Abstract
In mitosis, chromosome, cytoskeleton, and organelle dynamics must be coordinated for successful cell division. Here, we present evidence for a role for Arf1, a small GTPase associated with the Golgi apparatus, in the orchestration of mitotic Golgi breakdown, chromosome segregation, and cytokinesis. We show that early in mitosis Arf1 becomes inactive and dissociates from Golgi membranes. This is followed by the dispersal of numerous Arf1-dependent peripheral Golgi proteins and subsequent Golgi disassembly. If Arf1 is kept in an active state by treatment with the small molecule H89 or expression of its GTP-locked form, intact Golgi membranes with bound peripheral proteins persist throughout mitosis. These cells enter mitosis but exhibit gross defects in chromosome segregation and cytokinetic furrow ingression. These findings suggest that mitotic Golgi disassembly depends on Arf1 inactivation and is used by the cell to disperse numerous peripheral Golgi proteins for coordinating the behavior of Golgi membranes, chromosomes, and cytoskeleton during mitosis.
The mitotic cell must divide its nucleus and cytoplasm. Nuclear division involves several highly regulated processes including chromosome condensation, spindle maturation, chromosome alignment, and segregation (1). Cytoplasmic division is less precise. It relies on partitioning or breakdown/reassembly mechanisms for segregating protein assemblies, cytoskeletal elements, and membrane-bounded organelles (2). How nuclear and cytoplasmic division is coordinated is not known.
Many membrane-bound organelles, including endoplasmic reticulum (ER) and mitochondria, remain intact in mitosis. However the Golgi apparatus, which functions at the crossroads of many membrane trafficking pathways within cells (3), reversibly disassembles (2). One explanation for the disassembly is that it serves as a mechanism for equally partitioning the interconnected Golgi membrane system into daughter cells (4). A different explanation, explored in this study, is that disassembly provides a mechanism for regulating the activity and location of signaling/effector molecules that during interphase are peripherally associated with Golgi membranes.
The Golgi is known to be a platform for numerous cytoplasmic proteins with diverse cellular roles, including heterotrimeric G proteins, phosphatidylinositol 3-kinase, N-Ras, eNOS, tankyrase, cyclin B2, and Cdc42 (5-7). Golgi association of many of these molecules is regulated directly or indirectly by the small, Ras-like GTP binding protein known as Arf1 (6), whose activity is required for maintenance of Golgi structure and function. Arf1 is recruited to Golgi membranes by a guanine nucleotide exchange factor (GEF) that converts Arf1-GDP to its GTP-bound form (6, 8). Activated Arf1 then binds effector molecules that regulate protein sorting/trafficking within the Golgi (6-8) and that stimulate the recruitment of spectrin, actin, and other cytoplasmic proteins, including signaling molecules, onto Golgi membranes (5, 7). When Arf1 is inactivated in cells by brefeldin A (BFA) treatment or expression of dominant-negative Arf1 mutants, Arf1 dissociates from Golgi membranes and peripherally associated Golgi components are released (6), resulting in Golgi disassembly and retrograde transport of Golgi enzymes into the ER (9).
The fate and activity of Arf1 in mitosis is not known but could potentially play a role in mitotic Golgi breakdown, because persistent inactivation of Arf1 through BFA treatment leads to Golgi disassembly (9). Recently, Golgi disassembly caused by Arf1 inactivation in BFA-treated cells was shown to be blocked by pretreatment of cells with the small molecule H89, an inhibitor of PKA, and other kinases (10). This finding prompted us in this study to investigate whether H89 could also inhibit mitotic Golgi disassembly and, if it could, to determine whether this inhibition was mediated through effects on Arf1.
As detailed below, we found that H89 treatment blocked mitotic Golgi disassembly because of the failure of Arf1 to become constitutively inactive in mitosis (a change in Arf1 that normally occurs early in mitosis). Expressing a constitutively active Arf1 mutant (Arf1[Q71L]) (11) also blocked mitotic Golgi breakdown and, as found for H89 treatment, caused incomplete segregation of mitotic chromosomes and inhibition of furrow ingression during cytokinesis. In investigating the basis of these mitotic defects, we found that several proteins that participate in chromosome segregation and cytokinesis are Arf1-dependent, peripheral Golgi proteins, suggesting that mitotic Arf1 inactivation serves to release these peripheral Golgi proteins so they can perform mitotic functions.
Methods
All plasmids have been described (12, 13). Normal rat kidney (NRK) cells were synchronized with aphidicolin (see supporting information on the PNAS web site). Methods for time-lapse imaging, photobleaching, quantification, and fluorescence correlation spectroscopy are described in detail in the supporting information. ImmunoGold labeling was performed as reported (14).
Results
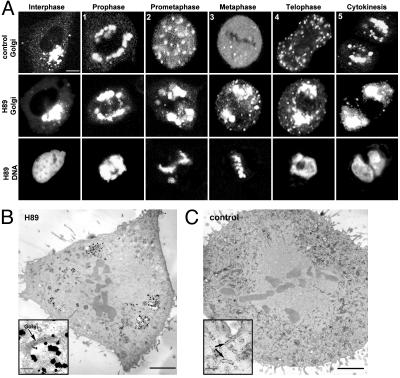
Golgi disassembly in BFA-treated, interphase cells can be blocked by the small molecule H89 (10). Given this effect, we asked whether H89 could also block the disassembly of the Golgi in mitosis. Synchronized NRK cells expressing the Golgi enzyme marker, galactosyltransferase-YFP (GalT-YFP), were treated with or without 50 μM H89 shortly before mitotic entry, and the fate of the Golgi was visualized by time-lapse confocal imaging (Fig. 1A).
