Abstract
We have visualized the movements of native mRNAs in living cells. Using nuclease-resistant molecular beacons, we imaged the transport and localization of oskar mRNA in Drosophila melanogaster oocytes. When the localization pattern was altered by genetic manipulation of the mRNA's 3′ untranslated region, or by chemical perturbation of the intracellular tubulin network, the distribution of the fluorescence signals changed accordingly. We tracked the migration of oskar mRNA in real time, from the nurse cells where it is produced to the posterior cortex of the oocyte where it is localized. Our observations reveal the presence of a transient, and heretofore elusive, stage in the transport of oskar mRNA. Direct visualization of specific mRNAs in living cells with molecular beacons will accelerate studies of intracellular RNA trafficking and localization, just as the use of green fluorescent protein has stimulated the study of specific proteins in vivo.
Cells respond to their internal genetic programs and external stimuli by modulating the synthesis of specific mRNAs. Some cells, such as fibroblasts, neurons, and oocytes, further refine these responses by sequestering specific mRNAs in different regions of the cell. The subcellular localization of mRNAs helps to restrict the activity of their encoded protein to where it is most needed. The mechanisms that cells employ to sequester mRNAs in subcellular zones without compartmentalization is the subject of intense investigation (1, 2). A particularly rich system for investigating this phenomenon occurs in Drosophila oocytes, where a number of different mRNAs are localized in distinct regions (1-3). Among these are bicoid and oskar mRNAs, which are sequestered, respectively, at the anterior and posterior ends of developing oocytes (4, 5). Another intensively studied mRNA, gurken, is initially localized at the posterior end, and later migrates to the dorsoanterior corner of the oocyte (6). The focus of this study is oskar mRNA, which is synthesized in nurse cells during the early stages of oogenesis and is then transported to the oocyte via interconnecting channels called ring canals (7). Once inside the oocyte, oskar mRNA traverses the cytoplasm to reach the posterior end where it is concentrated by binding to a localized anchor (8). An intact microtubule network (9), a functional kinesin I motor (10), and particular sequence motifs in the 3′ UTR of oskar mRNA (11) are all required for this transport, suggesting that either the motor proteins carry the mRNA directly (10, 12), or it is swept along by cytoplasmic flows that are generated by the motor-dependent transport of organelles (13). The organization and polarities of the oocyte microtubule network that control these transport processes may change during oogenesis and are still under debate (14, 15). To clarify the mechanism of transport of oskar mRNA, we have directly imaged its transport from the nurse cell to the posterior end of the oocyte.
Currently, the method of choice for mapping the intracellular distribution of mRNAs is in situ hybridization with labeled nucleic acid probes (16). In this method, the cells are fixed, and excess probes are removed by washing to detect the hybrids. Fixing leads to the denaturation and crosslinking of proteins and causes cell death, precluding the study of dynamic processes such as intracellular trafficking. To circumvent this problem, green fluorescent protein fused to bacteriophage MS2 RNA coat protein has been used to image engineered RNAs containing MS2 coat protein-binding sites in living cells (17). Recently, this approach was used to explore mechanism of nanos mRNA transport in Drosophila oocytes (18). The transport processes of a number of different mRNAs have also been dissected by injecting fluorescently labeled synthetic RNAs into live oocytes (13, 19-21). Through construction of mRNAs possessing deletions of different sequence motifs, and by injecting them into specific locations within the oocyte, valuable information has been gained about the requirements and nature of these processes. However, because naturally occurring mRNAs are processed and ”dressed” with protein factors (19) before being sent on their journey, it is important to be able to detect native mRNAs in live cells.
We have developed a general method for directly visualizing endogenous mRNAs in living cells by using hybridization probes called ”molecular beacons,” which generate fluorescence signals only when they hybridize to complementary nucleic acid target sequences (22). Molecular beacons are oligonucleotides that possess complementary sequences on either end of a probe sequence, enabling the molecule to assume a hairpin configuration in which a fluorophore and a quencher are held in close proximity (Fig. 1A). The formation of a probe-target hybrid disrupts the hairpin stem, removing the fluorophore from the vicinity of the quencher, restoring the probe's fluorescence. Because unbound molecular beacons are nonfluorescent, it is not necessary to remove excess probes to detect the hybrids. Moreover, these probes bind to their targets spontaneously at physiological temperatures. Therefore, the introduction of molecular beacons into cells is sufficient to illuminate a target mRNA. Furthermore, it is possible to detect different mRNAs simultaneously in the same cell by using molecular beacons that are specific for different targets and that are labeled with differently colored fluorophores (23). In this study, we use these probes to image the distribution and transport of specific mRNAs in living cells.
