Abstract
Adeno-associated virus (AAV) is a promising gene transfer vector tested in both animal studies and human clinical trials. However, current production methods are generally inefficient and require improvements to meet the increasing clinical need for economical, high titer and high quality rAAV vectors. The inefficiency of the current systems largely arises from the AAV helper function, which contains only the AAV coding region but lacks inverted terminal repeats. The terminal repeats were originally removed to prevent replication competent AAV contamination. Here we designed a novel and highly efficient rAAV helper function containing AAV terminal repeats. The new helper function not only mimics the wild-type AAV growth as it replicates along with the vector plasmid, but also restores the cis regulating function of the AAV terminal repeats. Addition of heterologous introns to the helper genome and use of a mutant AAV terminal repeat defective in packaging effectively controls the contamination of replication competent AAV particles. This new strategy also performs better in AAV producing cell lines than those based on non-replicating AAV rep and cap genome.
Keywords: AAV, adeno-associated virus, helper, replication competent AAV, hRCAAV
Introduction
Adeno-associated virus (AAV) is a defective nonpathogenic human parvovirus. Its single-stranded DNA genome consists of approximately 4.7 kb.1 Efficient replication and viral gene expression of AAV genome requires co-infection with a second helper virus, such as adenovirus or herpesvirus. In the absence of helper functions, AAV genomes integrate into the host chromosome to establish a latent infection. Such integration events have been shown to occur in a site-specific manner.2,3 In the case of the most studied AAV serotype 2, approximately 70% of integration occurs in a narrow region in human chromosome 19q13.3-qter.4,5 Upon superinfection of helper adenovirus, AAV provirus rescues from its latent state and enters its lytic life cycle. The AAV life cycle is regulated through a complicated system involving host factors, helper virus, genes encoded in the AAV genome, and cis element inverted terminal repeats (ITRs).6 ITRs are palindromic sequences which can assume a T shaped hairpin structure. This special configuration serves as the origin for viral DNA replication. In addition, the ITRs are essential for successful virus packaging, integration and rescue. The four non-structural proteins designated as Rep 78, Rep 68, Rep 52 and Rep 40 are the only trans elements in the AAV genome which are involved in the regulation of these events. At the 3′ end of AAV genome, the cap gene encodes three overlapping structural proteins VP1, VP2 and VP3. The ratio of VP1, VP2 and VP3 in the AAV virion is approximately 1:1:10.
The function of AAV ITRs in AAV replication, rescue, integration and packaging has been studied extensively.6,7 The Rep78 and Rep68 can both bind to the ITR at GAGY tandem repeats and nick duplex ITR in a site- and strand-specific manner.8,9 This cleavage reaction within the ITR allows the continuous replication of the AAV genome.7 The only non-palindromic sequence in the ITR is the 20-nucleotide D sequence. The D sequence is critical for AAV replication and packaging. Both the terminal resolution site (TRS) and AAV packaging signal localize to the D sequence. A single ITR with two D sequences permits the replication and packaging of the AAV genome.10 However, in the absence of the D sequence, the hairpin structures are insufficient for the encapsidation of the AAV genome.11 Although exact nucleotide sequences are not conserved among the identified AAV ITRs from various AAV serotypes, the overall T shaped hairpin structure is maintained.1,12-15 Generally, the Rep proteins from one AAV serotype can complement the ITRs from another AAV serotype. The only exception is AAV5, which has the least homology with any of the known AAV serotypes and its ITR has a unique TRS site.13,16
AAV-derived recombinant vectors represent an attractive gene delivery system for gene therapy due to their unique features such as lack of pathogenicity, the ability to transduce nondividing cells and stable long-term gene expression in a variety of tissues including skeletal muscle, photoreceptors, liver, and neurons from central nervous system.17-24 However, economical large-scale production of high quality rAAV remains a bottleneck for AAV vector applications. The traditional rAAV production system involves the transfection of an AAV vector plasmid and an AAV helper plasmid in the presence of a helper virus function.25 The vector plasmid contains AAV ITRs and a transgene cassette. The helper plasmid contains the AAV rep and cap gene, but not ITRs. Progress has been made to increase rAAV yield through regulating the AAV rep and cap gene expression, eliminating the cytolytic helper adenovirus and using the SV40 ori to increase helper plasmid copy number.26-29 Additional strategies to minimize the contamination of replication competent AAV particles (rcAAV) have also been proposed and studied, including replacement of intact ITRs in the vector construct with a truncated D sequence D10 and splitting rep and cap sequences to different helper plasmids when making an AAV packaging cell line.30,31 These various methods have been shown to be effective in previous studies. Recently, we developed the intronized AAV helper to increase rAAV yield, while concurrently eliminating the generation of rcAAV contamination.32 On the basis of this previous study, we designed a completely new strategy for the rAAV production. This system is based on a replication competent AAV helper function (hRCAAV). Unlike all previous AAV helper plasmids, the AAV ITR was retained in the helper plasmid. The contamination of rcAAV was controlled by using intronized AAV genome and ITRs deficient in packaging, but still capable of supporting AAV replication. The efficacy of this novel strategy was demonstrated in production systems based on both transfection and infection.
