properties of GCAP1. Furthermore, the Ca2+-induced structural changes in the EF-hands do not influence the structural environment that surrounds the sequestered myristoyl group. Unlike recoverin, the myristoyl group attached to GCAP1 remains buried inside the protein regardless of Ca2+ level and GCAP1 does NOT possess a functional Ca2+-myristoyl switch.
The Ca2+ dissociation constants (KEF2, KEF3 and KEF4) of wildtype GCAP1 are somewhat different from those of the mutants (Table 1), implying that the three Ca2+-binding sites must interact and possess some cooperativity. Indeed, the EF3− mutation in GCAP1 abolishes Ca2+ binding to EF4 (16), suggesting both ordered and cooperative Ca2+ binding at EF3 and EF4. In essence, Ca2+ must bind at EF3 first to facilitate subsequent binding at EF4. Such ordered Ca2+ binding is consistent with our measurement that EF3 has the highest binding affinity (KEF3 = 80 nM), whereas EF4 has intermediate affinity (KEF4 = 200 nM) and EF2 has lowest affinity (KEF2 = 0.9 μM). Cooperative Ca2+ binding between EF3 and EF4 is also consistent with their intimate structural contacts observed in the x-ray crystal structure (18), and concerted Ca2+-induced NMR spectral changes observed for Gly105 (EF3) and Gly149 (EF4) in Supplemental Fig. 4. This cooperativity seems at odds with the overall Ca2+-binding isotherm of GCAP1 having a Hill slope of 1.0 (16). The Hill coefficient in this case, however, is not always a reliable measure of cooperativity, particularly if the individual sites have different intrinsic affinities (45). The free energy of interaction between the two sites (ΔGinteract) is a more stringent definition of cooperativity and this interaction energy can be inferred by comparing how the dissociation constant of EF3 (KEF3 = exp(−ΔG°/RT)) changes when EF4 is disabled (EF4−). For GCAP1, KEF3 increases subtantially by disabling EF4 (Table 1), which means that ΔGinteract > 0 and the two sites have positive cooperativity. We propose that this positive cooperativity of Ca2+ binding between EF3 and EF4 could help explain how GCAP1 confers such a steep Ca2+-dependent activation of RetGC with a Hill coefficient = 2 (46). The cooperative cyclase activation may also arise in part by Ca2+-induced dimerization of RetGC (47).
The results of this study provide structural insights into the regulatory mechanism of RetGC. The activation of RetGC by Ca2+-free GCAP1 requires physiological Mg2+ levels (16). Our NMR structural analysis shows that the completely metal-free (apo) GCAP1 lacks organized tertiary structure, explaining why the apo state is physiologically inactive (Fig. 5A). Mg2+ binding induces structural changes in the EF-hands and stabilizes protein tertiary structure as evidenced by DSC (Fig. 4) and NMR (Fig. 5). Mg2+ binds most tightly to GCAP1 at EF2 (Table 2), which we suggest might constitute an important interaction site for cyclase activation. Indeed, recent biochemical and cell biology studies have shown that EF2 is required for the cyclase interaction (12, 17) and promotes docking of Mg2+-bound GCAP1 on RetGC (17). In addition, Mg2+ binding at EF3 appears to facilitate the docking of GCAP1 with RetGC, because cyclase binding to GCAP1 is weakened by a mutation that blocks Mg2+ binding at EF3 (12). Finally, Ca2+ binding at EF4 is necessary and sufficient for GCAP1 to inhibit RetGC when Ca2+ rises in the dark (12, 16). It seems likely that Ca2+ binding at EF4 may promote the docking of Ca2+ bound GCAP1 onto the cyclase (48, 49).