Fig. 1.
H89 treatment blocks Golgi fragmentation and dispersal in mitosis. (A) Confocal images of control and H89-treated synchronized NRK cells expressing GalT-YFP at different stages of mitosis. (Top and Middle) GalT-YFP distribution. (Bottom) Hoechst staining of DNA in the same H89-treated cell. H89 was added to synchronized cells ≈30-45 min before mitosis, when cells were in G2 as determined by the lack of DNA condensation and histone-H3 phosphorylation (not shown). (Bar, 5 μm.) (B) Silver-enhanced ImmunoGold labeling of GalT-YFP in a metaphase NRK cell treated with H89 showed an enrichment of gold particles in Golgi-like fragments. At higher magnification (Inset), gold particles were over stacked Golgi cisternae. (C) In a control metaphase cell, gold particles were distributed throughout the cell. At higher magnification (Inset), gold particles predominantly associated with tubular/vesicular profiles. (Bars, 5 μm at low magnification and 1 μm at high magnification.)
In untreated cells, mitotic Golgi breakdown and reassembly followed a series of steps (15, 16), including (i) lateral distension of the Golgi ribbon around the nuclear envelope in prophase, (ii) the appearance of scattered fragments in prometaphase, (iii) complete or nearly complete Golgi dispersal in metaphase, (iv) the reappearance of scattered fragments in telophase, and (ν) the coalescence of Golgi fragments in cytokinesis (Fig. 1 A, control). H89 treatment blocked this process before step ii, with minor scattering and no dispersal of Golgi membranes observed at any stage of mitosis. The Golgi membranes in H89-treated cells segregated with mother and daughter centrosomes to opposite spindle poles (Fig. 1 A, H89 Golgi), and cell entry and progression through mitosis occurred normally (86 ± 5% of n = 1,500 cells were in mitosis) (Fig. 1 A, H89 DNA; also see supporting information). The effect of H89 treatment was most striking during metaphase, in which ≈80% of cells (n = 50) exposed to H89 showed large intact Golgi membranes, whereas <10% of untreated cells (n = 50) had any visible Golgi-like elements at all. At metaphase, the Golgi membranes in H89-treated cells appeared as well organized stacks of flattened cisternae based on correlative light-electron microscopy (14) of GalT-YFP-expressing cells (Fig. 1B). No such structures were seen in electron micrographs of untreated metaphase cells; instead, GalT-YFP labeling was found within tubular vesicular membranes dispersed throughout the cell (Fig. 1C). These results indicate that H89 treatment inhibits mitotic Golgi breakdown, as it does BFA-induced Golgi breakdown (10), and this occurs without preventing cells from entering and progressing through mitosis.
Based on these findings, we followed two avenues of research. One involved an investigation into the molecular basis for the inhibition of mitotic Golgi breakdown by H89 treatment. The second involved an examination of the consequences of this inhibition for cell division.
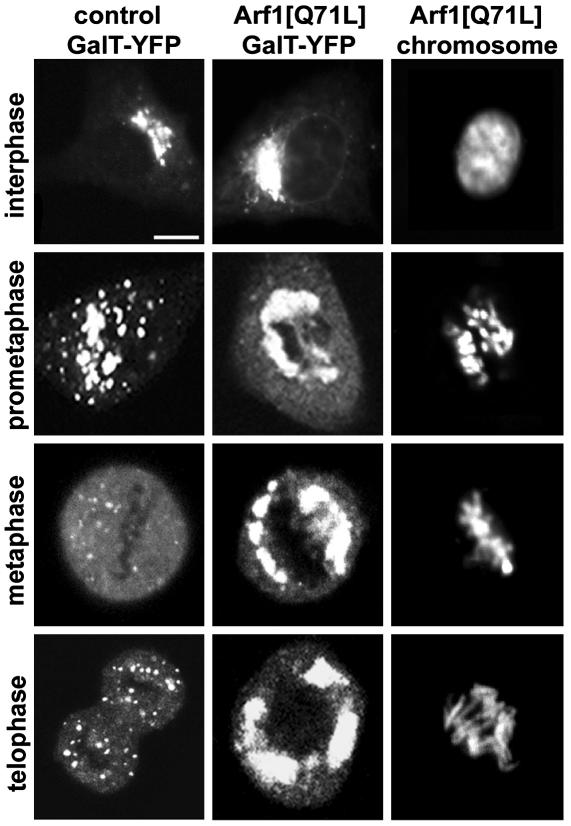
To gain insight into the molecular basis for H89 treatment's inhibition of mitotic Golgi breakdown, we first considered whether mitotic Golgi breakdown occurs by a similar mechanism to that of BFA-induced Golgi breakdown, because both processes are inhibited by H89 treatment. Golgi disassembly caused by BFA treatment results from the persistent inactivation of the Arf1 GTPase (6). This disassembly is due to BFA forming a noncompetitive complex with Arf1's guanine nucleotide exchange factor (GEF) (17), which normally recruits Arf1 to membranes by converting Arf1 to an active GTP-bound state. Because Arf1 cannot be recruited to and activated at Golgi membranes in the presence of BFA, Arf1 and its effectors accumulate in the cytoplasm, leading to Golgi disassembly (5, 6). Expression of the constitutively active form of Arf1, Arf1[Q71L] (11), which cannot hydrolyze its GTP, prevents BFA-induced Golgi disassembly by keeping activated Arf1 and its effectors on Golgi membranes. Given this effect, we tested whether Arf1[Q71L] expression could inhibit mitotic Golgi disassembly. Cells were cotransfected with Arf1[Q71L] and GalT-YFP, and Golgi membranes were examined during mitosis. Significantly, at all stages of mitosis, intact Golgi structures labeled with GalT-YFP were observed (Fig. 2). The phenotype was similar to that observed in H89-treated cells (Fig. 1 A), with Golgi membranes remaining as large structures that partitioned with centrosomes to opposite spindle poles. Thus, Golgi disassembly during mitosis is inhibited when Arf1 cannot undergo GTP hydrolysis and release from Golgi membranes.
