Abstract
Background
Melanoma metastasis status is highly associated with the overall survival of patients; yet, little is known about proteomic changes during melanoma tumor progression. To better understand the changes in protein expression involved in melanoma progression and metastasis, and to identify potential biomarkers, we conducted a global quantitative proteomic analysis on archival metastatic and primary melanomas.
Methodology and Findings
A total of 16 metastatic and 8 primary cutaneous melanomas were assessed. Proteins were extracted from laser captured microdissected formalin fixed paraffin-embedded archival tissues by liquefying tissue cells. These preparations were analyzed by a LC/MS-based label-free protein quantification method. More than 1500 proteins were identified in the tissue lysates with a peptide ID confidence level of >75%. This approach identified 120 significant changes in protein levels. These proteins were identified from multiple peptides with high confidence identification and were expressed at significantly different levels in metastases as compared with primary melanomas (q-Value<0.05).
Conclusions and Significance
The differentially expressed proteins were classified by biological process or mapped into biological system networks, and several proteins were implicated by these analyses as cancer- or metastasis-related. These proteins represent potential biomarkers for tumor progression. The study successfully identified proteins that are differentially expressed in formalin fixed paraffin-embedded specimens of metastatic and primary melanoma.
Introduction
The potential for primary melanomas to metastasize increases with the depth of invasion. The diagnosis of distant metastatic cutaneous melanoma is usually associated with poor prognosis. Currently, no single protein marker reliably predicts disease outcome in cutaneous melanoma. Understanding the mechanisms of metastasis and identifying proteomic biomarkers associated with melanoma progression and metastasis may aid in developing treatment strategies. In recent years, many studies have investigated gene expression signatures based on mRNA levels in melanomas [1], [2], [3]. However, changes at the mRNA level may not always directly correlate to changes at the protein level; translational and post-translational alterations also affect tumor progression and metastasis. Proteomics screening tools may identify relevant and significant changes in protein expression in relation to early and advance stage cancers.
Two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) coupled with tandem mass spectrometry (MS/MS) has been the principal tool for proteomic analysis of melanoma tissue [4], [5], but little data has been reported about the proteomic expression profiles of primary versus metastatic melanoma tissues. The ion intensity-based, label-free quantitative proteomics (LFQP) approach has gained popularity as mass spectrometer performance has significantly improved [6], [7], [8]. LFQP provides a powerful tool to resolve and identify thousands of proteins from a complex biological sample. Proteins are digested with a protease, and the peptide mixture is analyzed by liquid chromatography-mass spectrometry (LC/MS) and liquid chromatography with tandem mass spectrometry (LC/MS/MS); the relative protein abundance is determined by chromatographic peak intensity measurement. The LFQP approach is rapid and more sensitive than many other proteomic methods, and it increases the protein dynamic range 3- to 4-fold as compared to 2D-PAGE. This method can also be automated for large-scale proteome analysis.
To identify the mechanisms and proteomic biomarkers of melanoma metastasis, we used an LC/MS-based label-free protein quantification method to profile the global protein expression of microdissected primary and metastatic melanoma formalin fixed paraffin-embedded (FFPE) archival tissues. Proteins were extracted using reagents and protocols that yield a product suitable for MS analysis [9], [10]. The overall objective was to determine if proteomic profiling differences could be identified in FFPE tissue specimens of primary and metastatic cutaneous melanomas.
Methods
Tissue Processing
Formalin-fixed tissue blocks from 8 primary (five AJCC stage II and three AJCC stage III) and 16 metastatic melanomas (5 AJCC stage III lymph nodes and AJCC stage IV: 4 lung, 4 brain, 1 colon, 1 adrenal gland, and 1 extremity) were chosen for analysis. All cutaneous melanomas were identified by standard histopathology techniques. Five µm thick tissue sections were stained with hematoxylin and eosin to identify an 8-mm2 area containing defined melanoma cells for dissection. For tissue isolation and preparation of analyzable protein, the manufacturer's recommendations were followed (Expression Pathology, Inc., Gaithersburg, MD). In brief, a single 10-µm tissue section was cut from each tissue block, treated with xylene to remove paraffin, rehydrated through a series of graded ethanol solutions and distilled water, and stained with hematoxylin. An area 8 mm2 in size, corresponding to approximately 30,000 cells from the relevant region, was procured by microdissection for processing. Informed patient consents for all studies were approved by the human subjects IRB of Saint John's Health Center and JWCI. All of the consents are written and verbally explained to the patients. Patients' specimens were de-identified and run in a double-blinded manner.
Sample Preparation
Collected microdissected tissue was processed with the Liquid Tissue® MS Protein Prep Kit (Expression Pathology, Inc.). Briefly, the tissue was suspended in 20 µl of Liquid Tissue Buffer, incubated at 95°C for 90 min, and then cooled on ice for 2 min. Trypsin (1 µg) was added, and then the tissue was incubated at 37°C overnight. The sample was then heated for 5 min at 95°C to inactivate the trypsin. After the total amount of extracted protein was measured by a micro BCA (bicinchoninic acid) protein assay (PIERCE, Rockford, IL), DTT was added for a final concentration of 10 mM. The sample was stored at −20°C until LC/MS/MS analysis.
LC/MS/MS Analysis
Trypsin-digested samples (1 µg each) were injected onto an Agilent 1100 nano-HPLC system (Agilent Technologies, Inc., Santa Clara, CA) with a C18 capillary column in random order. Peptides were eluted with a linear gradient from 5 to 45% acetonitrile developed over 120 min at a flow rate of 500 nL/min, and effluent was electro-sprayed into the LTQ mass spectrometer (Thermo Fisher Scientific Inc., Waltham, MA). Data was collected in the “Triple-Play” (MS scan, Zoom scan, and MS/MS scan) mode. The acquired data were filtered and analyzed by a proprietary algorithm [11]. Database searches against the International Protein Index (IPI) human database (European Bioinformatics Institute, 2005) and the Non-Redundant-homo sapiens database (National Center for Biotechnology Information, 2005) were carried out using both the X!Tandem [12] and SEQUEST [13] algorithms. Protein quantification was carried out using the same algorithm described above [11]. Briefly, when the raw files were acquired from the LTQ mass spectrometer, all extracted ion chromatograms (XICs) were aligned by retention time. To be used in the protein quantification procedure, each aligned peak must have a matched precursor ion, charge state, fragment ions (MS/MS data) and retention time (within a one-minute window). After alignment, the area-under-the-curve (AUC) for each individually aligned peak from each sample was measured, normalized, and compared for relative abundance as previously described [11].
After obtaining the list of proteins that were differentially expressed in primary and metastatic samples from the LC/MS quantitative analysis, a corresponding gene list was created and input for bioinformatics analysis.
Biostatistics Analysis
Since the data has multiple sources of random variation, such as biological samples and replicates, a Linear Mixed Model (a generalization of an ANOVA, Analysis of Variance) was used for biostatistics analysis in this study. This model, especially when applied to complex experimental designs, cannot be handled by introductory methods such as t-tests. Also, the exact scale of the protein expression used in the model can make a difference in the sensitivity. In general, there is usually a large technical variation introduced by the act of ‘measurement’ in many ‘omics’ studies. Randomization of measurement order and normalization of the data will eliminate the technical bias. We used a statistically based quantile normalization method for data normalization [14]. Because ‘omics’ measures of expression are usually on an arbitrary scale, we chose to evaluate ratios or their equivalent differences on a log scale [15]. Log base 2 was chosen because a unit difference on the log scale is equivalent to a two-fold change.
