Abstract
Elongation of very long chain fatty acids (ELOVL)5 is one of seven mammalian fatty acid condensing enzymes involved in microsomal fatty acid elongation. To determine the in vivo substrates and function of ELOVL5, we generated Elovl5−/− mice. Studies using liver microsomal protein from wild-type and knockout mice demonstrated that the elongation of γ-linolenic (C18:3, n-6) to dihomo-γ-linolenic (C20:3, n-6) and stearidonic (C18:4, n-3) to ω3-arachidonic acid (C20:4, n-3) required ELOVL5 activity. Tissues of Elovl5−/− mice accumulated the C18 substrates of ELOVL5 and the levels of the downstream products, arachidonic acid (C20:4, n-6) and docosahexaenoic acid (DHA, C22:6, n-3), were decreased. A consequence of decreased cellular arachidonic acid and DHA concentrations was the activation of sterol regulatory element-binding protein (SREBP)-1c and its target genes involved in fatty acid and triglyceride synthesis, which culminated in the development of hepatic steatosis in Elovl5−/− mice. The molecular and metabolic changes in fatty acid metabolism in Elovl5−/− mice were reversed by dietary supplementation with arachidonic acid and DHA. These studies demonstrate that reduced ELOVL5 activity leads to hepatic steatosis, and endogenously synthesized PUFAs are key regulators of SREBP-1c activation and fatty acid synthesis in livers of mice.
Keywords: fatty acid synthesis, PUFAs, sterol regulatory element-binding protein, elongation of very long chain fatty acids, hepatic steatosis
Very long chain (⩾C18) saturated, monounsaturated, and PUFAs are synthesized by elongation and desaturation reactions that are carried out by enzymes located in the endoplasmic reticulum (ER) (1). Microsomal fatty acid elongation adds two-carbon units to pre-existing fatty acyl-CoAs using malonyl-CoA as the donor and NADPH as the reducing agent. Fatty acid elongation requires four sequential reactions: 1) condensation between fatty acyl-CoA and malonyl-CoA to generate 3-ketoacyl-CoA; 2) reduction of 3-ketoacyl-CoA using NADPH to form 3-hydroxyacyl-CoA; 3) dehydration of 3-hydroxyacyl-CoA to trans-2-enoyl-CoA; and 4) reduction of trans-2-enoyl-CoA to fully elongated acyl-CoA. Each reaction is catalyzed by different enzymes that are encoded by separate genes. Elongation of very long chain fatty acids (ELOVLs) are responsible for the rate-controlling and initial condensation reaction (2–4).
Seven ELOVL enzymes have been identified in mammals and are designated ELOVL1–7 (4–9). Each ELOVL has a distinct tissue distribution, and individual enzymes exhibit different fatty acid substrate preferences. ELOVL1 and ELOVL3 catalyze the condensation of very long chain (>C20) saturated or monounsaturated fatty acids (10). ELOVL2 elongates C20 and C22 PUFAs (4, 11) and ELOVL4 elongates fatty acids with chain lengths longer than C26 (12, 13). When expressed in yeast, mammalian ELOVL5 condenses a broad range of fatty acids including palmitoleic (C16:1, n-7), oleic (C18:1, n-9), γ-linolenic (C18:3, n-6), stearidonic (C18:4, n-3), arachidonic (C20:4, n-6) and eicosapentaenoic acid (EPA, C20:5, n-3) acids (8). ELOVL6 elongates long chain (C12–C16) saturated and monounsaturated fatty acids (4).
Very long chain PUFAs of the n-6 and n-3 series are key components of membrane phospholipids and are precursors of signaling molecules, such as prostanoids (14). In animal studies, feeding diets with high levels of EPA (C20:5, n-3) and docosahexaenoic acid (DHA) (C22:6, n-3) (fish oils) have multiple effects on lipid metabolism in liver, including the suppression of lipid synthesis and the activation of fatty acid oxidation (15, 16). The underlying mechanisms responsible for the metabolic actions of DHA and EPA are thought to be mediated by their ability to modulate the activity of key transcription factors that regulate lipid metabolism, including sterol regulatory element-binding protein (SREBP)-1c (17–25).
SREBP-1c is one of three SREBP family members, designated SREBP-1a, -1c, and -2 (26). All are membrane-bound transcription factors that regulate the gene expression of many genes encoding enzymes of lipid synthesis (27). The predominant forms present in liver are SREBP-1c and SREBP-2, which are specified by separate genes (28). SREBPs reside in the ER as inactive precursors; the active forms of SREBPs are released by sequential proteolytic cleavages that occur after transport to the Golgi. Once released from the membrane, the cleaved forms of SREBPs translocate to the nucleus where they activate the transcription of target genes (27). SREBP-1c preferentially activates genes involved in fatty acid and triglyceride synthesis, including ELOVL5 and ELOVL6, while SREBP-2 preferentially activates genes involved in cholesterol synthesis and the LDL receptor (27, 29).
SREBP-1c is transcriptionally induced by insulin. The induction of SREBP-1c transcription by insulin is dependent on the integrity of two liver X receptor α (LXRα) binding sites located in the promoter of SREBP-1c (22, 30, 31). Conversely, SREBP-1c transcription is suppressed by unsaturated fatty acids when they are added to the medium of cultured cells. In vitro, this suppression is a result of their ability to inhibit LXRα activity (21, 22, 32). Unsaturated fatty acids might also accelerate the degradation of SREBP-1c mRNAs and promote the degradation of the nuclear form of SREBP-1c through 26S proteosomes in rat hepatocytes (18, 33).
To determine whether endogenous very long chain PUFAs are involved in the regulation of SREBP-1c and fatty acid synthesis, Elovl5−/− mice were produced and characterized. Here, we demonstrate that the in vivo function of ELOVL5 is the elongation of C18:3, n-6 to C20:3, n-6 and C18:4, n-3 to C20:4, n-3 and that the reduction in PUFA synthesis associated with the loss of ELOVL5 derepresses SREBP-1c, resulting in the development of hepatic steatosis.
EXPERIMENTAL PROCEDURES
Materials and general methods
Plasma glucose and insulin were measured using an Autokit Glucose CII kit from Wako Chemicals USA, Inc. (Richmond, VA), and an Ultra Sensitive Rat Insulin ELISA Kit from Crystal Chem Inc. (Downers Grove, IL), respectively. Plasma free fatty acids were measured using a NEFA C kit from Wako Chemicals Inc. Plasma and liver cholesterol and triglycerides were measured as described (34).
Generation of Elovl5−/− mice
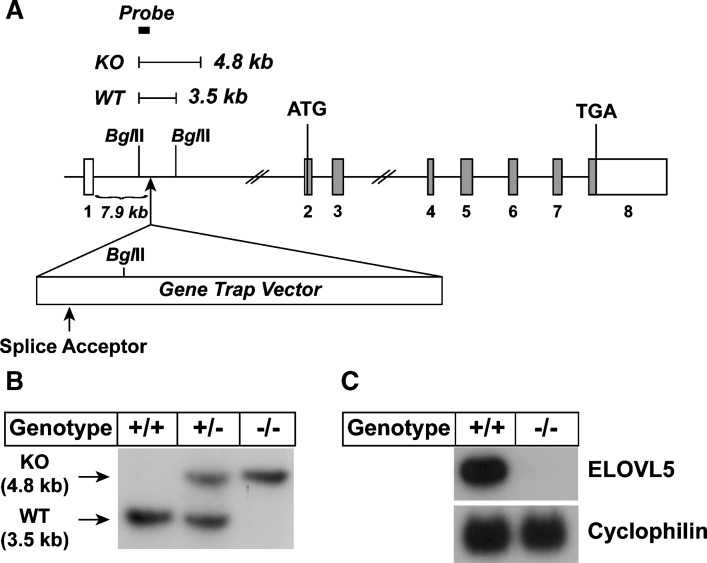
An ES cell clone (DTM098) was obtained from NIH/NHLBI-sponsored Bay Genomics ES cell gene trap resource (Mutant Mouse Regional Resource Center at UC Davis, Davis, CA). The ES cells were injected into C57BL/6J blastocysts, yielding chimeric mice as described (35). Chimeric male mice with over 90% agouti coat color were bred with C57BL/6J female mice (Jackson laboratory, ME) and germline transmission was confirmed by Southern blotting and PCR (35). Southern blotting was performed using BglII digested genomic DNA and the 32P-labeled probe generated by PCR using mouse genomic DNA as template and the following primers: 5′ primer, 5′-ATAGTCTGAACATAAGGCTTGTCTG-3′; and 3′ primer, 5′-ATAGAGGATGCTCCTTATGGAACCT-3′. The wild-type allele produced a 3.5 kb band, and the disrupted allele produced a 4.8 kb band. PCR was performed using genomic DNA prepared from tails with direct lysis kit (Viagen Biotech) and the following primers: 5′ primer, 5′-ACCACTAGTCATATGACGGC-3′; 3′ primer, 5′-TACAAAGCGTACAAAGGCTG-3′; primer 3, 5′-ATACAGTCCTCTTCACATCC-3′. The wild-type allele produced a 400 bp band, and the disrupted allele produced a 510 bp band.