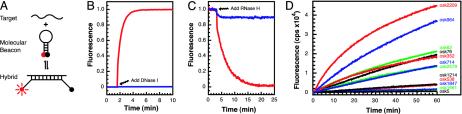
Fig. 1.
Design of the molecular beacon. (A) Principle of operation. When these probes hybridize to their mRNA targets, the fluorescence of an internally quenched fluorophore is restored, enabling target detection. (B and C) Substitution of 2′-O-methylribonucleotides for deoxyribonucleotides within the backbone of molecular beacons renders them resistant to cleavage by DNase I, and their hybrids with RNA become refractory to digestion by ribonuclease H. The fluorescence of molecular beacons possessing deoxyribonucleotide backbones (osk76-DNA) is shown in red, and the fluorescence of molecular beacons possessing 2′-O-methylribonucleotide backbones (osk76) is shown in blue. (D) Selection of molecular beacon-accessible target regions in oskar mRNA. The most accessible target sequences induce the fastest increase in fluorescence. The location of molecular beacon target sites within the predicted secondary structure of oskar mRNA is shown in Fig. 5, and the sequences of the molecular beacons specific for these sites are listed in Table 1.
Materials and Methods
Molecular Beacon Synthesis. Molecular beacons possessing a 2′-O-methylribonucleotide backbone were synthesized on an Applied Biosystems 394 DNA synthesizer by using 2′-O-methylcyanoethyl phosphoramidites (Glen Research, Sterling, VA). The controlled-pore glass columns used for these syntheses introduced either an amino group (Glen Research) or a BHQ2 group at the 3′ ends of the oligonucleotides (Biosearch). The last nucleotide added possessed a 5′-sulfhydryl group protected by a trityl moiety. Each oligonucleotide was then purified with the trityl moiety still attached. Either an iodoacetamide derivative of tetramethylrhodamine (TMR) or a maleimide derivative of the QSY7 quencher (Molecular Probes) was then coupled to the 5′-sulfhydryl group, and a succinimidyl ester derivative of Texas red was coupled to the 3′-amino group. The activated fluorophores were purchased from Molecular Probes. The completed molecular beacons were then purified by high-pressure liquid chromatography through a C-18 reverse-phase column, and their signal-to-background ratio was measured as described (23). A detailed protocol for molecular beacon synthesis is available at www.molecularbeacons.org.
Nuclease Sensitivity. To test the nuclease sensitivity of molecular beacons, the fluorescence of a 125-μl solution containing 40 mM Tris·HCl (pH 8.0), 10 mM MgSO4, 1 mM CaCl2, and 160 nM of either osk76 or osk76-DNA was measured as a function of time at 37°C. Five units of ribonuclease-free DNase I were added, and the subsequent change in fluorescence was recorded as a function of time. To test the susceptibility of molecular beacon-oskar mRNA hybrids to digestion by ribonuclease H, 20 pmol of either osk76 (a probe synthesized from 2′-O-methylribonucleotides) or osk76-DNA (a probe synthesized from 2′-deoxyribonucleotides) was incubated with 2.7 pmol of in vitro-transcribed oskar mRNA for 3 h at 37°C in 125 μl of the buffer described above. Full-length oskar mRNA was prepared by in vitro transcription from a plasmid containing oskar cDNA [which was a gift from R. Lehmann (24)]. Thereafter, 5 units of ribonuclease H were added, and the subsequent change in fluorescence was recorded as a function of time. For each enzyme, fluorescence intensity readings were normalized on a scale of 0 to 1.
In Vitro Hybridization. For determination of the accessibility of molecular beacon target sites on oskar mRNA, the fluorescence intensity of a 125-μl solution containing 20 mM Tris·HCl (pH 8.0), 1 mM MgCl2, and 80 nM molecular beacon was measured at 25°C until no change in fluorescence occurred; a 5-μl aliquot of 100 nM oskar mRNA was added to this solution, and the subsequent change in fluorescence intensity was measured as a function of time at 25°C. The time course of hybridization was recorded with the excitation wavelength set at 555 nm and the emission wavelength set at 575 nm. For the in vitro fluorescence resonance energy transfer (FRET) experiments, hybrids were formed with osk62, osk87, or both, by using twice the quantity of oskar mRNA. Emission spectra were recorded between 565 nm and 650 nm, while maintaining the excitation wavelength at 555 nm. All in vitro reactions were monitored by using a QuantaMaster spectrofluorometer (Photon Technology International, Lawrenceville, NJ).
