Abstract
Three paths are described to obtain crystals of reduced (II-E4Q/I-K258R) cytochrome ba3, and the structures of these are reported at ∼2.8 to 3.0 Å resolution. Microspectrophotometry of single crystals of Thermus ba3 oxidase at 100 K was used to show that crystals of the oxidized enzyme are reduced in an intense X-ray (beam line 7-1 at the Stanford Synchrotron Radiation Laboratory, U.S.A) being nearly complete in one minute. The previously reported structures of ba3 (PDB codes 1EHK and 1XME), having a crystallographically detectable water between the CuB and Fea3 metals of the dinuclear center, actually represent the X-ray radiation-reduced enzyme. Dithionite reduced crystals or crystals formed from dithionite reduced enzyme revealed the absence of the above mentioned water and an increase in the CuB - Fea3 distance of ∼0.3 A. The new structures are discussed in terms of enzyme function. An unexpected optical absorption envelope at ∼590 nm is also reported. This spectral feature is tentatively thought to arise from a 5-coordinate, low-spin, ferrous heme-a3 that is trapped in the frozen crystals.
In respiring organisms cytochrome c oxidase catalyzes proton translocation coupled to the reduction of O2 to water (1, 2). Cytochrome ba3 oxidase is one of two heme-copper oxidases isolated from T. thermophilus (3). It contains the dinuclear CuA center; a six-coordinated, low-spin, (6cLS) heme-B; and a (usually) five-coordinated, high-spin (5cHS) heme-As in close proximity to CuB ((3, 4) and references therein). The latter (heme-As and CuB) constitute the bimetallic site, and it is widely agreed that dioxygen binds to this site as the first step in O2-reduction and proton pumping. Two crystal structures of native and recombinant, oxidized ba3-type cytochrome c oxidase have been solved at <2.5 Å resolution (5, 6). However, the structure of reduced cytochrome ba3 oxidase has not been reported.
Oxidized and reduced forms of cytochrome c oxidase represent two important intermediates in its catalytic cycle, and one might expect differences in their structures. Particularly, the reduced enzyme should be structurally prepared to bind O2 (7, 8). However, such structures obtained so far for the A1-type (9) enzymes indicate very small, if any, changes near the redox centers (10-12). Reported structures of both redox states from the cytochrome aa3 of Paracoccus denitrificans also revealed essentially no changes at 3.0 to 3.3 Å resolution (10), while in the case of bovine heart cytochrome c oxidase (also of the aa3-type, and at a higher resolution), a redox-linked conformational change was observed in a surface loop (13) not present, for example, in ba3.
We recently reported the structure of a doubly-mutated form of cytochrome ba3 (II-E4Q/I-K258R) that forms robust crystals in just a few days due to a new lattice contact (14). This form of the enzyme is used throughout this work to examine properties and structures of the chemically-reduced enzyme. By recording optical absorption spectra from single crystals before and after exposure to the X-ray beam, we gained new information on the redox state of the two hemes. Small but significant, and possibly important structural changes were detected around the Fea3 - CuB dinuclear center. These changes, observed for defined spectroscopic states, support a direct, as opposed to an indirect (15), mechanism for the B-type oxidases (see Fee et al. (7)). The structures provide important new information on how the enzyme functions.
Experimental Procedure
Protein expression, purification and crystallization
The mutant (II-E4Q/I-K258R) cytochrome ba3 oxidase from Thermus thermophilus was expressed and purified as previously described (14, 16) and used throughout this study. Detergent exchange was conducted using DEAE resin (GE Science) in Econo-Column (Biorad)(14). Purified protein sample was eluted and concentrated in 13 mM n-Nonyl-β-D-glucopyranoside (2×CMC) in 10 mM Bis-Tris pH 7.0 with 0.1 M KCl. The oxidized protein was crystallized under the same conditions as the recombinant wild type (6). Crystals ranged from ∼250 to ∼400 microns along their largest dimension. The concentration of enzyme within the unit cell is ∼6 mM. In order to crystallize reduced mutant cytochrome ba3, 5 - 10 mM sodium dithionite (Sigma, La Jolla, CA, U.S.A) was used. Moreover, all solutions used in the experiment were degassed and the entire crystallization / archiving procedure was conducted in a glove box at < 1 ppm O2 (COY Laboratory Products Inc, U.S.A) filled with a gas mixture of 10 % hydrogen and 90 % nitrogen.
Single-crystal microspectrophotometry
Absorbance spectra of crystals mounted in an Oxford cryo-stream operating at ∼100 K were recorded using either an in-house system based on an Ocean Optics USB4000 spectrometer or a modified 4DX system (17) installed at SSRL on beamline 7-1, which utilizes an Ocean Optics QE65000 spectrometer. All crystals were exposed to degassed cryo-protectant, typically 50 % methylpentanediol, for ∼10 seconds before cryo-cooling and recording spectra. In the X-ray radiation-induced reduction experiments, crystal orientation was chosen to produce a satisfactory spectrum, and this was maintained in subsequent measurements. Spectral figures were prepared using Igor Pro software.
X-ray data collection
Where indicated, the cryo-protectant used previously (14) was supplemented with sodium dithionite (10 mM). This level of reductant was included to prevent oxidation of the enzyme during the time that the crystals were being transferred to liquid nitrogen. All datasets were collected on beamline 7-1 at SSRL (Stanford Synchrotron Radiation Laboratory, U.S.A) at ∼100 K, a wavelength of 0.979 Å, and an approximate total flux of 1 × 1013 photons/minute.