A byproduct of the Linear Mixed Model is a p-Value or measure of significance for an observed change in protein expression (signal). The p-Value estimates the proportion of times a change that large will be observed if in fact there is no real change (the False Positive Rate). All the p-Values were then transformed into q-Values that estimate the False Discovery Rate (FDR) [16].
For each protein a separate analysis of variance (ANOVA) model is fit:
 |
Log2(Intensity) is the protein intensity based on the weighted average of the quantile normalized log base 2 peptide intensities with the same protein identification. Group Effect refers to the fixed effects (not random) caused by the experimental conditions or treatments that are being compared. Sample Effect (nested within group) refers to the random effects from individual biological samples. It also includes the random effects from the individual sample preparations. Replicate Effect (nested within sample) refers to the random effects from replicate injections from the same sample preparation.
When an ANOVA model has two or more random effects (sample and replicate in this data) and at least one fixed effect (group in this data) it is referred to as a Mixed Model. For each protein these models were fit using PROC MIXED in SAS software (version 9) (SAS Institute Inc., Cary, NC). The REML method was used as a fit mechanism and degrees of freedom were computed using the Satterthwaite method. The RANDOM statement was used to model the covariance with the NOBOUND parameter option in the PROC statement.
Immunohistochemistry Analysis
Selective protein expressions were assessed by immunohistochemistry (IHC) on FFPE melanoma tissues. Five-µm sections were cut from FFPE archival tissue blocks and incubated overnight at 50°C for IHC preparation. The sections were then deparaffinized in xylene and rehydrated in ethanol. All IHC staining was conducted with the CSA II kit (CSA II, Biotin-Free Tyramide Signal Amplication System; Dakocytomation) as described previously [17]. To perform heat-induced antigen retrieval, sections were placed into 10 mM citrate buffer (pH 6.0) and heated to 100°C for 20 min. Endogenous peroxidase function was quenched with peroxidase blocking solution. Nonspecific protein binding sites were blocked with serum-free protein blocking solution. Sections were incubated with anti-STAT1 mAb (BD Transduction, San Jose, CA) at 1∶400 dilution in PBS for 30 min at room temperature, or anti-MIF mAb (Abcam, Cambridge, MA) at 1∶100 dilution in PBS, anti-pyruvate kinase (PKM1/M2) (Abcam) at 1∶50 dilution in antibody diluent (Dakocytomation) with 1% BSA, or prediluted anti-HSP90α (Hsp86 antibody, Abcam) overnight at 4°C. Negative control slides were incubated under the same conditions with universal negative control-mouse antibody or universal negative control-rabbit (Dakocytomation), respectively. After washing, tissue sections stained with mouse monoclonal primary antibodies were incubated for 15 min at room temperature with horseradish peroxidase conjugated anti-mouse Ig antibodies. CSA II Rabbit Link (DakoCytomation) was used for the detection of rabbit primary antibodies. The amplification system provided with the CSA II kit was applied as directed in the manufacturer's instructions. Slides were developed with DAB. Sections were counterstained with half-strength Gill's hematoxylin (Fisher Scientific) for 30 sec, then dehydrated and mounted. Sections were evaluated and photographed using a Nikon Eclipse Ti microscope system.
Pathway Networks Analysis
The differentially expressed proteins were mapped into biological networks by using a manually curated proprietary database (MetaCore™, GeneGo, St. Joseph, MI), a pathway analysis tool. Gene symbols of differentially expressed proteins were uploaded into the database. For enrichment analysis, gene IDs of the uploaded files were matched with gene IDs in functional ontologies in MetaCore™ [18], that included canonical pathway maps (GeneGo maps), GeneGo cellular processes, Gene Ontology (GO) cellular process and diseases categories. Canonical pathway maps represent a set of about 500 signaling and metabolic maps covering human biology. For network analysis, analyze networks (AN), transcription regulation and direct interactions algorithms were used. The analyze networks algorithm, a variant of the shortest paths algorithm, uses relative enrichment and relative saturation to generate subnetworks prioritized by their number of canonical pathways [19].
Results
Proteomic Profiling of FFPE Melanoma Tissue
Proteins were extracted from microdissected melanoma FFPE tissues and the global protein expression profile in metastases was compared to that in primary tissues. MS analysis results are summarized in Table 1 . Of the 1557 proteins identified and quantified, 555 proteins were identified with high confidence (Priority 1). The identified proteins were found in every cell compartment and appeared to cover a broad range of biological functions. Of the 555 Priority 1 proteins, 120 proteins were differentially expressed (q<0.05) in primary and metastatic melanomas. The q-value, an adjusted p-value, was used to estimate the False Discovery Rate (FDR) [20]. A False Discovery is a when protein is falsely declared to be changed. The significance threshold sets the FDR at less than 5%. The replicate median percentage coefficient of variation (%CV) for the Priority 1 proteins was 7.6%, and the combined replicate plus sample median %CV was 15%.
Table 1. LC/MS-based Label-free Protein Quantification Method.
| Protein Priority | Peptide ID Confidence | Multiple Sequences | Number of Proteins | Number Significant Changes | Maximum Foldchange* | Median %CV** | Median %CV*** |
| 1 | HIGH | YES | 555 | 120 | 2.8 | 7.6 | 15 |
| 2 | HIGH | NO | 480 | 133 | 6.8 | 14 | 25.3 |
| 3 | LOW | YES | 8 | 3 | 2 | 9.5 | 24.4 |
| 4 | LOW | NO | 514 | 227 | 7.1 | 13 | 32.9 |
| Overall | 1557 | 483 | 7.1 | 11.7 | 22.6 |
Absolute Fold Change.
replicate.
replicate+sample.
Characterization of Identified Proteins
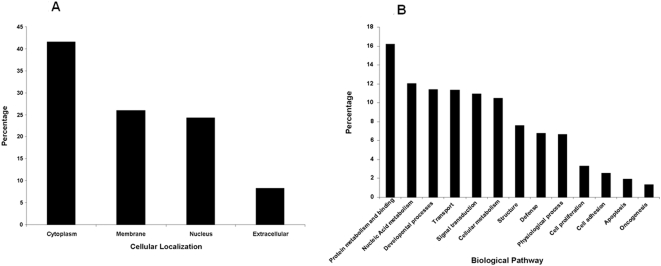
To investigate the properties of identified proteins, the 1557 identified proteins were classified based on the cellular location or biological process of the protein using the GO database ( Figure 1 ). Proteins that were not described or unspecified in the GO database were screened out and not further assessed for this report.
Figure 1. Location and functional distribution of proteins isolated and identified from melanoma FFPE tissue.
Proteins are categorized by A) cellular origin and, B) biological process, based on the Gene Ontology database.