The absence of an ELOVL5 transcript in Elovl5−/− mice was confirmed by Northern blot analysis using total RNA prepared from livers of wild-type and Elovl5−/− mice as described (34). A 32P-labeled ELOVL5 mouse cDNA that covered the entire coding region of ELOVL5 was used as the probe.
Mice were housed in colony cages and maintained on a 12-h light/12-h dark cycle and fed Teklad Mouse/Rat Diet 7002 from Harlan Teklad Premier Laboratory Diets (Madison, WI). All animal studies were approved by the UT Southwestern IACUC.
Diet studies
Oleic, arachidonic acid, and DHA were purchased from Nu-Chek prep (Elysian, MN) and were added to a chow diet (Teklad Mouse/Rat Diet 7001 from Harlan Teklad Premier Laboratory Diets, Madison, WI) as indicated. All diets were fed for 7 days. The first 4 days on the diet the mice were fed ad libitum, and the last 3 days they were given access to the food only during the dark cycle (7:00 PM–7:00 AM).
Measurement of liver free fatty acid and DAG concentrations
Frozen liver (100–200 mg) was homogenized in 4 ml Folch (choloforom/methanol, 2:1) and then mixed with 0.8 ml of saline. The organic phase was collected and evaporated under a N2 stream. The extracts were then dissolved in 0.5 ml of chloroform and immediately transferred to 500 mg aminopropyl Strata columns (Phenomenex, Torrance, CA) that were equilibrated with hexane. Nonpolar lipids were eluted with 6 ml of chloroform/isopropanol (2:1) and free fatty acids were eluted with 6 ml of diethyl ether/acetic acid (98:2). The eluates were dried under N2. Free fatty acids were dissolved in chloroform and the concentration measured using NEFA C kit. Diacylglycerols (DAG) were separated from the total lipid extracts using 500 mg-silica columns (Varian, Harbor City, CA). DAGs were eluted from the column with diethyl ether/hexane (30:70), dried under N2, and resuspended in chloroform. The concentration of DAG was measured as previously described for triglycerides (34).
Fatty acid elongation assay
ELOVL5 activity was measured in liver microsomes as described previously (4). Microsomes were prepared from wild-type and Elovl5−/− mice and the fatty acid elongation assay was performed using BSA-bound fatty acids as substrates and [14C]malonyl-CoA (American Radiolabeled Chemicals, Inc., St. Louis, MO) as described (4). Elongated fatty acids were extracted and incorporation of the radiolabeled malonyl-CoA was quantified.
Measurements of tissue fatty acid composition
Lipids from livers of individual mice were extracted and fractionated into cholesteryl esters, triglycerides, and phospholipids using 500 mg silica columns as described (36, 37). Fatty acids were methyl esterified and separated by gas-liquid chromatography (GLC) using a Hewlett Packard 6890 Series GC System (37). The identity of the fatty acid methyl esters was determined by comparing the retention times with fatty acid standards [Supelco 37 Component FAME Mix and PUFA-2, Animal Source (SUPELCO, Bellefonte, PA)]. The relative amount of each fatty acid was calculated in the fractionated lipid classes. To measure the concentration of fatty acids in total lipids, the lipid extracts were methyl esterified without fractionation. Pentadecanoic acid (C15:0) was added as an internal standard to the reaction, and the fatty acid concentration in total lipids was calculated based on the area of the C15:0 peak.
Immunoblot analyses
Membrane and nuclear proteins were prepared from frozen livers as described (38). Equal aliquots (8 μg) of protein from individual livers were pooled (total, 40 μg) and the proteins were subjected to SDS-PAGE on 8% gels and transferred to Hybond ECL membrane (GE Healthcare Bio-Sciences Corp., Piscataway, NJ). Immunoblot analyses were performed using polyclonal anti-mouse SREBP-1, SREBP-2, Insig-1, and Insig-2 antibodies, an anti-hamster SREBP cleavage-activating protein (SCAP) antibody, and an anti-human carbohydrate regulatory element-binding protein (ChREBP) antibody (Novus Biological, Inc., Littleton, CO) as described (34, 38, 39). Antibody-bound bands were detected using the SuperSignal CL-HRP substrate system (Pierce Biotechnology Inc., Rockford, IL). Anti-human CREB (cAMP response element binding protein, Invitrogen) and anti-human transferrin receptor (TFR) (Invitrogen) monoclonal antibodies were used as loading controls for liver nuclear and membrane proteins, respectively.
Quantitative real-time PCR
Total RNA was prepared from liver using RNA STAT-60 (TEL-TEST, Friendswood, TX). RNA was treated with DNase I using a DNA-free kit (Ambion Inc., Austin, TX), and first strand cDNA was prepared using a TaqMan Reverse Transcription Reagents kit (Roche Molecular Systems, Inc., Branchburg, NJ). Quantitative real-time PCR was performed as described (40). Specific primers for each gene were designed using Primer Express software (PE Biosystems) for the following genes: ELOVL1, 5′ primer, 5′-GTGGCCCAGCCCTACCTT-3′, and 3′ primer, 5′-TGTGCAGTGAGACCAGGACAA-3′; ELOVL2, 5′ primer, 5′-TCAATGCTTTCTTGGACAACATG-3′, and 3′ primer, 5′-GGTAAGAGTCCAGCAGGAACCA-3′; ELOVL3, 5′ primer, 5′-TTCTCACGCGGGTTAAAAATG-3′, and 3′ primer, 5′-GGGCCTTAAGTCCTGAAACGT-3′; ELOVL5, 5′ primer, 5′-ATGGACACCTTTTTCTTCATCCTT-3′, and 3′ primer, 5′-ATGGTAGCGTGGTGGTAGACATG-3′; GPAT, 5′ primer, 5′-CAACACCATCCCCGACATC-3′, and 3′ primer, 5′-GTGACCTTCGATTATGCGATCA-3′; AGPAT1, 5′ primer, 5′-GCTGGCTGGCAGGAATCAT-3′, and 3′ primer, 5′-GTCTGAGCCACCTCGGACAT-3′; AGPAT2, 5′ primer, 5′-TTTGAGGTCAGCGGACAGAA-3′, and 3′ primer, 5′-AGGATGCTCTGGTGATTAGAGATGA-3′; MGAT1, 5′ primer, 5′-GAGTAACGGGCCGGTTTCA-3′, and 3′ primer, 5′-AGACATTGCCACCTCCATCCT-3′; MGAT2, 5′ primer, 5′-GCCCATCGAGGTGCAGAT-3′, and 3′ primer, 5′-AGAGCTCCTTGATATAGCGCTGAT-3′; DGAT1, 5′ primer, 5′-GAGGCCTCTCTGCCCCTATG-3′, and 3′ primer, 5′-GCCCCTGGACAACACAGACT-3′; DGAT2, 5′ primer, 5′-CCGCAAAGGCTTTGTGAAG-3′, and 3′ primer, 5′-GGAATAAGTGGGAACCAGATCA-3′. Other primers for real time PCR were described previously (40–42).
Cholesterol and fatty acid synthesis in vivo
Rates of cholesterol and fatty acid synthesis were measured in wild-type and Elovl5−/− mice using 3H-labeled water as previously described (34). The rates of sterol and fatty acid synthesis were calculated by measuring incorporation of 3H-labeled water into fatty acids or digitonin-precipitable sterols.