Microinjection and Imaging. For microinjection, Drosophila melanogaster egg chambers were isolated from wild-type flies (Oregon R), osk3′-UTR-bcd flies (24), and from flies whose oocytes possess a kinesin I heavy chain null mutation, generated as described (10, 12). Well fed female flies were dissected 3 days after hatching. Their ovaries were teased out, and the egg chambers were separated under a dissecting microscope. Solutions of 100 ng/μl molecular beacons dissolved in water were microinjected into an oocyte or a nurse cell. In separate experiments, we estimated that 105 to 106 molecules of the molecular beacons are introduced in each injection. Images were captured at various time intervals, beginning immediately after microinjection. In some experiments, oskar mRNA localization was prevented by treatment with the microtubule-depolymerizing agent colcemid. For this purpose, wild-type flies were starved for one day after hatching and then fed fresh yeast paste containing 50 μg/ml colcemid overnight.
A Zeiss Axiovert 200M inverted fluorescence microscope, equipped with a Photometrics (Tucson, AZ) Coolsnap HQ camera, was used for imaging. The fluorescence of TMR was imaged by using excitation filter 546DF10, dichroic mirror 555DRLP, and emission filter 580DF30. The fluorescence of Texas red was imaged by using excitation filter 590DF10, dichroic mirror 610DRLP, and emission filter 630DF30. FRET from TMR-to-Texas red was imaged by using excitation filter 546DF10, dichroic mirror 610DRLP, and emission filter 630DF30. All of the filters and dichroic mirrors were purchased from Omega Optical (Brattleboro, VT). The images were acquired and analyzed by using OPENLAB and VOLOCITY software (Improvision, Lexington, MA). Confocal sectioning was performed by using a Leica (Deerfield, IL) laser scanning microscope and Leica TCS NT time-lapse acquisition software, and the images were analyzed by using the program NIH IMAGE, which is available at http://rsb.info.nih.gov/nih-image.
Results
Nuclease-Resistant Molecular Beacons. Ideal probes for targets in living cells should be stable inside the cell, should not induce the destruction or perturbation of their target, and should signal only the presence of their target when (and where) the target is present. Molecular beacons constructed from natural deoxyribonucleotides are not suitable for this purpose, because cellular nucleases can digest them and cellular ribonuclease H can digest the target RNA in the region where the probe is bound. The former process leads to the generation of nonspecific fluorescence, and the latter process leads to destruction of the targets and loss of signal due to reformation of the molecular beacon stems. Consequently, earlier attempts to use molecular beacons for the detection of mRNAs in living cells have yielded uncertain results (25-27). We therefore decided to modify the backbone of molecular beacons by substituting an oxymethyl group for the hydrogen located at the second position of the ribose sugar in each nucleotide (28). Although this modification does not adversely affect the spontaneity of their fluorogenic interaction with targets, it enables the molecular beacons to resist endonucleolytic cleavage (Fig. 1B). Furthermore, the mRNA in the hybrids that they form is not digested by ribonuclease H (Fig. 1C). Therefore, in all subsequent experiments, we used molecular beacons synthesized from 2′-O-methylribonucleotides.
Selection of Regions in mRNA That Are Accessible to Probes. For a molecular beacon to be able to find its target sequence in an mRNA, the target should not lie within a tight secondary structure or be bound to proteins. Using the RNA secondary structure prediction program MFOLD (29), we identified regions in oskar mRNA that were either single stranded or were paired with distant sequences in most of the thermodynamically favored foldings of oskar mRNA (see Fig. 5, which is published as supporting information on the PNAS web site). We considered all suboptimal structures in this analysis because folding programs are not reliable indicators of exact secondary structures, especially for large sequences. We then narrowed the choice of target regions to 12 sequences with the help of a second computer program, OLIGOWALK (30), which identifies probe sequences that bind most stably to their complements and cause the least disruption in RNA secondary structure on binding. Molecular beacons were synthesized for each of the 12 selected target regions, and then tested in vitro to determine their ability to bind to oskar mRNA. Because the rate of hybridization of a molecular beacon is influenced by the stability of its stem, we chose stem sequences that were either identical to one another or that possessed the same GC content and length (see Table 1, which is published as supporting information on the PNAS web site). Even though each molecular beacon was able to bind spontaneously to an oligonucleotide that was complementary to its probe sequence, only half were able to bind efficiently to full-length transcripts (Fig. 1D). Had the target sequences been selected randomly, only 5% of the probes would have been likely to bind (31). All of the molecular beacons that were able to bind to their target sites in vitro were also able to hybridize to oskar mRNA in live oocytes although detailed results are presented below only for probes osk62, osk76, and osk87, which are all designed to hybridize to the same region within oskar mRNA.