The replacement of bound Ca2+ by Mg2+ in GCAP1 is functionally important for switching GCAP1 from a Ca2+-bound inhibitor to a Mg2+-bound activator conformational state during light activation. The relatively low apparent Ca2+ binding affinity of EF2 measured by ITC is explained by its thermodynamic coupling to an unfavorable protein conformational change probed structurally by NMR (Fig. 5). The intrinsic Ca2+ binding to EF2 measured by a direct Ca2+ binding assay (Ka = 107 M−1) coupled to an unfavorable conformational change (Keq ∼ 10−1) yields an overall binding constant of Ktot = Ka*Keq ∼ 106 M−1, consistent with the overall KEF2 measured by ITC. Thus, the intrinsic Ca2+-binding constants inferred from the ITC analysis are consistent with those measured by direct equilibrium binding assays (16) and consistent with the functional behavior of GCAP1 in physiological analyses (16, 17). The relative binding affinities of Mg2+ (KEF2 = 0.7 mM) and Ca2+ (KEF2 = 0.9 μM) at EF2 suggest that a fraction of GCAP1 will be occupied with Mg2+ even in dark adapted rods (250 nM Ca2+ and 1 mM Mg2+ (35)) and virtually all of the EF2 sites are occupied by Mg2+ upon illumination, when the Ca2+ concentration decreases below 100 nM (50). For EF3 and EF4, the relatively weak Mg2+ binding (Kd = 2 mM) and strong Ca2+ binding (KEF3 = 80 nM and KEF4 = 200 nM) means that both of these sites are fully occupied by Ca2+ in the dark. Mg2+ can bind to these sites only under bright light illumination, when the free Ca2+ concentration in rod outer segment falls drastically to ∼25 nM (50). Mg2+ binding at EF3 is required for RetGC activation (12, 16, 17), whereas Mg2+ binding at EF4 is not essential (12, 16), suggesting that EF4 does not bind functionally to Mg2+.
Ca2+ binding to GCAP1 leads to protein conformational changes that cause inhibition of RetGC activity at high Ca2+ levels in the rod cell, which is necessary to maintain a steady cGMP level in the dark that otherwise may lead to retinal disease (8, 51). Our ITC and NMR studies suggest that GCAP1 binds cooperatively to Ca2+ at EF3 and EF4 that promotes a Ca2+-induced conformational change (Fig. 5). Indeed, a number of residues in EF3 and EF4 (e.g. Gly105 and Gly149) exhibit Ca2+-induced NMR spectral changes that occur in a concerted fashion (Suppl Fig. 4). Also, Ca2+ binding at EF3 and EF4 both display large negative ΔCp values (−379 cal mol−1 K−1 and −101 cal mol−1 K−1) suggesting that Ca2+ binding at both sites promotes an overall burial of hydrophobic residues (29). This seems somewhat at odds with Ca2+-dependent changes of intrinsic Trp fluorescence from GCAP1, especially for Trp21 in EF1 and Trp94 in EF3 (46, 52). Trp21 and Trp94 shift into polar environments in both the Mg2+-bound and Ca2+-bound GCAP1 relative to its apo-conformation, whereas Trp94 shifts back into a more hydrophobic environment when Ca2+ replaces Mg2+ at EF3 (16). Our ITC analysis detects an overall Ca2+-induced decrease in exposed hydrophobic surface area, because ITC detects signals from all residues of the protein (and not just Trp). Therefore, Ca2+ binding at EF3 and EF4 causes conformational changes leading to a net burial of hydrophobic residues in GCAP1 that presumably compensates for the exposure of Trp21 and Trp94.
