Abstract
We hypothesized that chlorophyllin (CHLN) would reduce BP-DNA adduct levels. Using NHMECs exposed to 4 μM BP for 24 hr in the presence or absence of 5 μM CHLN, we measured BP-DNA adducts by chemiluminescence immunoassay (CIA). The protocol included the following experimental groups: BP alone, BP given simultaneously with CHLN (BP+CHLN) for 24 hr, CHLN given for 24 hr followed by BP for 24 hr (preCHLN, postBP), and CHLN given for 48 hr with BP added for the last 24 hr (preCHLN, postBP+CHLN). Incubation with CHLN decreased BPdG levels in all groups, with 87 % inhibition in the preCHLN, postBP+CHLN group. To examine metabolic mechanisms, we monitored expression by Affymetrix microarray (U133A), and found BP-induced up-regulation of CYP1A1 and CYP1B1 expression, as well as up-regulation of groups of interferon-inducible, inflammation and signal transduction genes. Incubation of cells with CHLN and BP in any combination decreased expression of many of these genes. Using real time PCR (RT-PCR) the maximal inhibition of BP-induced gene expression, >85% for CYP1A1 and >70% for CYP1B1, was observed in the preCHLN, postBP+CHLN group. To explore the relationship between transcription and enzyme activity, the ethoxyresorufin-O-deethylase (EROD) assay was used to measure the combined CYP1A1 and CYP1B1 activities. BP exposure caused the EROD levels to double, compared to the unexposed controls. The CHLN-exposed groups all showed EROD levels similar to the unexposed controls. Therefore, the addition of CHLN to BP-exposed cells reduced BPdG formation and CYP1A1 and CYP1B1 expression, but EROD activity was not significantly reduced.
Keywords: polycyclic aromatic hydrocarbons, microarray, chemoprevention, chlorophyllin, NQO1, chemiluminescence immunoassay, interferon
INTRODUCTION
Polycyclic aromatic hydrocarbons (PAHs), ubiquitously present in the environment, are products of incomplete combustion of coal, wood, oil, gasoline, and organic substances (Phillips 1999; Sinha et al., 2005a; Sinha et al., 2005b). In the ambient environment PAHs are present as complex mixtures of chemicals that are a source of human exposure through inhalation of polluted air (e.g., tobacco smoke and diesel exhaust), and through ingestion of cooked food (Reeves et al., 2001; Rodgman et al., 2000). Firefighters, coke oven workers, aluminum and foundry workers and others may have occupational exposure to high levels of PAHs (Armstrong et al., 2004; Assennato et al., 1993; Boers et al., 2005; Caux et al., 2002; Ovrebo et al., 1995). These compounds are not mutagenic or carcinogenic until bioactivated by enzymes of the cytochrome P450 family (Mollerup et al., 2006; Whitlock et al., 1996) and epoxide hydrolases (Wood et al., 1976). Once metabolized, they become converted to highly reactive diol-epoxides that modify macromolecules. The covalent binding of activated PAHs to DNA, and subsequent DNA replication, may result in mutagenesis and tumorigenesis (Baird et al., 2005; Chakravarti et al., 1998; DeMarini et al., 2001; Denissenko et al., 1996; Gray et al., 2001; Hughes and Phillips 1990; Hughes and Phillips 1993; Pratt et al., 2007; Peltonen and Dipple 1995; Phillips et al., 1979).
The cytochrome P450 (CYP450) isozymes 1A1 and 1B1 are major CYP family enzymes responsible for metabolism of several PAHs, including BP, to DNA-reactive metabolites (Guengerich and Shimada 1991; Shimada and Fujii-Kuriyama 2004). CYP1A1 and 1B1 are expressed in many tissues, including liver, lung, kidney, breast, and blood leukocytes. CYP1B1 is constitutively expressed in several steroidogenic and steroid responsive tissues, including breast, while CYP1A1 is mostly inducible (Berge et al., 2004; Keshava et al., 2005; Shimada et al., 2003; Spink et al., 1998; Whyatt et al., 1998). Both these enzymes are highly induced in tissues of experimental animals that are exposed to dioxins or PAHs (Shimada and Fujii-Kuriyama 2004; Whitlock et al., 1989; Whyatt et al., 1998) through the aryl-hydrocarbon receptor (AhR) pathway (Tsuchiya et al., 2003). In addition, BP significantly induces CYP gene expression in normal human mammary epithelial cells (NHMECs) used in this study (Keshava et al., 2005).
Chlorophyllin (CHLN) is a semi-synthetic sodium/copper derivative of chlorophyll. Unlike chlorophyll, it is water-soluble, and like chlorophyll it has deodorizing activity. Chlorophyll and CHLN are antimutagenic in vitro in cultured cells exposed to mutagens such as BP, dibenzo(a,l) pyrene (DBP), 3-methylcholanthrene, N-methyl-N’-nitro-N’-nitrosoguanidine (MNNG), and aflatoxin B1 (AFB1) (Arimoto et al., 1993; Mata et al., 2004). Chlorophyll and CHLN also have anticarcinogenic effects in animal models exposed to carcinogens such as AFB1, 1,2-dimethylhydrazine, and DBP (Breinholt et al., 1995a; Breinholt et al., 1995b; Dashwood et al., 1998; Harttig and Bailey 1998; Hayashi et al., 1999). In China, where dietary AFB1 exposures are high and hepatocellular carcinoma is widespread, CHLN dietary supplementation was reported to substantially decrease AFB1-induced DNA damage (Egner et al., 2003). Therefore, we hypothesized that inhibition of PAH-DNA adduct formation would occur in human cells exposed simultaneously to BP and CHLN. In the current study we chose to use a strain of normal human mammary epithelial cells (NHMECs), reflecting a normal human, hormone-responsive tissue. We compared BPdG adduct formation, alterations in overall gene expression, alterations in CYP1A1 and 1B1 expression, and levels of EROD activity in NHMECs exposed to BP in the absence or presence of CHLN.
MATERIALS AND METHODS
Human cells and chemicals
NHMECs were isolated from normal breast tissue, collected at reduction mammoplasty, by a process involving mechanical and enzymatic disruption (Stampfer et al., 1980). The tissue was obtained through the Cooperative Human Tissue Network, which is sponsored by the National Cancer Institute and the National Disease Research Interchange. Human Studies Review Board approval was sought at NIOSH, and a waiver was granted as no unique identifiers accompanied the tissues obtained. BP (99 % purity) was purchased from the National Cancer Institute Chemical Carcinogen Reference Standard Repository, Midwest Research Institute (Kansas City, MO). Commercial grade CHLN, copper trisodium salt was obtained from Sigma-Aldrich (St. Louis, MO), and used without further purification or analysis to identify different copper chlorins. The manufacturer reported 4.0 % copper, and 6.0 % sodium in CHLN preparation. In a previous study, the CHLN from Sigma was reported to have ∼72 % copper chlorin e6 and ∼10 % copper chlorin e4, with little or no chlorin ethyl esters (Pratt et al., 2007). We made concentrated aqueous stock solutions of CHLN, diluted with medium, and included CHLN in cell culture studies at a final concentration of 5.0 μM CHLN.
Chemical Exposure and DNA preparation
To obtain a uniform culture of epithelial cells, NHMECs (strain 98013) were grown to passage 6 in serum free media (Clonetics™, Walkersville, MD). For all these experiments 4.0 μM BP and 5.0 μM CHLN were used. Based on previous experience with NHMECs (Keshava et al., personal communication), the doses of BP and CHLN were chosen because they were non-toxic but sufficiently high to give measurable DNA adduct levels. The exposure groups were as follows: (1) BP alone for 24 h; (2) BP+CHLN for 24 h; (3) preCHLN for 24 h, thorough washing and postBP for 24 h; (4) 24 h of preCHLN, and postBP + CHLN for an additional 24 h; (5) CHLN alone for 24 h; (6) CHLN alone for 48 h. A solvent only (acetone:ethanol, 1:22.5, final concentration <0.1 %) group was maintained throughout the experiment as the unexposed control. In the group exposed to preCHLN, postBP cell monolayers were washed 3 or more times with 20 ml of 1X PBS per T-75 flask to remove all of the green color. Experiments were performed twice, each time in duplicate, and specific assays were performed on each sample 6 times.