Imaging the Distribution of oskar mRNA in Drosophila Oocytes. Molecular beacons for microinjection into Drosophila oocytes were labeled with either TMR or Texas red to distinguish their orange and red fluorescence from the green autofluorescence that occurs in oocytes. In addition to preparing a TMR-labeled molecular beacon specific for oskar mRNA (osk76), we prepared a Texas red-labeled control molecular beacon that differed from the specific molecular beacon at several nucleotide positions within its probe sequence so that it could not bind to oskar mRNA (mismatch-osk76). We then microinjected an equimolar mixture of these two molecular beacons into stage 9 to 10 oocytes. The molecular beacons were distributed throughout the oocytes within 2 min. After 15 min, we observed an increase in TMR fluorescence at the posterior end of the oocytes whereas no change in Texas red fluorescence was detected in this region (Fig. 2 A and B). A detectable increase in fluorescence was also observed in the center of the oocytes, but the extent of the change was the same for both fluorophores, suggesting that this increase in fluorescence was a background signal due to the nonspecific binding of cellular components to both molecular beacons, causing the probes to change their conformation and fluorescence. This explanation is supported by computer-generated ratio images obtained by dividing the intensity of the TMR fluorescence by the intensity of the Texas red fluorescence at every pixel in the image (32). In these images, an enhanced ratio was observed only at the posterior end of the oocyte (Fig. 2C). In addition, the magnitude of the background fluorescence decreased substantially when fluorescence contributions from other focal planes were removed by deconvolution analysis of the images or by confocal sectioning (see Fig. 4).
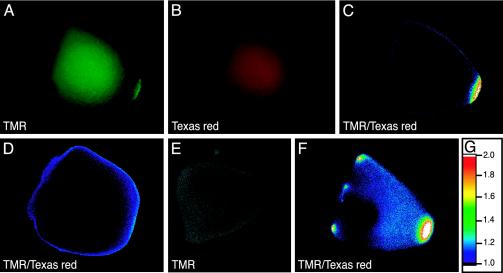
Fig. 2.
Imaging the distribution of oskar mRNA in Drosophila oocytes. A mixture containing a TMR-labeled molecular beacon that is specific for oskar mRNA (osk76) and a Texas red-labeled control molecular beacon that cannot bind to oskar mRNA (mismatched-osk76) was microinjected into stage 9 - 10 oocytes and imaged after 15 min. (A-C) A microinjected oocyte obtained from wild-type flies was imaged in the fluorescence emission range of TMR (A), in the fluorescence emission range of Texas red (B), and by ratio imaging (C). Ratio imaging was performed by dividing the TMR emission intensity by the Texas red emission intensity at every pixel in the image. (D) Ratio image of a microinjected oocyte obtained from a wild-type mother that was fed a microtubule depolymerizing agent to disrupt oskar mRNA translocation. (E) A microinjected oocyte that lacked a functional heavy chain component of the kinesin I motor protein, and was therefore unable to transport oskar mRNA to the posterior cortex, was imaged in the emission range of TMR with a confocal fluorescence microscope. (F) Ratio image of a microinjected oocyte from a transgenic fly possessing one copy of the oskar gene containing the 3′ UTR of bicoid mRNA in place of its own 3′ UTR, and another copy of the oskar gene containing its natural 3′ UTR. (G) Table of color codes used for depicting the ratios obtained in generating images C, D, and F.
Fig. 4.
Direct visualization of the transport of native oskar mRNA from a nurse cell to the posterior cortex of the oocyte. A TMR-labeled molecular beacon specific for oskar mRNA was injected into the cytoplasm of a nurse cell proximal to the oocyte. Images were acquired every 10 s over a period of 90 min, beginning immediately after microinjection, by using a confocal fluorescence microscope (the entire time course is shown in Movie 1). The anterior cortex (A) and the posterior cortex (P) of the oocyte and the location of the microinjected nurse cell are identified in the earliest image.
To confirm that the enhanced fluorescence observed at the posterior end of the oocytes was due to the binding of oskar mRNA-specific molecular beacons, and not to the nonspecific association of these molecular beacons with other structures present there, we prevented the localization of oskar mRNA at the posterior end. This result was accomplished either by disrupting the microtubules, which are the cytoskeletal fibers on which oskar mRNA is transported within the oocyte, or by using mutant oocytes that lack the functional heavy chain component of kinesin I, which is the motor protein responsible for the transport of oskar mRNA to the posterior pole (10). To disrupt the microtubular network in the oocytes, we added the microtubule depolymerizing agent colcemid to the diet of the mother flies 1 day before carrying out the experiment (12). When the mixture of molecular beacons was injected into oocytes from these flies, the fluorescence signal generated by ratio imaging did not appear at their posterior ends (Figs. 2D). Moreover, there was no localized increase in the fluorescence of oskar mRNA-specific molecular beacons in confocal sections of oocytes possessing the kinesin I heavy chain null mutation (Fig. 2E). The lack of a posterior signal in these two independent controls indicates that the localized signals seen in healthy wild-type oocytes are due to the specific binding of molecular beacons to oskar mRNA.