The melanosome is a tissue specific organelle containing melanin pigments. It is of interest to investigate how well the melanosomal proteins were extracted and identified in this study, since melanoma is associated with changes in pigmentation and melanosome biogenesis, and melanogenesis related proteins have been used as melanoma biomarkers [21], [22], [23]. Of the common melanosomal proteins 79 out of 100 were identified using our method. These common proteins are known as constituent and resident proteins of the melanosome. There are six well-known melanogenesis proteins that are also frequent signatures of melanomas: tyrosinase, TRP1/TYRP1, TRP2/DCT, gp100/pmel17, MART1, and GPR43/OA1. These proteins have been shown to play either an enzymatic or structural role in melanin synthesis [24], [25], [26]. Four out of six of these well-known melanosome-specific proteins found in cutaneous melanomas were identified in our proteomic profiling.
Differentially Expressed Proteins in Metastasis
Of the 120 differentially expressed (q<0.05) proteins in priority 1, 61 had expression levels that differed by more than 20% between metastatic and primary melanoma ( Table 2 ). A cut-off of 20% was selected based on a 15% median CV of the combined replicate and sample ( Table 1 ). Of the 61 differentially expressed proteins, 38 were up-regulated and 23 were down-regulated.
Table 2. Metastasis compared to Primary Melanoma.
| Accession number | Gene Symbol | Annotation | # of peptides sequenced | q-value |
| Summary of Upregulated Proteins * | ||||
| P62701 | RPS4X | 40S ribosomal protein S4, X isoform | 2 | 0.003 |
| P14786 | PKM2 | Pyruvate kinase isozymes M1/M2 | 20 | 0.003 |
| Q8TBK5 | RPL6 | 60S ribosomal protein L6 | 2 | 0.005 |
| Q02878 | GAPDHS | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific | 3 | 0.006 |
| Q59GX2 | GLUT1 | Solute carrier family 2, facilitated glucose transporter member 1 | 2 | 0.007 |
| Q14568 | HSP90AA2 | Heat shock protein HSP 90-alpha 2 | 17 | 0.008 |
| P07195 | LDHB | L-lactate dehydrogenase B chain | 12 | 0.008 |
| Q5RKT7 | RPS27A | 40S ribosomal protein S27a | 4 | 0.011 |
| P00354 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 24 | 0.012 |
| Q6FHV0 | MIF | Macrophage migration inhibitory factor | 4 | 0.012 |
| Q9BVK5 | PPIB | Peptidyl-prolyl cis-trans isomerase B precursor | 6 | 0.012 |
| Q3B872 | HIST1H2BI | Histone H2B type 1-C/E/F/G/I | 10 | 0.013 |
| Q3KQT6 | RPS2 | 40S ribosomal protein S2 | 3 | 0.014 |
| Q5J7W1 | PGK1 | Phosphoglycerate kinase 1 | 20 | 0.014 |
| Q8WV32 | TALDO1 | Transaldolase | 2 | 0.016 |
| Q99497 | DJ1 | Protein DJ-1 | 8 | 0.016 |
| Q9UBU5 | PRDX5 | Peroxiredoxin-5, mitochondrial precursor | 4 | 0.016 |
| Q76LA1 | CSTB | Cystatin-B | 3 | 0.016 |
| Q53X54 | HSPE1 | 10 kDa heat shock protein, mitochondrial | 3 | 0.019 |
| Q96DW8 | SERPINA3 | Alpha-1-antichymotrypsin precursor | 2 | 0.02 |
| Q9NTZ7 | CPNE1 | Copine-1 | 2 | 0.021 |
| Q53Y44 | PFN1 | Profilin-1 | 6 | 0.021 |
| Q8WU81 | MYST4 | Histone acetyltransferase MYST4 | 2 | 0.021 |
| P05141 | SLC25A5 | ADP/ATP translocase 2 | 7 | 0.021 |
| Q86WV2 | COX4I1 | Cytochrome c oxidase subunit 4 isoform 1, mitochondrial precursor | 2 | 0.023 |
| P12236 | SLC25A6 | ADP/ATP translocase 3 | 5 | 0.023 |
| P42224 | STAT1 | Signal transducer and activator of transcription 1-alpha/beta | 4 | 0.026 |
| P31945 | PRDX2 | Peroxiredoxin-2 | 8 | 0.028 |
| P07900 | HSP90AA1 | heat shock protein 90 kDa alpha | 4 | 0.029 |
| Q53ES3 | ARL6IP5 | PRA1 family protein 3 | 2 | 0.03 |
| P14866 | HNRPL | Heterogeneous nuclear ribonucleoprotein L | 2 | 0.034 |
| P13797 | PLS3 | Plastin-3 | 3 | 0.036 |
| Q99623 | PHB2 | Prohibitin-2 | 6 | 0.04 |
| Q9UDE9 | LDHA | L-lactate dehydrogenase A chain | 17 | 0.041 |
| P16402 | HIST1H1D | Histone H1.3 | 6 | 0.041 |
| Q9H4B7 | TUBB1 | tubulin, beta 1 | 6 | 0.043 |
| P02792 | FTL | Ferritin light chain | 5 | 0.047 |
| P32077 | PRDX6 | Peroxiredoxin-6 | 4 | 0.047 |
| Summary of Downregulated Proteins * : | ||||
| P33121 | ACSL1 | Long-chain-fatty-acid–CoA ligase 1 | 2 | 0.003 |
| Q5T0R7 | CAP1 | Adenylyl cyclase-associated protein 1 | 4 | 0.003 |
| P36957 | DLST | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial precursor | 2 | 0.003 |
| Q53FQ6 | CAPZA1 | F-actin capping protein subunit alpha-1 | 2 | 0.004 |
| P31930 | UQCRC1 | Ubiquinol-cytochrome-c reductase complex core protein 1, mitochondrial precursor | 3 | 0.005 |
| Q59EQ5 | CSRP1 | Cysteine and glycine-rich protein 1 | 2 | 0.006 |
| P07585 | DCN | Decorin precursor | 2 | 0.007 |
| P24941 | CDK2 | Cell division protein kinase 2 | 4 | 0.009 |
| Q6LBZ1 | APOE | Apolipoprotein E precursor | 4 | 0.009 |
| Q96QM7 | LUM | Lumican precursor | 6 | 0.009 |
| Q9Y4L1 | HYOU1 | 150 kDa oxygen-regulated protein precursor | 4 | 0.011 |
| Q504S5 | CKAP4 | Cytoskeleton-associated protein 4 | 3 | 0.012 |
| P62241 | RPS8 | 40S ribosomal protein S8 | 3 | 0.016 |
| P78528 | PRKDC | DNA-dependent protein kinase catalytic subunit | 2 | 0.016 |
| P05198 | EIF2S1 | Eukaryotic translation initiation factor 2 subunit 1 | 2 | 0.017 |
| P15586 | GNS | N-acetylglucosamine-6-sulfatase precursor | 2 | 0.021 |
| P41219 | PRPH | Peripherin-2 | 3 | 0.021 |
| Q9UKB0 | HMGA1 | High mobility group protein HMG-I/HMG-Y | 2 | 0.022 |
| Q3KQV4 | NACA | Nascent polypeptide-associated complex subunit alpha | 2 | 0.024 |
| Q4V324 | GAGE7 | G antigen 7 | 3 | 0.026 |
| P67809 | YBX1 | Nuclease sensitive element-binding protein 1 | 4 | 0.031 |
| P13667 | PDIA4 | Protein disulfide-isomerase A4 precursor | 4 | 0.034 |
| Q16208 | CD44 | CD44 antigen precursor | 2 | 0.042 |
Proteins with significant changes greater than 20%.