Structure determination and refinement
All diffraction data were processed with the programs MOSFLM and SCALA (CCP4 suite (18). The wild-type, recombinant structure (PDB code1XME) was the initial model for molecular replacement with PHASER (19). The solved structures were then refined using rigid refinement and further restrained refinement with REFMAC 5.0 (20). For all datasets, 5 % of the reflections were randomly selected to provide a test set for Rfree calculations (21). Water molecules were placed in ≥ 3 σ positive peaks in the Fo - Fc map and refined. Structure models and electron density maps were displayed with the program COOT (22).
Coordinates of Fea3 and CuB were refined against the anomalous difference data with MLPHARE (18) to independently determine the distance between them (f’ = 0.3 e-, f” =1.5 e- for Fe; f’ = 0.0 e- and f” = 2.2 e- for Cu at 0.979 Å). The SCALA mtz files containing |F +| and | F -| to the resolution limit of each data set were used with a 6σΔF cutoff. The observed standard deviation of the coordinate shift of the Fea3 - CuB sites is very small for each data set, with the estimated error for the metal-metal distance of ∼ ± 0.03 Å. However, the metal-metal distances vary from data set to data set, i.e., different crystals, and for this reason they are reported only to the first decimal place (see Table SI_1). Refinement of Xe and Kr positions within ba3 was carried out in a similar manner, see Ref. (8).
The program VOIDOO (23) was utilized to compute the O2-channel to include the cavity formed in the dinuclear site in the reduced structure (grid for plot files, 0.7 Å; probe radius, 1.0 Å). Figures were created using PyMOL (24). Details of data collection statistics and final refinement statistics are given in Table 1.
Table 1.
Data collection and refinement statistics
| A-path | B-path | C-path | |
|---|---|---|---|
| Beam line (SSRL) | 7-1 | 7-1 | 7-1 |
| Resolution (Å) | 3.10 (3.18-3.10) | 2.90 (2.98-2.90) | 2.80 (2.87-2.80) |
| P41212 cell dimensions (Å) | |||
| a = b | 119.75 | 120.85 | 115.63 |
| c | 150.81 | 150.40 | 149.17 |
| Solvent content (%) | 54.9 | 55.5 | 51.0 |
| Total no. of observations | 54047 | 96770 | 95862 |
| Unique reflection set | 19624 | 25168 | 25479 |
| Redundancy | 2.8 (2.3) | 3.8 (4.2) | 3.8 (4.0) |
| Completeness (%) | 96.3 (96.3) | 99.4 (99.4) | 99.6 (99.6) |
| <I/σI> | 5.9 (1.8) | 9.1 (1.8) | 7.1 (1.8) |
| Rsym* (%) | 7.4 (39.9) | 3.6 (35.4) | 4.4 (38.1) |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.024 | 0.022 | 0.020 |
| Bond angles (°) | 2.7 | 2.4 | 2.2 |
| Rwork/Rfree** (%) | 20.8/30.7 | 20.8/27.8 | 20.9/27.1 |
| PDB code | 3EH3 | 3EH4 | 3EH5 |
Values in parentheses are for the last shell.
Rsym = Σ |I - < I >|/ Σ I, where I is the integrated intensity for a reflection.
Rfree is the conventional crystallographic R factor, Rwork, calculated on 5% of the data exclude from the refinement.
Results
Crystals of reduced cytochrome ba3 suitable for X-ray diffraction studies can be obtained (A-path) by treatment of crystals of oxidized, as-isolated protein with a cryoprotectant solution containing excess dithionite followed by plunging into liquid nitrogen, (B-path) by X-ray irradiation of crystals of oxidized protein maintained in a cryostream, and (C-path) by growth of crystals of dithionite-reduced protein in an anaerobic environment prior to freezing in liquid nitrogen. Optical absorption spectra of crystals so-obtained are shown in Fig. 1(A- , B-, and C-paths), and spectra of a solution of ba3 are shown in the lower set of Fig. 1C-path; more extensive solution spectra are presented in (3) and (25).
Fig. 1.