To eliminate the possibility that the fluorescence signals stem from other posteriorly localized components, we repeated the experiments with oocytes in which the localization pattern of oskar mRNA was genetically altered. These oocytes were obtained from transgenic flies in which the 3′ UTR of oskar mRNA was replaced with the 3′ UTR of bicoid mRNA (24). In addition to this chimeric oskar gene, these flies possessed a normal oskar gene encoding its natural 3′ UTR. Because the disparate localization of oskar mRNA and bicoid mRNA within the oocyte is determined by the characteristic sequence within each mRNA's 3′ UTR, the localization pattern of oskar mRNA in these transgenic oocytes should have the character of both oskar mRNA and bicoid mRNA. As expected, the results show that enhanced TMR fluorescence occurred at both poles of these oocytes (Fig. 2F). The signal at the posterior pole was from oskar mRNAs possessing a normal oskar 3′ UTR, and the signal at the anterior pole was from chimeric oskar mRNAs possessing a bicoid 3′ UTR. The localization of the anterior signal into dorsal and ventral regions, as well as into regions near the ring canal openings, which is a characteristic of bicoid mRNA distribution in stage 9-10 oocytes (12), demonstrates the high specificity of detection achieved with this technique. Furthermore, the ”normal” oskar gene in these transgenic flies (possessing a natural oskar 3′ UTR) harbors a premature termination codon that results in poorly localized oskar mRNA (24). Consistent with this genetic background, the posterior signal in these oocytes was more diffuse than in corresponding images obtained from wild-type flies (compare Fig. 2 F and C).
Binary Molecular Beacons. Because molecular beacons generated a detectable background fluorescence in the body of the oocytes (Fig. 2 A and B), we used a novel molecular beacon configuration to prove that fluorescence signals at the posterior end of the oocyte are due to the hybridization of the probe to oskar mRNA and are not merely correlated with the presence of the probe. In this approach, two different molecular beacons, each possessing a differently colored fluorophore, were designed to bind to the same target at nearly adjacent positions so that, on hybridization, their fluorophores would interact via FRET (33). Because the efficiency of FRET decreases with the sixth power of the distance between the fluorophores, molecular beacons that are bound nonspecifically to cellular constituents fluoresce in their own characteristic emission colors but are not able to participate in the generation of a FRET signal (Fig. 3A). To maximize the efficiency of target-mediated FRET, we placed the donor fluorophore (TMR) at the 5′ end of one molecular beacon (osk62) and the acceptor fluorophore (Texas red) at the 3′ end of the other molecular beacon (osk87). The fluorophore-bearing arms of these molecular beacons (as well as the loop sequences) were designed to be complementary to their target sequences (Fig. 3A). Furthermore, the sequences of the two probes were chosen so that, when they both bind to the same oskar mRNA, their fluorophores are separated from each other by seven intervening target nucleotides. At this distance, the intensity of the FRET signal is at a maximum, optimally balancing the negative effect of mutual fluorescence quenching and the positive effect of resonance energy transfer for these particular fluorophores. Fig. 3B shows that, when both molecular beacons are bound to oskar mRNA (black curve), TMR fluorescence is depressed and Texas red fluorescence is enhanced, compared with the intensity of their fluorescence when they are bound individually (green and red curves).
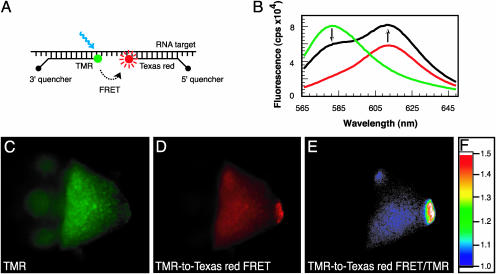
Fig. 3.
Use of binary molecular beacons to confirm that signals at the posterior pole of oocytes stem from hybridization to oskar mRNA. (A) The fluorophores of two appropriately designed molecular beacons can undergo FRET. Binding of differently colored molecular beacons to nearly adjacent positions on a target mRNA enables their two fluorophores to interact by FRET, which generates a uniquely detectable signal, whereas the nonspecific association of these molecular beacons with cellular components that elicit fluorescence does not place the pair of probes in a position that enables FRET to occur. (B) Detection of oskar mRNA in vitro with FRET, by using molecular beacon osk62 (labeled at its 5′ end with TMR) and molecular beacon osk87 (labeled at its 3′ end with Texas red), both of which bind to oskar mRNA at locations seven nucleotides apart from each other. The fluorescence spectrum of osk62 bound alone to oskar mRNA is shown by the green curve, and the fluorescence spectrum of osk87 bound alone to oskar mRNA is shown by the red curve. When they both bind to oskar mRNA, TMR fluorescence is depressed and Texas red fluorescence is elevated (black curve). (C and D) Imaging a wild-type oocyte 30 min after microinjection of a mixture of osk62 and osk87, by using either a filter set designed to stimulate the fluorescence of TMR and record the emission of TMR (C)ora filter set designed to highlight the FRET signal (D), which illuminates the specimen in the optimal excitation range of TMR and records the image in the optimal emission range of Texas red. (E) A ratio image, generated by dividing the intensity of the FRET signal in image D by the intensity of TMR fluorescence in image C at every pixel in the image. (F) Table of color codes used for depicting the ratios obtained in generating image E.