Culture of mouse primary hepatocytes and measurement of TG synthesis
Hepatocytes were isolated from wild-type and Elovl5−/− mice and cultured as described previously (43). After a 2 h attachment period, the cells were incubated with 1.5 mM [14C]glycerol for 3 h. The cells were harvested in 1 ml of PBS and lipids were extracted in 4 ml of chloroform/methanol (2:1). The extracts were centrifuged for 10 min at 500 g, and the upper aqueous phase was removed. The organic phase was washed twice with 0.1 M NaCl/methanol/chloroform (47:45:3) and the lower organic phase was evaporated. The extracted lipids were dissolved in 40 μl of chloroform, and separated by TLC using hexane/diethylether/acetic acid (80:20:1). The TLC plate was stained with I2 and a band corresponding to triglycerides was scraped. [14C]radioactivity was measured using a β-counter. Protein concentrations of the samples were measured using an aliquot of the harvested cells with the BCA protein assay kit (Thermo Scientific, Rockford, IL).
Statistical Analysis
Statistical analyses were performed using SigmaStat 3.0 software (SPSS, Inc., Chicago, IL). One-way ANOVA, followed by multiple pairwise comparisons (Holm-Sidak method), was used for multigroup comparisons. P values of <0.05 were considered statistically significant.
RESULTS
Elovl5−/− mice were generated using ES cells obtained from NIH/NHLBI-sponsored Bay Genomics ES cell gene trap resource (44). These ES cells have an insertion of a gene trap vector that contains a splice acceptor site immediately upstream of a promoter-less β-geo (the fusion of β-galactosidase and neomycin phosphotransferase gene) and that gives rise to a fusion transcript composed of upstream coding sequence of the trapped gene linked to the β-geo sequences (44, 45). The ES cell clone (DTM 098) harbored an insertion of the gene trap vector 7.9 kb downstream of exon 1 of Elovl5 (Fig. 1A). The genotype of mice generated using these ES cells was confirmed by Southern blot analysis (Fig. 1B). Northern blot analysis using total RNA isolated from livers of wild-type and Elovl5−/− mice and a 32P-labeled cDNA probe corresponding to the coding region of Elovl5 showed a ∼99% reduction in ELOVL5 transcript levels in homozygous knockout mice (Fig. 1C). Matings between mice heterozygous for the allele harboring the gene trap vector (Elovl5+/−) produced Elovl5−/− mice in the expected Mendelian ratios. The majority of Elovl5−/− female mice were not fertile while Elovl5−/− male mice were fertile. The mechanism responsible for the infertility was not investigated further.
Fig. 1.
Gene structure of Elovl5 and generation of Elovl5−/− mice. A: Gene structure of the Elovl5 wild-type and disrupted allele. Elovl5 contains 8 exons and 7 introns. The first ATG and the stop (TGA) codon reside on exon 2 and exon 8, respectively. Coding regions are shown by gray boxes. The insertion site of the gene trap vector and the splice acceptor in the vector are indicated with arrows. BglII restriction sites and the expected DNA fragments produced by BglII digestion are shown. The location of the probe for Southern blotting with BglII digestion is shown by a filled box. B: Southern blot result of BglII digested genomic DNA from wild-type (+/+), heterozygous (+/−), and Elovl5−/− (−/−) mice. C: Total RNA prepared from livers of wild-type (WT) (+/+) and Elovl5−/− (−/−) mice were subjected to Northern blot analysis for elongation of very long chain fatty acids (ELOVL)5 and cyclophilin as described under Experimental Procedures. KO, knockout.
The phenotypic parameters of Elovl5−/− mice are listed in Table 1. Body weight and liver weight were not significantly different in Elovl5−/− mice compared with wild-type controls of either sex fed a standard chow diet. Similarly, plasma triglycerides and cholesterol (Table 1), and plasma free fatty acids, glucose, and insulin levels (data not shown), were not altered in Elovl5−/− mice of either sex. Disruption of Elovl5 resulted in the development of hepatic steatosis. Liver triglyceride concentrations were 1.6-fold higher in Elovl5−/− male mice and 2.4-fold higher in Elovl5−/− in female mice. Liver free fatty acid concentrations in wild-type and Elovl5−/− mice were not significantly different (0.4 ± 0.1 vs. 0.5 ± 0.0 mg/g, respectively). Liver DAG concentrations in Elovl5−/− mice were increased 2-fold compared with those of wild-type mice (1.9 ± 0.2 vs. 1.1 ± 0.1 mg/g, respectively).
TABLE 1.
Phenotypic comparison of wild-type and Elovl5−/− mice
| Parameter | Elovl5+/+ | Elovl5−/− | Elovl5+/+ | Elovl5−/− |
|---|---|---|---|---|
| Sex | Male | Male | Female | Female |
| Age (weeks) | 8–9 | 8–9 | 8–9 | 8–9 |
| Number of mice | 10 | 10 | 10 | 10 |
| Body weight (g) | 26.7 ± 1.4 | 24.9 ± 0.6 | 21.6 ± 0.5 | 20.8 ± 0.7 |
| Liver weight (g) | 1.53 ± 0.13 | 1.34 ± 0.05 | 1.08 ± 0.04 | 1.09 ± 0.05 |
| Liver weight/Body weight (%) | 5.66 ± 0.25 | 5.37 ± 0.16 | 5.00 ± 0.11 | 5.27 ± 0.15 |
| Liver cholesterol (mg/g) | 2.5 ± 0.1 | 2.6 ± 0.1 | 2.9 ± 0.1 | 3.9 ± 0.2a |
| Liver triglyceride (mg/g) | 7.1 ± 0.3 | 11.4 ± 1.4a | 14.5 ± 1.1 | 34.2 ± 3.6a |
| Plasma cholesterol (mg/dl) | 97 ± 4 | 100 ± 7 | 88 ± 5 | 79 ± 6 |
| Plasma triglycerides (mg/dl) | 106 ± 9 | 102 ± 16 | 93 ± 8 | 91 ± 9 |
Mice were fed a chow diet ad libitum. Each value represents the mean ± SE.
P < 0.05 using Student's t-test.
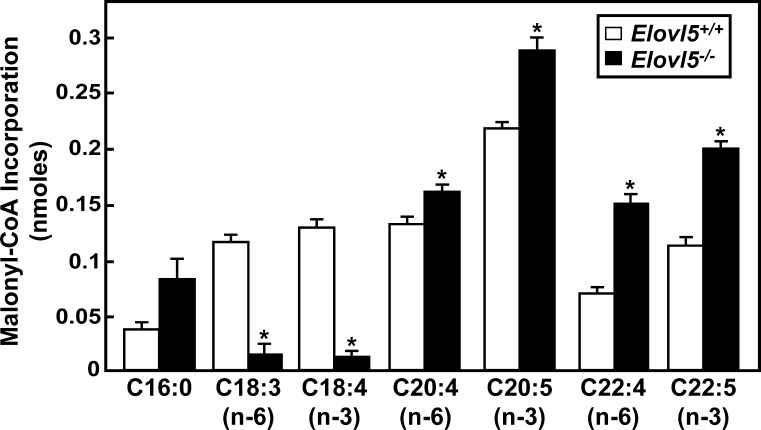
To identify in vivo fatty acid substrates of ELOVL5, a fatty acid elongation assay was performed using microsomes prepared from livers of wild-type and Elovl5−/− mice. Fatty acids of various chain lengths and degrees of desaturation were used as substrates in the reaction. As shown in Fig. 2, γ-linolenic (C18:3, n-6) and stearidonic (C18:4, n-3) acids were the only fatty acids tested in which there was a ∼88–90% reduction in elongation activity in microsomes from Elovl5−/− mice. Conversely, the elongation activities for palmitic (C16:0) acid, [a substrate of ELOVL6 (4)], arachidonic (C20:4, n-6), eicosapentaenoic (C20:5, n-3), docosatetraenoic (C22:4, n-6), and docosapentaenoic (C22:5, n-3) acid, [substrates of ELOVL2 (4, 11)], were significantly elevated (1.3 to 2.2-fold) in microsomes from Elovl5−/− mice. Elongation of palmitoleic (C16:1, n-9), stearic (C18:0), oleic (C18:1, n-9), linoleic (C18:2, n-6), linolenic (C18:3, n-3), and arachidic (C20:0) acids was undetectable in microsomes from livers of both wild-type and Elovl5−/− mice (data not shown). These results demonstrate that microsomes from Elovl5−/− mouse livers have a specific defect in the elongation of γ-linolenic (C18:3, n-6) to dihomo-γ-linolenic (C20:3, n-6) acid and stearidonic (C18:4, n-3) to eicosatetraenoic (C20:4, n-3) acid. The increased elongation activity of ELOVL6 and ELOVL2 substrates suggested that these enzymes might be induced in livers of Elovl5−/− mice.