For DNA preparation, two of the replicates were washed with PBS, lysed with 5 ml lysis buffer (100 mM Tris pH 8.5, 5 mM EDTA, 0.2 % SDS, 200 mM NaCl) and incubated with RNase A (250 μg, Qiagen, Valencia, CA) for 20 min at 37 °C followed by proteinase K (500 μg, Qiagen, Valencia, CA) for 3 h at 37 °C. At the end of proteinase K digestion, an equal volume of isopropanol was added to each sample to precipitate DNA. The DNA precipitate was washed twice with ethanol (70 %), dissolved in 200 μl of molecular biology grade water (Laird et al., 1991), and stored at -70 °C for BPdG adduct analysis.
Cytotoxicity
NHMEC strain 98013 cells at passage 6 were exposed on two separate occasions, and cell survival was evaluated using the Cell Titer Glo Luminescent Cell Viability Assay (Promega, Madison, WI). In brief, cells were seeded in 12-well plates (75,000 cells/well) in triplicate. After attaching, cells were exposed to 4 μM BP and 5 μM CHLN in the same exposure groups 1-6 as described above. Following exposure, 1x RIPA Lysis Buffer (Upstate Cell Signaling, Temecula, CA) was added to lyse the cells. A diluted sample of homogeneous cell lysate was transferred to a 96-well plate and combined with an equivalent volume of Cell Titer Glo. Luminescence was measured using Tropix 717 Luminometer (Applied Biosystems, Foster City, CA). Cells exposed to the vehicle (acetone: ethanol [1:22.5]) constituted the 100 % viable control, and cell viability was expressed as a percentage of this control.
Preparation of RNA and High Density Oligonucleotide Array Expression Analysis
The total RNA was isolated from the remaining two replicates using the RNeasy kit (Qiagen, Valencia, CA) as per manufacturer’s protocol. The RNA was resuspended in 50 μl of nuclease-free water, and the concentration, purity, and quality were measured by spectrophotometry and gel electrophoresis, respectively. The RNA was converted to cRNA for the purpose of microarray analyses as per the manufacturer’s (Affymetrix, Santa Clara, CA) protocol. Briefly, total RNA (12 μg) was used for the preparation of double stranded cDNA using an oligonucleotide (dT)24 primer with a T7 RNA polymerase promoter sequence at its 5′ end. A labeled cRNA transcript was generated from the cDNA in an in vitro transcription reaction using the Enzo BioArray high yield RNA transcript labeling kit (Enzo Diagnostics Inc., Farmingdale, NY). The labeled antisense cRNA was purified using RNeasy kit (Qiagen) and each cRNA sample (20 μg) was fragmented (94 °C for 35 min) in a buffer containing Tris-acetate (40 mM, pH 8.1), potassium acetate (100 mM), and magnesium acetate (30 mM).
The fragmented cRNA samples (15 μg) were mixed with eukaryotic hybridization control oligonucleotides (20X; BioB, BioC, BioD, cre at 1.5, 5, 25, 100 pM, respectively), control oligonucleotide B2, herring sperm DNA (10 mg/ml), acetylated BSA (50 mg/ml), and hybridization buffer (2X) to form the hybridization mix. The hybridization mix was heated (99 °C for 5 min; and 45 °C for 5 min) prior to hybridization to the U133A human genome microarrays. Hybridization was allowed to proceed (45 °C in a rotary hybridization oven set at 60 rpm) for 16 h, and subsequently the arrays were washed and stained using the GeneChip™ fluidics station protocol EukGE-WS2 V4. Following washing and staining, probe arrays were scanned using GeneArray® 2500 scanner (Affymetrix). All experiments were performed in duplicate.
Data Analysis Using Affymetrix Software
Global gene expression profiles in response to BP and/or CHLN exposures were generated using the Affymetrix human genome U133A microarray. Each array was examined individually for quality of hybridization prior to comparative analysis. For pairwise comparison, increases or decreases in transcription of ≥2.0-fold with p<0.05 were reported. Image files obtained from the scanner were analyzed with the Affymetrix Microarray Suite (MAS) 5.0 software and normalized by global scaling to 1500 and to the average fluorescence intensity for the entire microarray. Absolute analysis was performed for each array prior to comparative analysis. To identify differentially expressed transcripts, pairwise comparison analyses were carried out with MAS 5.0 (Affymetrix). Statistical significance (p-values) were determined by the Wilcoxon’s signed rank test and denoted as increase, decrease, or no change. A transcript was considered significantly altered in relative abundance when p<0.05. Analysis using MAS 5.0 provides a signal log ratio (SLR), which estimates the magnitude and direction of change of a transcript when two arrays are compared (experimental versus control). The SLR output was converted into fold-change as recommended by Affymetrix. Furthermore, stringent criteria that were used to identify robust signals included a software call of ‘present’ and ≥2.0-fold change or SLR 1.0 in both replicates. Average and standard deviations were calculated for all the fold-change values. In general, only transcripts induced or suppressed by ≥2 fold were considered as differentially expressed.
Reverse Transcription Real-time PCR (RT-PCR)
Relative quantitation of gene transcripts was performed by RT-PCR using Taq-Man™ probe technology (Applied Biosystems). The cDNA obtained from the total RNA for microarray analysis was used as a template to perform real-time PCR after dilution (1:50) in a 50 μl reaction mixture containing 2X Taq-Man Universal PCR Master Mix and 20X Assays-on-Demand Gene Expression Primers and Probes for both CYP1A1 (Hs00153120_m1/X02612), CYP1B1 (Hs00164383_m1/U03688) and GAPDH (Hs99999905_m1/NM002046) (Applied Biosystems/GenBank). A two step PCR reaction was performed using the ABI 7700 Sequence Detection System. Each sample was assayed in duplicate, Cycle Threshold (CT) values were normalized to the housekeeping gene GAPDH and the fold change was calculated using the 2-ΔΔCT method (Livak and Schmittgen 2001).
Quantification of BP-DNA adducts
The BPDE-DNA chemiluminescence immunoassay (BPDE-CIA) was performed as described (Divi et al., 2002) with minor modifications. The CIA-specific reagents, including streptavidin-alkaline phosphatase, I block (casein), and CDP-Star with Emerald II, were obtained from PE Applied Biosystems (Foster City, CA). Biotinylated anti-IgG was from Jackson ImmunoResearch Laboratories, Inc., (West Grove, PA). Reacti-Bind DNA coating solution (Pierce Biotechnology, Inc., Rockford, IL) was used to coat opaque 96-well high binding polystyrene microtiter plates (Greiner Bio-one, Longwood, FL). Unless otherwise indicated, incubations were for 90 min at 37 °C and washes were conducted with PBS-Tween 20 (PBST) using an automated plate washer (Ultrawash Plus, Dynex Technologies, Guernsey, UK).
Microtiter plates, coated with 100 pg of sonicated calf-thymus DNA, either unmodified or containing 0.33 % BPdG, were stored frozen until use. Thawed plates were washed, incubated with I-block (0.25 %) and washed again. Unknown sample DNA, or standard DNA modified with BPdG to 0.3 adducts/108 nucleotides, was sonicated (20 sec, 20 % amplitude, Ultrasonic Processor, Sonics & Materials, Inc., Newtown, CT), denatured (4 min at 95 °C), and cooled (10 min on ice) before being mixed with an equal volume of anti-BPDE-DNA antiserum (rabbit # 31, bleed 8/16/1978) diluted 1:3,000,000 in I-block. Serial dilutions of DNA containing BPdG, plus carrier calf-thymus DNA, were prepared such that each well contained an equal quantity of total DNA and 0-5 fmol BPdG adduct/well. After incubation, plates were washed and incubated with biotinylated anti-rabbit antibody (1:2,500; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) in I-block solution. Plates were subsequently incubated with streptavidin alkaline phosphatase in I-block at room temperature (60 min) and washed with PBST, distilled water, and Tris buffer (20 mM Tris, 1 mM MgCl2, pH 9.5), before adding CDP Star-Emerald and incubating at room temperature for 20 min and at 4 °C overnight. Luminescence was measured using a TR717 Microplate Luminometer (PE Applied Biosystems, Foster City, CA). The lower limit of detection in assays that used 10 μg DNA was 0.3 adducts/108 nucleotides. DNA isolated from BP-unexposed NHMEC strains and BP-exposed (4.0 μM) human lymphoblastoid cells (MCL-5) served as negative and positive controls, respectively. The BPDE-DNA standard curve in the CIA showed 50 % inhibition at 0.60 ± 0.08 fmol BPdG (mean ± SE, n=30).