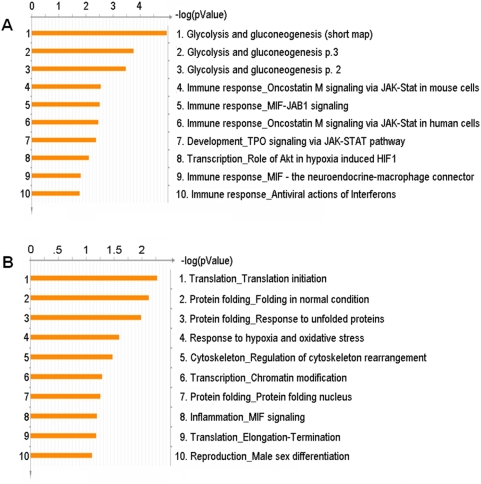
To further interpret the likely roles of differentially-expressed proteins involved in melanoma metastasis, we used the MetaCore™ pathway mapping tool to analyze and build the biological networks related to these proteins. First, we investigated the enrichment of the 61 differentially expressed proteins across two different ontological categories using GeneGo canonical pathway maps and GeneGo processes by calculating the hypergeometric distribution p-value of this list with respect to each category. Figure 2A shows the top ten GeneGo canonical pathway maps associated with the differentially expressed proteins and Figure 2B shows the top ten GeneGo cellular process networks. Glycolysis and gluconeogenesis, which involve PGK1, GAPDH, GAPDHS, PKM2, LDHA, and LDHB proteins, are suggested to have the highest significance in the pathway maps. Translation initiation and protein folding are the two most significant cellular process networks. Individual mapping of up-regulated and down-regulated proteins showed that glycolysis and gluconeogenesis were the most up-regulated pathways, while DNA damage-induced response (cell cycle), nucleocytoplasmic transport of CDK/cyclins (cell cycle), and DNA-damage-induced apoptosis were the most down-regulated (data not shown).
Figure 2. Representation of two ontological categories of differentially expressed proteins in metastasis by GeneGo pathways analysis.
The results are ordered by −log10 of the p-value of the hypergeometric distribution. A) GeneGo canonical pathway maps, and B) GeneGo cellular process.
Next, we analyzed direct interaction between the 61 differentially expressed proteins. STAT1 directly interacted with HSP90AA1, SERPINA3, and APOE, whereas HMGA1 interacted with CD44. We then analyzed the transcription factors regulating 61 differentially expressed proteins. The most prominent regulatory proteins were c-MYC, SP1, and P53, interacting with 21, 20, and 11 differentially expressed proteins, respectively. Proteins directly activated by c-MYC included HSPE1, HSP90AA1, LDHA, CDK2, HMGA1, EIF2S1, and YBX1. Finally, we used the analyze network (AN) algorithm to reveal the most relevant networks associated with the 61 differentially expressed proteins. The networks were prioritized based on the number of fragments of the canonical pathways in the network. As a result, the top-scored AN network brought 11 proteins together: up-regulated proteins were MYST4, HIST1H2BI, PPIB, ARL6IP5, CPNE1, PFN1, and DJ1; downregulated proteins were YBX1, NACA, PRKDC, and CDK2.
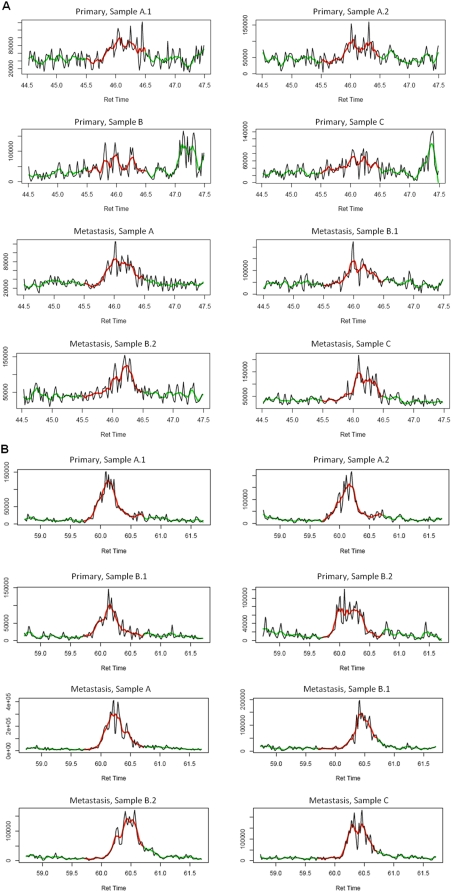
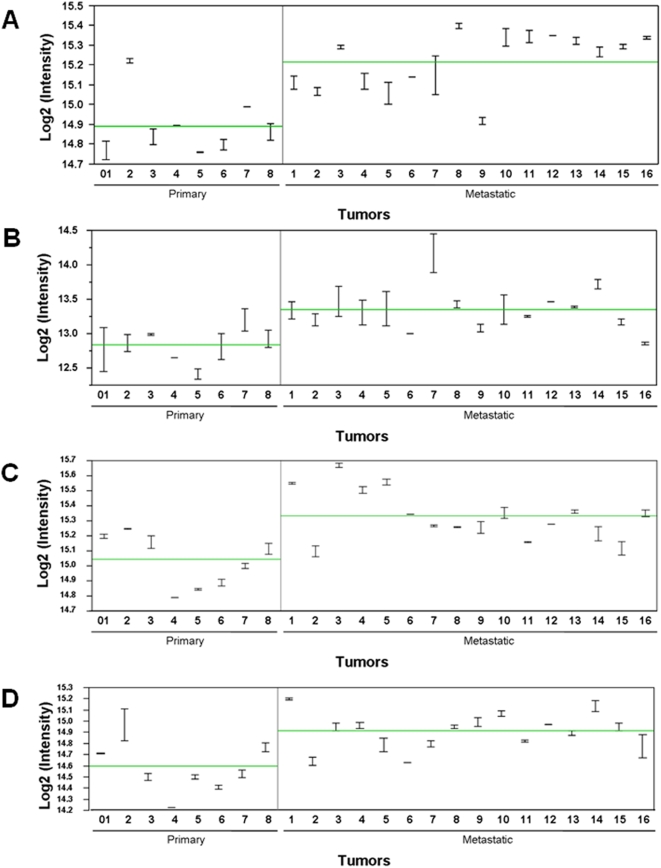
To identify the potential proteomic markers, our first objective was to focus on the up-regulated proteins that are categorized in the biological processes related to melanoma progression or metastasis ( Table 3 ). These categories include cell structure and motility, cell adhesion, cell defense, cell cycle, cell proliferation and differentiation, apoptosis, metabolic process, and melanosome-related. Interestingly, all of the listed proteins have been previously implicated as cancer-related, and some have been suggested to be related to metastasis, including pyruvate kinase isozymes M1/M2 (PKM2), glucose transporter isoform 1 (GLUT1), macrophage migration inhibitory factor (MIF), peroxiredoxin-2 (PRDX2), L-lactate dehydrogenase A chain (LDHA), and heat shock protein HSP 90 alpha 2 (HSP90AA2). Furthermore, GLUT1, HSP90AA2, prohibitin-2 (PHB2), peptidyl-prolyl cis-trans isomerase B precursor (PPIB), and L-lactate dehydrogenase A chain (LDHA) have been identified in the melanosome. In Figure 3 we provide representative MS tracings of XIC for specific peptides in PKM2 and HSP90AA2. Figure 4 shows representative plots of protein intensities of the four most significant proteins are shown: PKM2 (q value = 0.003), GLUT1 (q = 0.007), HSP90AA2 (q = 0.008) and L-lactate dehydrogenase B chain (LDHB, q = 0.008) in individual samples of primary and metastatic melanomas.