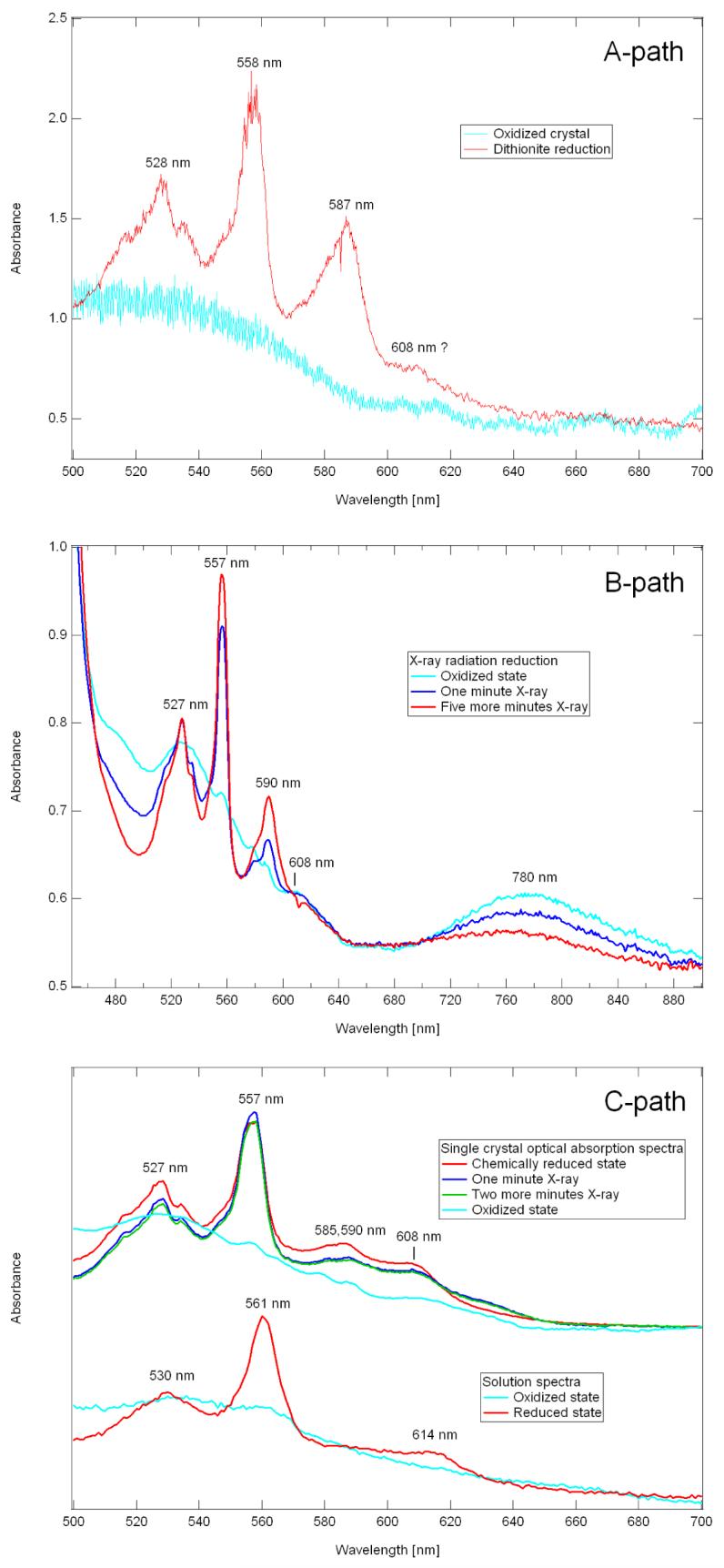
A-path. Optical absorption spectra recorded from two different crystals of cytochrome ba3 from T. thermophilus. Spectrum of a crystal grown from oxidized (which is the same as the as-isolated enzyme) cytochrome ba3, cryoprotected in the absence of dithionite and cooled to 100 K in a cryostream (cyan trace). Spectrum of a crystal grown from oxidized, as-isolated cytochrome ba3, cryoprotected in the presence of ∼10 mM dithionite and cooled to 100 K in a cryostream (red trace), i.e., Path-A. Spectra were recorded at TSRI using the Ocean Optics USB4000 spectrophotometer. B-path. Effects of 1 Å X-ray radiation on the absorption spectra of an oxidized crystal of cytochrome ba3 from T. thermophilus at 100 K. The cyan trace shows the optical absorption spectrum of this crystal prior to exposure to X-radiation. The blue trace shows the optical absorption spectrum of the same crystal after one minute of exposure to an X-ray beam (Path-B). The red trace, recorded after an additional five minutes of radiation indicates maximum radiation-induced reduction. Spectra were recorded using the Ocean Optics QE65000 spectrometer mounted at beamline 7.1 of SSRL. C-path. Effect of X-ray radiation on the 100 K optical absorption spectrum of a single crystal of reduced cytochrome ba3 from T. thermophilus (top set). The cyan trace is that of the oxidized crystal shown in Fig. 1B-path (top). The red trace was recorded from a different crystal grown from reduced protein (C-path, see text). The nearly overlapping blue and green traces were obtained after additional exposure to radiation. (lower set) Optical absorption spectra of oxidized (cyan) and dithionite reduced (red) recorded from detergent solubilized cytochrome ba3. Spectra were recorded as described in the legend to Fig. 1A-path using the Ocean Optics USB4000 spectrometer. Spectral figures were created with Igor Pro software.
Optical absorption spectra of single crystals
Crystals formed from the as-isolated enzyme and maintained at 100 K in a cryostream, exhibit a broad, generally featureless optical absorption spectrum (cyan traces in Fig. 1(A- , B-, and C-path)) comparable to spectra recorded in solution at room temperature (cyan trace, lower set, Fig. 1C-path). The broad absorption band at ∼780 nm, cyan trace in Fig. 1B-path, probably arises from the oxidized form of the weakly absorbing CuA center, although this feature is not observed in all crystals of as-isolated enzyme that we have examined.
(A-path)
When a crystal of oxidized enzyme at room-temperature is exposed to a cryoprotectant solution containing ∼10 mM dithionite for ∼10 seconds prior to being frozen in liquid nitrogen, its optical spectrum shows distinct features that are reasonably assigned to the 6cLS ferrous heme-b, ∼528 nm and ∼558 nm (Fig. 1A-path). In addition, an intense but unexpected absorption envelope appears near ∼587 nm.