CYP1A1 and 1B1 activity determined in intact cells by EROD assay
EROD assays were conducted on cells cultured in 12-well plates with 1 ml medium/well. Confluent cells were exposed to BP and/or CHLN for 24-48 hr as described above, and after incubation the medium was removed and the wells were washed three times with Dulbecco’s Phosphate-Buffered Saline (DPBS). EROD activity was measured as previously described (Ciolino and Yeh, 1999) with slight modifications (Radenac et al., 2004). Intact cells were incubated in 250 μl of DPBS containing 5 μM ethoxyresorufin and 1.5 mM salicylamide for 20 min at 37 °C. At the end of incubation, duplicate aliquots of 100 μl were transferred to wells of an opaque 96 well plate. The resorufin formed was measured using an Infinite 200 fluorescence reader (Teacan, Männedorf, Switzerland) set at excitation and emission wavelengths of 560 nm and 592 nm, respectively. Values (fmol/min/mg protein) for exposure groups were calculated using a resorufin standard curve generated using the same 96 well plates. Protein was measured in each well of the 12-well plate by Bicinchoninic Acid protein assay as per the manufacturers’ protocol (Thermo Scientific, Rockford, IL).
Statistical Analysis
The statistical significance of the resulting changes was analyzed using SigmaStat 3.11 (Systat Software Inc., San Jose, CA). Results from the different exposure groups were compared to data from the unexposed controls and when the equality of the variance and the normality of the data were confirmed, they were further analyzed by ANOVA. Comparison of the group exposed to BP alone with the other groups, and pairwise multiple comparisons, were performed using the Holm-Sidak test (statistical significance at p< 0.05). In addition, correlation of adduct level reduction with fold decrease in CYP1A1 and CYP1B1 expression in the presence or absence of CHLN was determined using the Pearson product moment correlation with correlation coefficient values of (-1) to (+1). The pair of variables with positive correlation coefficients and p<0.05 increased together. For analysis of the EROD assay, groups were compared by Student’s t test.
RESULTS
Cell survival
Cells exposed according to the protocol outlined for exposure groups 1-5 were harvested at the appropriate time points and subjected to analysis by Cell Titer Glo cytotoxicity test. Cell survival, expressed as percent of the solvent control (mean ± range for 2 experiments), was 96.7 ± 7.6, 85.2 ± 1.5, 81.4 ± 0.2, 73.2 ± 0.3 and 96.7 ± 8.4 for groups 1, 2, 3, 4, and 5, respectively.
BPdG adduct levels were reduced in the presence of chlorophyllin
The BPDE-DNA CIA was used to examine the effect of CHLN on BP-induced DNA adduct formation in NHMECs. The data showed a CHLN-mediated reduction in BPdG levels (Figure 1). Exposure of NHMECs to BP alone resulted in formation 23 BPdG adducts/108 nucleotides. Co-incubation of BP with CHLN resulted in formation of 14 adducts/108 nucleotides (p≤ 0.001 compared to BP alone). Similarly, the preCHLN group, that was washed thoroughly and subsequently exposed to BP had 12 adducts/108 nucleotides (p≤ 0.001 compared to BP alone). Finally, a further decrease in BPdG formation was observed in the preCHLN, postBP+CHLN group, with a level of 3 adducts/108 nucleotides, 87% lower than for BP alone.
Figure 1.
Reduction of BPdG levels in NHMECs exposed to BP with and without CHLN. Cells were exposed on 2 separate occasions with 2 replicates for each exposure, and each replicate was assayed 6 times by BPDE-DNA CIA. Bars show mean ± range for 2 experiments and asterisks indicate values that are significantly different from the group exposed to BP alone (p < 0.05).
Gene expression profiles determined by microarray analysis
In order to explore mechanisms that might contribute to the CHLN-modulation of BPdG levels, we subjected RNA from each exposure group to Affymetrix microarray. Of the 22,283 probe sets on the array, about 45 % were detected, and overall analysis indicated significant, ≥2 signal-log ratio (SLR) alteration of 111 genes. We were primarily interested in examining a subset of genes that was significantly increased in response to BP exposure and reduced by coexposure to CHLN and BP (Tables I, II, III). These are grouped as: genes related to interferon production or activity (Table I), genes related to inflammation and signal transduction (Table II), and genes related to xenobiotic metabolism (Table III).
TABLE I.
Microarray fold-change values (mean ± SD, p < 0.05) for interferon-associated genes that are induced ≥2 fold in response to BP exposure and mitigated by chlorophyllin
| Accession No | Gene Symbol | Gene Description | BP alonea | BP+CHLNb | Pre CHLN and Post BPc | Pre CHLN, Post BP+CHLNd |
|---|---|---|---|---|---|---|
| NM_002462 | MX1 | Interferon-inducible protein p78 (mouse) | 7.5 ± 0.58 | 1.5 ± 0.34 | 0.9 ± 0.01 | 0.7 ± 0.03 |
| NM_005101 | G1P2 | Interferon, alpha-inducible protein | 7.2 ± 0.96 | 1.1 ± 0.38 | 0.5 ± 0.02 | 0.4 ± 0.08 |
| NM_001548 | IFIT1 | Interferon-induced protein with tetratricopeptide repeats 1 | 6.3± 0.38 | 0.9 ± 0.53 | 0.9 ± 0.54 | 0.3 ± 0.03 |
| NM_003641 | IFITM1 | Interferon induced transmembrane protein 1 | 4.0 ± 0.03 | 1.0 ± 0.06 | 1.4 ± 0.17 | 1.0 ± 0.06 |
| NM_001549 | IFIT4 | Interferon-induced protein with tetratricopeptide repeats 4 | 2.4 ± 0.37 | 0.7 ± 0.04 | 0.8 ± 0.12 | 0.8 ± 0.04 |
| NM_005532 | IFI27 | Interferon, alpha-inducible protein 27 | 3.3 ± 0.58 | 1.3 ± 0.10 | 0.6 ± 0.23 | 1.0 ± 0.21 |
| NM_006332 | IFI30 | Interferon, gamma-inducible protein 30 | 2.2 ± 0.09 | 1.3 ± 0.07 | 1.2 ± 0.11 | 1.2 ± 0.05 |
| NM_022873 | G1P3 | Interferon, alpha-inducible protein | 3.3 ± 0.06 | 1.1 ± 0.37 | 0.7 ± 0.02 | 0.7 ± 0.05 |
BP Alone = cells were exposed to BP alone for 24 h.
BP+CHLN = cells were exposed to BP along with CHLN for 24 h; differences in expression levels between BP alone and BP + CHLN were statistically significant.
Pre CHLN and Post BP = cells were exposed to CHLN for 24 h, washed and then incubated with BP for 24 h.
Pre CHLN and Post BP+CHLN= cells were exposed to CHLN for 24 h, washed and then exposed to BP and CHLN for 24 h.
TABLE II.