Table 3. Selected Up-regulated Proteins in Melanoma Metastasis.
| Category | Gene Symbol |
| Cell Structure and Motility, Cell Adhesion | TUBB1, PLS3 |
| Cell Cycle | MYST4, PHB2, TUBB1 |
| Cell proliferation and differentiation | DJ1, MYST4, PHB2 |
| Cell Defense | MIF, PPIB, DJ1, PRDX5, PRDX2, PRDX6, HSP90AA2 |
| Carbohydrate metabolic process | PKM2, GLUT1,LDHB, LDHA, PGK1 |
| Protein metabolic process | HSP90AA1, HSP90AA2, HSPE1, MYST4, PPIB |
| Nucleic acid metabolic process | DJ1, MYST4, PHB2, STAT1 |
| Melanosome-Associated | PKM2, GLUT1, HSP90AA1, PPIB, PHB2, LDHA, PLS3 |
Figure 3. MS tracings of representative specific peptides.
X-axis: retention time. Y-axis: peak intensity. A) Extracted ion chromatogram (XIC) for peptide GDYPLEAVR from pyruvate kinase isozymes M1/M2 of representative samples. Primary melanoma samples A.1 and A.2 are duplicate samples. Metastatic melanoma samples B.1 and B.2 are duplicates. Peak intensities were calculated by smoothing (as indicated by the green and red trace lines) and integrating the AUC of smoothed peaks for all samples within the same one-minute window (RT from 45.5 min to 46.5 min as indicated by the red trace line). B) Extracted ion chromatogram (XIC) for peptide ALLFVPR from heat shock protein HSP90AA2 of representative samples. Primary melanoma samples A.1 and A.2, and samples B.1 and B.2, are duplicate samples, respectively. Metastatic melanoma samples B.1 and B.2 are duplicates. RT from 59.7 min to 60.7 min as indicated by the red trace line.
Figure 4. Representative plots of protein intensities in the individual samples of primary and metastatic melanoma.
The individual protein intensities for A) PKM2, B) GLUT1, C) HSP90AA2, and D) LDHB Are plotted on a log2 scale. The horizontal line is the group mean, and duplicate intensities of individual samples are joined by a vertical line.
IHC Analysis of Proteins
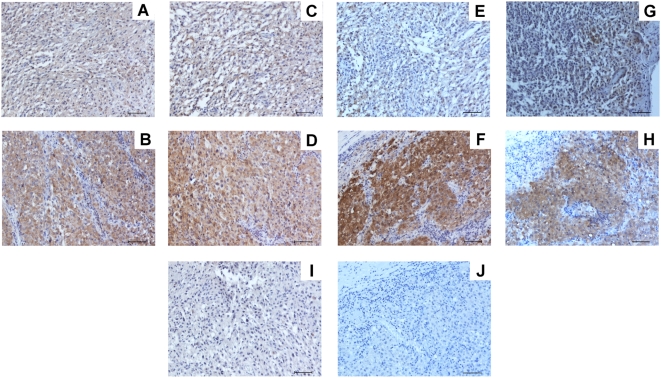
We examined several key proteins that were significantly upregulated in metastasis compared to primary melanoma. IHC staining was limited to availability of antibodies for FFPE tissues. In Figure 5 we show representative examples of IHC staining of PKM2, HSP90 alpha, MIF, and STAT1 in both primary and metastatic melanomas. These studies confirmed the MS analysis of proteins in melanomas.
Figure 5. Representative IHC staining of significant proteins found in melanomas.
Anti-PKM1/M2 (rabbit) Ab: A) primary and B) brain metastasis. Anti-HSP90 (rabbit) Ab: C) primary and D) brain metastasis. Anti-MIF (mouse) Ab: E) primary and F) brain metastasis. Anti-STAT1 (mouse) Ab: G) primary and H) brain metastasis. I) Universal negative control rabbit Ig on primary melanoma tissue. J) Universal negative control mouse Ig on brain metastasis tissue. All magnifications were ×100. Scale bars represent 100 µm.
Discussion
Quantitative proteome analysis by MS has become a useful tool for understanding biological functions. While FFPE tissues offer great opportunity for retrospective proteomic analysis, isolating proteins from FFPE tissues suitable for MS analysis has been a challenge. Quality control and uniformity of preparation across, or even within, institutions remain problematic [27]. The reproducibility of results requires further validation on a large set of specimens.
In the present study, the global protein expression profile in FFPE tissue lysates of melanoma metastases was compared with that of primary tumors. Throughout this study, we have addressed several questions regarding FFPE melanoma tissues: (1) Is this methodology useful for profiling the protein expression of archival tissue? (2) How many proteins can be identified from small quantities of FFPE melanoma tissue using Liquid Tissue® reagents and high resolution LC-ESI/MS/MS? (3) Do specific identified proteins discriminate between metastatic and primary tissue? (4) Can known melanoma-related biomarkers be identified? (5) Are any known metastasis-related markers identified? (6) Can the identified protein be validated by other methods?
We have demonstrated that proteins can be extracted from microdissected FFPE tissues and utilized for MS analysis. Using LC/MS/MS analysis, over 1500 proteins can be identified in a small amount of cells (the equivalent of approximately 5,000 cells per injection). Moreover, identified proteins appear to cover a wide variety of biological functions and appear in diverse cellular compartments. The proteomic profiling of FFPE tissue demonstrated the detection of the major known melanogenesis pathway proteins that are known to be expressed in melanomas. Over 70% of previously known common melanosomal proteins can be identified, including four of the six well known melanoma specific markers. MART1 and TRP2 were not identified, which may either be due to their low abundance or interference from melanin [28], [29]. Melanin has been shown to bind to proteins, causing difficulties in protein solubility and binding affinity to chromatography columns, which impairs protein characterization using LC/MS analysis [30], [31]. It has been demonstrated that MART1 can be identified only if melanin is removed before 2D-LC/MS [29]. Lack of detection of some proteins may be due to the level of protein in the tissues. Of the 120 differentially expressed proteins identified with multiple peptides, 61 showed differences exceeding 20%. Many of these proteins have been reported to be associated with characteristic steps of metastasis including motility, adhesion, migration and tumor progression including cellular defense, apoptosis, and proliferation.
Several of the up-regulated proteins ( Table 3 ) identified in this study have been described previously and implicated as cancer-related. Some proteins are implicated as metastasis-related proteins (e.g. LDH, HSP90, GLUT1, MIF, DJ1, PKM2 and PHB2). HSP90 and GLUT1 have been previously described in melanoma metastasis tissues as up-regulated [32], [33]. LDH protein levels have been described in serum from melanoma patients [34]. The expression of pyruvate kinase, HSP90 alpha, MIF, and STAT1 in primary and metastatic melanomas could be demonstrated by IHC staining. Upregulated expression levels of these proteins in melanoma metastasis compared to primary melanomas were demonstrated, confirming MS analysis.