(Path-B)
As shown by the blue and red traces of Fig. 1B-path, a crystal of as-isolated, oxidized enzyme, maintained in a cryostream at ∼100 K, becomes reduced upon exposure to the X-ray beam. Exposure for as little as one minute (see Experimental) caused the appearance of the sharp absorption bands (∼527 nm and ∼557 nm) due to the 6cLS ferrous heme-b, and an additional five minutes of exposure to the beam caused only a modest (≈ 20 %) further increase in these absorption bands. Here again, the unexpected ∼590 nm band is also seen, but probably because the bands appear sharper in this configuration (see Experimental), it has a shoulder at ∼585 nm.
(C-path)
Spectra of crystals obtained from dithionite reduced enzyme, cryo-cooled in liquid nitrogen and examined at 100 K, also showed strong absorption bands due to reduced heme-b (∼527 nm and ∼557 nm), relatively weak absorption near ∼585,590 nm, and yet a new and stronger band near ∼608 nm (Fig. 1C-path).
We were concerned that additional exposure to X-ray radiation would cause further changes in the optical spectrum. However, beyond the possible emergence of a broad band at ∼630 nm, this did not occur, indicating that one-electron reduction is not followed by, for example, further reduction of the porphyrin π-system (Fig. 1C-path). With this concern allayed, we determined the structures of the two, chemically-reduced forms of cytochrome ba3, i.e., those obtained by A- and C-paths, and compared them to the structure resulting from the (B-path).
X-ray crystallographic structures
The rms differences between the structure of ba3 in chemically reduced crystals (A-path), crystals of the as-isolated protein (B-path), and crystallized reduced protein (C-path) are of the order, ∼0.3 Å. A few residues located on the surface of the protein show slightly larger deviations, because they reside in flexible loops. No significant change was observed at the CuA, the CuB, or the heme-b metals. However, small changes do occur in and around the dinuclear site (Fig. 2). These include movement of the Fea3-atom as well as possible changes in the interaction between the guanidinium group of I-Arg449 and the D-ring propionate of heme-a3 while the interaction between the A-ring propionate O1D to ND1 of I-His376 is unchanged. Further work is required to determine if these small changes are important to the enzyme’s mechanism.
Fig. 2.
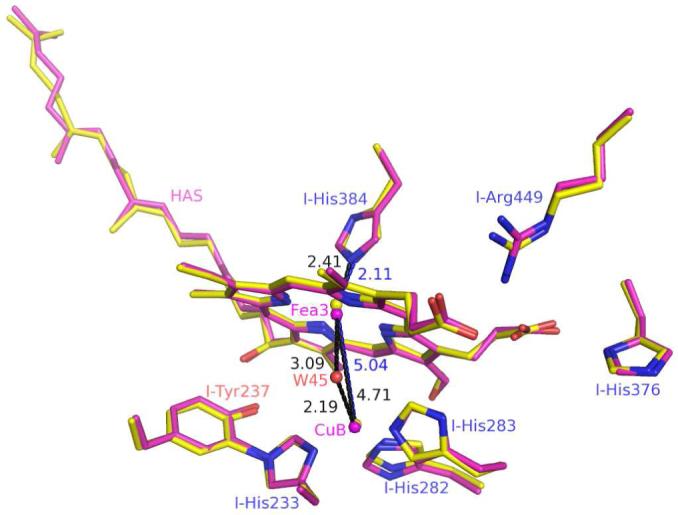
Superimposition of binuclear site structures of T. thermophilus cytochrome ba3 in oxidized, as-isolated crystals (magenta, reduced by the B-path) and in crystals grown from chemically reduced protein (C-path, yellow). The two structures were superimposed using COOT and then displayed with PyMOL. Additional metric data are given in Table SI_1.
In order to avoid bias in restrained refinement and more accurately determine the distance between iron and copper atoms, anomalous difference maps were created (Figs. 3a - c). At high contour levels (4 σ), these illustrate the shift in metal positions. The Fea3 - CuB distances were derived by refining coordinates of the two atoms against the anomalous scattering data (see Methods). Metric comparisons of the dinuclear center in the chemically reduced structure with that in the as-isolated cytochrome ba3 structure are presented in Table SI_1. The distance between Fea3 and CuB, which is 4.7 Å in crystals of as-isolated protein (Path-B), increases to 5.1 Å and 5.0 Å, respectively, in reduced crystals (Path-A) and in crystals grown from chemically reduced protein (Path-C). The shorter 4.7 Å distance of the B-path structure is comparable to the 4.4 Å distance obtained previously from higher-resolution structures that were studied under different conditions (Table SI_1, and PDB codes 1XME and 1EHK).