What is the structural and functional role of the N-terminal myristoyl group of GCAP1? Myristoylation of GCAP1 is thought to be important for the regulation of RetGC (53, 54), and the recent crystal structure of GCAP1 shows the myristate to be buried inside the protein hydrophobic core (18). In this study, our NMR analyses confirmed that the myristoyl group in GCAP1 is indeed sequestered inside a protein environment in the native conformation of the protein in solution. Upfield chemical shifts and NOE patterns of the myristoyl resonances (Fig. 6, Table 3) are consistent with ring-currents imposed by aromatic residues (F39, F43, F63 and Y76) shown to be near the myristate in the crystal structure. Ca2+- and/or Mg2+-binding to GCAP1 has very little influence on the chemical shifts and NOE patterns of the myristoyl group (Fig. 6), suggesting that the myristoyl group remains in a similar environment in both the Mg2+-bound activator and Ca2+-bound inhibitor forms of GCAP1. Surprisingly, however, myristoylation of GCAP1 has only a small effect on the overall protein folding stability (Fig. 4) and structure of the EF-hands (Fig. 5). Thus, it seems that myristoylation interacts most strongly with residues outside the EF-hand regions; namely, the N- and C-terminal helices that contact the myristate in the crystal structure (italicized in Fig. 1). Interestingly, the NMR resonances of these N- and C-terminal residues and those of myristate are all spectrally quite broad (linewidth, Δν = 50−80 Hz) due to exchange processes on the millisecond time scale (τex ∝ 1/Δν), suggesting a highly dynamic environment around the myristate. These millisecond exchange processes (activation energy ∼30 kJ/mol) occur at physiological temperatures but are essentially frozen out under conditions for x-ray crystallography (77 K), explaining why this dynamical behavior apparently was not observed in the crystal structure. We propose that conformational fluctuations at the myristoyl binding site might regulate the cyclase by enabling specific recognition of RetGC or by facilitating secondary interactions required for RetGC activation.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Dr. Jeff de Ropp for help with NMR experiments, Dr. Frits Abildgaard for providing NMR pulse-sequence programs, and Frank Delaglio for writing computer software for NMR data processing and analysis. A.M.D. is The Martin and Florence Hafter Chair Professor of Pharmacology.
1Abbreviations
- Ca2+
calcium ion
- Mg2+
magnesium ion
- DSC
differential scanning calorimetry
- GCAP1
guanylyl cyclase activating protein 1
- EDTA
ethylenediaminetetraacetic acid
- HMQC
heteronuclear multiple quantum coherence
- HSQC
heteronuclear single quantum coherence
- IPTG
isopropylthiogalactoside
- ITC
isothermal titration calorimetry
- NOE
nuclear Overhauser effect
- NOESY
nuclear Overhauser effect spectroscopy
- RetGC
retinal guanylyl cyclase
- SDS PAGE
sodium dodecylsulfate polyacrylamide gel electrophoresis
Footnotes
This work was supported by NIH Grants NS045909, EY012347 (J.B.A.) and RR11973 (UC Davis NMR).
SUPPORTING INFORMATION AVAILABLE