Fig. 2.
ELOVL5 activities in liver microsomes from wild-type and Elovl5−/− mice. Microsomes were prepared from livers of five wild-type and five Elovl5−/− mice. Microsomal proteins (30 μg) were incubated with a reaction mixture containing 150 μM [14C]malonyl-CoA and 20 μM of the indicated fatty acids in the presence of 100 μM CoA, 1 mM ATP, and 1 mM NADPH at 37°C for 10 min. Fatty acids were extracted from the reaction mixture and the total incorporation of [14C]malonyl-CoA to fatty acid substrates was measured. Each bar represents the mean ± SE of the values from five mice. * P < 0.01.
To determine if the loss of ELOVL5 altered the cellular fatty acid composition, hepatic lipids were extracted and fatty acids quantified by GLC (Table 2). The fatty acid composition of the Teklad diet used is presented in supplementary Table I. Normally, the γ-linolenic (C18:3, n-6), a substrate of ELOVL5, is present at very low concentrations in mouse liver; however, in Elovl5−/− livers the concentration of this fatty acid was increased ∼24-fold. The absolute concentration of the predominant downstream n-6 product of γ-linolenic (C18:3, n-6) elongation and desaturation, arachidonic acid (C20:4, n-6), was reduced by 36%. The Elovl5−/− mouse livers also contained 1.3-fold more linoleic acid (C18:2, n-6) than wild-type mice. Inasmuch as the concentrations of n-3 fatty acids that are ELOVL5 substrates are normally very low, we were not able to detect stearidonic acid (C18:4, n-3); however, the concentration of the downstream n-3 product, DHA, was reduced by 32% in Elovl5−/− mouse livers. When Elovl5−/− mice were fed AIN-93G (TestDiet, Richmond, IN), in which the α-linolenic acid (C18:3, n-3) content is five times higher than that in the Teklad chow (see supplementary Table I), the livers accumulated measurable n-3 substrate; stearidonic acid (C18:4, n-3); and both downstream products, EPA (C20:5, n-3) and DHA, were reduced by 60% and 20%, respectively (data not shown). The residual PUFAs with chain lengths ⩾C20 present in Elovl5−/− mouse livers are likely derived from fatty acids ingested from the diet, although it is also possible that they are produced by low level redundant activities associated with other elongases.
TABLE 2.
Fatty acid composition of major lipid classes in livers of wild-type and Elovl5−/− mice
| Fatty Acid Compositions |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Lipid class | Genotype | C16:0 | C16:1 | C18:0 | C18:1, n-9 | C18:2, n-6 | C18:3, n-6 | C20:4, n-6 | C22:6, n-3 |
| Total lipids (μg/mg) | +/+ | 10 ± 0.2 | 0.6 ± 0.1 | 6.1 ± 0.3 | 9.5 ± 0.6 | 5.7 ± 0.3 | 0.05 ± 0.1 | 6.4 ± 0.2 | 1.9 ± 0.1 |
| −/− | 15 ± 1.1a | 2.7 ± 0.5a | 5.5 ± 0.4 | 23 ± 2.8a | 7.6 ± 0.5 | 1.2 ± 0.1a | 4.1 ± 0.1a | 1.3 ± 0.0a | |
| Total lipids (% of total) | +/+ | 24 ± 0.5 | 1.4 ± 0.2 | 15 ± 0.9 | 23 ± 1.5 | 14 ± 0.6 | 0.1 ± 0.1 | 16 ± 0.8 | 4.7 ± 0.3 |
| −/− | 24 ± 0.4 | 4.3 ± 0.6a | 9.2 ± 1.0a | 36 ± 1.6a | 13 ± 0.3 | 2.0 ± 0.1a | 6.9 ± 0.6a | 2.1 ± 0.1a | |
| Phospho-lipids (%) | +/+ | 26 ± 0.3 | 0.5 ± 0.2 | 22 ± 0.7 | 11 ± 0.4 | 13 ± 0.3 | ND | 20 ± 0.3 | 5.4 ± 0.2 |
| −/− | 26 ± 0.2 | 1.8 ± 0.2a | 18.± 0.7a | 16 ± 0.4a | 17 ± 0.2a | 2.6 ± 0.1a | 13 ± 0.6a | 3.7 ± 0.2a | |
| Triglycerides (%) | +/+ | 26 ± 0.6 | 3.1 ± 0.4 | 2.0 ± 0.1 | 45 ± 0.4 | 17 ± 1.3 | 0.5 ± 0.0 | 1.9 ± 0.1 | 0.5 ± 0.1 |
| −/− | 23 ± 0.6a | 7.4 ± 1.0a | 1.4 ± 0.1a | 50 ± 1.1a | 12 ± 0.5a | 3.1 ± 0.3a | 0.1 ± 0.1a | ND | |
| Cholesteryl esters (%) | +/+ | 33 ± 0.9 | 2.5 ± 0.3 | 4.1 ± 0.5 | 47 ± 1.0 | 7.9 ± 0.6 | ND | 1.7 ± 0.2 | ND |
| −/− | 27 ± 0.7a | 5.5 ± 0.6a | 2.5 ± 0.2a | 55 ± 0.9a | 6.3 ± 0.4 | 0.6 ± 0.1 | 0.2 ± 0.0a | ND | |
ND, not detected. Livers from 8- to 9-wk-old female mice fed a chow diet were extracted and the major classes of lipids were separated on a silica column. Lipid fractions were methyl esterified and quantified by gas-liquid chromatography (GLC). Each value represents the mean ± SE from five mice.
Levels of statistical significance of P < 0.05 (Student's t-test).
The concentrations of monounsaturated fatty acids, such as palmitoleic (C16:1, n-7) and oleic acid (C18:1, n-9) from de novo fatty acid synthesis were increased by 2.4 to 4.5-fold, respectively in livers from Elovl5−/− mice. Vaccenic acid (C18:1, n-7), a minor fatty acid generated from the elongation of palmitoleic acid, was 1.6-fold increased in livers of Elovl5−/− mice (data not shown).
We next determined whether the changes measured in the total lipid fatty acid compositions were also present in the major lipid classes: phospholipids, triglycerides, and cholesteryl esters (Table 2). The relative amount of the ELOVL5 n-6 substrate, γ-linolenic (C18:3, n-6), was increased from undetectable levels in wild-type livers to 2.6% of all fatty acids in the phospholipid lipid fraction. The hepatic phospholipids of Elovl5−/− mice also contained 1.4-fold more linoleic acid (C18:2, n-6). ELOVL5 downstream products, arachidonic acid (C20:4, n-6) and DHA (C22:6, n-3), were reduced by 37% and 32% of those measured in wild-type liver, respectively. Similar but less pronounced changes were measured in the fatty acid compositions of triglycerides and cholesteryl esters. All lipid fractions from Elovl5−/− livers were enriched with monounsaturated fatty acids, palmitoleic (C16:1), and oleic (C18:1) acids.