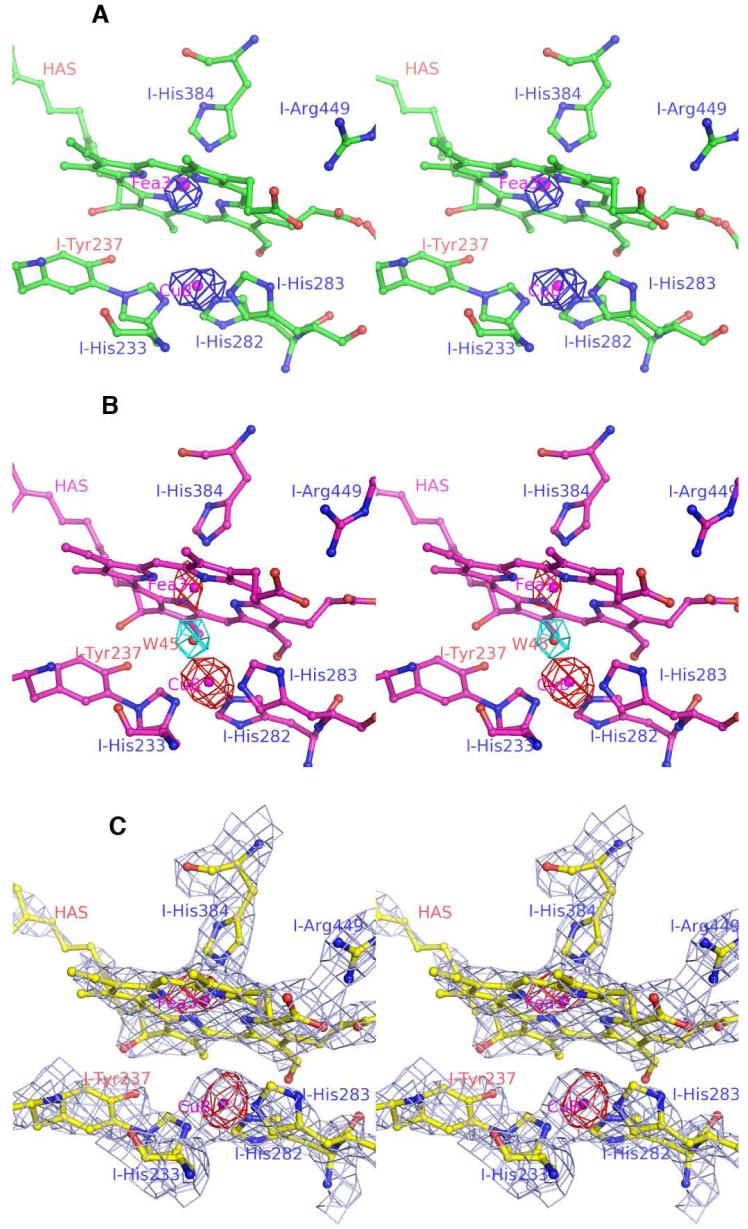
Fig 3.
Stereo-views (cross-eye) of the binuclear sites of three T. thermophilus cytochrome ba3 oxidase structures: (A) the crystallized, chemically-reduced (see Fig. 1A-path for spectra, Path-A); (B) the crystallized, X-ray radiation-reduced corresponding to previously reported structures considered at the time to be “oxidized” (see Fig. 1B-path for spectra). One water molecule at the binuclear site (cyan Fo - Fc ‘omit’ map contoured at 4-sigma) is present in Path-B crystals; (C) chemically-reduced, crystallized enzyme (see Fig. 1C-path for spectra). The 2Fo - Fc map illustrates the clean absence of electron density (2 σ) between Fea3 and CuB in the C-path structure. The anomalous difference maps are contoured at 4-sigma for the CuB and Fea3.
In the structures reported here, the greater Fea3 - CuB separation results mainly from movement of the Fea3-atom out of the plane of the porphyrin toward the NE2-atom of I-His384, while the CuB atom remains in its original position (Fig. 2). This movement of the Fea3-atom is accommodated and perhaps even caused by shortening of the I-His384 NE2 to Fea3 interaction ∼ 0.3 Å in the path-A and -C structures, suggesting that I-His384 and CuB are anchored in protein-space while the Fea3-atom can move along the normal to the heme plane. Such differences may find their origin in the chemistry that is occurring in the reaction chamber (see for example (7)). Note that in the structure of cytochrome aa3 of Rhodobacter sphaeroides reported by Qin et al. (26) there appear to be two O-atoms in the as-isolated enzyme.
The other significant finding is that the water molecule residing between CuB and Fea3 in the B-path structure (Fig. 3b and PDB codes 1EHK and 1XME) is no longer present in either structure of chemically-reduced ba3 (Paths A and C, Fig. 3a and 3c). A 2Fo - Fc map (at 1 σ, not shown) indicates no connection between the two metals. Fig. 3b shows the |Fo|- |Fc | map (at 4 σ) indicating the presence of a strong water peak in the dinuclear center of the X-ray radiation-reduced structure (i.e. Path-B). The |Fo|- |Fc | maps for the Path-A and Path-C structures also show no significant features between the Fea3 and the CuB sites. These data indicate that an O-atom resides between CuB and Fea3 in the oxidized, as-isolated structure, but they do not reveal to which metal this atom is coordinately bonded prior to radiation reduction (see Discussion).
Discussion
Recent studies with other heme proteins (see (27-29) and references therein) have provided evidence for spectral and/or structural changes induced by intense X-ray radiation. The chemical, spectral and structural results described here for Thermus cytochrome ba3 can be understood in terms of the following: First, crystalline, oxidized cytochrome ba3 can be fully reduced by a short incubation in buffered cryo-solution containing ∼10 mM Na2S2O4, or alternatively, crystals can be formed from reduced protein in dithionite-containing mother liquor and maintained O2-free. Second, because electrons formed during X-ray photolysis of water (27) are strongly reducing, it might have been expected that synchrotron derived X-ray beams, with their high photon flux, would result in reduction of the metal centers of cytochrome c oxidases (30-32). However, as shown in Fig. 1C-path, the X-ray beam does not significantly alter the heme chromophores beyond the level of reduction already effected by dithionite. Third, atom motions that might normally accompany reduction of the enzyme are likely to be resisted by the protein itself and possibly(?) crystal lattice forces, both of which will be less inclined to undergo change at 100 K. This is particularly important following Path-B. If the radiation-induced reduction of metal-centers leads to unstable reduced states, e.g., those having high affinity for protons (see Fee et al. (7) for review), then movement of these within the protein may create forces that are resisted under these conditions, thereby resulting in non-equilibrium structures. In contrast, when exposing crystals to dithionite at room temperature, the metal centers may relax to the states observed along (Paths A- or C-). Applying these facts and concepts allows a tentative interpretation of both spectral and structural data.