Summary of DSC data of myristoylated and unmyristoylated recoverin; fluorescence spectra of myristoylated and unmyristoylted GCAP1; 13C(F3)-filtered/13C(F1)-edited NOESY-HMQC spectrum of GCAP1 containing a 13C-labeled myristoyl group; and a plot of the relative NMR intensities of amide 1H resonances from Gly 105 (EF3) and Gly 149 (EF4) vs. Ca2+ are available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Burgoyne RD. Neuronal calcium sensor proteins: generating diversity in neuronal Ca2+ signalling. Nat. Rev. Neurosci. 2007;8:182–193. doi: 10.1038/nrn2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burgoyne RD, Weiss JL. The neuronal calcium sensor family of Ca2+-binding proteins. Biochem. J. 2001;353:1–12. [PMC free article] [PubMed] [Google Scholar]
- 3.Ames JB, Tanaka T, Stryer L, Ikura M. Portrait of a myristoyl switch protein. Curr. Opin. Struct. Biol. 1996;6:432–438. doi: 10.1016/s0959-440x(96)80106-0. [DOI] [PubMed] [Google Scholar]
- 4.Dizhoor AM, Lowe DG, Olsevskaya EV, Laura RP, Hurley JB. The human photoreceptor membrane guanylyl cyclase, RetGC, is present in outer segments and is regulated by calcium and a soluble activator. Neuron. 1994;12:1345–1352. doi: 10.1016/0896-6273(94)90449-9. [DOI] [PubMed] [Google Scholar]
- 5.Palczewski K, Subbaraya I, Gorczyca WA, Helekar BS, Ruiz CC, Ohguro H, Huang J, Zhao X, Crabb JW, Johnson RS. Molecular cloning and characterization of retinal photoreceptor guanylyl cyclase-activating protein. Neuron. 1994;13:395–404. doi: 10.1016/0896-6273(94)90355-7. [DOI] [PubMed] [Google Scholar]
- 6.Palczewski K, Polans AS, Baehr W, Ames JB. Ca(2+)-binding proteins in the retina: structure, function, and the etiology of human visual diseases. Bioessays. 2000;22:337–350. doi: 10.1002/(SICI)1521-1878(200004)22:4<337::AID-BIES4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 7.Stephen R, Filipek S, Palczewski K, Sousa MC. Ca2+ -dependent regulation of phototransduction. Photochem. Photobiol. 2008;84:903–910. doi: 10.1111/j.1751-1097.2008.00323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Payne AM, Downes SM, Bessant DA, Taylor R, Holder GE, Warren MJ, Bird AC, Bhattacharya SS. A mutation in guanylate cyclase activator 1A (GUCA1A) in an autosomal dominant cone dystrophy pedigree mapping to a new locus on chromosome 6p21.1. Hum. Mol. Genetics. 1998;7:273–277. doi: 10.1093/hmg/7.2.273. [DOI] [PubMed] [Google Scholar]
- 9.Baylor D. How photons start vision. Proc. Natl. Acad. Sci. USA. 1996;93:560–565. doi: 10.1073/pnas.93.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koch KW, Stryer L. Highly cooperative feedback control of retinal rod guanylate cyclase by calcium ions. Nature. 1988;334:64–66. doi: 10.1038/334064a0. [DOI] [PubMed] [Google Scholar]
- 11.Olshevskaya EV, Calvert PD, Woodruff ML, Peshenko IV, Savchenko AB, Makino CL, Ho YS, Fain GL, Dizhoor AM. The Y99C mutation in guanylyl cyclase-activating protein 1 increases intracellular Ca2+ and causes photoreceptor degeneration in transgenic mice. J. Neurosci. 2004;24:6078–6085. doi: 10.1523/JNEUROSCI.0963-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peshenko IV, Dizhoor AM. Activation and inhibition of photoreceptor guanylyl cyclase by guanylyl cyclase activating protein 1 (GCAP-1): the functional role of Mg2+/Ca2+ exchange in EF-hand domains. J. Biol. Chem. 2007;282:21645–21652. doi: 10.1074/jbc.M702368200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dizhoor AM, Olshevskaya EV, Henzel WJ, Wong SC, Stults JT, Ankoudinova I, Hurley JB. Cloning, sequencing and expression of a 24-kDa Ca2+-binding protein activating photoreceptor guanylyl cyclase. J. Biol. Chem. 1995;270:25200–25206. doi: 10.1074/jbc.270.42.25200. [DOI] [PubMed] [Google Scholar]