In addition, LDHA, HSP90, GLUT1, MIF, DJ1, PKM2 PHB2, PGK1 and PPIB are secreted from cancer cells and can be detected in the blood, offering great potential as biomarkers of tumor progression. It may be interesting to investigate whether these proteins are involved in melanoma progression. Many of these proteins are poorly-documented, and their role in metastasis requires further investigation (e.g. MYST4, PLS3, and TUBB1). We were able to identify several proteins implicated in melanoma progression and metastasis; their biological functions and deregulations in tumorigenesis and metastasis are further discussed below.
Plastins are actin-binding proteins that have three cell-type specific isoforms: T-plastin, L-plastin and I-plastin. L-plastin is expressed in hemapoietic cells, while T-plastin is in cells of solid tissue, including fibroblasts, epithelial cells, and melanocytes. Upregulation of L-plastin has been well documented in many types of cancer [35] and has been implicated in metastasis [36]. However, the role of T-plastin in tumor progression is not known and overexpression of T-plastin in cultured fibroblasts showed increased mobility and altered cellular architecture. Although PLS3 (T-plastin) has not been reported to be specifically present in the melanosome organelle, it has been shown to be expressed in melanocytes [37].
HSP90 has been a potential target for cancer therapy in the last decade, and a HSP90 inhibitor, 17-allylamino-17 demethoxygeldanamycin (17-AAG), is currently in clinical trials [38]. HSP90 is involved in maintaining the conformation, stability, activity, and cellular localization of several key oncogenic client proteins, including ERBB2, C-RAF, CDK4, AKT/PKB mutated P53, hypoxia-inducible factor-1α (HIF-1α), survivin telomerase hTERT, and steroid hormone receptors [39]. Inhibition of HSP90 simultaneously targets multiple oncogenes and pathways, as well as characteristic traits of malignancy. HSP90α, an isoform of HSP90, is stress-inducible and overexpressed in many cancer cells [40]. Furthermore, HSP90α has been shown to be related to invasiveness and found to be secreted [40]. HSP90 mRNA levels are up-regulated in primary and metastatic melanoma when compared to melanocytic nevi [32]. Our analysis showed significantly higher levels of HSP90 protein in metastatic than in primary melanomas.
DJ1 protein was initially discovered as a putative oncogene that promotes cell survival by negatively regulating tumor suppressor PTEN [41]. DJ1 has also been implicated in cell protection against stresses including chemotherapy and oxidative stresses, [42]. Overexpression of DJ1 has been shown to be associated with tumorigenesis. Interestingly, DJ1 has been detected in serum from uveal malignant melanoma patients [43].
MYST4, a member of the MYST histone acetyltransferase family, may be involved in both upregulation and downregulation of tumorigenesis. MYST4 reportedly fuses to the transcription co-activators CBP, p300, and TIF2 in acute myeloid leukemia. MYST4 was shown to be required for RUNX2-dependent transcriptional activation, and to interact with P53 in cancer [44], [45]. There is little information about the expression profile of MYST4 in cancer. Further investigation is needed to determine whether MYST4 is associated with brain metastasis, since MYST4 has been implicated in neural development and maintenance of neural stem cells, and in this study we found MYST4 expression is higher in brain melanoma metastases when compared to primary melanoma.
Proteins of the prohibitin family (PHB1 and PHB2) are localized in many cellular compartments (e.g. mitochondria, nucleus, and cell-surface) and may have diverse functions in different locations. PHB appears to function as a chaperone in the stabilization of mitochondrial proteins [46], yet the functions of nuclear PHB are still controversial. PHB has been proposed to play a role as a potential tumor suppressor, but higher levels of PHB protein have been reported in a variety of cancers [47]. Prohibitin was demonstrated to play an unexpected role in the activation of the Ras-Raf signaling pathway and in modulating epithelial cell adhesion and migration, indicating that PHB may play a role in metastasis formation [47].
Several of the most significant proteins identified that increased in metastasis were those involved in the aerobic glycolysis pathway in cancer cells (Warburg effect). Particular upregulated proteins were GLUT1, PKM1/M2, PGK1, and LDHA/B. Most of these proteins have been shown to be upregulated in cancer, including melanomas. Interestingly, in our analysis these proteins of the glycolysis pathway were quite significantly elevated in metastatic tissues. LDHA and LDHB are subunits of lactate dehydrogenase (LDH) and associate to generate five tetrameric forms of LDH isoenzymes. LDHA is involved in metabolic activities, particularly the redox reaction at the end of glycolysis. An enhanced level of LDHA is observed in cancer cells and a higher level of LDHA is associated with proliferation [48]. Elevated serum levels of LDH indicate progression of AJCC stage IV melanoma; LDH is a biomarker for AJCC staging of melanomas [49]. Our results support the finding that upregulation of LDH in cells is related to aggressive metastatic melanoma tumors.
GLUT1 may be responsible for constitutive or basal glucose uptake. Overexpression of GLUT1 is associated with increased glucose metabolism and has been correlated with progression of several types of cancer [50]. Studies have shown that GLUT1 is modulated by hypoxia-responsive elements and upregulated by overexpression of HIF-1 [51]. GLUT1 protein is expressed in most melanoma metastases with a broad range of expression levels as shown by Western immunoblot analysis [33]. Our results confirm these findings.
PKM2 is an M2 type of pyruvate kinase (PK) which plays a critical role in aerobic glycolysis regulation in tumorigenesis. During oncogenesis, the PK isoform M1 is converted to PKM2 and is present as a less active dimer form. PKM2 is up-regulated in tumor cells, and is under control of Ras and transcription factors HIF-1, SP1, and SP2. Interestingly, tumor M2-PK is expressed at higher levels in metastases than in primary tumors, and the level of PKM2 in plasma of various cancer types is correlated with tumor size and stage [52]. Recent studies have shown that PKM2 isoform is highly necessary for aerobic glycolysis, which provides a selective growth advantage for tumor cells [53].
PGK1 (phosphoglycerate kinase), which is involved in the glycolytic pathway, is regulated by HIF-1α and is enhanced in states of hypoxia due to increased demand for glycolysis. PGK1 also affects DNA replication and repair in the nucleus. Extracellular secretion of PGK1 by tumor cells increases angiostatin activity, causing inhibition of angiogenesis. In several types of cancer, PGK1 has been shown to be up-regulated [54]. In pancreatic cancers, PGK1 is secreted into the bloodstream, and studies have shown increased levels of serum PGK1. Therefore, this protein shows promise as a potential serum marker for cancer [54].