Results
Strategy for generating replication competent AAV helper
Our strategy is based on the common observation that wild-type AAV growth is much more efficient than current recombinant AAV production systems. We hypothesize that the main deficiency of rAAV production is due to the fact that the AAV ITRs are removed from the AAV helper. This deletion results in the loss of the regulating function of AAV ITRs in AAV gene expression. Moreover, it also leads to the major difference between wildtype AAV growth and rAAV production in terms of the copies of rep and cap genes during the AAV growth process. In neither transfection-based systems nor systems with integrated AAV helper genomes can the copies of rep and cap genes synchronize to the replication of vector sequences. Although at the early stage of wild-type AAV growth only a limited number of templates for transcription are available, it is sufficient to supply necessary Rep proteins for massive replication of the AAV genome. In the late stage of AAV growth, when large amounts of AAV Cap proteins are required for the packaging process, the already replicated AAV genomes allow the high level of cap expression to meet the demand of efficient encapsidation. In contrast, in the transfection-based rAAV production system, there are large numbers of AAV helper genomes available right after the transfection procedure. This results in the oversupply of Rep proteins in the early stage of rAAV production. However, in the late stage of AAV packaging, AAV templates start to decrease because of cell division. This imbalance between rep and cap may contribute to the inefficiency of the current AAV production systems. The modulation of AAV helper rep and cap gene expression was very efficient at improving rAAV packaging.26 In the case of integrated AAV helpers, reports showed that cell lines harboring rep and cap sequence could undergo inducible amplification by adenovirus infection.33-40 Cell lines with this property worked better in producing rAAV vectors. However, they also appear to be dependent on integration loci. For rep sequence carried by other replicating virus or vectors, it will not be able to mimic wild-type AAV growth.41-43 Since ITRs are critical for AAV replication, they are removed from current AAV helpers in order to prevent the generation of rcAAV contamination. However, recent progress in identification of AAV ITR components required for AAV packaging and our invention of an intronized AAV helper has allowed us to design a new class of AAV helpers containing AAV ITRs.11,31,32 An illustration of these helper constructs is shown in Figure 1. These AAV helpers may achieve high efficiency rAAV packaging by mimicking wild-type AAV growth. The rcAAV contamination was controlled by increasing the AAV helper genome through heterologous intron insertion and utilizing an AAV mutant ITR that was defective for encapsidation, but still supported AAV replication.
Figure 1.
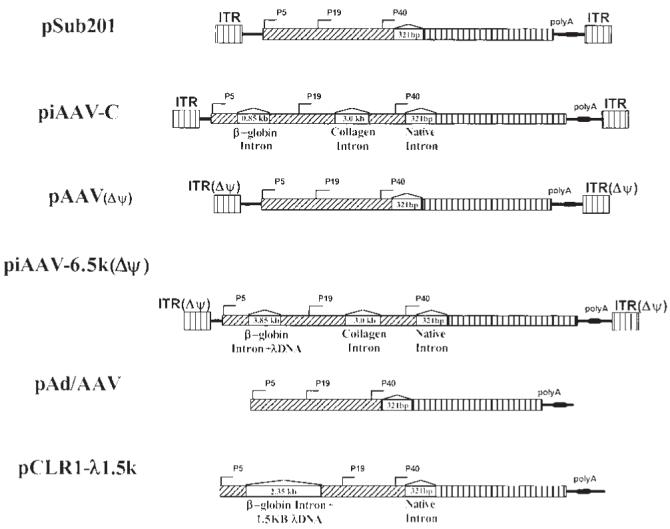
An illustration of components of AAV helper constructs. Key elements are identified in the figure. Note that the sizes of the plasmids are not to scale. Δψ indicates mutation in ITR that affects AAV packaging, but still supports helper replication.
Replication competent helpers support wild type free rAAV production in transfection systems
To test whether the replication competent AAV helpers support AAV production, all constructs listed in Figure 1 were used to make rAAV-CAG-EGFP. The rAAV vector yield and wtAAV contamination are shown in Table 1. The plasmid pSub201 is an AAV infectious clone. It is very efficient in replicating and packaging itself and therefore, produces large amounts of wild-type AAV particles. Wild-type AAV could be detected even after 1:106 dilution. However, the rAAV vector yield from using pSub201 was about two to three logs lower than those from other AAV helpers. A simple heterologous intron insertion to the AAV genome was able to increase the rAAV titer to the level of pAd/AAV, because its genome was oversized to 160% of the wtAAV genome (piAAVC). However, the replication competent AAV contamination was still a problem. On the other hand, replacing the ITR in pSub201 with a packaging-defective, replication-permissive AAV ITR mutant, pAAV (Δψ), could greatly improve rAAV yield and also minimize the contamination of replication competent AAV. The replication of this form of mutant ITR which lacks the 15 nucleotides distal to the AAV terminal repeat hairpin has been demonstrated by Srivastava’s group (refer to Figures 2 and 3 in Ref. 44).11,31,45 Plasmid piAAV-6.5k(Δψ) carries two heterologous introns and AAV packaging-defective ITRs. The combination of these two approaches could further improve the rAAV yield dramatically over our previous intronized helper pCLR1-λ1.5k without detectable rcAAV contamination.
Table 1.
A comparison of performance of various AAV helper constructs in a triple transfection AAV production method
| Helper | Size % of wtAAV | AAV ITR | Vector yield (units/ml) | wtAAV titer (dilution) |
|---|---|---|---|---|
| pSub201 | 100 | Yes | (3.58 ± 1.71) × 104 | 106 |
| piAAV-C | 165 | Yes | (1.7 ± 0.4) × 106 | 104 |
| pAAV(Δψ) | 100 | Yes* | (6.1 ± 0.6) × 107 | 102 |
| piAAV-6.5k (Δψ) | 240 | Yes* | (5.1 ± 0.6) × 107 | ND |
| pAd/AAV | 100 | No | (3.3 ± 1.0) × 106 | 10 |
| pCLR1-λ1.5k | 150 | No | (8.6 ± 2.2) × 106 | ND |
Helper plasmids listed in the table were co-transfected along with pAAV-CAG-EGFP and mini adenovirus plasmid pFΔ13 at a ratio 1:1:2 to approximately 2 × 106 293 cells, respectively. The cells were harvested 4 days after transfection and resuspended in 1 ml 10 mM Tris.Cl (pH 8.0), frozen and thawed three times. The vector yield was determined by monitoring GFP expression in 293 cells. The wild-type AAV titer was determined by serial dilution of 2 μl cell lysate and infecting 293 cells in the presence of adenovirus infection at MOI of 5. The total cellular DNA was isolated and subjected to real-time PCR amplification of the rep sequence. At least 10-fold difference between the samples in the presence or absence of adenovirus was scored as rep replication competent.