The increase in hepatic triglyceride concentration and the enrichment of monounsaturated fatty acids in livers of Elovl5−/− mice suggested that rates of de novo fatty acid and triglyceride synthesis might be higher in the knockout livers. To evaluate this possibility, the mRNA levels of genes involved in lipid metabolism were quantified by real-time PCR in wild-type and knockout livers. As shown in Table 3, the mRNA levels of genes involved in monounsaturated fatty acid synthesis, acetyl-CoA carboxylase 1 (ACC-1), fatty acid synthase (FAS), ELOVL6, and stearoyl-CoA desaturase 1 (SCD-1), were increased 1.9- to 5.5-fold in livers of Elovl5−/− mice. The mRNAs encoding glucose-6-phosphate dehydrogenase (G6PD) and malic enzyme, enzymes required for NAPDH production, were increased 3.9- to 5.9-fold in Elovl5−/− livers. The mRNA level of glycerol-3-phosphate acyl transferase (GPAT), the enzyme responsible for the first step in triglyceride synthesis, was increased 3.1-fold. No significant changes in the mRNA levels were measured in 1-acylglycerol-3-phosphate-O-acyltransferase (AGPAT) or diacylglycerol acyltransferase (DGAT). Small increases in the hepatic mRNA levels of ELOVL2, Δ6-desaturase, and Δ5 desaturase, enzymes involved in PUFA elongation and desaturation, were also measured in Elovl5−/− mice. The fatty acid biosynthetic genes are transcriptionally regulated by SREBP-1c and ChREBP (46, 47); however the mRNA level of SREBP-1c was unchanged and the mRNA level of ChREBP was only slightly increased in livers of the Elovl5−/− mice. In addition, the mRNA levels of PPARα regulated genes involved in fatty acid oxidation were either unchanged or slightly increased.
TABLE 3.
Expression of mRNAs in livers of Elovl5−/− mice
| mRNA | Relative Expression | |
|---|---|---|
| Elovl5 genotype | +/+ | −/− |
| SREBP pathway | ||
| SREBP-1c | 1.0 ± 0.3 | 1.1 ± 0.1 |
| SREBP-1a | 1.0 ± 0.1 | 1.1 ± 0.1 |
| SREBP-2 | 1.0 ± 0.1 | 1.2 ± 0.2 |
| Insig-1 | 1.0 ± 0.2 | 1.6 ± 0.1 |
| Insig-2 | 1.0 ± 0.1 | 2.0 ± 0.1a |
| Fatty acid synthesis | ||
| ACC-1 | 1.0 ± 0.0 | 1.9 ± 0.2a |
| FAS | 1.0 ± 0.2 | 3.0 ± 0.5a |
| ELOVL6 | 1.0 ± 0.1 | 2.3 ± 0.5 |
| SCD-1 | 1.0 ± 0.3 | 5.5 ± 0.9a |
| NADPH production | ||
| G6PD | 1.0 ± 0.3 | 5.9 ± 1.1a |
| Malic enzyme | 1.0 ± 0.2 | 3.9 ± 0.7a |
| TG synthesis | ||
| GPAT | 1.0 ± 0.2 | 3.1 ± 0.4a |
| AGPAT1 | 1.0 ± 0.1 | 0.9 ± 0.1 |
| AGPAT2 | 1.0 ± 0.1 | 1.4 ± 0.1 |
| MGAT1 | 1.0 ± 0.5 | 2.1 ± 0.5 |
| MGAT2 | 1.0 ± 0.1 | 1.4 ± 0.3 |
| DGAT1 | 1.0 ± 0.1 | 1.5 ± 0.3 |
| DGAT2 | 1.0 ± 0.2 | 1.4 ± 0.0 |
| PUFA synthesis | ||
| ELOVL1 | 1.0 ± 0.1 | 1.0 ± 0.0 |
| ELOVL2 | 1.0 ± 0.1 | 2.1 ± 0.1a |
| ELOVL3 | 1.0 ± 0.3 | 1.7 ± 0.3 |
| Δ6 desaturase | 1.0 ± 0.1 | 2.2 ± 0.0a |
| Δ5 desaturase | 1.0 ± 0.1 | 1.9 ± 0.1a |
| LXRα target genes | ||
| ABCG5 | 1.0 ± 0.2 | 1.5 ± 0.2 |
| ABCG8 | 1.0 ± 0.2 | 1.8 ± 0.2a |
| PLTP | 1.0 ± 0.2 | 5.3 ± 1.0a |
| Lipoprotein lipase | 1.0 ± 0.3 | 0.7 ± 0.1 |
| PPAR-α target genes | ||
| Acyl-CoA oxidase | 1.0 ± 0.2 | 1.4 ± 0.1 |
| Cyp4a10 | 1.0 ± 0.3 | 1.3 ± 0.1 |
| L-PBE | 1.0 ± 0.1 | 1.9 ± 0.2a |
| CD-36 | 1.0 ± 0.1 | 1.2 ± 0.1 |
| LCAD | 1.0 ± 0.1 | 1.5 ± 0.1a |
| CPT-1 | 1.0 ± 0.2 | 1.3 ± 0.1 |
| ChREBP | 1.0 ± 0.1 | 1.4 ± 0.1a |
ACC, acetyl-CoA carboxylase; AGPAT, 1-acylglycerol-3-phosphate-O-acyltransferase; ChREBP, carbohydrate regulatory element-binding protein; DGAT, diacylglycerol acyltransferase; ELOVL, elongation of very long chain fatty acids; FAS, fatty acid synthase; G6PD, glucose-6-phosphate dehydrogenase; GPAT, glycerol-3-phosphate acyl transferase; MGAT, monoacylglycerol acyltransferase; SREPB, sterol regulatory element-binding protein; SCD-1, stearoyl-CoA desaturase 1. Four female wild-type and four Elovl5−/− mice were used for the experiment. Total RNA from individual livers were subjected to quantitative real-time PCR as described in Experimental Procedures. Cyclophilin was used as the invariant control. Values represent the mean ± SE amount of mRNA from four mice relative to that of wild-type mice fed chow, which is defined as 1.
Levels statistically different than wild-type mice fed chow (P < 0.05, Student's t-test). Similar results were obtained from three independent experiments.
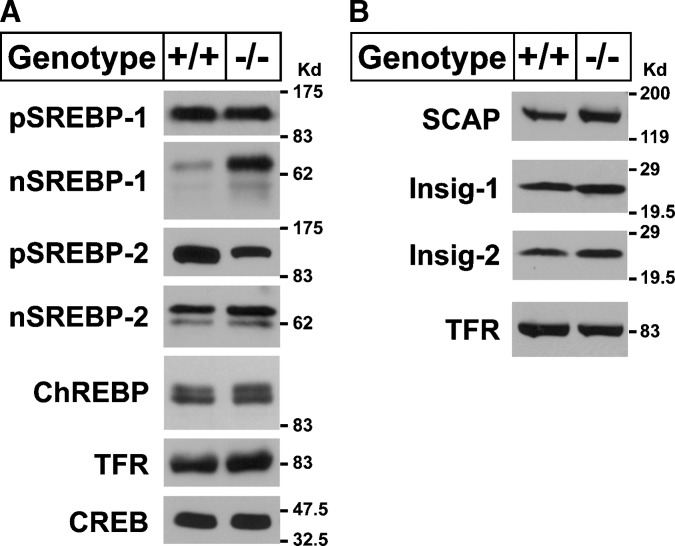
To determine whether the loss of ELOVL5 selectively altered the abundance of the cleaved and transcriptionally active nuclear form of SREBP-1 or nuclear ChREBP, immunoblot analyses of liver nuclear extracts and membranes was performed. As shown in Fig. 3A, the nuclear form of SREBP-1 was ∼4-fold higher in livers from Elovl5−/− mice, whereas, the nuclear form of SREBP-2 was unchanged. The nuclear abundance of ChREBP was not increased in Elovl5−/− mouse livers, suggesting that SREBP-1c was responsible for the activation of the lipogenic genes.
Fig. 3.
Immunoblot analysis of sterol regulatory element-binding proteins (SREBPs), SCAP, Insig-1, and Insig-2 in livers of wild-type and Elovl5−/− mice. Membrane and nuclear proteins were prepared from livers of five wild-type and Elovl5−/− female mice and equal aliquots from each were pooled (total, 30 μg) and subjected to SDS-PAGE. A, Immunoblotting of SREBP-1, SREBP-2, transferrin receptor (TFR), and cAMP response element binding protein (CREB) was performed using anti-mouse SREBP-1, SREBP-2, anti-human CREB, and TFR antibodies as described in Experimental Procedures. B: Immunoblotting of SREBP cleavage-activating protein (SCAP), Insig-1, Insig-2, and TFR was performed using anti-hamster SCAP, anti-mouse Insig-1, Insig-2, and anti-human TFR antibodies. Similar results were obtained in five independent experiments. ChREBP, carbohydrate regulatory element-binding protein.