Fig. 2.
Arf1[Q71L] expression inhibits mitotic Golgi breakdown. NRK cells were transfected with GalT-YFP (Left) or Arf1[Q71L] and GalT-YFP (Center), and mitotic stages were identified by using Hoechst 33342 (Right). Shown are representative mitotic states illustrating the distribution of Golgi membranes. Arf1[Q71L]-expressing cells were easily identifiable based on their unique Golgi morphology and lack of sensitivity to BFA. (Bar, 10 μm.)
These findings raised the possibility that mitotic Golgi breakdown resembled BFA-induced Golgi breakdown in involving the persistent inactivation of Arf1. During BFA treatment, Arf1's steady-state distribution on Golgi membranes (as a GTP-bound, active form) and in the cytoplasm (as a GDP-bound, inactive form) rapidly shifts to a single inactive population in the cytoplasm (8, 12). This shift occurs because GTP-bound Arf1 can hydrolyze its GTP and dissociate from the Golgi but cannot exchange GTP/GDP and rebind to the Golgi in the presence of BFA (6, 8, 12, 17). To investigate whether a similar phenomenon occurs in mitotic cells, we looked for changes in the Golgi and cytoplasmic pool sizes of Arf1 by using a GFP-tagged version of Arf1 (Arf1-GFP) whose activity mirrors that of the endogenous protein (12).
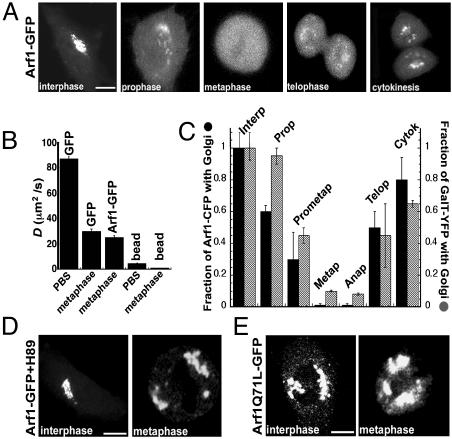
Significantly, the steady-state distribution of Arf1-GFP on Golgi membranes and in the cytoplasm began to change as soon as cells entered prophase, with Arf1-GFP's pool on the Golgi decreasing and its pool in the cytoplasm increasing (Fig. 3A, interphase and prophase). By metaphase, all of Arf1 was found dispersed in the cytoplasm (Fig. 3A, metaphase). In telophase, Arf1-GFP began reassociating with Golgi membranes, and by cytokinesis, it had nearly reestablished its steady-state Golgi and cytoplasmic levels (Fig. 3A, telophase and cytokinesis). These changes in distribution were not due to de novo synthesis of Arf1-GFP, because no fluorescence recovered when all cellular fluorescence was photobleached (not shown). Thus, between prophase and metaphase, the subcellular distribution of Arf1-GFP shifts to a single population in the cytoplasm, suggesting that Arf1 is persistently inactive during metaphase.
Fig. 3.
Arf1 distribution in mitosis. (A) Confocal images of mitotic NRK cells expressing Arf1-GFP. (B) Diffusion coefficients, D, of Arf1-GFP and free GFP in metaphase cells (n = 10 for each) and of free GFP and 40-nm fluorescent beads in PBS measured by using fluorescence correlation spectroscopy (FCS). The estimated D for beads in metaphase cells was extrapolated from the D for beads in PBS by using the difference in D observed between GFP in PBS and GFP in a metaphase cell. (C) Individual synchronized NRK cells (n = 10) coexpressing Arf1-CFP and GalT-YFP were followed as they advanced through mitosis. A simple image-processing method involving two regions of interest (ROI) was used to quantify Golgi and non-Golgi pools of Arf1-CFP and GalT-YFP. One ROI was drawn around GalT-YFP-labeled structures and fragments (representing Golgi labeling), and the other ROI was drawn around the rest of the cell (representing non-Golgi labeling). During each stage of mitosis, the mean fluorescence intensity associated with the Golgi and non-Golgi ROIs was measured for both Arf1-CFP and GalT-YFP. The total Arf1-CFP fluorescence associated with the Golgi was expressed as a fraction of the total cellular fluorescence (i.e., the sum of the Golgi and non-Golgi contributions) and normalized to the initial interphase value. This procedure was repeated for GalT-YFP, and both were plotted. (D) Arf1-GFP distribution in H89-treated interphase and metaphase cells that were treated before entry into mitosis. (E) Arf1[Q71L]-GFP expression in interphase and metaphase cells. (Bar, 10 μm.)