Spectral changes
Upon chemical reduction of the enzyme in solution, all sites will be reduced: CuA, heme-b, and the CuB - Fea3 dinuclear center (3, 16, 25, 33), and each heme will exhibit a unique absorption spectrum. Heme-b is a typical 6cLS electron carrier, known to be in rapid redox equilibrium with CuA (25). Its unique optical spectrum serves as a sensitive indicator of enzyme reduction, and its features are similar in crystals obtained along each path as well as in solution (Fig. 1C-path).
By contrast, heme-a3 contributes unique, path-dependent spectra in the form of the ∼585, 590 nm envelope in all crystalline forms, and the ∼608 nm peak observed in path-C crystals. Absorption of light at ∼590 nm is commonly observed with the 6cLS forms of ferrous heme-a3 such as its CO (34) and CN--complexes (35). However, all the structures reported here clearly demonstrate the absence of a 6th ligand to Fea3. Hence, the ∼590 nm envelope cannot be due to a 6cLS ferrous heme-a3. Although the reason for a spin change is not known, we tentatively propose that a 5cLS electronic configuration of ferrous heme-a3 is responsible for the ∼585,590 nm envelope in all the crystal forms. Such an unusual electronic configuration could result from structural trapping at 100 K. It is important to note that these optical transitions have been observed only at low temperature in the crystalline state.
Crystals from the C-path, however, show rather little ∼585,590 nm absorption compared to paths A and B crystals but instead have a significant band at ∼608 nm. We suggest this represents the 5cHS electronic configuration of ferrous heme-a3 and would correspond to the low-temperature, crystal-shifted version of the solution absorption band (Fig. 1C-path).
Structural changes
Each path for obtaining reduced, crystalline ba3 leads to overall, similar structures. However, each form is spectrally distinct and therefore, at some level, it must also be structurally different. Heme-a3 and CuB, forming the O2-reduction / H+-pumping site must pass through numerous electronation / protonation states during catalysis (7, 36, 37) Small structural changes will be outside the current resolution, but they are likely to be of importance in enzyme function. Two real differences between chemical- and radiation-reduction are movement of Fea3 away from CuB by ∼0.3 Å and loss of the intermetal O-atom. These changes are likely to be mechanistically important because they prepare the reaction chamber to bind an O2 molecule. Reduction of the metals leads to an increase in the volume of the reaction chamber and causes the appearance of apposing, open-coordination positions on those metals, which appear perfectly evolved to bind O2 and reduce it to a bridging peroxo dianion (see Ref. 100 of (7)). As summarized in Table SI_1, a similar CuB - Fea3 distance change also occurs in A1-type oxidases.
As might be expected, the O2-channel (8), should extend to include the space between Fea3 and CuB, which is the ultimate binding site for dioxygen. Fig. 4 shows the VOIDOO representation of this extended channel with a blue ball to mark the penultimate O2-binding site (The Xe1 position described in Ref. (8)).
Fig. 4.
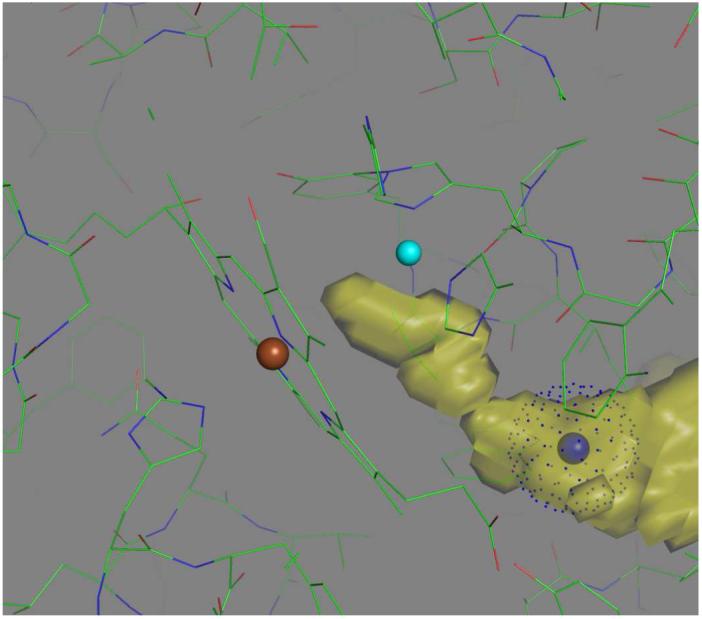
Extension of the O2-channel (yellow, see Luna et al. (8)) in chemically reduced, Thermus ba3 oxidase, as determined using VOIDOO (see Methods). Colored balls mark the positions of CuB (cyan), Fea3 (brown), and Xe1 (blue) Dots associated with the latter indicate the van der Waal radius of Xe. Figure created using PyMOL.