The ER-bound precursor forms of SREBP protein must be cleaved to release the forms that subsequently enter the nucleus. SCAP and Insig proteins play crucial regulatory and permissive roles in this process (48). SCAP escorts the precursor forms of SREBPs from the ER to Golgi where proteolytic processing occurs. Insig proteins prevent the movement of SREBP:SCAP complexes from ER to Golgi by binding to SCAP and preventing SREBP processing (49, 50). To determine whether changes in the amount of SCAP or Insig proteins were responsible for the increased nuclear SREBP-1c levels, immunoblot analysis of these proteins was performed using liver membranes of wild-type and Elovl5−/− mice. As shown in Fig. 3B, SCAP, Insig-1, and Insig-2 proteins were not significantly changed in livers of Elovl5−/− mice and thus do not explain the observed increase in nuclear SREBP-1.
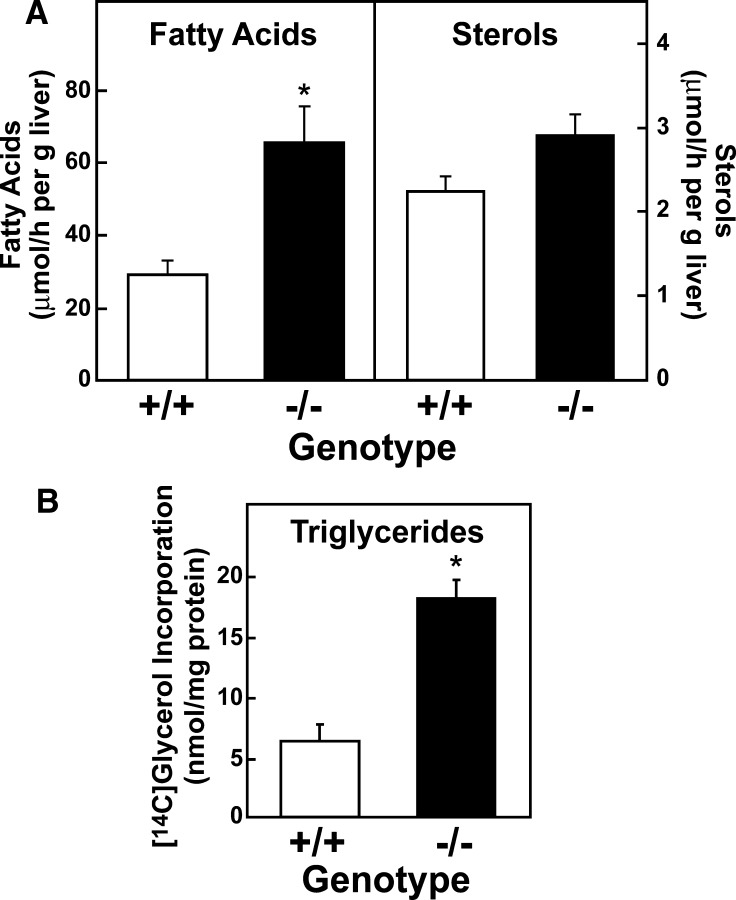
To confirm that the changes in mRNA levels measured for the genes encoding the fatty acid biosynthetic enzymes in livers of Elovl5−/− mice reflected a change in flux through the pathway, the rates of fatty acid and cholesterol synthesis were determined in vivo using [3H]H2O (34). Consistent with the changes measured in mRNA levels, synthesis of fatty acids was more than 2-fold higher in livers of Elovl5−/− mice, while cholesterol synthesis was unchanged (Fig. 4A). To determine if the higher rates of fatty acid synthesis translated into higher rates of triglyceride synthesis, freshly isolated hepatocytes from wild-type and Elovl5−/− mice were incubated with [14C]glycerol and incorporation into newly synthesized triglycerides was measured. As shown in Fig. 4B, triglyceride synthesis was also 2.9-fold higher in Elovl5−/− hepatocytes.
Fig. 4.
Synthetic rates of fatty acids, sterols, and triglycerides in livers of wild-type and Elovl5−/− mice. A: In vivo rates of fatty acids and sterol synthesis in liver. Five wild-type mice and five Elovl5−/− mice were injected intraperitoneally with [3H]H2O (50 mCi in 0.2 ml of saline). One h after injection, livers were removed and fatty acids and digitonin-precipitable sterols were extracted. Incorporation of 3H into newly synthesized fatty acids and sterols were measured as described in Experimental Procedures. Each bar represents the mean ± SE of the values from five mice. * P < 0.05 using Student's t-test. Similar results were obtained in two additional independent experiments. B: Triglyceride synthesis in primary hepatocytes isolated from WT and Elovl5−/− mice. Primary hepatocytes were isolated from three WT and three Elovl5−/− mice and attached on collagen coated dishes for 2 h in duplicate. After the attachment period, the cells were incubated with 1.5 mM [14C]glycerol for 3 h. Lipids were extracted from the cells and triglycerides were separated by TLC as described in Experimental Procedures. Incorporation of radiolabeled glycerol to triglycerides was measured. Each bar represents the mean ± SE.
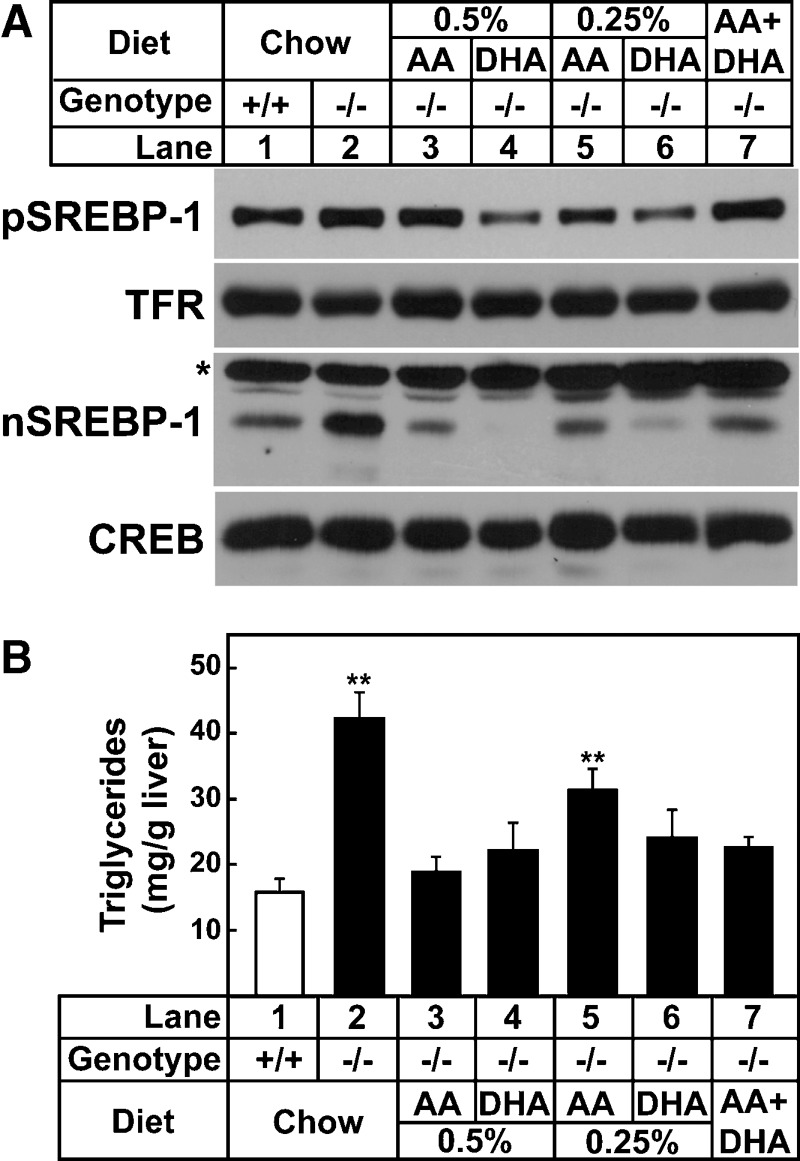
Arachidonic acid (C20:4, n-6) and DHA (C22:6, n-3) are synthesized by a series of elongation and desaturation reactions from the essential fatty acids linoleic acid (C18:2, n-6) and α-linolenic acid (C18:3, n-3), respectively. To determine if the molecular and metabolic changes measured in Elovl5−/− livers were due to a lack of arachidonic acid and/or DHA, mice were fed a standard chow diet supplemented with varying amounts of arachidonic acid and/or DHA for 7 days. Diets supplemented with <0.5% (w/w) of arachidonic acid or <0.25% (w/w) of DHA for 7 days did not normalize triglycerides nor nuclear SREBP-1 protein levels in livers of Elovl5−/− mice (data not shown). Elovl5−/− mice fed diets supplemented with 0.5% (w/w) of arachidonic acid or 0.25% (w/w) of DHA for 7 days did normalize nuclear SREBP-1 protein levels (Fig. 5A, lanes 1, 3, and 5) and liver triglycerides (Fig. 5B). However, this degree of supplementation resulted in 1.5-fold higher arachidonic acid and 3-fold higher DHA liver concentrations compared with wild-type mice fed chow (Table 4).