- 14.Imanishi Y, Li N, Sokal I, Sowa ME, Lichtarge O, Wensel TG, Saperstein DA, Baehr W, Palczewski K. Characterization of retinal guanylate cyclase-activating protein 3 (GCAP3) from zebrafish to man. Eur. J. Neurosci. 2002;15:63–78. doi: 10.1046/j.0953-816x.2001.01835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imanishi Y, Yang L, Sokal I, Filipek S, Palczewski K, Baehr W. Diversity of guanylate cyclase-activating proteins (GCAPs) in teleost fish: characterization of three novel GCAPs (GCAP4, GCAP5, GCAP7) from zebrafish (Danio rerio) and prediction of eight GCAPs (GCAP1−8) in pufferfish (Fugu rubripes) J. Mol. Evol. 2004;59:204–217. doi: 10.1007/s00239-004-2614-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peshenko IV, Dizhoor AM. Ca2+ and Mg2+ binding properties of GCAP-1. Evidence that Mg2+-bound form is the physiological activator of photoreceptor guanylyl cyclase. J. Biol. Chem. 2006;281:23830–23841. doi: 10.1074/jbc.M600257200. [DOI] [PubMed] [Google Scholar]
- 17.Peshenko IV, Olshevskaya EV, Dizhoor AM. Binding of guanylyl cyclase activating protein 1 (GCAP1) to retinal guanylyl cyclase (RetGC1). The role of individual EF-hands. J. Biol. Chem. 2008;283:21747–21757. doi: 10.1074/jbc.M801899200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stephen R, Bereta G, Golczak M, Palczewski K, Sousa MC. Stabilizing function for myristoyl group revealed by the crystal structure of a neuronal calcium sensor, guanylate cyclase-activating protein 1. Structure. 2007;15:1392–1402. doi: 10.1016/j.str.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ames JB, Dizhoor AM, Ikura M, Palczewski K, Stryer L. Three-dimensional structure of guanylyl cyclase activating protein-2, a calcium-sensitive modulator of photoreceptor guanylyl cyclases. J Biol Chem. 1999;274:19329–19337. doi: 10.1074/jbc.274.27.19329. [DOI] [PubMed] [Google Scholar]
- 20.Ames JB, Ishima R, Tanaka T, Gordon JI, Stryer L, Ikura M. Molecular mechanics of calcium-myristoyl switches. Nature. 1997;389:198–202. doi: 10.1038/38310. [DOI] [PubMed] [Google Scholar]
- 21.Ames JB, Ikura M, Stryer L. Molecular structure of membrane-targeting calcium sensors in vision: recoverin and guanylate cyclase-activating protein 2. Methods Enzymol. 2000;316:121–132. doi: 10.1016/s0076-6879(00)16720-5. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka T, Ames JB, Harvey TS, Stryer L, Ikura M. Sequestration of the membrane-targeting myristoyl group of recoverin in the calcium-free state. Nature. 1995;376:444–447. doi: 10.1038/376444a0. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka T, Ames JB, Kainosho M, Stryer L, Ikura M. Differential isotype labeling strategy for determining the structure of myristoylated recoverin by NMR spectroscopy. J Biomol NMR. 1998;11:135–152. doi: 10.1023/a:1008212316986. [DOI] [PubMed] [Google Scholar]
- 24.Valentine K, Mesleh M, Ikura M, Ames JB, Opella S. Structure, topology and dynamics of myristoylated recoverin bound to phospholipid bilayers. Biochemistry. 2003;42:6333–6340. doi: 10.1021/bi0206816. [DOI] [PubMed] [Google Scholar]
- 25.Osawa M, Dace A, Tong KI, Valiveti A, Ikura M, Ames JB. Mg2+ and Ca2+ differentially regulate DNA binding and dimerization of DREAM. J Biol Chem. 2005;280:18008–18014. doi: 10.1074/jbc.M500338200. [DOI] [PubMed] [Google Scholar]
- 26.Ames JB, Hamasaki N, Molchanova T. Structure and calcium-binding studies of a recoverin mutant (E85Q) in an allosteric intermediate state. Biochemistry. 2002;41:5776–5787. doi: 10.1021/bi012153k. [DOI] [PubMed] [Google Scholar]