To demonstrate that the cytoplasmic pool of Arf1-GFP in metaphase is, in fact, persistently inactive (as observed in BFA-treated cells) and unable to bind to dispersed mitotic membranes (e.g., vesicles), we performed FCS (ref. 18; see supporting information). This technique measures the effective diffusion coefficient, D, of individual fluorescent molecules in living cells (18). If a fraction of Arf1-GFP at metaphase were active and bound to vesicles, FCS would detect this as a species of Arf1 that diffused slower (with a D characteristic of vesicles) than Arf1 that was inactive and unable to bind to membranes (which would diffuse with a D characteristic of a small cytoplasmic molecule). Our measurements revealed that >98% of Arf1-GFP molecules within metaphase cells had a D similar to that of free GFP expressed in these cells (Fig. 3B). The D for a 40-nm bead (corresponding to the smallest vesicle in a cell) measured in PBS or extrapolated to the cytoplasm were ≈5 and ≈25 times smaller, respectively, than that of Arf1-GFP (Fig. 3B). We concluded, therefore, that Arf1-GFP in metaphase cells is persistently inactive (i.e., GDP-bound) and unable to associate with Golgi membranes.
Conversion of Arf1 to a persistently inactive species in BFA-treated cells causes Arf1 effectors that are necessary for Golgi maintenance to redistribute into the cytoplasm, with Golgi disassembly occurring after this redistribution. To determine in mitotic cells whether conversion of Arf1 to a persistently inactive species, likewise, precedes mitotic Golgi disassembly, we coexpressed Arf1-CFP and GalT-YFP in NRK cells and monitored their distribution by time-lapse microscopy in cells progressing through mitosis (Fig. 3C). The use of GalT-YFP, an integral Golgi membrane protein, allowed us to identify Golgi membranes independently of Arf1-CFP fluorescence and thereby to quantify Arf1-CFP's shift from an active form on Golgi membranes to an inactive form in the cytoplasm. Notably, 50% of the Golgi pool of Arf1-CFP shifted to the cytoplasm by the end of prophase, even though there was little change in the Golgi levels of GalT-YFP at this time. At metaphase and anaphase, both Arf1-CFP and GalT-YFP were dispersed throughout cells. In telophase and cytokinesis, GalT-YFP-enriched membranes reappeared and Arf1-CFP increased its association with these membranes gradually to levels found in interphase. These data indicate that in mitosis, inactive pools of Arf1 in the cytoplasm have significantly increased before Golgi disassembly occurs.
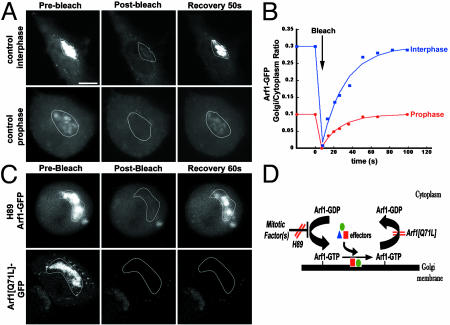
We next focused on the mechanism leading to the increase in Arf1's inactive cytoplasmic pool during prophase. This increase could be due to a decrease in Arf1's binding rate, an increase in Arf1's dissociation rate, or both. To distinguish between these possibilities, we looked for differences between the rates of Golgi association and dissociation of Arf1-GFP in prophase and interphase cells by using photobleaching and kinetic modeling techniques (see supporting information). Golgi fluorescence in Arf1-GFP-expressing cells was selectively photobleached, and the extent of fluorescence recovery onto Golgi membranes was measured over time in either prophase or interphase cells. We found that Golgi fluorescence recovered rapidly and completely in both interphase and prophase cells (Fig. 4A), but the ratio of Golgi to cytoplasmic fluorescence of Arf1 that was reestablished after recovery in prophase cells was only one-third of that observed in interphase cells (Fig. 4B). Using this data together with data on the relative abundances of Arf1-GFP in cytoplasmic and Golgi pools during prophase and interphase, we then calculated Arf1's rates of membrane binding and dissociation in prophase and interphase by kinetic modeling techniques. The estimated rate constants in NRK cells were 0.0051%·sec-1 for binding of cytoplasmic Arf1 to Golgi in interphase, 0.0018%·sec-1 for binding in prophase, 0.0203%·sec-1 for dissociation in interphase, and 0.0227%·sec-1 for dissociation in prophase. These values were not affected by the degree of expression of exogenous Arf1-GFP over levels of endogenous Arf1 (see supporting information). Because Arf1's binding rate constant decreased 2.8-fold in prophase, whereas its dissociation rate constant remained essentially unchanged, the data suggest that the increase in Arf1's inactive, cytoplasmic pool in prophase is due to a reduced rate of Arf1 membrane binding. This binding rate presumably decreases still further as cells progress into metaphase, because Arf1 binding to Golgi membranes was completely inhibited by metaphase, with all Arf1 existing free in the cytoplasm (Fig. 3A).
Fig. 4.
Arf1 dynamics in untreated, H89-treated, and Arf1[Q71L]-expressing cells. (A) Photobleaching the Golgi pool of Arf1-GFP in interphase and prophase cells and monitoring recovery of fluorescence. (B) An example of recoveries observed for one interphase cell and one prophase cell. The Golgi to cytoplasm ratio of Arf1-GFP was plotted through the course of the experiment. (C) Photobleaching the Golgi pool of Arf1-GFP in an H89-treated metaphase cell or an Arf1[Q71L]-GFP-expressing cell. In H89-treated cells, Arf1-GFP recovered rapidly upon photobleaching the Golgi pool of its fluorescence (Upper), whereas Arf1[Q71L]-GFP-expressing cells showed negligible recovery over the same time period (Lower). (D) Model for Arf1 inactivation during mitosis and the effects of H89 and Arf1[Q71L]. Recruitment of Arf1 to Golgi membranes is inhibited early in mitosis by a mechanism that is sensitive to H89. This leads to the accumulation of Arf1-GDP in the cytoplasm, the inability to recruit Arf1 effector proteins onto Golgi membranes, and the disassembly of the Golgi. These mitotic effects can be inhibited either by expression of Arf1[Q71L] or by H89 treatment. (Bar, 10 μm.)