Figure 2.
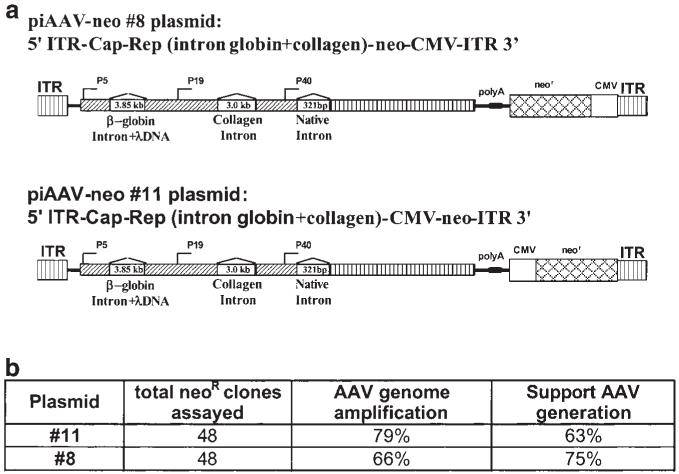
Establishment of hRCAAV cell lines. (a) Illustration of plasmids for the establishment of stable cell lines. (b) List of cell clones obtained from hRCAAV helper. The column ‘AAV genome amplification’ indicates the clones that allow the amplification of AAV genome in the presence of adenovirus infection assayed by quantitative PCR.
Figure 3.
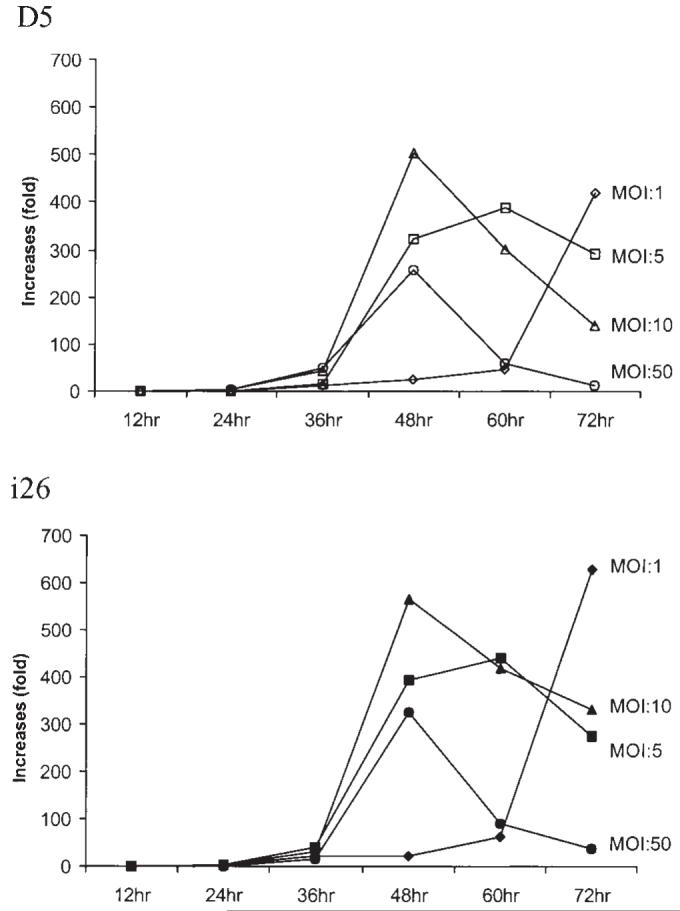
Characterization of AAV gene amplification in i26 and D5. Cell line D5 (with integrated wild-type AAV genome) and hRCAAV helper cell clone i26 were infected by adenovirus at various MOI. The control group was not infected. Cells were harvested at the indicated time after adenovirus infection. Total cellular DNA was isolated and AAV DNA was quantitated by real-time PCR. The Y-axis shows amplification of AAV sequence relative to that of the control group without infection by adenovirus.
Establishment and characterization of replication competent AAV helper cell lines
The establishment of highly efficient AAV packaging cell lines expressing rep and cap is often a challenge and very labor-intensive, because AAV Rep proteins are highly cytotoxic when overexpressed in mammalian cells. For example, the B50 cell line reported by Gao et al46 was obtained by screening hundreds of cell clones. Given AAV’s biological feature of establishing an efficient latent infection in the absence of helper virus, we hypothesized that the replication competent AAV helper could resemble this feature of wild-type AAV. Upon the presence of helper virus, the integrated AAV genome can be rescued from the host chromosome. Therefore, the integration loci should no longer remain a major concern for the performance of the helpers. The constructs illustrated in Figure 2 were used to establish the cell lines. After G418 selection, approximately 70% of neomycin-resistant clones were identified to be AAV replication competent and capable of supporting rAAV growth (Figure 2). The two different constructs were similarly efficient at generating hRCAAV cell lines. These cell clones remained stable after consecutive passages.
One of the hRCAAV cell clones, i26, was chosen to compare its helper replication to that of Detroit 5 (D5), a cell line with integrated wild-type AAV genome. Both D5 and i26 were infected with helper adenovirus at various MOI. The increase of AAV copy number was determined by quantitative PCR. As shown in Figure 3, D5 and i26 exhibited similar amplification curves under similar conditions.
We also characterized the performance of i26 in supporting rAAV production. To avoid the discrepancy of transfection, the cis element was supplied by infecting i26 and D5 cell lines with rAAV-CAG-EGFP. The vectors were harvested at 72 h after adenovirus infection. As shown in Figure 4a, the AAV vector yield from i26 cell lines was several logs higher than that of the D5 cell line at various MOI of adenovirus infection. The rcAAV contamination was well controlled in the i26 cell line as shown in Figure 4b.
Figure 4.