- 27.Tsien RY. A new generation of Ca2+ indicators with greatly improved fluorescence properties. Methods Cell Biol. 1989;30:127–156. [PubMed] [Google Scholar]
- 28.Wingard JN, Chan J, Bosanac I, Haeseleer F, Palczewski K, Ikura M, Ames JB. Structural analysis of Mg2+ and Ca2+ binding to CaBP1, a neuron-specific regulator of calcium channels. J Biol Chem. 2005;280:37461–37470. doi: 10.1074/jbc.M508541200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gomez J, Hilser VJ, Xie D, Freier E. The heat capacity of proteins. Proteins. 1995;22:404–412. doi: 10.1002/prot.340220410. [DOI] [PubMed] [Google Scholar]
- 30.Gomez J, Freier E. Thermodynamic mapping of the inhibitor site of the aspartic protease endothiapepsin. J. Mol. Biol. 1995;252:337–350. doi: 10.1006/jmbi.1995.0501. [DOI] [PubMed] [Google Scholar]
- 31.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 32.Ames JB, Tanaka T, Ikura M, Stryer L. Nuclear magnetic resonance evidence for Ca(2+)-induced extrusion of the myristoyl group of recoverin. J. Biol. Chem. 1995;270:30909–30913. doi: 10.1074/jbc.270.52.30909. [DOI] [PubMed] [Google Scholar]
- 33.Ames JB, Porumb T, Tanaka T, Ikura M, Stryer L. Amino-terminal myristoylation induces cooperative calcium binding to recoverin. J Biol Chem. 1995;270:4526–4533. doi: 10.1074/jbc.270.9.4526. [DOI] [PubMed] [Google Scholar]
- 34.Maune JF, Klee CB, Beckingham K. Ca2+ binding and conformational change in two series of point mutations to the individual Ca(2+)-binding sites of calmodulin. J Biol Chem. 1992;267:5286–5295. [PubMed] [Google Scholar]
- 35.Chen C, Nakatani K, Koutalos Y. Free magnesium concentration in salamander photoreceptor outer segments. J. Physiol. 2003;553:125–135. doi: 10.1113/jphysiol.2003.053280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flaherty KM, Zozulya S, Stryer L, McKay DB. Three-dimensional Structure of Recoverin, a Calcium Sensor in Vision. Cell. 1993;75:709–716. doi: 10.1016/0092-8674(93)90491-8. [DOI] [PubMed] [Google Scholar]
- 37.Zhou W, Qian Y, Kunjilwar K, Pfaffinger PJ, Choe S. Structural insights into the functional interaction of KChIP1 with Shal-type K(+) channels. Neuron. 2004;41:573–586. doi: 10.1016/s0896-6273(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 38.Ames JB, Hendricks KB, Strahl T, Huttner IG, Hamasaki N, Thorner J. Structure and calcium-binding properties of Frq1, a novel calcium sensor in the yeast Saccharomyces cerevisiae. Biochemistry. 2000;39:12149–12161. doi: 10.1021/bi0012890. [DOI] [PubMed] [Google Scholar]
- 39.Yamniuk AP, Nguyen LT, Hoang TT, Vogel HJ. Metal ion binding properties and conformational states of calcium- and integrin-binding protein. Biochemistry. 2004;43:2558–2568. doi: 10.1021/bi035432b. [DOI] [PubMed] [Google Scholar]
- 40.Gombos Z, Durussel I, Ikura M, Rose DR, Cox JA, Chakrabarti A. Conformational coupling of Mg2+ and Ca2+ on the three-state folding of calexcitin B. Biochemistry. 2003;42:5531–5539. doi: 10.1021/bi034047j. [DOI] [PubMed] [Google Scholar]
- 41.Qi XF, Bagby S, Gombos Z, Ikura M, Chakrabarti A. Alternate routes to conformational specificity in a Greek key beta barrel protein. Eur. J. Biochem. 2001;268:4653–4663. doi: 10.1046/j.1432-1327.2001.02388.x. [DOI] [PubMed] [Google Scholar]
- 42.Altieri AS, Hinton DP, Byrd RA. Association of biomolecular systems via pulsed field gradient NMR self-diffusion measurements. J. Am. Chem. Soc. 1995;117:7566–7567. [Google Scholar]