An equimolar mixture of these two molecular beacons was microinjected into oocytes, and fluorescence was recorded, either by using a TMR filter set, or by using a TMR-to-Texas red FRET filter set. In the FRET filter set, the excitation wavelength range was optimal for TMR, and the emission wavelength range was optimal for Texas red. When viewed with the TMR filter (Fig. 3C), the specific signal at the posterior end of the oocyte was not clearly distinguishable from background fluorescence in the body of the oocyte because the fluorescence from TMR-labeled molecular beacons at the posterior end was depressed by energy transfer to Texas red-labeled molecular beacons. However, when TMR was optimally excited and the optimal emission of Texas red was recorded by using the FRET filter set, the posterior signal was clearly detectable (Fig. 3D). This signal was even more distinct in a ratio image obtained by dividing the intensity of the FRET signal by the intensity of the TMR signal (Fig. 3E). When this experiment was repeated with a pair of nonspecific TMR and Texas red molecular beacons, no FRET signal was detectable.
Monitoring the Transport of oskar mRNA. Buoyed by the unambiguous detection of oskar mRNA in living oocytes, we set out to follow the migration of this RNA from its birthplace in the nurse cells to its final destination in the posterior cortex of the oocyte. We microinjected oskar mRNA-specific molecular beacons (osk76) into a nurse cell proximal to the oocyte and monitored their fluorescence by using confocal laser scanning microscopy. Optical sectioning obviated the need for ratio imaging because we found that the intensity of the background signals from the body of the oocytes was insignificant. We followed the time course of fluorescence in the focal plane that included the nucleus of the microinjected nurse cell, the ring canal that links the nurse cell to the oocyte, and the posterior cortex of the oocyte. Stage 7 to 8 oocytes were used for this experiment because the rate of migration of oskar mRNA into oocytes is at a peak in this stage of maturation. As shown in Fig. 4, the nucleus of the nurse cell first becomes fluorescent, due to the tendency of the free molecular beacons to become sequestered in nuclei [a process that occurs with all oligonucleotides (34)]. Later, fluorescence dissipates from the nucleus and spreads into the cytoplasm of the nurse cell, simultaneously acquiring a speckled character. The fluorescent speckles then funnel through the ring canal into the oocyte. The speckles linger in the center of the oocyte for ≈25 min, before clearing the center and migrating to the posterior cortex (see Fig. 4 and Movie 1, which is published as supporting information on the PNAS web site). We interpret these speckles to be ribonucleoprotein particles that may contain as many as 100 oskar mRNA molecules in each particle (13). Detection of these particles may have been facilitated by the tendency of molecular beacons to become sequestered in the nucleus where they have access to target sequences in nascent mRNAs during the assembly of the ribonucleoprotein particles. These events were not seen when a molecular beacon that cannot bind to oskar mRNA (mismatch-osk76) was microinjected into nurse cells.
This unprecedented observation of a native mRNA moving in its living context may help to resolve a lively debate that is waging about the nature of the transport mechanism of this intensively studied mRNA. Earlier studies suggested that the microtubule network in stage 7-8 oocytes is organized such that the minus ends of the microtubules are situated within the anterior cortex and the plus ends are concentrated in the vicinity of the posterior cortex (7). Thus, oskar mRNA was predicted to travel from the anterior cortex to the posterior cortex (10). However, recent observations suggest that the minus ends of the microtubules may be present throughout the cortex, and the plus ends may be concentrated in the center of stage 7-8 oocytes (12). Consequently, oskar mRNAs should transiently accumulate in the center of the oocyte. It is now hypothesized that the pool of oskar mRNA suspended in the center of the oocyte ultimately reaches the posterior cortex as a consequence of a developmentally programmed shrinking (or dissolution) of the posterior microtubules (12, 14). This alternative model predicts the existence of a transient stage in which oskar mRNA is present in the center of the oocyte. Just such a transient pool of oskar mRNA is visible in Fig. 4 (from 45 to 65 min). The existence of this pool is so ephemeral that it cannot be detected when molecular beacons are injected directly into stage 9-10 oocytes (Figs. 2 and 3). These real-time observations of moving endogenous oskar mRNA in live oocytes provide support for the latter model.