To gain insight into the molecular basis for the inhibition of Arf1 membrane binding in mitosis and to relate this to mitotic Golgi breakdown, we investigated the behavior of Arf1 in mitotic cells treated with H89 or expressing Arf1[Q71L], because these cells do not disassemble their Golgi in mitosis. We found that in H89-treated cells, Arf1-GFP remained associated with Golgi membranes throughout mitosis and showed no noticeable redistribution into the cytoplasm (Fig. 3D, metaphase). The same result was observed in mitotic cells expressing Arf1[Q71L]-GFP (Fig. 3E, metaphase) where Arf1 GTP hydrolysis and release from membranes is inhibited.
To determine the basis for Arf1 retention on Golgi membranes in H89-treated, mitotic cells, we selectively photobleached Golgi fluorescence in Arf1-GFP-expressing, metaphase cells treated with H89 and then monitored fluorescence recovery into the Golgi region over time (Fig. 4C, H89 Arf1-GFP). Rapid recovery of Arf1-GFP into the photobleached area was observed, indicating that Arf1-GFP continued to exchange between Golgi and cytoplasmic pools in these cells. By contrast, similar photobleaching experiments performed in Arf1[Q71L]GFP-expressing cells showed no fluorescence recovery into the Golgi region after photobleaching (Fig. 4C, Arf1Q71L-GFP). This finding can be explained by Arf1[Q71L]-GFP being locked on Golgi membranes because of an inability to hydrolyze GTP. Thus, H89 treatment and Arf1[Q71L] expression use different mechanisms to maintain activated Arf1 and its effectors on mitotic Golgi membranes (Fig. 4D). Whereas Arf1[Q71L] expression prevents Arf1 loss from the Golgi by locking activated Arf1 onto Golgi membranes, H89 treatment permits continued recruitment and release of Arf1 at the Golgi, suggesting that H89 treatment interferes with a mechanism in mitosis that normally inhibits Arf1 recruitment to Golgi membranes. Because this leads to the presence of activated Arf1 and its effectors on Golgi membranes, Golgi membranes do not disassemble during mitosis.
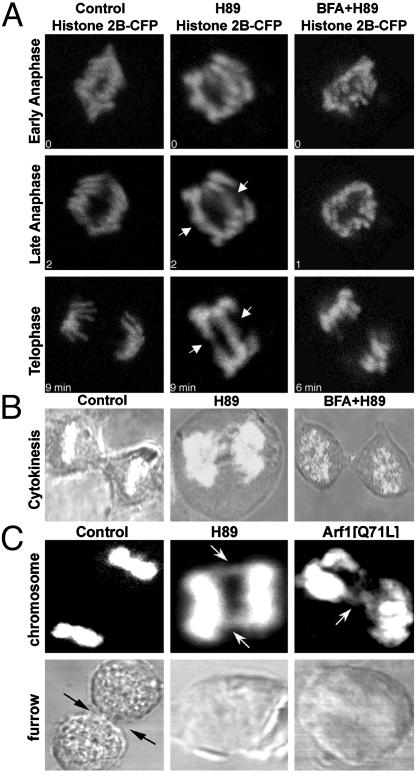
What are the consequences for cell division when Golgi disassembly is blocked by H89 treatment or Arf1[Q71L] expression in this manner? Under both conditions, Golgi structures can still segregate with centrosomes to opposite spindle poles (Figs. 1 and 2). This finding indicates that Golgi breakdown is not necessary for Golgi partitioning into daughter cells. To determine whether the presence of intact Golgi structures in mitosis has effects on other mitotic processes, we examined mitotic cells treated with H89 or expressing Arf1[Q71L] (Fig. 5). In these cells, most mitotic processes remained normal, including entry into mitosis, chromosome condensation/decondensation, spindle formation, nuclear envelope breakdown, and endoplasmic reticulum (ER) exit site dissolution (see supporting information). However, in telophase the resolved sister chromatid pairs (data not shown) were pulled apart but remained attached at their ends in 84 ± 4% of H89-treated mitotic cells (n = 91), compared with 11 ± 2% of untreated cells (Fig. 5A). These cells also did not go on to form a contractile furrow; therefore, cytokinesis was inhibited (Fig. 5B). Significantly, similar mitotic defects were observed in Arf1[Q71L]-expressing cells (75 ± 12%, n = 12) (Fig. 5C).
Fig. 5.
Inhibiting Arf1 inactivation in mitotic cells causes defects in chromosome segregation and cytokinesis. (A) Synchronized NRK cells expressing histone 2B-CFP were either untreated (control), treated with H89 (50 μM) 30 - 45 min before mitotic entry (H89), or treated with BFA (5 μg/ml) 15 min before the addition of H89 (BFA+H89) while cells were in G2 phase. Chromosomes were imaged by confocal microscopy as cells exited mitosis. In H89-treated cells, chromosome pairs did not completely separate in telophase (arrows), whereas in cells treated with BFA to disassemble the Golgi before H89, no chromosome bridging was observed. (B) Cells were examined by bright-field microscopy to observe cytokinesis. Control cells (Left) and cells treated with BFA before H89 (Right) completed furrow ingression, whereas H89-treated cells (Center) did not form a contractile furrow. (C) Untransfected, Arf1[Q71L]-transfected or H89-treated mitotic cells were stained with Hoechst 33342 to visualize telophase chromosomes. Bright-field images were collected from each cell to assess cytokinetic furrow ingression.