- 43.Vogel A, Schroder T, Lange C, Huster D. Characterization of the myristoyl lipid modification of membrane-bound GCAP-2 by 2H solid-state NMR spectroscopy. Biochim. Biophys. Acta. 2007;1768:3171–3181. doi: 10.1016/j.bbamem.2007.08.024. [DOI] [PubMed] [Google Scholar]
- 44.Wingard JN, Ladner J, Vanarotti M, Fisher AJ, Buchanan KT, Engman DM, Ames JB. Structural insights into membrane targeting by the flagellar calcium-binding protein (FCaBP), a myristoylated and palmitoylated calcium sensor in Trypanosoma cruzi. J. Biol. Chem. 2008;283:23388–23396. doi: 10.1074/jbc.M803178200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Forsen S, Linse S. Cooperativity: over the Hill. Trends. Biochem. Sci. 1995;20:495–497. doi: 10.1016/s0968-0004(00)89115-x. [DOI] [PubMed] [Google Scholar]
- 46.Peshenko IV, Dizhoor AM. Guanylyl cyclase-activating proteins (GCAPs) are Ca2+/Mg2+ sensors: implications for photoreceptor guanylyl cyclase (RetGC) regulation in mammalian photoreceptors. J. Biol. Chem. 2004;279:16903–16906. doi: 10.1074/jbc.C400065200. [DOI] [PubMed] [Google Scholar]
- 47.Ramamurthy V, Tucker C, Wilkie SE, Daggett V, Hunt DM, Hurley JB. Interactions within the coiled-coil domain of RetGC-1 guanylyl cyclase are optimized for regulation rather than for high affinity. J. Biol. Chem. 2001;276:26218–26229. doi: 10.1074/jbc.M010495200. [DOI] [PubMed] [Google Scholar]
- 48.Dizhoor AM, Boikov SG, Olshevskaya EV. Constitutive activation of photoreceptor guanylate cyclase by Y99C mutant of GCAP-1. Possible role in causing human autosomal dominant cone degeneration. J. Biol. Chem. 1998;273:17311–17314. doi: 10.1074/jbc.273.28.17311. [DOI] [PubMed] [Google Scholar]
- 49.Krylov DM, Niemi GA, Dizhoor AM, Hurley JB. Mapping sites in guanylyl cyclase activating protein-1 required for regulation of photoreceptor membrane guanylyl cyclases. J. Biol. Chem. 1999;274:10833–10839. doi: 10.1074/jbc.274.16.10833. [DOI] [PubMed] [Google Scholar]
- 50.Woodruff ML, Sampath AP, Mathews HR, Krasnoperova NV, Lem J, Fain GL. Measurement of cytoplasmic calcium concentration in the rods of wild-type and transducin knock-out mice. J. Physiol. 2002;542:843–854. doi: 10.1113/jphysiol.2001.013987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woodruff ML, Janisch KM, Peshenko IV, Dizhoor AM, Tsang SH, Fain GL. Modulation of phosphodiesterase6 turnoff during background illumination in mouse rod photoreceptors. J. Neurosci. 2008;28:2064–2074. doi: 10.1523/JNEUROSCI.2973-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sokal I, Otto-Bruc AE, Surgucheva I, Verlinde CL, Wang CK, Baehr W, Palczewski K. Conformational changes in guanylyl cyclase-activating protein 1 (GCAP1) and its tryptophan mutants as a function of calcium concentration. J. Biol. Chem. 1999;274:19829–19837. doi: 10.1074/jbc.274.28.19829. [DOI] [PubMed] [Google Scholar]
- 53.Hwang JY, Koch KW. Calcium- and myristoyl-dependent properties of guanylate cyclase-activating protein-1 and protein-2. Biochemistry. 2002;41:13021–13028. doi: 10.1021/bi026618y. [DOI] [PubMed] [Google Scholar]
- 54.Hwang JY, Lange C, Helten A, Hoppner D, Duda T, Sharma RK, Koch KW. Regulatory modes of rod outer segment membrane guanylate cyclase differ in catalytic efficiency and Ca(2+)-sensitivity. Eur. J. Biochem. 2003;270:3814–3821. doi: 10.1046/j.1432-1033.2003.03770.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.