Microarray fold-change values (mean ± SD, p < 0.05) for inflammatory and signal transduction genes that were induced ≥ 2 fold in response to BP exposure, and significantly mitigated by chlorophyllin
| Accession No. | Gene Symbol | Gene Description | BP alonea | BP+CHLNb | Pre CHLN and Post BPc | Pre CHLN, and Post BP+CHLNd |
|---|---|---|---|---|---|---|
| NM_000575 | IL1A | Interleukin 1, alpha | 2.1 ± 0.07 | 1.5 ± 0.49 | 0.9 ± 0.28 | 1.7 ± 0.01 |
| NM_021258 | IL22RA1 | Interleukin 22 receptor, alpha 1 | 2. 6 ± 0.23 | 1.3 ± 0.20 | 2.0 ± 0.30 | 1.4 ± 0.01 |
| NM_000598 | IGFBP3 | Insulin-like growth factor binding protein 3 | 3.8 ± 0.46 | 3.4 ± 1.47 | 1.9 ± 0.16 | 2.5 ± 0.10 |
| NM_014070 | C6orf15 | Chromosome 6 open reading frame 15 | 17.6 ± 2.09 | 8.7 ± 5.30 | 10.6 ± 1.67 | 12.8 ± 2.65 |
| NM_172087 | TNFSF13 | Tumor necrosis factor (ligand) superfamily, member 13 | 21.0 ± 0.51 | 7.2 ± 1.63 | 7.5 ± 2.07 | 4.9 ± 0.03 |
| NM_006301 | MAP3K12 | Mitogen-activated protein kinase kinase kinase 12 | 6.8 ± 0.07 | 4.9 ± 1.42 | 3.9 ± 0.26 | 4.2 ± 0.69 |
| NM_007068 | DMC1 | DMC1 dosage suppressor of mck1 homolog | 4.7 ± 1.21 | 3.0 ± 0.18 | 3.1 ± 1.13 | 1.4 ± 0.36 |
| NM_004795 | KL | Klotho | 4.1 ± 0.26 | 2.9 ± 0.06 | 1.0 ± 0.17 | 0.8 ± 0.03 |
| NM_018476 | BEX1 | Brain expressed, X-linked 1 | 3.8 ± 0.11 | 2.7 ± 0.23 | 2.7 ± 0.04 | 1.9 ± 0.15 |
| NM_018260 | FLJ10891 | Hypothetical protein FLJ10891 | 3.6 ± 0.45 | 2.3 ± 0.29 | 1.3 ± 0.10 | 1.7 ± 0.04 |
| NM_016632 | LOC51326 | ARF protein | 3.5 ± 0.49 | 1.6 ± 0.24 | 2.2 ± 0.46 | 1.8 ± 0.12 |
| NM_002878 | RAD51L3 | RAD51-like 3 (S. cerevisiae) | 2.8 ± 0.01 | 1.6 ± 0.27 | 1.4 ± 0.05 | 1.8 ± 0.17 |
| NM_014634 | PPM1F | Protein phosphatase 1F (PP2C domain containing) | 2.3 ± 0.19 | 1.2 ± 0.01 | 1.2 ± 0.19 | 1.2 ± 0.06 |
| NM_139266 | STAT1 | Signal transducer and activator of transcription 1 | 2.3 ± 0.10 | 0.9 ± 0.02 | 0.8 ± 0.11 | 0.7 ± 0.02 |
| NM_001565 | CXCL10 | Chemokine (C-X-C motif) ligand 10 | 2.1 ± 0.10 | 1.0 ± 0.02 | 1.0 ± 0.29 | 1.0 ± 0.06 |
a,b,c,dExposure conditions were as described in Methods and Table I.
TABLE III.
Microarray fold-change (mean ± SD, p < 0.05) values for metabolism genes that were induced in response to BP exposure and mitigated by chlorophyllin
| Accession No. | Gene Symbol | Gene Description | BP alonea | BP+CHLNb | Pre CHLN and Post BPc | Pre CHLN, and Post BP+CHLNd |
|---|---|---|---|---|---|---|
| NM_000104 | CYP1B1 | Cytochrome P450, family 1, subfamily B1 | 16.5 ± 1.53 | 9.3 ± 1.42 | 7.8 ± 0.59 | 6.4 ± 0.58 |
| NM_000499 | CYP1A1 | Cytochrome P450, family 1, subfamily A1 | 12.2 ± 0.12 | 6.3 ± 0.96 | 6.8 ± 1.47 | 5.1 ± 0.22 |
| NM_025163 | SMP3 | SMP3 mannosyl-transferase | 7.7 ± 1.46 | 3.2 ± 0.20 | 2.3 ± 0.57 | 2.1 ± 0.92 |
| NM_000903 | NQO1* | NAD(P)H dehydrogenase,quinone1 | 3.0 ± 0.19 | 2.4 ± 0.28 | 1.9 ± 0.03 | 1.9 ± 0.11 |
| NM_017436 | A4GALT | Alpha 1,4-galactosyltransferase | 7.0 ± 0.12 | 4.3 ± 1.27 | 4.1 ± 0.95 | 5.4 ± 0.37 |
| NM_002534 | OAS1 | 2′,5′-oligoadenylate synthetase 1, | 3.0 ± 0.01 | 0.7 ± 0.15 | 0.9 ± 0.03 | 0.6 ± 0.16 |
| NM_002535 | OAS2 | 2′-5′-oligoadenylate synthetase 2, | 3.2 ± 0.25 | 0.5 ± 0.17 | 0.8 ± 0.09 | 1.5 ± 0.59 |
a,b,c,dExposure conditions were as described in Methods and Table I.
CHLN alone induced NQO1 2.0 ± 0.19 (by RT-PCR).
Table I shows changes in interferon-inducible genes that were up-regulated (2- to 7-fold) in the presence of BP alone and substantially reduced in the groups containing BP plus CHLN. Table II shows a group of inflammatory and signal transduction genes that were up-regulated 2- to 21-fold in response to BP, and decreased in the cells exposed to both BP and CHLN. For example, the expression of tumor necrosis factor superfamily member 13 (TNFSF13) was up-regulated 21-fold with BP alone, 7-fold with BP+CHLN or preCHLN, post-BP, and only 5-fold in the preCHLN, postBP+CHLN group (Table II).
Significant gene expression changes in enzymes related to xenobiotic metabolism are presented in Table III. Microarray analysis revealed that CYP1A1 was induced ∼12-fold in cells exposed to BP alone, and in the presence of CHLN the BP-mediated CYP1A1 induction was 5.1- to 6.3-fold, a reduction of 40-58 % (Table III). Similarly, CYP1B1 expression was induced ∼16 fold in cells exposed to BP alone, but only 6.4- to 9.3-fold in any group of cells exposed to CHLN and BP, a reduction of 44-62 %. Therefore, CHLN mitigates BP-related induction of CYP1A1 and CYP1B1 expression.
Microarray analysis also showed that expression of NAD(P)H dehydrogenase (quinone1) (NQO1), a Phase II enzyme, was induced 3-fold in cells exposed to BP alone, and that incubation of cells with BP and CHLN in any combination reduced the BP-induced expression of NQO1 (Table III). In addition, expression levels of other metabolism genes, such as mannosyl-transferase (SMP3), α1,4-galactosyltransferase (A4GALT), and oligoadenylate synthetase 1 and 2 (OAS1, OAS2) were significantly increased in the presence of BP alone and mitigated by exposure to BP in the presence of CHLN.
Confirmation of microarray data by RT-PCR
Because CYP1A1and CYP1B1 are directly involved in the metabolism of BP on the pathway to DNA adduct formation, and because the microarray data indicated that induction of these genes is reduced in the presence of CHLN, it was of interest to confirm the microarray data using RT-PCR. For the RT-PCR experiment, but not the microarray, we included two groups with CHLN alone, one taken at 24 hr and one taken at 48 hr, as controls for the BP-exposed groups. For CYP1A1, at 24 hr of CHLN exposure there was no change from the unexposed control, but at 48 hr there was a significant 2.4-fold increase. For CYP1B1, at 24 hr of CHLN exposure there was no change from the unexposed control, but at 48 hr there was a significant 1.7-fold increase. Figure 2A shows RT-PCR for CYP1A1expression for each exposure group. CYP1A1 was up-regulated 66-fold in cells exposed to BP alone, and reductions of 76-88 % were found in groups exposed to BP plus CHLN. The microarray data (Table III) correlated well with the RT-PCR CYP1A1 expression data (Figure 2A) (r = 0.99, P<0.01). By RT-PCR, CYP1B1 was up-regulated approximately 40-fold in cells exposed to BP alone, and reductions of 55-70 % were found in groups exposed to BP plus CHLN (Figure 2B). For CYP1B1, the data from RT-PCR and microarray analysis were also highly correlated (r = 1.00, P<0.01).
Figure 2.