Discussion
We have described two approaches for distinguishing the target-specific fluorescence signals from the background fluorescence signals due to the interactions of molecular beacons with non-specific cellular constituents. In the first approach, we used a control molecular beacon that could not bind to the target RNA and was labeled with a differently colored fluorophore than the fluorophore that was used to label the target-specific molecular beacon. A pixel-by-pixel ratio (32) of the fluorescence intensities of the two images revealed the locations in the cells where the target was present. In the second approach, we also used two molecular beacons, but, in this case, they both could bind to the target at adjacent sites, giving rise to a uniquely detectable FRET signal. Although, the second method provides a better level of confidence on the nature of signals, it requires that a target site be found that is sufficiently long to accommodate both molecular beacons, and that the fraction of targets that are bound at any given moment to only one member of the molecular beacon pair be low. In practice, we found that both methods provide similar signal-to-background ratios and dynamic ranges.
Real-time monitoring of the transport of oskar mRNA clarified the mechanism of its transport. Similar analyses of the movements of other mRNAs in living cells will improve the understanding of how they reach their destinations. A case in point is gurken mRNA, which is seen at the posterior cortex in stage 6 oocytes and at the dorsoanterior corner in stage 8 oocytes (6). When a synthetic gurken mRNA is microinjected into the center of stage 8 oocytes, it first moves toward the anterior, and then switches direction, moving toward the dorsoanterior corner above the nucleus of the oocyte (21). It is not clear how this RNA is able to make these abrupt changes in the direction of its movement. Studies with molecular beacons will allow the natural migratory pattern of gurken mRNA to be deciphered. The success of these studies will depend on the abundance of the target mRNAs. However, if low level signals are encountered, it will be possible to enhance them by simultaneously targeting multiple sites on the same RNA with many molecular beacons.
By monitoring the transport of several different mRNAs at the same time, it will be possible to study how the transport of various mRNAs is coordinated. The different mRNAs can be distinguished from one another by using molecular beacons that are specific for different mRNAs and are labeled with differently colored fluorophores, and that are microinjected together into oocytes or nurse cells. Furthermore, concomitant labeling of microtubules (35) or their ends may allow the relationship between microtubule remodeling and RNA transport to be explored. Such studies may uncover new points of control that developmental programs exercise. Moreover, this method will be useful for the study of RNA trafficking in other contexts, such as the export of RNAs from the nucleus to the cytoplasm, and the transport of mRNAs within axons.
Direct visualization of mRNAs in living cells will also be useful for the study of RNAs that are not localized, but change their concentration as a function of time. It will be possible to observe cyclical changes in gene expression, such as those associated with cell division and cellular circadian rhythms. Because prior techniques for analyzing gene expression resulted in cell death, it was not feasible to ask questions about a cell's fate after measurements were taken. Thus, the use of molecular beacons for the detection, imaging, and quantification of specific mRNAs in living cells should have broad application in cell biology research.
Supplementary Material
Acknowledgments
We thank Ruth Lehmann and William Theurkauf for fly strains and plasmids, and Salvatore Marras for synthesis of the molecular beacons. This work was supported by National Institutes of Health Grants ES-10536 and EB-00277. B.-J.C. is grateful for support from the Harold Whitworth Pierce Charitable Trust and the American Cancer Society (CSM-101560).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TMR, tetramethylrhodamine; FRET, fluorescence resonance energy transfer.