Finally, the nature of the intermetal O-atom in the oxidized protein is potentially of great mechanistic importance because its position and protonation state (O2-, HO-, or H2O°) will affect the overall charge and the distribution of charge throughout the active site (7).
Supplementary Material
Acknowledgements
We thank Dr. Elizabeth Getzoff for the use of her microscope objectives, crystal mounting equipment, and cryo control system and Dr. David Goodin for the use of his Ocean Optics USB4000 spectrophotometer.
Supported by NIH grants GM35342 (JAF). Part of this research was carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the US Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences.
Abbreviations
- 5cLS
five-coordinate low-spin
- 5cHS
five-coordinate high-spin
- 6cLS
six-coordinate low-spin electronic configuration
References
- (1).Richter O-MH, Ludwig B. Cytochrome c oxidase - structure, function, and physiology of a redox driven molecular machine. Rev. Physiol. Biochem. Pharmacol. 2003;147:47–74. doi: 10.1007/s10254-003-0006-0. [DOI] [PubMed] [Google Scholar]
- (2).Brunori M, Giuffre A, Sarti P. Cytochrome c oxidase, ligands and electrons. J. Bioinorg. Chem. 2005;99:324–326. doi: 10.1016/j.jinorgbio.2004.10.011. [DOI] [PubMed] [Google Scholar]
- (3).Zimmermann BH, Nitsche CI, Fee JA, Rusnak F, Munck E. Properties of a copper-containing cytochrome ba3: a second terminal oxidase from the extreme thermophile Thermus thermophilus. Proc. Nat’l. Acad. Sci. U. S. A. 1988;85:5779–5783. doi: 10.1073/pnas.85.16.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Oertling WA, Surerus KK, Einarsdottir O, Fee JA, Dyer RB, Woodruff WH. Spectroscopic characterization of cytochrome ba3, a terminal oxidase from Thermus thermophilus: Comparison of the a3 / CuB site to that of bovine cytochrome aa3. Biochemistry. 1994;33:3128–3141. doi: 10.1021/bi00176a048. [DOI] [PubMed] [Google Scholar]
- (5).Soulimane T, Buse G, Bourenkov GB, Bartunik HD, Huber R, Than ME. Structure and mechanism of the aberrant ba3-cytochrome c oxidase from Thermus thermophilus. EMBO J. 2000;19:1766–1776. doi: 10.1093/emboj/19.8.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Hunsicker-Wang LM, Pacoma RL, Chen Y, Fee JA, Stout CD. A novel cryoprotection scheme for enhancing the diffration of crystals of the recombinant, integral-membrane cytochrome ba3 from Thermus thermophilus. Acta Cryst. 2005;D61:340–343. doi: 10.1107/S0907444904033906. [DOI] [PubMed] [Google Scholar]
- (7).Fee JA, Case DA, Noodleman L. Toward a chemical mechanism of proton pumping by the B-type cytochrome c oxidases: Application of density functional theory to cytochrome ba3 of Thermus thermophilus. J. Am. Chem. Soc. 2008;130:15002–15021. doi: 10.1021/ja803112w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Luna VMM, Chen Y, Fee JA, Stout CD. Crystallographic studies of Xe and Kr binding within the large internal cavity of cytochrome ba3 from Thermus thermophilus: Structural analysis and role of oxygen transport channels in the heme-Cu oxidases. Biochemistry. 2008;47:4657–4665. doi: 10.1021/bi800045y. [DOI] [PubMed] [Google Scholar]
- (9).Pereira MM, Santana M, Teixeira M. A novel scenario for the evolution of haem-copper oxygen reductases. Biochim. Biophys. Acta. 2001;1505:185–208. doi: 10.1016/s0005-2728(01)00169-4. [DOI] [PubMed] [Google Scholar]
- (10).Harrenga A, Michel H. The cytochrome c oxidase from Paracoccus denitrificans does not change the metal center ligation upon reduction. J. Biol. Chem. 1999;274:33296–33299. doi: 10.1074/jbc.274.47.33296. [DOI] [PubMed] [Google Scholar]
- (11).Tsukihara T, Shimokata K, Katayama Y, Shimada H, Muramoto K, Aoyama H, Mochizuki M, Shinzawa-Itoh K, Yamashita E, Yao M, Ishimura Y, Yoshikawa S. The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process. Proc. Natl. Acad. Sci. USA. 2003;100:15304–15309. doi: 10.1073/pnas.2635097100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Yoshikawa S, Shinzawa-Itoh K, Tsukihara T. Crystal structure of bovine heart cytochrome c oxidase at 2.8 A resolution. Journal of Bioenergetics & Biomembranes. 1998;30:7–14. doi: 10.1023/a:1020595108560. [DOI] [PubMed] [Google Scholar]
- (13).Tsukihara T, Shimokata K, Katayama Y, Shimada H, Muramoto K, Aoyama H, Mochizuki M, Shinzawa-Itoh K, Yamashita E, Yao M, Ishimura Y, Yoshikawa S. The Low-Spin Heme of Cytochrome C Oxidase as the Driving Element of the Proton-Pumping Process. Proc. Nat. Acad. Sci. U.S.A. 2003;100:15304–15309. doi: 10.1073/pnas.2635097100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Liu B, Luna VM, Chen Y, Stout CD, Fee JA. An unexpected outcome of surface engineering an integral membrane protein: improved crystallization of cytochrome ba3 from Thermus thermophilus. Acta Cryst. 2007;F63:1029–1034. doi: 10.1107/S1744309107054176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Michel B, Bosshard HR. Oxidation of cytochrome c by cytochrome c oxidase: Spectroscopic binding studies and steady-state kinetics support a conformational transition mechanism. Biochemistry. 1989;28:244–252. doi: 10.1021/bi00427a034. [DOI] [PubMed] [Google Scholar]