A and B. Reduction in BP-induced CYP1A1 expression (A) and CYP1B1 expression (B) determined by RT-PCR in primary NHMECs exposed to CHLN. Results are expressed as mean ± range of 2 experiments, each with replicate samples, where every replicate was assayed 6 times. Asterisks indicate expression levels that are significantly different from the group exposed to BP alone (p < 0.05).
BPdG levels correlated with CHLN-modulation of CYP1A1 and 1B1 expression
Correlation of BPdG adduct values, determined by BPDE-DNA CIA, with expression levels of CYP1A1 and CYP1B1, determined by RT-PCR, are shown in Figures 3A and B, respectively. Figure 3 shows excellent correlations between the CHLN-modulated reduction in BPdG adduct levels and decreases in expression levels of CYP1A1 (Figure 3A; r = 0.882, p≤ 0.05) and CYP1B1 (Figure 3B; r = 0.948, p≤ 0.014). Therefore, BPdG levels decreased in direct correlation with the CYP1A1 and CYP1B1 expression levels in the CHLN-exposed groups.
Figure 3.
A and B. Correlation of CYP1A1 expression (A) and CYP1B1 expression (B) determined by RT-PCR, and BPdG level determined by BPDE-DNA CIA, in NHMECs. Points represent mean ± range of 2 experiments, each with replicates exposures, where every replicate was assayed 6 times. The exposure groups are as follows: (△) Solvent alone (Control); (◆) BP alone; (○) BP + CHLN; (■) preCHLN, post BP; and (●) preCHLN, postBP+CHLN. (A) The CHLN-mediated decrease in BP-induced CYP1A1 expression is associated with reduction in BPdG level (r = 0.868, p< 0.001). (B) The CHLN-mediated decrease in BP-induced CYP1B1 expression is associated with reduction in BPdG level (r = 0.913, p< 0.001).
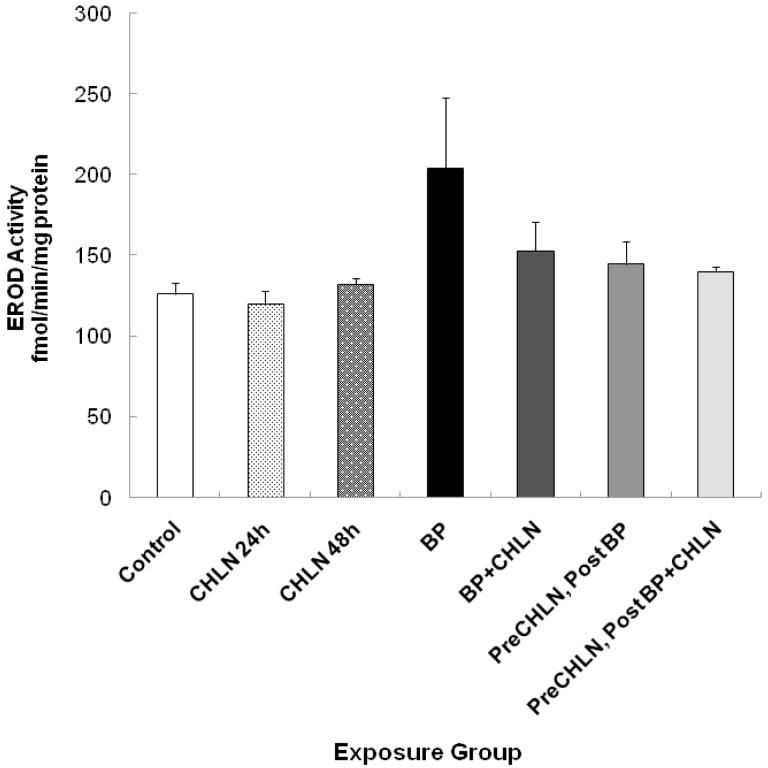
Enzyme activity of combined CYP1A1 and CYP1B1 (EROD) in NHMECs
Whereas the CHLN-modulated decreases in BP-induced CYP1A1 and CYP1B1 expression levels correlated well with BPdG values, the only functional indicator of actual enzyme activity is the EROD assay, which cannot distinguish between activity of CYP1A1 and CYP1B1 but measures both together. This precludes a fine analysis of differential modulation, but allows for an overall indication of enzyme activity. In these experiments EROD activity was 204.1 fmol/min/mg protein in the presence of BP, and 124.0 fmol/min/mg protein in the unexposed control. EROD activity, in the groups exposed to CHLN alone, was 121.0 and 137.0 fmol/min/mg protein at 24 and 48 hr, respectively. These values are similar to the 119.8 and 126.0 fmol/min/mg protein found in the unexposed controls. Though EROD values appeared higher in the group exposed to BP alone (204.2 fmol/min/mg protein), the differences were not statistically significant. By Student’s t test, p values for reduction in EROD activity in the preCHL, post BP group, and the preCHL, postBP+CHL group, compared to the BP group, were 0.090 and 0.066, respectively. The data suggest that, in spite of the strong correlation between CYP1A1 and CYP1B1 gene expression and DNA adducts, the enzyme activity, measured at this time point, does not correlate tightly with BPdG adduct formation.
DISCUSSION
In this study we showed that in NHMEC strain 98013 the soluble chlorophyll derivative CHLN effectively reduced BPdG adduct formation and the expression of the BP-metabolizing genes CYP1A1 and CYP1B1. However, specific activity of EROD, a measure of the combined CYP1A1 and CYP1B1 enzyme activities, was not correlated with adduct formation. The CHLN inhibition of BPdG and gene expression suggest that there may be direct and indirect mechanisms involved, because in cells exposed first to CHLN, then washed extensively and exposed to BP, both BPdG adducts and CYP1A1 and CYP1B1 expression were reduced. This set of pilot studies indicates that further exploration of the mechanisms by which these events occur is warranted. These studies were not designed to address the finer points of mechanism with regard to CHLN activity, but rather to determine if we could correlate DNA damage, expression changes and protein levels in a relevant biological system. As it turns out, the enzyme activities were not significantly inhibited at the same time point as the DNA adducts and expression changes, but it is possible that kinetics play a role here, and future experiments should examine the time course for EROD activity in order to obtain a better evaluation of the correlation between transcript level and enzyme level. In short, the experiments presented here are preliminary and limited, but these results can be used to design studies that will elucidate mechanisms underlying CHLN activity in this relevant biologically system.
In the literature a number of mechanisms have been proposed for the chemopreventive activity of CHLN. Scavenging of reactive oxygen species (Hernaez et al., 1997; Kumar et al., 2001), induction of phase 2 cytoprotective enzymes (Fahey et al., 2005) and inhibition of xenobiotic transport (Guo and Dashwood 1994; Mata et al., 2004) are possible mechanisms. Furthermore, formation of direct complexes between the xenobiotic and chlorophyll or CHLN through interactions between their planar unsaturated cyclic rings would decrease the bioavailability of the xenobiotic agent (Breinholt et al., 1995b). CHLN-mediated down regulation of CYP gene expression may be accomplished through several mechanisms. CHLN may sequester BP (Tachino et al., 1994), and thereby reduce bioavailability and subsequent AhR-mediated CYP gene expression. CHLN binds to RNA (Marty et al., 2004), and may inhibit translation or alter the stability of the transcript. CHLN binds to DNA through intercalation into G-C and A-T rich regions (Tajmir-Riahi et al., 2004), and may perturb DNA structure via binding to CYP genes, XRE elements upstream of CYP genes, or transcription factor binding sites having G-C rich sequences. The end result would be interference in the AhR pathway and reduction of CYP gene expression, presumably reducing levels of active CYP enzymes.