In this report, Liquid Tissue® reagents effectively solubilized proteins and peptides from microdissected formalin-fixed melanoma tissues and enabled proteomics analysis by ESI-LC MS/MS. Global protein profiling using label-free protein quantitative analysis revealed potential metastasis-related proteins that are differentially expressed between metastatic and primary melanoma. Up-regulation of some of these proteins has been previously implicated in tumor progression by other methodologies and is in accordance with our findings. Hence, the method described here is extremely useful for proteomics studies in a high-throughput setting. Moreover, in the search for potential biomarker, candidates can be confirmed in a larger set of samples to validate the findings. The advantage of this approach is that it allows analysis of retrospective tumor tissue specimens with known disease outcomes. This provides a unique strategy to develop new molecular prognostic biomarkers of melanoma.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported in part by NIH, National Cancer Institute P0 grants CA029605 and CA012582, the Leslie and Susan Gonda (Goldschmied) Foundation, and the Martin H. Weil Research Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, et al. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000;406:536–540. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- 2.Haqq C, Nosrati M, Sudilovsky D, Crothers J, Khodabakhsh D, et al. The gene expression signatures of melanoma progression. Proc Natl Acad Sci U S A. 2005;102:6092–6097. doi: 10.1073/pnas.0501564102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winnepenninckx V, Lazar V, Michiels S, Dessen P, Stas M, et al. Gene expression profiling of primary cutaneous melanoma and clinical outcome. J Natl Cancer Inst. 2006;98:472–482. doi: 10.1093/jnci/djj103. [DOI] [PubMed] [Google Scholar]
- 4.Pardo M, Garcia A, Antrobus R, Blanco MJ, Dwek RA, et al. Biomarker discovery from uveal melanoma secretomes: identification of gp100 and cathepsin D in patient serum. J Proteome Res. 2007;6:2802–2811. doi: 10.1021/pr070021t. [DOI] [PubMed] [Google Scholar]
- 5.Rivera J, Megias D, Bravo J. Proteomics-based strategy to delineate the molecular mechanisms of the metastasis suppressor gene BRMS1. J Proteome Res. 2007;6:4006–4018. doi: 10.1021/pr0703167. [DOI] [PubMed] [Google Scholar]
- 6.Zhang B, VerBerkmoes NC, Langston MA, Uberbacher E, Hettich RL, et al. Detecting differential and correlated protein expression in label-free shotgun proteomics. J Proteome Res. 2006;5:2909–2918. doi: 10.1021/pr0600273. [DOI] [PubMed] [Google Scholar]
- 7.Wang G, Wu WW, Zeng W, Chou CL, Shen RF. Label-free protein quantification using LC-coupled ion trap or FT mass spectrometry: Reproducibility, linearity, and application with complex proteomes. J Proteome Res. 2006;5:1214–1223. doi: 10.1021/pr050406g. [DOI] [PubMed] [Google Scholar]
- 8.Ono M, Shitashige M, Honda K, Isobe T, Kuwabara H, et al. Label-free quantitative proteomics using large peptide data sets generated by nanoflow liquid chromatography and mass spectrometry. Mol Cell Proteomics. 2006;5:1338–1347. doi: 10.1074/mcp.T500039-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Hood BL, Conrads TP, Veenstra TD. Mass spectrometric analysis of formalin-fixed paraffin-embedded tissue: unlocking the proteome within. Proteomics. 2006;6:4106–4114. doi: 10.1002/pmic.200600016. [DOI] [PubMed] [Google Scholar]
- 10.Hood BL, Darfler MM, Guiel TG, Furusato B, Lucas DA, et al. Proteomic analysis of formalin-fixed prostate cancer tissue. Mol Cell Proteomics. 2005;4:1741–1753. doi: 10.1074/mcp.M500102-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Higgs RE, Knierman MD, Gelfanova V, Butler JP, Hale JE. Comprehensive label-free method for the relative quantification of proteins from biological samples. J Proteome Res. 2005;4:1442–1450. doi: 10.1021/pr050109b. [DOI] [PubMed] [Google Scholar]
- 12.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 13.Eng J, McCormack A, Yates RR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 14.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 15.Limpert E, Stahel WA, Abbt M. Log-normal distributions across the sciences: keys and clues. BioScience. 2001;51:341–352. [Google Scholar]
- 16.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 17.Goto Y, Ferrone S, Arigami T, Kitago M, Tanemura A, et al. Human high molecular weight-melanoma-associated antigen: utility for detection of metastatic melanoma in sentinel lymph nodes. Clin Cancer Res. 2008;14:3401–3407. doi: 10.1158/1078-0432.CCR-07-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ekins S, Nikolsky Y, Bugrim A, Kirillov E, Nikolskaya T. Pathway mapping tools for analysis of high content data. Methods Mol Biol. 2007;356:319–350. doi: 10.1385/1-59745-217-3:319. [DOI] [PubMed] [Google Scholar]
- 19.Ekins S, Bugrim A, Nikolsky Y, Nikolskaya T. Systems biology: applications in drug discovery. In: Gad SC, editor. Drug Discovery Handbook. New York: Wiley; 2005. pp. 123–183. [Google Scholar]
- 20.Fitzpatrick DPG, You J-S, Bemis KG, Wery J-P, Ludwig JR, et al. Searching for potential biomarkers of cisplatin resistance in human ovarian cancer using a label-free LC/MS-based protein quantification method. PROTEOMICS - Clinical Applications. 2007;1:246–263. doi: 10.1002/prca.200600768. [DOI] [PubMed] [Google Scholar]
- 21.Takeuchi H, Kuo C, Morton DL, Wang HJ, Hoon DS. Expression of differentiation melanoma-associated antigen genes is associated with favorable disease outcome in advanced-stage melanomas. Cancer Res. 2003;63:441–448. [PubMed] [Google Scholar]
- 22.Koyanagi K, O'Day SJ, Gonzalez R, Lewis K, Robinson WA, et al. Microphthalmia transcription factor as a molecular marker for circulating tumor cell detection in blood of melanoma patients. Clin Cancer Res. 2006;12:1137–1143. doi: 10.1158/1078-0432.CCR-05-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez S, Takeuchi H, Hoon D. The clinical use of molecular markers as predictors of disease outcome and response to therapy in malignant melanoma. In: Hearing V, Leong S, editors. Melanocytes to melanoma: The progression to malignancy. Humana Press; 2005. pp. 551–576. [Google Scholar]
- 24.Huang SK, Okamoto T, Morton DL, Hoon DS. Antibody responses to melanoma/melanocyte autoantigens in melanoma patients. J Invest Dermatol. 1998;111:662–667. doi: 10.1046/j.1523-1747.1998.00354.x. [DOI] [PubMed] [Google Scholar]