Fig. 5.
Levels of SREBP-1 proteins and triglyceride concentration in livers of wild-type and Elovl5−/− mice fed a chow diet or the chow diet supplemented with various fatty acids. A, Arachidonic acid (AA) and docosahexaenoic acid (DHA) were added to a chow diet (Teklad Mouse/Rat Diet 7001 from Harlan Teklad Premier Laboratory Diets) at 0.5%, 0.25% (w/w) individually, or both arachidonic acid (0.2%) and DHA (0.05%) were added. Mice were fed the diets for 7 days. Membrane and nuclear proteins were prepared from livers of five mice of the indicated group. Equal aliquots of protein from each liver were pooled (total, 40 μg) and subjected to SDS-PAGE and immunoblotting using anti-mouse SREBP-1 as described under Experimental Procedures. * indicates a nonspecific band. B: Liver triglyceride contents were measured from the livers shown in A. Each bar represents the mean ± SE of the values from five mice. ** P < 0.05 of one-way ANOVA, followed by multiple pairwise comparisons (Holm-Sidak method).
TABLE 4.
Fatty acid composition of livers from wild-type and Elovl5−/− mice fed diets supplemented with arachidonic acid and docosahexaenoic acid (DHA)
| Fatty Acid Concentration (μg/mg liver) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Diet | Genotype | C16:0 | C16:1 | C18:0 | C18:1, n-9 | C18:2, n-6 | C18:3, n-6 | C20:4, n-6 | C22:6, n-3 |
| Chow (4% fat) | +/+ | 11.4 ± 0.9 | 0.8 ± 0.1 | 5.7 ± 0.3 | 13.8 ± 1.5 | 6.3 ± 0.4 | ND | 6.1 ± 0.2 | 2.5 ± 0.1 |
| −/− | 15.8 ± 0.9a | 3.3 ± 0.5a | 4.9 ± 0.2 | 27.8 ± 2.6a | 8.0 ± 0.4a | 1.5 ± 0.1a | 4.1 ± 0.1a | 1.9 ± 0.1a | |
| 0.5% Arachidonic acid | +/+ | 8.4 ± 0.2a | 0.3 ± 0.0a | 5.7 ± 0.3 | 6.6 ± 0.5a | 4.6 ± 0.2a | ND | 8.9 ± 0.2a | 2.4 ± 0.1 |
| −/− | 11.2 ± 0.5 | 1.0 ± 0.1 | 5.6 ± 0.1 | 13.1 ± 1.7 | 5.0 ± 0.6 | 0.6 ± 0.1a | 9.5 ± 0.3a | 1.2 ± 0.1a | |
| 0.25% DHA | +/+ | 9.0 ± 0.5 | 0.2 ± 0.1a | 6.2 ± 0.2 | 6.2 ± 0.7a | 6.0 ± 0.3 | ND | 4.7 ± 0.1a | 7.2 ± 0.3a |
| −/− | 12.7 ± 1.2 | 1.1 ± 0.2 | 5.6 ± 0.3 | 14.0 ± 2.0 | 8.0 ± 0.6a | 0.6 ± 0.1a | 3.0 ± 0.2a | 7.7 ± 0.4a | |
| 0.2% Arachidonic acid + 0.05% DHA | +/+ | 8.3 ± 0.4a | 0.4 ± 0.0a | 5.4 ± 0.3 | 6.8 ± 0.5a | 5.0 ± 0.2 | ND | 6.9 ± 0.2a | 3.3 ± 0.2a |
| −/− | 12.4 ± 0.7 | 1.3 ± 0.1a | 5.6 ± 0.4 | 14.2 ± 0.8 | 6.0 ± 0.3 | 0.7 ± 0.1a | 6.9 ± 0.3a | 3.2 ± 0.1a | |
Livers from 11- to 15-week-old female mice fed the indicated diets for 7 days were extracted. Fatty acids were methyl esterified and quantified by GLC. Each value represents the mean ± SE from five mice.
Levels statistically different than wild-type mice fed chow (P < 0.05, Student's t-test).
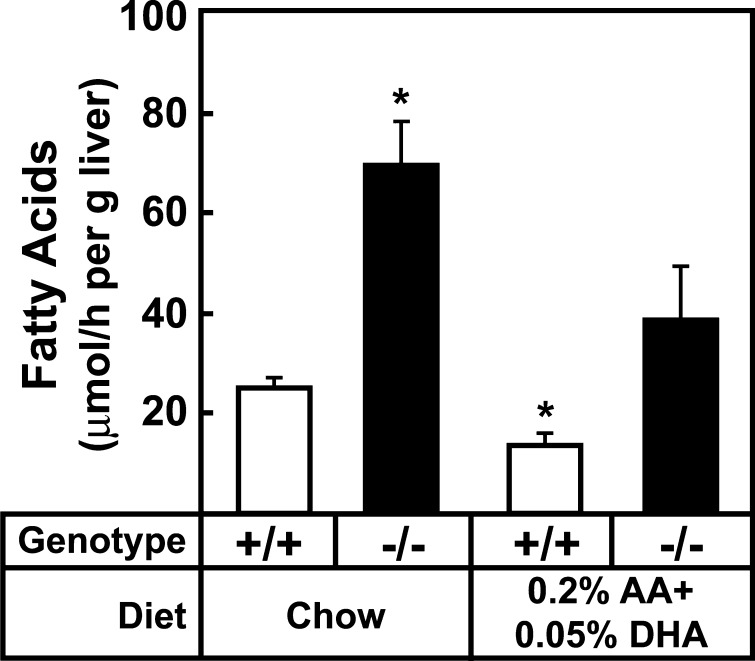
To determine whether supplementing both arachidonic and DHA was more effective than either alone in reversing the molecular and metabolic changes resulting from ELOVL5 deficiency, diets were supplemented with various concentrations of both fatty acids for 7 days. As shown in Table 4, 0.2% (w/w) arachidonic acid and 0.05% (w/w) DHA added to chow resulted in concentrations of these fatty acids in the lipid extracts of Elovl5−/− mouse livers that only slightly exceeded levels measured in livers of wild-type mice fed the normal chow diet. Combined fatty acid supplementation normalized nSREBP-1 protein levels, triglyceride concentrations, and the amount of palmitic, palmitoleic, and oleic acids in Elovl5−/− livers to levels measured in wild-type mice fed chow (Table 4, Fig. 5). Consistent with the normalization of nSREBP-1 levels, the mRNA levels of SREBP-1 regulated genes involved in fatty acid synthesis, fatty acid elongation and desaturation were reduced to levels found in livers of wild-type mice fed chow (Table 5). The supplementation of 0.25% oleic acid to the diet had no effect on the fatty acid composition, triglyceride content, nSREBP-1c level or the gene mRNA levels in Elovl5−/− livers (data not shown).
TABLE 5.