In spite of strong correlations between the CHLN-modulated decrease in CYP1A1 and CYP1B1 expression and BPdG adducts, the EROD activities did not correlate well with DNA adduct levels. EROD levels in groups exposed to CHLN were all numerically lower than the EROD activity in the BP-alone group, but the differences were not statistically significant at the p<0.05 level. A comparison of EROD values between cells exposed to BP alone, and the preCHLN, postBP+CHLN group, yielded marginal significance (p=0.066). The documented coordinate regulation of CYP1A1 and CYP1B1 enzymes suggests that measuring EROD is a reasonable approach, but the EROD assay cannot specifically measure the activities of CYP1A1 or CYP1B1. Therefore, if activity of one or the other of these enzymes was altered preferentially in the presence of CHLN it could account for the lack of complete correlation between the gene expression data and the enzyme assays. In addition, it is possible that in these normal human breast cells intermediates of the estrogen metabolism pathway may also induce CYP1A1 or CYP1B1 and confound these correlations (Singhal et al., 2008). Part of the design of future studies using these cells will include analysis of steroid hormone metabolites to specifically measure the individual CYP450s.
Several xenobiotic metabolism genes were up-regulated in BP exposed cells, and the extent of up-regulation was reduced in the groups exposed to BP and CHLN in any combination. Arguably the most important of these are CYP1A1 and CYP1B1. The increased expression of these genes in response to PAH exposure may result in increased DNA damage both directly and indirectly through activation of estradiol (Belous et al., 2007; Hayes et al., 1996; Spink et al., 1998; Spink et al., 1997). In this study we intentionally excluded estradiol from the medium to avoid hormonal activation. However, in vivo it is possible that both mechanisms may occur. An additional up-regulated xenobiotic metabolism gene was NQO1, a Phase II enzyme, that decreases the formation of the BP-3,6-quinone-induced DNA adduct (Joseph and Jaiswal 1994). In this study, BP induced NQO1 expression by 3-fold, but incubation with BP plus CHLN in any combination produced less NQO1 induction than incubation with BP alone.
A second important aspect of this study is the survey of gene expression changes conducted by Affymetrix microarray, indicating that BP-exposure alone induces up-regulation of 8 genes involved in interferon and inflammation-related pathways, and that all of these genes are down-regulated during exposure to CHLN plus BP. One of the functions of interferons is inhibition of cell growth, and therefore, up-regulation of interferon-inducible genes may constitute a general protective response with cell cycle arrest taking place to allow for genome repair. In addition, 15 inflammatory and signal transduction genes were up-regulated by BP, and 14 of these were down-regulated in the groups exposed to BP plus CHLN. These genes may be considered to participate in cellular stress-response pathways, and as such may provide specific elucidation of the CHLN effect observed here.
These investigations were performed largely because of previous studies in which CHLN supplementation was reported to reduce AFB1-DNA adduct formation (Breinholt et al., 1995a; Breinholt et al., 1995b; Dashwood et al., 1998), and to decrease excreted AFB1-DNA adduct levels in human urine (Egner et al., 2003). We hypothesized that CHLN might reduce BP-DNA adduct formation in NHMECs, and indeed, we found a reduction in BPdG formation accompanied by down-regulation of CYP1A1 and CYP1B1 gene expression levels. Incubation of BP-exposed cells in the presence of CHLN reduced the formation of BPdG adducts in direct correlation with the reduction in CYP1A1 and CYP1B1 gene expression, but the concomitant reduction in EROD activity was lower in magnitude and not statistically significant. As an attempt to elucidate factors responsible for BP-induced DNA adduct formation, this study reveals that many events may contribute. The fact that BPdG adducts were substantially lowered by CHLN, even though EROD was only partially affected, suggests a number of possibilities. Among other things, CHLN may be: inhibiting xenobiotic transport (Guo and Dashwood 1994; Mata et al., 2004); complexing with BP (Breinholt et al., 1995b); binding to RNA (Marty et al., 2004) to alter the stability of the transcript; or, binding to DNA causing perturbation of transcription factor binding sites (Tajmir-Riahi et al., 2004). In addition, longer incubation of NHMECs with CHLN might be necessary for EROD levels to drop to significance as a consequence of the drop in expression. Also, levels of detoxification enzyme activity might be altered by CHLN such that adduct removal by that mechanism is enhanced. All of these possibilities would not be reflected in enzyme levels as measured by EROD, and will be the subject of future investigations in this area.
Figure 4.
EROD (combined CYP1A1 and CYP1B1) activity. Each bar represents mean ± range from 2 separate exposures with 2 replicates each, from which each replicate was assayed 6 times. Statistical significance for comparison between: the BP alone group vs. the preCHLN, postBP group, p=0.090; and the BP alone group vs. the preCHLN, postBP+CHLN group, p=0.066.
Acknowledgments and Disclaimers
This work was supported in part by the Intramural Research Programs of the US Centers for Disease Control (CDC) National Institute for Occupational Safety and Health (NIOSH), and the US National Institutes of Health (NIH) National Cancer Institute (NCI) Center for Cancer Research. The mention of commercial products in this report does not constitute an endorsement by NIOSH, NCI, or the US Environmental Protection Agency (EPA). The findings and conclusions in this report are those of the authors and do not necessarily represent the views or policies of NIOSH, NCI or the EPA.
Abbreviations
- AFB1
aflatoxin B1
- AhR
aryl hydrocarbon receptor
- BP
benzo[a]pyrene
- BPDE
r7, t8-dihydroxy-t-9, 10-oxy-7, 8, 9, 10-tetrahydrobenzo[a]pyrene
- BPDE-DNA CIA
BPDE-DNA chemiluminescence immunoassay
- BPdG
r7, t8, t9-trihydroxy-c-10-(N2deoxyguanosyl)-7, 8, 9, 10-tetrahydro-benzo[a]pyrene
- CHLN
chlorophyllin
- CIA
chemiluminescence immunoassay
- CYP1A1
cytochrome P450 1A1
- CYP1B1
cytochrome P450 1B1
- DBP
dibenzo[a,l]pyrene
- DPBS
Dulbecco’s Phosphate Buffered Saline
- EROD
ethoxyresorufin-O-deethylase
- MNNG
N-methyl-N’-nitro-N’-nitrosoguanidine
- NQO1
NAD(P)H:Quinone Oxidoreductase 1
- NHMEC
normal human mammary epithelial cell
- OA
oligonucleotide array
- PAH
polycyclic aromatic hydrocarbon
- PBST
1x Phosphate buffered saline containing 0.05% Tween 20
- RT-PCR
reverse transcription real-time polymerase chain reaction
- SLR
signal-log ratio
REFERENCES
- Arimoto S, Fukuoka S, Itome C, Nakano H, Rai H, Hayatsu H. Binding of polycyclic planar mutagens to chlorophyllin resulting in inhibition of the mutagenic activity. Mutat Res. 1993;287:293–305. doi: 10.1016/0027-5107(93)90022-8. [DOI] [PubMed] [Google Scholar]