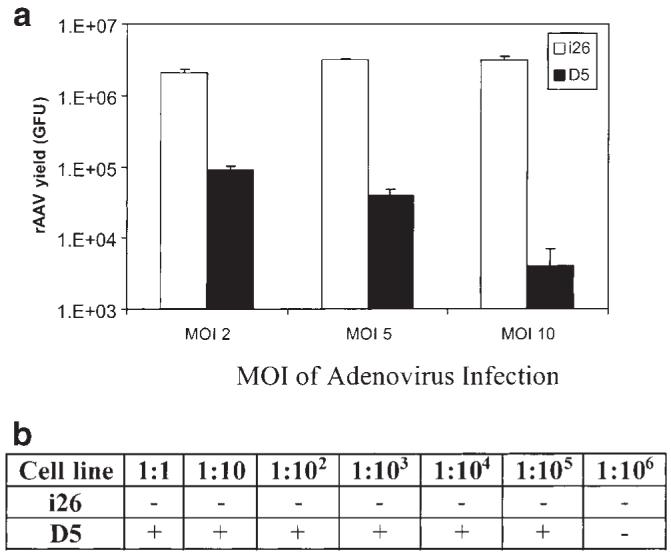
hRCAAV cell lines support rAAV production. Approximately 1.5 × 105 D5 or i26 cells were infected with adenovirus at MOI 2, 5 and 10, respectively. At 24 h after adenovirus infection, cells were further infected with rAAV-CAG-GFP and harvested 48 h after rAAV infection. The rAAV yield and replication competent AAV contamination were determined as described in Table 1. The graph shows the vector yield and the table summarizes the rcAAV contamination detected at various dilutions.
The performance of the hRCAAV cell line was studied under a variety of conditions. One of the major factors affecting the vector yield was the adenovirus infection. For example, the time interval between rAAV infection and adenovirus infection needed to be optimized. When rAAV and adenovirus were used to infect i26 cells simultaneously, the vector yield was low (Figure 5). The vector yield was significantly increased when adenovirus infection took place 36 h before rAAV infection, which is consistent with the occurrence of high-level amplification of the AAV genome (Figure 3). However, very few if any vectors can be obtained if the adenovirus infection occurs 48 h before rAAV infection because the severe cytotoxicity derived from the long-term exposure to adenovirus infection does not allow for efficient rAAV production before harvesting.
Figure 5.
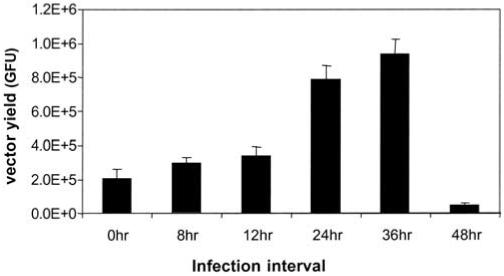
Timing of adenovirus infection affects the performance of hRCAAV cell line. Approximately 1.5 × 105 i26 cells were infected with adenovirus at the MOI of 5. Cells were further infected with rAAV-CAGGFP at the indicated time after adenovirus infection and harvested 72 h after adenovirus infection. The rAAV yield was determined as described in Table 1. The X-axis shows the time of rAAV infection relative to the infection of adenovirus.
We compared the gene expression profile between the replication competent AAV cell line i26, D5 and transfection of 293 cells with pAd/AAV. Equal amounts of D5, i26 cells and pAd/AAV transfected 293 cells were infected with adenovirus at MOI of 10. Total RNA was isolated at various time points and subjected to quantitative RT-PCR analysis. The rep transcription and cap transcription in D5 and i26 showed similar dynamics (Figure 6). Transcription in both cells remained at a low level until the late stage and reached their peak at 48 h following adenovirus infection. However, the transcription in 293 cells transfected with pAd/AAV was significantly different. The rep transcription continued to go up and reached a peak at 48 h after adenovirus infection. Its relative expression level during the early stage was much higher than that of i26 or D5. On the other hand, the cap expression quickly reached its peak at 12 h after transfection followed by the drop in transcription, with only 15% of the peak level retained at 48 h after infection. Such dramatic differences in the expression pattern of rep and cap genes may be a reason that the transfection-based AAV production systems are not as efficient as that of wild-type AAV growth.
Figure 6.
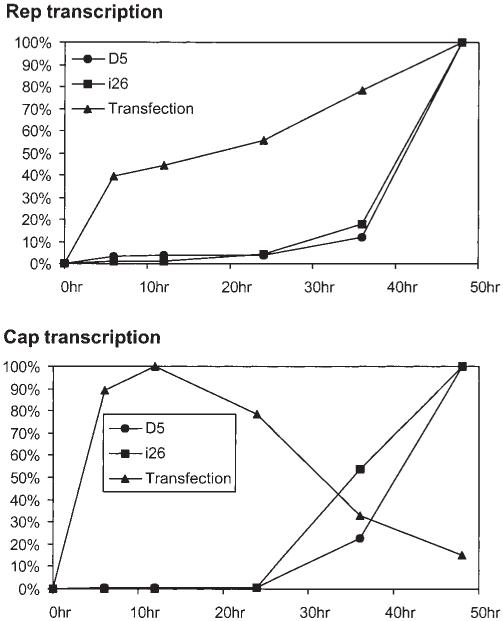
hRCAAV helper cell line can mimic wt AAV gene expression. Helper plasmid pAd/AAV was transfected into 293 cells. The transfected 293 cells, untransfected i26 or D5 cells were then infected with adenovirus at the MOI of 10. Cells were harvested at the indicated time points after Ad infection. Total RNA were isolated and subjected to quantitative RTPCR using primers specific to rep and cap transcript, respectively. The figure shows the amount of rep and cap transcripts at various time points relative to its peak level.
Discussion
In our current study, we have demonstrated a new class of AAV helpers that is capable of mimicking wild-type AAV growth and, moreover, possesses the potential of maximum efficiency in providing AAV transgene products for AAV packaging. These AAV helpers carry AAV ITRs and are capable of replicating themselves. The selfpackaging of such helpers was disabled through dramatic oversizing of the helper’s genome by the insertion of heterologous introns, as well as the use of encapsidationdefective mutant AAV ITRs. The data in this study suggest that this system can not only efficiently produce rAAV vectors, but also limit rcAAV contamination.