These defects were directly due to the maintenance of activated Arf1 on intact Golgi structures in mitosis, rather than to independent effects of H89 treatment or Arf1[Q71L] expression. If Arf1 was inactivated and the Golgi was disassembled through BFA treatment before H89 treatment in late G2, subsequent observations of the cells revealed that 76 ± 3% of cells (n = 50) had normal chromosome segregation and 76 ± 5% of this population had normal furrow ingression (Fig. 5 A and B, BFA+H89). These results suggest that unless Arf1 is persistently inactive and the Golgi is disassembled during mitosis, chromosome segregation and cytokinesis cannot properly occur.
We next investigated why it is that in mitosis Arf1 needs to become persistently inactive and the Golgi needs to become disassembled for normal chromosome segregation and cytokinesis to proceed. A wide variety of cytoplasmic proteins with roles in interphase and mitosis are known to associate with Golgi membranes, including: coatomer (COPI), ankyrin, spectrin, tankyrase, cullin family members, some polo-like kinases, Nir2, myosinIIA, and cdc42 (19-25). Many of these proteins have mitotic activities on chromatin, the spindle, and the cytoskeleton that require them to be in the cytoplasm during mitosis (19-24). Given this, one potential explanation for the observed mitotic defects is that persistent inactivation of Arf1 and Golgi disassembly are necessary for dispersing these proteins into the cytoplasm so they can perform their mitotic functions.
To explore this idea, we studied the changes in localization of several peripheral Golgi proteins during interphase and mitosis under different experimental conditions. In interphase, these proteins were concentrated at the Golgi (Fig. 6A, control interphase), as evidenced by their codistribution with Golgi markers (not shown) and their cytoplasmic release when the Golgi was disassembled by BFA treatment (Fig. 6A, BFA interphase). During metaphase, the peripheral proteins redistributed into the cytoplasm (Fig. 6A, control metaphase) and were no longer membrane-associated (not shown).
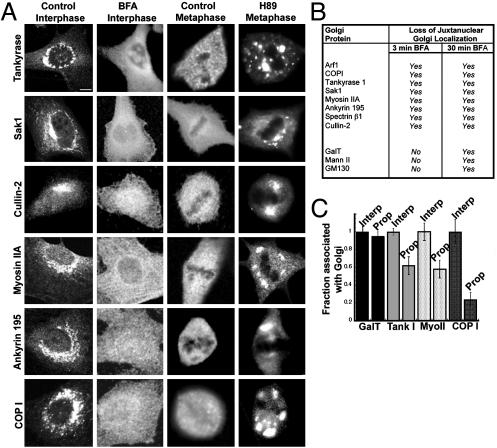
Fig. 6.
Inhibiting Arf1 inactivation blocks the mitotic dispersion of a wide variety of peripheral Golgi proteins. (A) Interphase cells treated without or with BFA (5 μg/ml, 30 min) and mitotic cells in metaphase treated with or without H89 (50 μM) were fixed and stained with antibodies against tankyrase 1 (3T3-L1), Sak1 (HeLa), Cullin-2 (EA.hy926), myosin IIA (NRK), ankyrin 195 (NRK), and εCOPI (NRK). (Bar, 5 μm.) (B) Table summarizing the effect of short and long BFA treatments on Golgi proteins. (C) Peripheral Golgi proteins start to disperse in prophase before Golgi membranes fragment. Interphase and prophase cells (staged with Hoechst 33342) were labeled for Golgi peripheral (myosin IIA, tankyrase, and COP1) and integral membrane (GalT) proteins. ROIs were drawn around the Golgi and the whole cell. The Golgi fluorescence for each protein was background subtracted, expressed as a fraction of the total cell fluorescence, and normalized to the interphase values for 10 cells.
To determine whether these peripheral Golgi proteins depended on Arf1 activity for their normal Golgi association, we examined whether they dissociated from the Golgi before Golgi disassembly during BFA treatment, because Arf1 inactivation and membrane dissociation precedes Golgi disassembly under these conditions. All of the proteins dissociated from Golgi membranes after short BFA treatment (3 min, i.e., before Golgi disassembly), suggesting that their membrane association depended on Arf1 GTPase activity (Fig. 6B).
We next tested whether Arf1 inactivation is responsible for the cytoplasmic redistribution of proteins in mitosis. Two observation were consistent with this possibility. First, we observed that in prophase the fraction of these proteins (including Myo II, COPI, and tankyrase) associated with the Golgi decreased, in contrast to resident enzymes [GalT and Mann II (not shown)] (Fig. 6C). This decrease occurred at the same time that Arf1 became persistently inactive and redistributed into the cytoplasm during mitosis. Second, and more significantly, when Arf1 redistribution into the cytoplasm was inhibited during mitosis through H89 treatment, the peripheral Golgi proteins remained associated with Golgi membranes throughout mitosis (Fig. 6A, H89)
Discussion
Our data suggest that mitotic Golgi disassembly is regulated by Arf1 and serves to disperse peripheral Golgi proteins, which, in turn, assist in chromosome segregation and cytokinesis. In this model (Fig. 4D), Arf1 recruitment to Golgi membranes is inhibited early in mitosis by interference in its conversion to an active, GTP-bound state. Arf1-dependent peripheral proteins are then unable to be recruited from the cytoplasm on to Golgi membranes and, therefore, the Golgi disassembles. A consequence is the cytoplasmic redistribution of other peripheral Golgi proteins that help coordinate mitotic processes, including chromosome segregation and cytokinesis. As discussed below, this model and its supporting data shed new light on the molecular mechanisms of mitotic Golgi breakdown, the roles of Arf1 effectors during mitosis, and the mitotic functions of different Golgi inheritance strategies.