- Armstrong B, Hutchinson E, Unwin J, Fletcher T. Lung cancer risk after exposure to polycyclic aromatic hydrocarbons: a review and meta-analysis. Environ Health Perspect. 2004;112:970–8. doi: 10.1289/ehp.6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assennato G, Ferri GM, Tockman MS, Poirier MC, Schoket B, Porro A, Corrado V, Strickland PT. Biomarkers of carcinogen exposure and cancer risk in a coke plant. Environ Health Perspect. 1993;99:237–9. doi: 10.1289/ehp.9399237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird WM, Hooven LA, Mahadevan B. Carcinogenic polycyclic aromatic hydrocarbon-DNA adducts and mechanism of action. Environ Mol Mutagen. 2005;45:106–14. doi: 10.1002/em.20095. [DOI] [PubMed] [Google Scholar]
- Belous AR, Hachey DL, Dawling S, Roodi N, Parl FF. Cytochrome P450 1B1-mediated estrogen metabolism results in estrogen-deoxyribonucleoside adduct formation. Cancer Res. 2007;67:812–7. doi: 10.1158/0008-5472.CAN-06-2133. [DOI] [PubMed] [Google Scholar]
- Berge G, Mollerup S, S OV, Hewer A, Phillips DH, Eilertsen E, Haugen A. Role of estrogen receptor in regulation of polycyclic aromatic hydrocarbon metabolic activation in lung. Lung Cancer. 2004;45:289–97. doi: 10.1016/j.lungcan.2004.02.014. [DOI] [PubMed] [Google Scholar]
- Boers D, Zeegers MP, Swaen GM, Kant I, van den Brandt PA. The influence of occupational exposure to pesticides, polycyclic aromatic hydrocarbons, diesel exhaust, metal dust, metal fumes, and mineral oil on prostate cancer: a prospective cohort study. Occup Environ Med. 2005;62:531–7. doi: 10.1136/oem.2004.018622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breinholt V, Hendricks J, Pereira C, Arbogast D, Bailey G. Dietary chlorophyllin is a potent inhibitor of aflatoxin B1 hepatocarcinogenesis in rainbow trout. Cancer Res. 1995a;55:57–62. [PubMed] [Google Scholar]
- Breinholt V, Schimerlik M, Dashwood R, Bailey G. Mechanisms of chlorophyllin anticarcinogenesis against aflatoxin B1: complex formation with the carcinogen. Chem Res Toxicol. 1995b;8:506–14. doi: 10.1021/tx00046a004. [DOI] [PubMed] [Google Scholar]
- Caux C, O’Brien C, Viau C. Determination of firefighter exposure to polycyclic aromatic hydrocarbons and benzene during fire fighting using measurement of biological indicators. Appl Occup Environ Hyg. 2002;17:379–86. doi: 10.1080/10473220252864987. [DOI] [PubMed] [Google Scholar]
- Chakravarti D, Mailander P, Franzen J, Higginbotham S, Cavalieri EL, Rogan EG. Detection of dibenzo[a,l]pyrene-induced H-ras codon 61 mutant genes in preneoplastic SENCAR mouse skin using a new PCR-RFLP method. Oncogene. 1998;16:3203–10. doi: 10.1038/sj.onc.1201853. [DOI] [PubMed] [Google Scholar]
- Ciolino HP, Yeh GC. Inhibition of aryl hydrocarbon-induced cytochrome P-450 1A1 enzyme activity and CYP1A1 expression by resveratrol. Mol. Pharmacol. 1999;56:760–767. [PubMed] [Google Scholar]
- Dashwood R, Negishi T, Hayatsu H, Breinholt V, Hendricks J, Bailey G. Chemopreventive properties of chlorophylls towards aflatoxin B1: a review of the antimutagenicity and anticarcinogenicity data in rainbow trout. Mutat Res. 1998;399:245–53. doi: 10.1016/s0027-5107(97)00259-5. [DOI] [PubMed] [Google Scholar]
- DeMarini DM, Landi S, Tian D, Hanley NM, Li X, Hu F, Roop BC, Mass MJ, Keohavong P, Gao W. Lung tumor KRAS and TP53 mutations in nonsmokers reflect exposure to PAH-rich coal combustion emissions. Cancer Res. 2001;61:6679–81. others. [PubMed] [Google Scholar]
- Denissenko MF, Pao A, Tang M, Pfeifer GP. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science. 1996;274:430–2. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- Divi RL, Beland FA, Fu PP, Von Tungeln LS, Schoket B, Camara JE, Ghei M, Rothman N, Sinha R, Poirier MC. Highly sensitive chemiluminescence immunoassay for benzo[a]pyrene-DNA adducts: validation by comparison with other methods, and use in human biomonitoring. Carcinogenesis. 2002;23:2043–9. doi: 10.1093/carcin/23.12.2043. [DOI] [PubMed] [Google Scholar]
- Egner PA, Munoz A, Kensler TW. Chemoprevention with chlorophyllin in individuals exposed to dietary aflatoxin. Mutat Res. 2003;523-524:209–16. doi: 10.1016/s0027-5107(02)00337-8. [DOI] [PubMed] [Google Scholar]
- Gray DL, Warshawsky D, Xue W, Nines R, Wang Y, Yao R, Stoner GD. The effects of a binary mixture of benzo(a)pyrene and 7H-dibenzo(c,g)carbazole on lung tumors and K-ras oncogene mutations in strain A/J mice. Exp Lung Res. 2001;27:245–53. doi: 10.1080/019021401300054000. [DOI] [PubMed] [Google Scholar]
- Guengerich FP, Shimada T. Oxidation of toxic and carcinogenic chemicals by human cytochrome P-450 enzymes. Chem Res Toxicol. 1991;4:391–407. doi: 10.1021/tx00022a001. [DOI] [PubMed] [Google Scholar]
- Guo D, Dashwood R. Inhibition of 2-amino-3-methylimidazo[4,5-f]quinoline (IQ)-DNA binding in rats given chlorophyllin: dose-response and time-course studies in the liver and colon. Carcinogenesis. 1994;15:763–6. doi: 10.1093/carcin/15.4.763. [DOI] [PubMed] [Google Scholar]
- Harttig U, Bailey GS. Chemoprotection by natural chlorophylls in vivo: inhibition of dibenzo[a,l]pyrene-DNA adducts in rainbow trout liver. Carcinogenesis. 1998;19:1323–6. doi: 10.1093/carcin/19.7.1323. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Schimerlik M, Bailey G. Mechanisms of chlorophyllin anticarcinogenesis: dose-responsive inhibition of aflatoxin uptake and biodistribution following oral co-administration in rainbow trout. Toxicol Appl Pharmacol. 1999;158:132–40. doi: 10.1006/taap.1999.8695. [DOI] [PubMed] [Google Scholar]
- Hayes CL, Spink DC, Spink BC, Cao JQ, Walker NJ, Sutter TR. 17 beta-estradiol hydroxylation catalyzed by human cytochrome P450 1B1. Proc Natl Acad Sci U S A. 1996;93:9776–81. doi: 10.1073/pnas.93.18.9776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernaez J, Xu M, Dashwood R. Effects of tea and chlorophyllin on the mutagenicity of N-hydroxy-IQ: studies of enzyme inhibition, molecular complex formation, and degradation/scavenging of the active metabolites. Environ Mol Mutagen. 1997;30:468–74. doi: 10.1002/(sici)1098-2280(1997)30:4<468::aid-em12>3.0.co;2-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes NC, Phillips DH. Covalent binding of dibenzpyrenes and benzo[a]pyrene to DNA: evidence for synergistic and inhibitory interactions when applied in combination to mouse skin. Carcinogenesis. 1990;11:1611–9. doi: 10.1093/carcin/11.9.1611. [DOI] [PubMed] [Google Scholar]
- Hughes NC, Phillips DH. 32P-postlabelling analysis of the covalent binding of benzo[ghi]perylene to DNA in vivo and in vitro. Carcinogenesis. 1993;14:127–33. doi: 10.1093/carcin/14.1.127. [DOI] [PubMed] [Google Scholar]
- Joseph P, Jaiswal AK. NAD(P)H:quinone oxidoreductase1 (DT diaphorase) specifically prevents the formation of benzo[a]pyrene quinone-DNA adducts generated by cytochrome P4501A1 and P450 reductase. Proc Natl Acad Sci U S A. 1994;91:8413–7. doi: 10.1073/pnas.91.18.8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshava C, Divi RL, Whipkey DL, Frye BL, McCanlies E, Kuo M, Poirier MC, Weston A. Induction of CYP1A1 and CYP1B1 and formation of carcinogen-DNA adducts in normal human mammary epithelial cells treated with benzo[a]pyrene. Cancer Lett. 2005;221:213–24. doi: 10.1016/j.canlet.2004.08.038. [DOI] [PubMed] [Google Scholar]