The inclusion of ITRs in the packaging system allowed the replication of an AAV helper genome. However, the amplification of helper sequences may not be the only advantage for improving rAAV packaging. The presence of ITRs in the AAV helper may also restore the critical cis element for efficient temporal and spatial regulation of AAV gene expression.47 As reported by Weger et al,47 the AAV ITR is required for the enhancement of p40 gene expression in the presence of helper virus. The p40 gene products are generally considered to be one of the limiting factors for AAV packaging. This new modification may have improved rAAV packaging efficiency through this mechanism, even though we did not observe an obvious increase in cap expression by Western blot (data not shown). As demonstrated in Table 1, utilizing of hRCAAV helper functions could increase the vector yield up to 20-fold without any modulation of rep gene expression, which is in contrast to the ACG helper generated by Li et al.26
Another advantage of the replication competent helper is that the stable cell lines harboring hRCAAV helpers are much easier to establish as shown in Figure 2b. As wild-type AAV is able to establish latent infection by the integration of its genome into the host chromosome, the hRCAAV helpers would be likely to reach similar efficiency since they carry functional ITRs. In addition, the hRCAAV helpers should be capable of rescuing from their integration loci and provide the rep and cap expression. Therefore, the performance of hRCAAV cell lines may be relatively independent of the integration loci. This is a unique feature in contrast to other established AAV helper cell lines whose performances are completely determined by the integration loci such as B50 and H32. It is worth noting that certain AAV cell clones devoid of ITRs could also undergo AAV genome amplification upon adenovirus infection.33-40,48 This property has been shown to improve their efficiency in producing rAAV vectors. It may be due to the integration loci in host chromosomes where some elements resemble AAV ITRs in function. One known site is the human chromosome 19 preintegration region AAVS1, which has been shown to be amplified upon adenovirus infection and rep expression. The hRCAAV helper, however, will be able to rescue and replicate much more frequently because the majority of clones possessed this property (see Figure 2b).
The key to the application of hRCAAV helper is to prevent the packaging of hRCAAV helper itself. As a proof of principle, we used the same type of ITR as in the vector plasmid. Given the homology between the helper constructs and the vector plasmids, there may remain the risk of generating fully functional ITRs attached to the helper constructs by homologous recombination. The use of AAV5 ITRs may help to overcome this problem because it shares little homology with AAV2 (Dr Richard Jude Samulski, UNC at Chapel Hill, NC, USA, personal communication). The modulation of AAV packaging via heterologous intron insertion can be exploited as a second safety measure to prevent hRCAAV contamination.32 In our current report, only two introns were inserted into the coding region of the rep gene. Theoretically, a variety of combinations of intron insertions and other types of sequence insertions to make the helper extraordinarily large49-51 can potentially eliminate the possibility of hRCAAV contamination derived from revertants arisen from nonhomologous recombination events.
Replication competent helpers have been studied extensively for gutted adenovirus vector production systems which generally employ cre-loxP recombination to control helper virus contamination.52-54 Based on published data, it is very difficult to achieve vector stocks completely devoid of helper virus. Our new hRCAAV helper scheme appears to be more flexible and has more room for manipulation to meet the requirement for large scale AAV vector production. Although the cre-loxP system cannot be readily adopted for AAV production, the heterologous intron insertion would be a universal method to exploit the packaging limit of DNA viruses and reduce the contamination of wild-type virus, which could also be readily adopted in the gutted adenovirus production system. Another immediate application is to use it for autonomous parvovirus vector generation, which closely resembles rAAV production.55,56
Materials and methods
Cell culture
Human 293 cells, Detroit 5 cells and HeLa cells were maintained as monolayer cultures in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal calf serum, 100 μg of penicillin/ml, 100 U of streptomycin/ml as recommended by the manufacturer (GIBCO, Gaithersburg, MD, USA).
Plasmid construction
Plasmids pCLR1-λ1.5k and piAAV-C containing insertions of heterologous introns in the AAV coding region were described elsewhere.32 Plasmid pAAV (Δψ) was made by the following procedures. First, plasmid pSub201 was digested with MscI and XhoI. The fragment with hairpin sequence of ITR at both ends was then recovered and ligated to the annealed oligos of DS1 and DA2 (DS1: 5′ CCA ACT CCG 3′; DA2: 5′ AAT TCG GAG TTG 3′). After ligation, the fragment obtained was filled-in with Klenow DNA polymerase and further ligated with the fragment containing the AAV genome without ITRs, which was obtained from pAd/AAV by XbaI digestion and subsequently filling in. The obtained ITR lacks the last 15 nucleotides distal to the hairpin structure. A similar method was used to construct piAAV-6.5k (Δψ). Plasmid pCMV-neo was made by self-ligating the large fragment from pcDNA3.1(-) (Invitrogen, Carlsbad, CA, USA) digested with EcoRV and SmaI. The plasmid pCMV-neo was digested with SalI and then ligated to the backbone of pSub201 which had been digested with SnaBI before obtaining the pAM/CMV-neo-SV40 polyA. To make plasmids piAAV-neo (Nos 8 and 11) for generation of the stable cell line (see Figure 2), the XbaI fragment containing the AAV genome including two heterologous introns (globin intron + lambda DNA sequence and collagen intron) was inserted into the Acc65I site (filled in with Klenow) in the pAM/CMV-neo-SV40 polyA. In Nos 8 and 11, the neo cassette was inserted in the opposite orientation.
Recombinant AAV production by triple-transfection
Helper plasmids were co-transfected with vector plasmids and mini adenovirus plasmid pFΔ13 at the ratio 1:1:2 to 2 × 106 293 cells, respectively. The cells were harvested 4 days after transfection. The cells were resuspended in 1 ml 10 mM Tris pH 8.0, frozen and thawed three times. The vectors were then purified by CsCl gradient or used directly.