- (16).Chen Y, Hunsicker-Wang LM, Pacoma RL, Luna E, Fee JA. A homologous expression system for obtaining engineered cytochrome ba3 from Thermus thermophilus HB8. Protein Expression& Purification. 2005;40:299–318. doi: 10.1016/j.pep.2004.11.014. [DOI] [PubMed] [Google Scholar]
- (17).Hadfield AT, Hajdu J. A fast and portable microspectrophotometer for time-resolved X-ray diffraction experiments. J. Appl. Cryst. 1993;26:839–842. [Google Scholar]
- (18).Bailey S. Acta Crystallographica Section D-Biological Crystallography. 1994. pp. 760–763. [DOI] [PubMed] [Google Scholar]
- (19).Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Cryst. D. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- (20).Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. D. 1999;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- (21).Brunger AT. X-PLOR Version 3.0. Yale University; New Haven, C.T: 1992. [Google Scholar]
- (22).Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- (23).Kleywegt GJ, Jones TA. Detection, delineation, measurement, and display of cavities in macromolecular structures. Acta Cryst. 1994;D50:178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- (24).DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- (25).Farver O, Chen Y, Fee JA, Pecht I. Electron transfer among the CuA-, heme b- and heme a3-centers of Thermus thermophilus cytochrome ba3. FEBS Lett. 2006;580:3417–3421. doi: 10.1016/j.febslet.2006.05.013. [DOI] [PubMed] [Google Scholar]
- (26).Qin L, Hiser C, Mulichak A, Garavito RM, Ferguson-Miller S. Identification of conserved lipid/detergent-binding sites in a high-resolution structure of the membrane protein cytochrome c oxidase. Proc. Nat. Acad. Sci. U.S.A. 2006;103:16117–16122. doi: 10.1073/pnas.0606149103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Garman EF, Owen RL. Cryocooling and radiation damage in macromolecular crystalography. Acta Cryst. 2006;D62:32–47. doi: 10.1107/S0907444905034207. [DOI] [PubMed] [Google Scholar]
- (28).Unno M, Chen H, Kusama S, Shaik S, Ikeda-Saito M. Structural characterization of the fleeting ferric peroxo species in myoglobin, Experiment and theory. J. Am. Chem. Soc. 2007;129:13394–13395. doi: 10.1021/ja076108x. [DOI] [PubMed] [Google Scholar]
- (29).Denisov IG, Victoria DC, Sligar SG. Cryoradiolytic reduction of heme proteins: Maximizing dose-dependent yield. Rad. Phys. Chem. 2007;76:714–721. doi: 10.1016/j.radphyschem.2006.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272:1136–44. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- (31).Iwata S, Ostermeier C, Ludwig B, Michel H. Structure at 2.8 A resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995;376:660–9. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- (32).Svensson-Ek M, Abramson J, Larsson G, Törnath S, Brzezinski P, Iwata S. The X-ray crystal structures of wild-type and EQ(I-286) mutant cytochrome c oxidases from Rhodobacter sphaeroides. J. Mol. Biol. 2002;321:329–339. doi: 10.1016/s0022-2836(02)00619-8. [DOI] [PubMed] [Google Scholar]
- (33).Sousa FL, Verissimo AF, Baptista AM, Soulimane T, Teixeira M, Pereira MM. Redox properties of Thermus thermophilus ba3: different electron-proton coupling in oxygen reductases? Biophys. J. 2008;94:2434–2441. doi: 10.1529/biophysj.107.122614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Goldbeck RA, Dawes TD, Einarsdottir O, Woodruff WH, Kliger DS. Time-resolved magnetic circular dichroism spectroscopy of photolyzed carbonmonoxy cytochrome c oxidase. Biophys. J. 1991;60:125–134. doi: 10.1016/S0006-3495(91)82036-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Surerus KK, Oertling WA, Fan C, Gurbiel RJ, Einarsdottir O, Antholine WE, Dyer RB, Hoffman BM, Woodruff WH, Fee JA. Reaction of cyanide with cytochrome ba3 from Thermus thermophilus: spectroscopic characterization of the Fe(II)a3-CN.Cu(II)B-CN complex suggests four 14N atoms are coordinated to CuB. Proc. of the Nat’l. Acad. Sci. U. S. A. 1992;89:3195–3199. doi: 10.1073/pnas.89.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Siletsky S, Soulimane T, Azarkina N, Vygodina TV, Buse G, Kaulen A, Konstantinov A. Time-resolved generation of a membrane potential by ba3 cytochrome oxidase from Thermus thermophilus. FEBS Lett. 1999;457:98–102. doi: 10.1016/s0014-5793(99)01019-4. [DOI] [PubMed] [Google Scholar]
- (37).Smirnova IA, Zaslavsky D, Fee JA, Gennis RB, Brzezinski P. Electron and proton transfer in the ba3 oxidase from Thermus thermophilus. J. Bioenerg Biomembr. 2008;40:281–287. doi: 10.1007/s10863-008-9157-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.