- Kumar SS, Devasagayam TP, Bhushan B, Verma NC. Scavenging of reactive oxygen species by chlorophyllin: an ESR study. Free Radic Res. 2001;35:563–74. doi: 10.1080/10715760100301571. [DOI] [PubMed] [Google Scholar]
- Laird PW, Zijderveld A, Linders K, Rudnicki MA, Jaenisch R, Berns A. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Marty R, Ouameur AA, Neault JF, Tajmir-Riahi HA. RNA adducts with chlorophyll and chlorophyllin: stability and structural features. J Biomol Struct Dyn. 2004;22:45–50. doi: 10.1080/07391102.2004.10506979. [DOI] [PubMed] [Google Scholar]
- Mata JE, Yu Z, Gray JE, Williams DE, Rodriguez-Proteau R. Effects of chlorophyllin on transport of dibenzo(a, l)pyrene, 2-amino-1-methyl-6-phenylimidazo-[4,5-b]pyridine, and aflatoxin B(1) across Caco-2 cell monolayers. Toxicology. 2004;196:117–25. doi: 10.1016/j.tox.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Mollerup S, Berge G, Baera R, Skaug V, Hewer A, Phillips DH, Stangeland L, Haugen A. Sex differences in risk of lung cancer: Expression of genes in the PAH bioactivation pathway in relation to smoking and bulky DNA adducts. Int J Cancer. 2006;119:741–4. doi: 10.1002/ijc.21891. [DOI] [PubMed] [Google Scholar]
- Ovrebo S, Haugen A, Hemminki K, Szyfter K, Drablos PA, Skogland M. Studies of biomarkers in aluminum workers occupationally exposed to polycyclic aromatic hydrocarbons. Cancer Detect Prev. 1995;19:258–67. [PubMed] [Google Scholar]
- Peltonen K, Dipple A. Polycyclic aromatic hydrocarbons: chemistry of DNA adduct formation. J Occup Environ Med. 1995;37:52–8. doi: 10.1097/00043764-199501000-00008. [DOI] [PubMed] [Google Scholar]
- Phillips DH. Polycyclic aromatic hydrocarbons in the diet. Mutat Res. 1999;443:139–47. doi: 10.1016/s1383-5742(99)00016-2. [DOI] [PubMed] [Google Scholar]
- Phillips DH, Grover PL, Sims P. A quantitative determination of the covalent binding of a series of polycylic hydrocarbons to DNA in mouse skin. Int J Cancer. 1979;23:201–8. doi: 10.1002/ijc.2910230211. [DOI] [PubMed] [Google Scholar]
- Pratt MM, Reddy AP, Hendricks JD, Pereira C, Kensler TW, Bailey GS. The importance of carcinogen dose in chemoprevention studies: quantitative interrelationships between, dibenzo[a,l]pyrene dose, chlorophyllin dose, target organ DNA adduct biomarkers and final tumor outcome. Carcinogenesis. 2007;28:611–24. doi: 10.1093/carcin/bgl174. [DOI] [PubMed] [Google Scholar]
- Radenac G, Coteur G, Danis B, Dubois Ph, Warnau M. Measurement of EROD Activity: Caution on Spectral Properties of Standards Used. Mar. Biotechnol. 2004;6:307–311. doi: 10.1007/s10126-004-3014-4. [DOI] [PubMed] [Google Scholar]
- Reeves WR, Barhoumi R, Burghardt RC, Lemke SL, Mayura K, McDonald TJ, Phillips TD, Donnelly KC. Evaluation of methods for predicting the toxicity of polycyclic aromatic hydrocarbon mixtures. Environ Sci Technol. 2001;35:1630–6. doi: 10.1021/es001689a. [DOI] [PubMed] [Google Scholar]
- Rodgman A, Smith CJ, Perfetti TA. The composition of cigarette smoke: a retrospective, with emphasis on polycyclic components. Hum Exp Toxicol. 2000;19:573–95. doi: 10.1191/096032700701546514. [DOI] [PubMed] [Google Scholar]
- Shimada T, Fujii-Kuriyama Y. Metabolic activation of polycyclic aromatic hydrocarbons to carcinogens by cytochromes P450 1A1 and 1B1. Cancer Sci. 2004;95:1–6. doi: 10.1111/j.1349-7006.2004.tb03162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Sugie A, Shindo M, Nakajima T, Azuma E, Hashimoto M, Inoue K. Tissue-specific induction of cytochromes P450 1A1 and 1B1 by polycyclic aromatic hydrocarbons and polychlorinated biphenyls in engineered C57BL/6J mice of arylhydrocarbon receptor gene. Toxicol Appl Pharmacol. 2003;187:1–10. doi: 10.1016/s0041-008x(02)00035-2. [DOI] [PubMed] [Google Scholar]
- Singhal R, Shankar K, Badger TM, Ronis MJ. Estrogenic status modulates aryl hydrocarbon receptor--mediated hepatic gene expression and carcinogenicity. Carcinogenesis. 2008;29:227–236. doi: 10.1093/carcin/bgm288. [DOI] [PubMed] [Google Scholar]
- Sinha R, Kulldorff M, Gunter MJ, Strickland P, Rothman N. Dietary benzo[a]pyrene intake and risk of colorectal adenoma. Cancer Epidemiol Biomarkers Prev. 2005a;14:2030–4. doi: 10.1158/1055-9965.EPI-04-0854. [DOI] [PubMed] [Google Scholar]
- Sinha R, Peters U, Cross AJ, Kulldorff M, Weissfeld JL, Pinsky PF, Rothman N, Hayes RB. Meat, meat cooking methods and preservation, and risk for colorectal adenoma. Cancer Res. 2005b;65:8034–41. doi: 10.1158/0008-5472.CAN-04-3429. [DOI] [PubMed] [Google Scholar]
- Spink DC, Spink BC, Cao JQ, DePasquale JA, Pentecost BT, Fasco MJ, Li Y, Sutter TR. Differential expression of CYP1A1 and CYP1B1 in human breast epithelial cells and breast tumor cells. Carcinogenesis. 1998;19:291–8. doi: 10.1093/carcin/19.2.291. [DOI] [PubMed] [Google Scholar]
- Spink DC, Spink BC, Cao JQ, Gierthy JF, Hayes CL, Li Y, Sutter TR. Induction of cytochrome P450 1B1 and catechol estrogen metabolism in ACHN human renal adenocarcinoma cells. J Steroid Biochem Mol Biol. 1997;62:223–32. doi: 10.1016/s0960-0760(97)00024-1. [DOI] [PubMed] [Google Scholar]
- Stampfer M, Hallowes RC, Hackett AJ. Growth of normal human mammary cells in culture. In Vitro. 1980;16:415–25. doi: 10.1007/BF02618365. [DOI] [PubMed] [Google Scholar]
- Tachino N, Guo D, Dashwood WM, Yamane S, Larsen R, Dashwood R. Mechanisms of the in vitro antimutagenic action of chlorophyllin against benzo[a]pyrene: studies of enzyme inhibition, molecular complex formation and degradation of the ultimate carcinogen. Mutat Res. 1994;308:191–203. doi: 10.1016/0027-5107(94)90154-6. [DOI] [PubMed] [Google Scholar]
- Tajmir-Riahi HA, Neault JF, Diamantoglou S. DNA adducts with chlorophyll and chlorophyllin as antimutagenic agents: synthesis, stability, and structural features. Methods Mol Biol. 2004;274:159–71. doi: 10.1385/1-59259-799-8:159. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Nakajima M, Yokoi T. Critical enhancer region to which AhR/ARNT and Sp1 bind in the human CYP1B1 gene. J Biochem (Tokyo) 2003;133:583–92. doi: 10.1093/jb/mvg075. [DOI] [PubMed] [Google Scholar]
- Whitlock JP, Jr., Denison MS, Fisher JM, Shen ES. Induction of hepatic cytochrome P450 gene expression by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Mol Biol Med. 1989;6:169–78. [PubMed] [Google Scholar]
- Whitlock JP, Jr., Okino ST, Dong L, Ko HP, Clarke-Katzenberg R, Ma Q, Li H. Cytochromes P450 5: induction of cytochrome P4501A1: a model for analyzing mammalian gene transcription. Faseb J. 1996;10:809–18. doi: 10.1096/fasebj.10.8.8666157. [DOI] [PubMed] [Google Scholar]
- Whyatt RM, Bell DA, Jedrychowski W, Santella RM, Garte SJ, Cosma G, Manchester DK, Young TL, Cooper TB, Ottman R. Polycyclic aromatic hydrocarbon-DNA adducts in human placenta and modulation by CYP1A1 induction and genotype. Carcinogenesis. 1998;19:1389–92. doi: 10.1093/carcin/19.8.1389. others. [DOI] [PubMed] [Google Scholar]
- Wood AW, Levin W, Lu AY, Yagi H, Hernandez O, Jerina DM, Conney AH. Metabolism of benzo(a)pyrene and benzo (a)pyrene derivatives to mutagenic products by highly purified hepatic microsomal enzymes. J Biol Chem. 1976;251:4882–90. [PubMed] [Google Scholar]