Recombinant AAV production by co-infection
In cell lines in which rep and cap genes have been integrated into the host chromosome, the rAAV vectors were made by infecting the host cells with purified rAAV vectors and wild-type adenovirus at MOI 5. The cells were harvested at 72 h after infection when full CPE appeared. The cells were resuspended in 10 mM Tris pH 8.0, frozen and thawed three times followed by incubation at 56°C for 30 min to inactivate the adenovirus. The rAAV yield and replication competent AAV contamination were determined as described elsewhere in this section. Experiments comparing the titer of different AAV helpers were repeated at least twice using different preparations of plasmids.
AAV titer determination
The rAAV infectious titer was determined by measuring GFP expression in 293 cells. Each green cell under the fluorescence microscope represents one IU (or GFU). Positive cells in the different dilutions were counted using a calibrated microscope (ocular diameter, 3.1 mm2) and then multiplied by the area of the well and the dilution of the virus. Data were represented as the average number of positive cells in a minimum of 10 fields per well.
Replication competent AAV determination
The titer of wild-type AAV or replication competent AAV was determined by serial dilution of 2 μl cell lysate and infecting 293 cells in the absence or presence of adenovirus infection at MOI of 5. The total cellular DNA was isolated 48 h after infection. Real-time PCR was then used to detect the amplification of the rep sequence. At least 10-fold difference between the samples in the presence or absence of adenovirus infection was scored as rep replication competent.
Establishment of replication competent AAV (hRCAAV) cell line
The two piAAV-neo plasmids were transfected to HeLa cells using LipofectAmine (GIBCO BRL) according to the manufacturer’s instructions, respectively. Cells were passed at 48 h after transfection and selected with 600 μg/ml Genecitin (GIBCO BRL). Single cell clones were selected and expanded for further experiments. The established cell clones were no longer maintained in selection medium.
Assay of the rescue and amplification of AAV genome sequence in cell lines
Cell clones were seeded in 24-well plates. Each well contained approximately 5 × 104 cells. One set was infected by adenovirus at MOI of 10 and the other was not infected to serve as control. Cells were harvested when full CPE occurred. Total cellular DNA was isolated and subjected to real-time PCR amplification of the rep sequence using primers (AAV-F, AAC AAG GTG GTG GAT GAG TGC; AAV-R, ACG CCC ACT GGA GCT CAG). The quantitative PCR was carried out using PRISM/7700 Sequence Detector (PE Applied Biosystems, Foster City, CA, USA). The reactions were performed according to the instructions of the manufacturer using the SYBR Green PCR Core Reagent Kit (PE Biosystems). The cell clones demonstrating at least 10-fold difference between the samples in the presence or absence of adenovirus were scored as positive for AAV genome amplification and selected for further experiments.
Assay of AAV gene expression
Helper plasmid pAd/AAV was transfected to 2 × 106 293 cells using LipofectAmine. The transfected 293 cells, untransfected hRCAAV cell clone i26 or Detroit 5 (D5) cells were then infected with adenovirus at the MOI of 10. Cells were harvested at various times after Ad infection. Total RNA was isolated using TRIzol LS Reagent (GIBCO BRL) according to the manufacturer’s instructions. Reverse transcription reaction was performed using TaqMan Reverse Transcription Reagents (PE Biosystems). The cDNA obtained from each sample was subjected to quantitative PCR using primers specific to rep and cap transcript, respectively, (rep-F, 5′ CGG CAA GAG GAA CAC CAT CT 3′, rep-R, 5′ CCT CCG CGA TGT TGG TCT 3′; cap-F, 5′ GGG CCC TGC CCA CCT 3′, cap-R, 5′ TGT CGT TCG AGG CTC CTG A 3′). Taqman β-actin Control Reagents (PE Biosystems) were used as endogenous control. All quantitation results were normalized to a β-actin endogenous control. Data are shown as the percentage of their maximum expression level in each cell line.
References
- 1.Srivastava A, Lusby EW, Berns KI. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J Virol. 1983;45:555–564. doi: 10.1128/jvi.45.2.555-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Samulski RJ, et al. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 1991;10:3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kotin RM, et al. Site-specific integration by adeno-associated virus. Proc Natl Acad Sci USA. 1990;87:2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang CC, et al. Cellular recombination pathways and viral terminal repeat hairpin structures are sufficient for adeno-associated virus integration in vivo and in vitro. J Virol. 1997;71:9231–9247. doi: 10.1128/jvi.71.12.9231-9247.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rutledge EA, Russell DW. Adeno-associated virus vector integration junctions. J Virol. 1997;71:8429–8436. doi: 10.1128/jvi.71.11.8429-8436.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muzyczka N. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr Topic Microbiol Immunol. 1992;158:97–129. doi: 10.1007/978-3-642-75608-5_5. [DOI] [PubMed] [Google Scholar]
- 7.Berns KI. Parvoviridae: the viruses and their replication. In: Fields FN, Knipe DM, Howley PM, editors. Fundamental Virology. Lippincott-Raven; Philadelphia, PA: 1995. pp. 1007–1041. [Google Scholar]