Mechanistic studies of mitotic Golgi breakdown up until now have focused on mitosis-dependent phosphorylation of Golgi matrix and structural proteins by MEK1 and Cdc2 kinases (26, 27). In in vitro experiments, these kinases have been suggested to act sequentially in orchestrating mitotic Golgi breakdown, with MEK1 activity involved in the initial fragmentation of the Golgi and Cdc2 kinase activity involved in the conversion of fragments into smaller membrane elements and vesicles (27, 28). The data in this paper provide evidence for an additional mechanism for controlling mitotic Golgi breakdown that involves the inactivation of Arf1, a GTPase that is a key regulator of Golgi structure (6).
Evidence for a role of Arf1 inactivation in Golgi disassembly was threefold. First, direct observations of Arf1-GFP in living cells progressing through mitosis revealed that Arf1-GFP shifted its distribution into the cytoplasm between prophase and metaphase, and this redistribution began before Golgi structures had disassembled and dispersed. Second, FCS measurements demonstrated that >98% of Arf1-GFP molecules in the cytoplasm of metaphase cells were freely diffusing as small molecules rather than attached to membranes, as expected if Arf1-GFP existed in an inactive, GDP-bound form. Third, and most significantly, preventing Arf1 from being persistently inactive in mitosis, either by H89 treatment or Arf[Q71L] expression, blocked the mitotic breakdown of the Golgi apparatus.
Given these results, how does the role of Arf1 relate to that of MEK1 or Cdc2 kinases in regulating mitotic Golgi breakdown? H89 treatment or Arf1[Q71L] expression inhibited the scattering and subsequent cytoplasmic dispersal of Golgi membranes, but Golgi membranes still segregated by coordinated movement with mother and daughter centrosomes to opposite spindle poles. Because downstream effects of MEK1 and Cdc2 kinases appeared normal under these conditions (e.g., cells entered mitosis, and the spindle apparatus formed), the observed ordered Golgi movement in association with centrosomes could be orchestrated by the activities of these kinases. This possibility remains to be tested. The fact that Golgi membranes in these cells did not undergo further breakdown (i.e., dispersal) implies that MEK1 and Cdc2 kinase activities are insufficient to mediate later steps in the overall mitotic Golgi breakdown process when activated Arf1 still associates with Golgi membranes.
How Arf1 becomes persistently inactive in mitotic cells is an important topic for future investigations. Our data suggest that it involves the inhibition of GTP exchange on Arf1 at Golgi membranes because Arf1's association (but not dissociation) rate was inhibited at the time Arf1 redistributed off Golgi membranes in prophase. Because Arf1's association and dissociation rates remained unchanged between interphase and metaphase in H89-treated cells, it is possible that H89 interferes directly with the mitotic factor(s) involved in preventing Arf1 binding to membranes.
Associated with Golgi membranes are a wide variety of cytoplasmic proteins whose functions are necessary for DNA replication, chromosome condensation, segregation, and cytokinesis (19-25). The inactivation of Arf1 early in mitosis could serve to release such components into the cytoplasm in a timely way so that they can carry out their respective mitosis-related activities (Fig. 4D). Consistent with this, when we blocked Arf1 inactivation by H89 treatment or Arf1[Q71L] expression, membrane dissociation of many peripheral Golgi proteins was prevented and two key events of mitosis, chromosome segregation and cytokinesis, were abnormal. These abnormalities were directly dependent on Arf1 activity and its downstream effects because BFA addition, which specifically inactivates Arf1, before H89 treatment prevented these defects.
In mitosis, organelles partition between daughter cells either by an ordered or a stochastic process (4). Our ability to inhibit mitotic Golgi disassembly allowed us to demonstrate that the Golgi can follow either pathway in mammalian cells. Under normal conditions, a stochastic inheritance process is followed with dispersed Golgi membranes partitioning between daughter cells (15, 16). Under conditions in which Golgi disassembly is inhibited, however, an ordered pathway is followed involving the tracking of Golgi elements with centrosomes. The latter pathway resembles that found in plant and simple eukaryotic cells, which do not disassemble their Golgi in mitosis (29, 30).
Given that the Golgi readily partitions into daughter cells without undergoing disassembly, why do mammalian and other higher eukaryotic cells disassemble their Golgi in mitosis? Our data suggest they do so not for Golgi partitioning, but for the coordination of other events in mitosis, including chromosome segregation and cytokinesis. The membranes of the Golgi serve as a scaffold for a diverse population of cytoplasmic proteins. Many of these proteins, as shown here, are regulated by Arf1. The inactivation of Arf1 in mitosis, therefore, provides a means for redistributing these proteins into the cytoplasm so that they can perform their mitotic functions.
Supplementary Material
Acknowledgments
We thank Grégoire Altan-Bonnet for collaboration on FCS and discussion, and the Lippincott-Schwartz lab, Cathy Jackson, and Julie Donaldson for stimulating discussions.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BFA, brefeldin A; FCS, fluorescence correlation spectroscopy; NRK, normal rat kidney.