References
- 1.Tekotte, H. & Davis, I. (2002) Trends Genet. 18, 636-642. [DOI] [PubMed] [Google Scholar]
- 2.Jansen, R. P. (2001) Nat. Rev. Mol. Biol. 2, 247-256. [DOI] [PubMed] [Google Scholar]
- 3.Dubowy, J. & Macdonald, P. M. (1998) Mech. Dev. 70, 193-195. [DOI] [PubMed] [Google Scholar]
- 4.Berleth, T., Burri, M., Thoma, G., Bopp, D., Richstein, S., Frigerio, G., Noll, M. & Nusslein-Volhard, C. (1988) EMBO J. 7, 1749-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim-Ha, J., Smith, J. L. & Macdonald, P. M. (1991) Cell 66, 23-35. [DOI] [PubMed] [Google Scholar]
- 6.Thio, G. L., Ray, R. P., Barcelo, G. & Schupbach, T. (2000) Dev. Biol. 221, 435-446. [DOI] [PubMed] [Google Scholar]
- 7.Grunert, S. & St. Johnston, D. (1996) Curr. Opin. Genet. Dev. 6, 395-402. [DOI] [PubMed] [Google Scholar]
- 8.Erdelyi, M., Michon, A. M., Guichet, A., Glotzer, J. B. & Ephrussi A. (1995) Nature 377, 524-527. [DOI] [PubMed] [Google Scholar]
- 9.Pokrywka, N. J. & Stephenson, E. C. (1995) Dev. Biol. 167, 363-370. [DOI] [PubMed] [Google Scholar]
- 10.Brendza, R. P., Serbus, L. R., Duffy, J. B. & Saxton, W. M. (2000) Science 289, 2120-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bashirullah, A., Cooperstock, R. L. & Lipshitz, H. D. (1998) Annu. Rev. Biochem. 67, 335-394. [DOI] [PubMed] [Google Scholar]
- 12.Cha, B.-J., Serbus, L. R., Koppetsch, B. S. & Theurkauf, W. E. (2002) Nat. Cell Biol. 4, 592-598. [DOI] [PubMed] [Google Scholar]
- 13.Glotzer, J. B., Saffrich, R., Glotzer, M. & Ephrussi, A. (1997) Curr. Biol. 7, 326-337. [DOI] [PubMed] [Google Scholar]
- 14.Pellettieri, J. & Seydoux, G. (2002) Science 298, 1946-1950. [DOI] [PubMed] [Google Scholar]
- 15.Cohen, R. S. (2002) Curr. Biol. 12, 797-799. [Google Scholar]
- 16.Levsky, J. M. & Singer, R. H. (2003) J. Cell Sci. 116, 2833-2838. [DOI] [PubMed] [Google Scholar]
- 17.Bertrand, E., Chartrand, P., Schaefer, M., Shenoy, S. M., Singer, R. H. & Long, R. M. (1998) Mol. Cell 2, 437-445. [DOI] [PubMed] [Google Scholar]
- 18.Forrest, K. M. & Gavis E. R. (2003) Curr. Biol. 13, 1159-1168. [DOI] [PubMed] [Google Scholar]
- 19.Cha, B.-J., Koppetach, B. S. & Theurkauf, W. E. (2001) Cell 106, 35-46. [DOI] [PubMed] [Google Scholar]
- 20.Bullock S. L. & Ish-Horowicz D. (2001) Nature 414, 611-616. [DOI] [PubMed] [Google Scholar]
- 21.MacDougall, N., Clark, A., MacDougall, E. & Davis, I. (2003) Dev. Cell 4, 307-319. [DOI] [PubMed] [Google Scholar]
- 22.Tyagi, S. & Kramer, F. R. (1996) Nat. Biotechnol. 14, 303-308. [DOI] [PubMed] [Google Scholar]
- 23.Tyagi, S., Bratu, D. P. & Kramer, F. R. (1998) Nat. Biotechnol. 16, 49-53. [DOI] [PubMed] [Google Scholar]
- 24.Ephrussi, A. & Lehmann, R. (1992) Nature 358, 387-392. [DOI] [PubMed] [Google Scholar]
- 25.Sokol, D. L., Zhang, X., Lu, P. & Gewirtz, A. M. (1998) Proc. Natl. Acad. Sci. USA 95, 11538-11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuo, T. (1998) Biochim. Biophys. Acta 1379, 178-184. [DOI] [PubMed] [Google Scholar]
- 27.Dirks, R. W., Molenaar, C. & Tanke, H. J. (2001) Histochem. Cell Biol. 115, 3-11. [DOI] [PubMed] [Google Scholar]
- 28.Cummins, L. L., Owens, S. R., Risen, L. M., Lesnik, E. A., Freier, S. M., McGee, D., Guinosso, C. J. & Cook, P. D. (1995) Nucleic Acids Res. 23, 2019-2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathews, D. H., Sabina, J., Zuker, M. & Turner, D. H. (1999) J. Mol. Biol. 288, 911-940. [DOI] [PubMed] [Google Scholar]
- 30.Mathews, D. H., Burkard, M. E., Freier, S. M., Wyatt, J. R. & Turner, D. H. (1999) RNA 5, 1458-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luebke, K. J., Balog, R. P. & Garner, H. R. (2003) Nucleic Acids Res. 31, 750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dunn, K. & Maxfield, F. R. (1998) Methods Cell Biol. 56, 217-236. [PubMed] [Google Scholar]
- 33.Cardullo, R. A., Agrawal, S., Flores, C. Zamecnick, P. C. & Wolf, D. E. (1988) Proc. Natl. Acad. Sci. USA 85, 8790-8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lorenz, P., Misteli, T., Baker, B. F., Bennett, C. F. & Spector, D. L. (2000) Nucleic Acids Res. 28, 582-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grieder, N. C., de Cuevas, M. & Spradling, A. C. (2000) Development 127, 4253-4264. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.