- 8.Brister JR, Muzyczka N. Rep-mediated nicking of the adenoassociated virus origin requires two biochemical activities, DNA helicase activity and transesterification. J Virol. 1999;73:9325–9336. doi: 10.1128/jvi.73.11.9325-9336.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weitzman MD, Kyostio SR, Kotin RM, Owens RA. Adeno-associated virus (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc Natl Acad Sci USA. 1994;91:5808–5812. doi: 10.1073/pnas.91.13.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao X, Xiao W, Li J, Samulski RJ. A novel 165-base-pair terminal repeat sequence is the sole cis requirement for the adeno-associated virus life cycle. J Virol. 1997;71:941–948. doi: 10.1128/jvi.71.2.941-948.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang XS, Qing K, Ponnazhagan S, Srivastava A. Adeno-associated virus type 2 DNA replication in vivo: mutation analyses of the D sequence in viral inverted terminal repeats. J Virol. 1997;71:3077–3082. doi: 10.1128/jvi.71.4.3077-3082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao W, et al. Gene therapy vectors based on adeno-associated virus type 1. J Virol. 1999;73:3994–4003. doi: 10.1128/jvi.73.5.3994-4003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiorini JA, Kim F, Yang L, Kotin RM. Cloning and characterization of adeno-associated virus type 5. J Virol. 1999;73:1309–1319. doi: 10.1128/jvi.73.2.1309-1319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rutledge EA, Halbert CL, Russell DW. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. J Virol. 1998;72:309–319. doi: 10.1128/jvi.72.1.309-319.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muramatsu S, Mizukami H, Young NS, Brown KE. Nucleotide sequencing and generation of an infectious clone of adeno-associated virus 3. Virology. 1996;221:208–217. doi: 10.1006/viro.1996.0367. [DOI] [PubMed] [Google Scholar]
- 16.Chiorini JA, Afione S, Kotin RM. Adeno-associated virus (AAV) type 5 Rep protein cleaves a unique terminal resolution site compared with other AAV serotypes. J Virol. 1999;73:4293–4298. doi: 10.1128/jvi.73.5.4293-4298.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samulski RJ. Adeno-associated virus: integration at a specific chromosomal locus. Curr Opin Genet Dev. 1993;3:74–80. doi: 10.1016/s0959-437x(05)80344-2. [DOI] [PubMed] [Google Scholar]
- 18.Xiao X, Li J, Samulski RJ. Efficient long-term gene transfer into muscle tissue of immunocompetent mice by adeno-associated virus vector. J Virol. 1996;70:8098–8108. doi: 10.1128/jvi.70.11.8098-8108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jooss K, Yang Y, Fisher KJ, Wilson JM. Transduction of dendritic cells by DNA viral vectors directs the immune response to transgene products in muscle fibers. J Virol. 1998;72:4212–4223. doi: 10.1128/jvi.72.5.4212-4223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herzog RW, et al. Long-term correction of canine hemophilia B by gene transfer of blood coagulation factor IX mediated by adeno-associated viral vector. Nat Med. 1999;5:56–63. doi: 10.1038/4743. [DOI] [PubMed] [Google Scholar]
- 21.Kay MA, et al. Evidence for gene transfer and expression of factor IX in haemophilia B patients treated with an AAV vector. Nat Genet. 2000;24:257–261. doi: 10.1038/73464. [DOI] [PubMed] [Google Scholar]
- 22.During MJ, Leone P. Adeno-associated virus vectors for gene therapy of neurodegenerative disorders. Clin Neurosci. 1995;3:292–300. [PubMed] [Google Scholar]
- 23.During MJ, et al. An oral vaccine against NMDAR1 with efficacy in experimental stroke and epilepsy. Science. 2000;287:1453–1460. doi: 10.1126/science.287.5457.1453. [DOI] [PubMed] [Google Scholar]
- 24.Flotte TR, Carter BJ. Adeno-associated virus vectors for gene therapy. Gene Therapy. 1995;2:357–362. [PubMed] [Google Scholar]
- 25.Samulski RJ, Chang LS, Shenk T. Helper-free stocks of recombinant adeno-associated viruses: normal integration does not require viral gene expression. J Virol. 1989;63:3822–3828. doi: 10.1128/jvi.63.9.3822-3828.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Samulski RJ, Xiao X. Role for highly regulated rep gene expression in adeno-associated virus vector production. J Virol. 1997;71:5236–5243. doi: 10.1128/jvi.71.7.5236-5243.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrari FK, Xiao X, McCarty D, Samulski RJ. New developments in the generation of Ad-free, high-titer rAAV gene therapy vectors. Nat Med. 1997;3:1295–1297. doi: 10.1038/nm1197-1295. [DOI] [PubMed] [Google Scholar]
- 28.Vincent KA, Piraino ST, Wadsworth SC. Analysis of recombinant adeno-associated virus packaging and requirements for rep and cap gene products. J Virol. 1997;71:1897–1905. doi: 10.1128/jvi.71.3.1897-1905.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flotte TR, et al. An improved system for packaging recombinant adeno-associated virus vectors capable of in vivo transduction. Gene Therapy. 1995;2:29–37. [PubMed] [Google Scholar]
- 30.Allen JM, Halbert CL, Miller AD. Improved adeno-associated virus vector production with transfection of a single helper adenovirus gene, E4orf6. Mol Ther. 2000;1:88–95. doi: 10.1006/mthe.1999.0010. [DOI] [PubMed] [Google Scholar]
- 31.Wang XS, et al. Characterization of wild-type adeno-associated virus type 2-like particles generated during recombinant viral vector production and strategies for their elimination. J Virol. 1998;72:5472–5480. doi: 10.1128/jvi.72.7.5472-5480.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao L, Liu Y, During MJ, Xiao W. High-titer, wild-type free recombinant adeno-associated virus vector production using intron-containing helper plasmids. J Virol. 2000;74:11456–11463. doi: 10.1128/jvi.74.24.11456-11463.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clark KR, Voulgaropoulou F, Fraley DM, Johnson PR. Cell lines for the production of recombinant adeno-associated virus. Hum Gene Ther. 1995;6:1329–1341. doi: 10.1089/hum.1995.6.10-1329. [DOI] [PubMed] [Google Scholar]