References
- 1.Nasmyth, K., Peters, J. M. & Uhlmann, F. (2000) Science 288, 1379-1384. [DOI] [PubMed] [Google Scholar]
- 2.Warren, G. & Wickner, W. (1996) Cell 84, 395-400. [DOI] [PubMed] [Google Scholar]
- 3.Marsh, B. J. & Howell, K. E. (2002) Nat. Rev. Mol. Cell Biol. 3, 789-795. [DOI] [PubMed] [Google Scholar]
- 4.Shorter, J. & Warren, G. (2002) Annu. Rev. Cell Dev. Biol. 18, 379-420. [DOI] [PubMed] [Google Scholar]
- 5.Donaldson, J. G. & Lippincott-Schwartz, J. (2000) Cell 101, 693-696. [DOI] [PubMed] [Google Scholar]
- 6.Donaldson, J. G. & Jackson, C. L. (2000) Curr. Opin. Cell Biol. 12, 475-482. [DOI] [PubMed] [Google Scholar]
- 7.De Matteis, M. A. & Morrow, J. (2000) J. Cell Sci. 113, 2331-2343. [DOI] [PubMed] [Google Scholar]
- 8.Donaldson, J. G. & Klausner, R. D. (1994) Curr. Opin. Cell Biol. 6, 527-532. [DOI] [PubMed] [Google Scholar]
- 9.Lippincott-Schwartz, J., Donaldson, J. G., Schweizer, A., Berger, E. G., Hauri, H. P., Yuan, L. C. & Klausner, R. D. (1990) Cell 60, 821-836. [DOI] [PubMed] [Google Scholar]
- 10.Lee, T. H. & Linstedt, A. (2000) Mol. Biol. Cell 11, 2577-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dascher, C. & Balch, W. E. (1994) J. Biol. Chem. 269, 1437-1448. [PubMed] [Google Scholar]
- 12.Presley, J. F., Ward, T. H., Pfeifer, A. C., Siggia, E. D., Phair, R. D. & Lippincott-Schwartz, J. (2002) Nature 417, 187-193. [DOI] [PubMed] [Google Scholar]
- 13.Ward, T. H., Polishchuk, R. S., Caplan, S., Hirschberg, K. & Lippincott-Schwartz, J. (2001) J. Cell Biol. 155, 557-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polishchuk, R. S., Polishchuk, E. V., Marra, P., Alberti, S., Buccione, R., Luini, A. & Mironov, A. A. (2000) J. Cell Biol. 148, 45-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shima, D. T., Cabrera-Poch, N., Pepperkok, R. & Warren, G. (1998) J. Cell Biol. 114, 955-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaal, K. J. M., Smith, C. L., Polishchuk, R. S., Altan, N., Cole, N. B., Ellenberg, J., Hirschberg, K., Presley, J. F., Roberts, T. H., Siggia, E., et al. (1999) Cell 99, 589-601. [DOI] [PubMed] [Google Scholar]
- 17.Peyroche, A., Antonny, B., Robineau, S., Acker, J., Cherfils, J. & Jackson, C. L. (1999) Mol. Cell 3, 275-285. [DOI] [PubMed] [Google Scholar]
- 18.Krichevsky, O. & Bonnet, G. (2002) Rep. Prog. Physics 65, 251-297. [Google Scholar]
- 19.Litvak, V., Tian, D., Carmon, S. & Lev, S. (2002) Mol. Cell. Biol. 22, 5064-5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hudson, J. W., Kozarova, A., Cheung, P., MacMillan, J. C., Swallow, C. J., Cross, J. C. & Dennis, J. W. (2001) Curr. Biol. 11, 441-446. [DOI] [PubMed] [Google Scholar]
- 21.Feng, H., Zhong, W., Punkosdy, G., Gu, S., Zhou, L., Seabolt, E. K. & Kipreos, E. T. (1999) Nat. Cell Biol. 1, 486-492. [DOI] [PubMed] [Google Scholar]
- 22.Drechsel, D. N., Hyman, A. A., Hall, A. & Glotzer, M. (1997) Curr. Biol. 7, 12-23. [DOI] [PubMed] [Google Scholar]
- 23.Smith, S. & de Lange, T. (1999) J. Cell Sci. 112, 3649-3656. [DOI] [PubMed] [Google Scholar]
- 24.Singer, J. D., Gurian-West, M., Clurman, B. & Roberts, J. M. (1999) Genes Dev. 13, 2375-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sutterlin, C., Lin, C. Y., Feng, Y., Ferris, D. K., Erikson, R. L. & Malhotra, V. (2001) Proc. Natl. Acad. Sci. USA 98, 9128-9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lowe, M., Rabouille, C., Nakamura, N., Watson, R., Jackman, M., Jamsa, E., Rahman, D., Pappin, D. & Warren, G. (1998) Cell 94, 783-793. [DOI] [PubMed] [Google Scholar]
- 27.Acharya, U., Mallabiabarrena, A., Acharya, J. K. & Malhotra, V. (1998) Cell 92, 183-192. [DOI] [PubMed] [Google Scholar]
- 28.Colanzi, A., Suetterlin, C. & Malhotra, V. (2003) Curr. Opin. Cell Biol. 15, 1-6. [DOI] [PubMed] [Google Scholar]
- 29.Pelletier, L., Stern, C. A., Pypaert, M., Sheff, D., Ngo, H. M., Roper, N., He, C. Y., Hu, K., Toomre, D., Coppens, I., et al. (2002) Nature 418, 548-552. [DOI] [PubMed] [Google Scholar]
- 30.Nebenfuhr, A., Frohlick, J. A. & Staehelin, L. A. (2000) Plant Physiol. 124, 135-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.