- 25.Okamoto T, Irie RF, Fujii S, Huang SK, Nizze AJ, et al. Anti-tyrosinase-related protein-2 immune response in vitiligo patients and melanoma patients receiving active-specific immunotherapy. J Invest Dermatol. 1998;111:1034–1039. doi: 10.1046/j.1523-1747.1998.00411.x. [DOI] [PubMed] [Google Scholar]
- 26.Yamaguchi Y, Brenner M, Hearing VJ. The regulation of skin pigmentation. J Biol Chem. 2007;282:27557–27561. doi: 10.1074/jbc.R700026200. [DOI] [PubMed] [Google Scholar]
- 27.Blow N. Tissue preparation: Tissue issues. Nature. 2007;448:959–963. doi: 10.1038/448959a. [DOI] [PubMed] [Google Scholar]
- 28.Basrur V, Yang F, Kushimoto T, Higashimoto Y, Yasumoto K, et al. Proteomic analysis of early melanosomes: identification of novel melanosomal proteins. J Proteome Res. 2003;2:69–79. doi: 10.1021/pr025562r. [DOI] [PubMed] [Google Scholar]
- 29.Kushimoto T, Basrur V, Valencia J, Matsunaga J, Vieira WD, et al. A model for melanosome biogenesis based on the purification and analysis of early melanosomes. Proc Natl Acad Sci U S A. 2001;98:10698–10703. doi: 10.1073/pnas.191184798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chi A, Valencia JC, Hu ZZ, Watabe H, Yamaguchi H, et al. Proteomic and bioinformatic characterization of the biogenesis and function of melanosomes. J Proteome Res. 2006;5:3135–3144. doi: 10.1021/pr060363j. [DOI] [PubMed] [Google Scholar]
- 31.Wakamatsu K, Ito S. Advanced chemical methods in melanin determination. Pigment Cell Res. 2002;15:174–183. doi: 10.1034/j.1600-0749.2002.02017.x. [DOI] [PubMed] [Google Scholar]
- 32.McCarthy MM, Pick E, Kluger Y, Gould-Rothberg B, Lazova R, et al. HSP90 as a marker of progression in melanoma. Ann Oncol. 2008;19:590–594. doi: 10.1093/annonc/mdm545. [DOI] [PubMed] [Google Scholar]
- 33.Wachsberger PR, Gressen EL, Bhala A, Bobyock SB, Storck C, et al. Variability in glucose transporter-1 levels and hexokinase activity in human melanoma. Melanoma Res. 2002;12:35–43. doi: 10.1097/00008390-200202000-00006. [DOI] [PubMed] [Google Scholar]
- 34.Meyers ML, Balch CM. Diagnosis and Treatment of Metastatic Melanoma. In: Balch CM, Houghton AN, Sober AJ, Soong S, editors. Cutaneous Melanoma. 3rd Ed. ed. St. Louis, Missouri: Quality Medical Publishing, Inc; 1998. pp. 325–372. [Google Scholar]
- 35.Park T, Chen ZP, Leavitt J. Activation of the leukocyte plastin gene occurs in most human cancer cells. Cancer Res. 1994;54:1775–1781. [PubMed] [Google Scholar]
- 36.Otsuka M, Kato M, Yoshikawa T, Chen H, Brown EJ, et al. Differential expression of the L-plastin gene in human colorectal cancer progression and metastasis. Biochem Biophys Res Commun. 2001;289:876–881. doi: 10.1006/bbrc.2001.6047. [DOI] [PubMed] [Google Scholar]
- 37.Lin CS, Park T, Chen ZP, Leavitt J. Human plastin genes. Comparative gene structure, chromosome location, and differential expression in normal and neoplastic cells. J Biol Chem. 1993;268:2781–2792. [PubMed] [Google Scholar]
- 38.Ramanathan RK, Trump DL, Eiseman JL, Belani CP, Agarwala SS, et al. Phase I pharmacokinetic-pharmacodynamic study of 17-(allylamino)-17-demethoxygeldanamycin (17AAG, NSC 330507), a novel inhibitor of heat shock protein 90, in patients with refractory advanced cancers. Clin Cancer Res. 2005;11:3385–3391. doi: 10.1158/1078-0432.CCR-04-2322. [DOI] [PubMed] [Google Scholar]
- 39.Maloney A, Workman P. HSP90 as a new therapeutic target for cancer therapy: the story unfolds. Expert Opin Biol Ther. 2002;2:3–24. doi: 10.1517/14712598.2.1.3. [DOI] [PubMed] [Google Scholar]
- 40.Picard D. Hsp90 invades the outside. Nat Cell Biol. 2004;6:479–480. doi: 10.1038/ncb0604-479. [DOI] [PubMed] [Google Scholar]
- 41.Kim RH, Peters M, Jang Y, Shi W, Pintilie M, et al. DJ-1, a novel regulator of the tumor suppressor PTEN. Cancer Cell. 2005;7:263–273. doi: 10.1016/j.ccr.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 42.Taira T, Saito Y, Niki T, Iguchi-Ariga SM, Takahashi K, et al. DJ-1 has a role in antioxidative stress to prevent cell death. EMBO Rep. 2004;5:213–218. doi: 10.1038/sj.embor.7400074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pardo M, Garcia A, Thomas B, Pineiro A, Akoulitchev A, et al. The characterization of the invasion phenotype of uveal melanoma tumour cells shows the presence of MUC18 and HMG-1 metastasis markers and leads to the identification of DJ-1 as a potential serum biomarker. Int J Cancer. 2006;119:1014–1022. doi: 10.1002/ijc.21942. [DOI] [PubMed] [Google Scholar]
- 44.Yang XJ, Ullah M. MOZ and MORF, two large MYSTic HATs in normal and cancer stem cells. Oncogene. 2007;26:5408–5419. doi: 10.1038/sj.onc.1210609. [DOI] [PubMed] [Google Scholar]
- 45.Troke PJ, Kindle KB, Collins HM, Heery DM. MOZ fusion proteins in acute myeloid leukaemia. Biochem Soc Symp. 2006:23–39. doi: 10.1042/bss0730023. [DOI] [PubMed] [Google Scholar]
- 46.Nijtmans LG, de Jong L, Artal Sanz M, Coates PJ, Berden JA, et al. Prohibitins act as a membrane-bound chaperone for the stabilization of mitochondrial proteins. Embo J. 2000;19:2444–2451. doi: 10.1093/emboj/19.11.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rajalingam K, Wunder C, Brinkmann V, Churin Y, Hekman M, et al. Prohibitin is required for Ras-induced Raf-MEK-ERK activation and epithelial cell migration. Nat Cell Biol. 2005;7:837–843. doi: 10.1038/ncb1283. [DOI] [PubMed] [Google Scholar]
- 48.Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 49.Deichmann M, Benner A, Bock M, Jackel A, Uhl K, et al. S100-Beta, melanoma-inhibiting activity, and lactate dehydrogenase discriminate progressive from nonprogressive American Joint Committee on Cancer stage IV melanoma. J Clin Oncol. 1999;17:1891–1896. doi: 10.1200/JCO.1999.17.6.1891. [DOI] [PubMed] [Google Scholar]
- 50.Kunkel M, Reichert TE, Benz P, Lehr HA, Jeong JH, et al. Overexpression of Glut-1 and increased glucose metabolism in tumors are associated with a poor prognosis in patients with oral squamous cell carcinoma. Cancer. 2003;97:1015–1024. doi: 10.1002/cncr.11159. [DOI] [PubMed] [Google Scholar]
- 51.Krishnamachary B, Berg-Dixon S, Kelly B, Agani F, Feldser D, et al. Regulation of colon carcinoma cell invasion by hypoxia-inducible factor 1. Cancer Res. 2003;63:1138–1143. [PubMed] [Google Scholar]
- 52.Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin Cancer Biol. 2005;15:300–308. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 53.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 54.Hwang TL, Liang Y, Chien KY, Yu JS. Overexpression and elevated serum levels of phosphoglycerate kinase 1 in pancreatic ductal adenocarcinoma. Proteomics. 2006;6:2259–2272. doi: 10.1002/pmic.200500345. [DOI] [PubMed] [Google Scholar]