- 34.Clark KR, Voulgaropoulou F, Johnson PR. A stable cell line carrying adenovirus-inducible rep and cap genes allows for infectivity titration of adeno-associated virus vectors. Gene Therapy. 1996;3:1124–1132. [PubMed] [Google Scholar]
- 35.Liu XL, Clark KR, Johnson PR. Production of recombinant adeno-associated virus vectors using a packaging cell line and a hybrid recombinant adenovirus. Gene Therapy. 1999;6:293–299. doi: 10.1038/sj.gt.3300807. [DOI] [PubMed] [Google Scholar]
- 36.Tamayose K, Hirai Y, Shimada T. A new strategy for large-scale preparation of high-titer recombinant adeno-associated virus vectors by using packaging cell lines and sulfonated cellulose column chromatography. Hum Gene Ther. 1996;7:507–513. doi: 10.1089/hum.1996.7.4-507. [DOI] [PubMed] [Google Scholar]
- 37.Chadeuf G, et al. Efficient recombinant adeno-associated virus production by a stable rep-cap HeLa cell line correlates with adenovirus-induced amplification of the integrated rep-cap genome. J Gene Med. 2000;2:260–268. doi: 10.1002/1521-2254(200007/08)2:4<260::AID-JGM111>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 38.Inoue N, Russell DW. Packaging cells based on inducible gene amplification for the production of adeno-associated virus vectors. J Virol. 1998;72:7024–7031. doi: 10.1128/jvi.72.9.7024-7031.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu X, et al. Selective Rep-Cap gene amplification as a mechanism for high-titer recombinant AAV production from stable cell lines. Mol Ther. 2000;2:394–403. doi: 10.1006/mthe.2000.0132. [DOI] [PubMed] [Google Scholar]
- 40.Salvetti A, et al. Factors influencing recombinant adeno-associated virus production. Hum Gene Ther. 1998;9:695–706. doi: 10.1089/hum.1998.9.5-695. [DOI] [PubMed] [Google Scholar]
- 41.Conway JE, et al. High-titer recombinant adeno-associated virus production utilizing a recombinant herpes simplex virus type I vector expressing AAV-2 Rep and Cap. Gene Therapy. 1999;6:986–993. doi: 10.1038/sj.gt.3300937. [DOI] [PubMed] [Google Scholar]
- 42.Conway JE, et al. Recombinant adeno-associated virus type 2 replication and packaging is entirely supported by a herpes simplex virus type 1 amplicon expressing Rep and Cap. J Virol. 1997;71:8780–8789. doi: 10.1128/jvi.71.11.8780-8789.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X, et al. High-titer recombinant adeno-associated virus production from replicating amplicons and herpes vectors deleted for glycoprotein H. Hum Gene Ther. 1999;10:2527–2537. doi: 10.1089/10430349950016861. [DOI] [PubMed] [Google Scholar]
- 44.Wang XS, Ponnazhagan S, Srivastava A. Rescue and replication of adeno-associated virus type 2 as well as vector DNA sequences from recombinant plasmids containing deletions in the viral inverted terminal repeats: selective encapsidation of viral genomes in progeny virions. J Virol. 1996;70:1668–1677. doi: 10.1128/jvi.70.3.1668-1677.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang XS, Ponnazhagan S, Srivastava A. Rescue and replication signals of the adeno-associated virus 2 genome. J Molec Biol. 1995;250:573–580. doi: 10.1006/jmbi.1995.0398. [DOI] [PubMed] [Google Scholar]
- 46.Gao GP, et al. High-titer adeno-associated viral vectors from a Rep/Cap cell line and hybrid shuttle virus. Hum Gene Ther. 1998;9:2353–2362. doi: 10.1089/hum.1998.9.16-2353. [DOI] [PubMed] [Google Scholar]
- 47.Weger S, Wistuba A, Grimm D, Kleinschmidt JA. Control of adeno-associated virus type 2 cap gene expression: relative influence of helper virus, terminal repeats and Rep proteins. J Virol. 1997;71:8437–8447. doi: 10.1128/jvi.71.11.8437-8447.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tessier J, et al. Characterization of adenovirus-induced inverted terminal repeat-independent amplification of integrated adeno-associated virus rep-cap sequences. J Virol. 2001;75:375–383. doi: 10.1128/JVI.75.1.375-383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hermonat PL, Muzyczka N. Use of adeno-associated virus as a mammalian DNA cloning vector: transduction of neomycin resistance into mammalian tissue culture cells. Proc Natl Acad Sci USA. 1984;81:6466–6470. doi: 10.1073/pnas.81.20.6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dong JY, Fan PD, Frizzell RA. Quantitative analysis of the packaging capacity of recombinant adeno-associated virus. Hum Gene Ther. 1996;7:2101–2112. doi: 10.1089/hum.1996.7.17-2101. [DOI] [PubMed] [Google Scholar]
- 51.McLaughlin SK, Collis P, Hermonat PL, Muzyczka N. Adenoassociated virus general transduction vectors: analysis of proviral structures. J Virol. 1988;62:1963–1973. doi: 10.1128/jvi.62.6.1963-1973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou HA, et al. Cre-expressing cell line and an E1/E2a doubledeleted virus for preparation of helper-dependent adenovirus vector. Mol Ther. 2001;3:613–622. doi: 10.1006/mthe.2001.0288. [DOI] [PubMed] [Google Scholar]
- 53.Parks RJ, et al. A helper-dependent adenovirus vector system: removal of helper virus by Cre-mediated excision of the viral packaging signal. Proc Natl Acad Sci USA. 1996;93:13565–13570. doi: 10.1073/pnas.93.24.13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mitani K, Graham FL, Caskey CT, Kochanek S. Rescue, propagation, and partial purification of a helper virus-dependent adenovirus vector. Proc Natl Acad Sci USA. 1995;92:3854–3858. doi: 10.1073/pnas.92.9.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maxwell IH, et al. Recombinant LuIII autonomous parvovirus as a transient transducing vector for human cells. Hum Gene Ther. 1993;4:441–450. doi: 10.1089/hum.1993.4.4-441. [DOI] [PubMed] [Google Scholar]
- 56.Brandenburger A, Russell S. A novel packaging system for the generation of helper-free oncolytic MVM vector stocks. Gene Therapy. 1996;3:927–931. [PubMed] [Google Scholar]