Relative amounts of mRNAs in livers from wild-type and Elovl5−/− mice on diets supplemented with unsaturated fatty acids
| Diets | Chow | 0.2% Arachidonic + 0.05% DHA | ||
|---|---|---|---|---|
| Elovl5 genotype | +/+ | −/− | +/+ | −/− |
| SREBP pathway | ||||
| SREBP-1c | 1.0 ± 0.2 | 1.3 ± 0.2 | 0.7 ± 0.1 | 1.1 ± 0.2 |
| SREBP-1a | 1.0 ± 0.1 | 1.3 ± 0.1 | 1.0 ± 0.1 | 1.1 ± 0.0 |
| SREBP-2 | 1.0 ± 0.2 | 1.0 ± 0.1 | 1.1 ± 0.1 | 0.8 ± 0.1 |
| Insig-1 | 1.0 ± 0.1 | 1.3 ± 0.1 | 0.7 ± 0.1 | 0.7 ± 0.1 |
| Insig-2 | 1.0 ± 0.1 | 1.7 ± 0.1a | 0.9 ± 0.0 | 1.0 ± 0.0 |
| Fatty acid synthesis | ||||
| ACC-1 | 1.0 ± 0.2 | 2.3 ± 0.4a | 0.8 ± 0.1 | 1.1 ± 0.1 |
| FAS | 1.0 ± 0.3 | 2.8 ± 0.4a | 0.5 ± 0.1 | 0.9 ± 0.2 |
| SCD-1 | 1.0 ± 0.2 | 4.6 ± 0.8a | 0.6 ± 0.2 | 1.1 ± 0.1 |
| G6PD | 1.0 ± 0.2 | 7.8 ± 1.4a | 0.7 ± 0.1 | 1.3 ± 0.2 |
| Malic enzyme | 1.0 ± 0.2 | 2.7 ± 0.3a | 0.5 ± 0.1 | 1.4 ± 0.2 |
| ELOVL2 | 1.0 ± 0.1 | 1.6 ± 0.1a | 0.6 ± 0.0a | 0.8 ± 0.1 |
| ELOVL6 | 1.0 ± 0.2 | 2.8 ± 0.2a | 0.5 ± 0.1 | 1.0 ± 0.2 |
| Δ6 desaturase | 1.0 ± 0.1 | 2.4 ± 0.3a | 0.8 ± 0.1 | 0.9 ± 0.1 |
| Δ5 desaturase | 1.0 ± 0.1 | 1.8 ± 0.2a | 0.8 ± 0.1 | 0.9 ± 0.1 |
Total RNA from each liver of five mice used in Table 4 was subjected to quantitative real-time PCR as described in Experimental Procedures. Values represent the mean ± SE amount of mRNA from five mice relative to that of wild-type mice fed chow, which is defined as 1. Cyclophilin was used as the invariant control.
Levels statistically different than wild-type mice fed chow (P < 0.05, Student's t-test).
To confirm that 0.2% (w/w) arachidonic acid and 0.05% (w/w) DHA also normalized rates of fatty acid synthesis in Elovl5−/− livers, mice fed chow or the arachidonic and DHA supplemented diet for 7 days were injected with [3H]H2O and incorporation into newly synthesized fatty acids was quantified (Fig. 6). As was found in the experiments shown in Fig. 4, rates of fatty acid synthesis in Elovl5−/− livers were increased ∼3-fold on the chow diet. Dietary supplementation with 0.2% arachidonic acid and 0.05% DHA resulted in a modest but significant reduction in fatty acid synthesis in wild-type mouse liver and lowered rates of fatty acid synthesis in livers of knockout mice to those that were not significantly different from wild-type mice fed chow. These results suggest that activation of SREBP-1, the resulting higher rates of fatty acid synthesis, and excessive triglyceride accumulation in livers of Elovl5−/− mice resulted from a deficiency of arachidonic acid and DHA.
Fig. 6.
In vivo synthetic rates of fatty acids in livers of wild-type and Elovl5−/− mice. Five female mice from the indicated groups were fed a chow (4% fat) or the chow diet supplemented with 0.2% arachidonic acid and 0.05% DHA for 7 days and then injected intraperitoneally with 3H-labeled water (40 mCi in 0.2 ml of water). One h after injection, livers were removed and fatty acids were extracted. Incorporation of 3H-labeled water into the newly synthesized fatty acids was measured as described in Experimental Procedures. Each bar represents the mean ± SE of the values from five mice. * P < 0.05 of Student's t-test.
DISCUSSION
Elovl5−/− mice were produced to determine the in vivo fatty acid substrates of ELOVL5 and the physiologic importance of the enzyme's products in regulating lipid metabolism. ELOVL5-deficient microsomes were unable to support the elongation of C18:3, n-6 to C20:3, n-6 and C18:4, n-3 to C20:4, n-3 in vitro. Elongation of C18 PUFAs is an essential step for the generation of cellular arachidonic acid and DHA using dietary linoleic (18:2, n-6) and α-linolenic acid (C18:3, n-3) as substrates and the defect in C18 PUFA elongation in Elovl5−/− mice resulted in significantly lower cellular levels of arachidonic acid and DHA in liver. The metabolic consequence of this reduction was derepression of SREBP-1c, which led to the transcriptional activation of lipogenic genes, increased lipogenesis, and hepatic steatosis. The molecular and metabolic changes were normalized when hepatic arachidonic acid and DHA levels of Elovl5−/− mice were restored to those of wild-type mice by dietary supplementation. We conclude that the metabolic changes present in Elovl5−/− mice result from insufficient arachidonic acid and DHA, and that cellular levels of these fatty acids produced endogenously play an important role in the tonic regulation of nuclear SREBP-1 and fatty acid synthesis.
Several potential mechanisms have been proposed for PUFA-mediated regulation of SREBP-1c expression including transcriptional (21, 22), posttranscriptional (18), and posttranslational (17, 33). In vivo, the mRNA level for SREBP-1c was markedly reduced in rats fed diets containing a very high concentration of fish oils (51). The unsaturated fatty acids oleic (C18:1, n-9), linoleic (C18:2, n-6), α-linolenic (C18:3, n-3), arachidonic (C20:4, n-6), EPA (C20:5, n-3), and DHA (C22:6, n-3) suppress SREBP-1c transcription by inhibition of LXRα in HEK-293 cells, FTO-2B cells, and in primary rat hepatocytes when added to the medium at 100 μM concentrations (17, 21, 22). Elovl5−/− livers contained higher concentrations of oleic acid and C18 PUFAs than wild-type mice, which suggest that these fatty acids do not normally regulate nuclear SREBP-1 levels in vivo.
Based on the reported effects of PUFAs on LXR activity, we initially hypothesized that the deletion of Elovl5 and the subsequent reduction in PUFA concentrations might be explained by increased LXRα activity, which in turn would increase SREBP-1c transcription. In fact, some of the LXRα target genes, such as ABCG5, ABCG8, and PLTP were modestly induced in livers of Elovl5−/− mice (Table 3), suggesting possible activation of LXRα; however, the mRNA levels of SREBP-1c and the precursor form of SREBP-1 were not increased in Elovl5−/− mouse livers, suggesting that LXRα activation of SREBP-1c transcription was not the major mechanism responsible for increased nuclear SREBP-1 protein levels.
The absence of significant changes in the steady-state mRNA levels of SREBP-1c suggests that posttranslational mechanisms are responsible for the accumulation of nuclear SREBP-1 in Elovl5−/− livers, either by activation of proteolytic processing or by increased stability of nuclear SREBP-1. Various unsaturated fatty acids inhibit SREBP-1 cleavage in cultured cells lines (17), and several proteins are involved in the cleavage of SREBPs and thus may be the regulatory targets of arachidonic acid and DHA. The critical role of SCAP, Insig-1, and Insig-2 in the regulation of SREBP cleavage was shown previously in cultured cells and in animal models (43, 49, 50, 52, 53). In the current studies, neither SCAP nor Insig protein levels were changed in Elovl5−/− livers, suggesting that alterations in the expression of these proteins do not explain the increase in nSREBP-1. It is possible that changes in fatty acid composition affect the stability of the Insig:SREBP:SCAP complex in the ER; however, it is not possible to quantify the strength of this interaction with currently available tools.
A second possibility is that reduced cellular arachidonic acid and DHA concentrations in Elovl5−/− livers inhibit the degradation of nuclear SREBP-1. Botolin et al. (33) demonstrated that degradation of nuclear SREBP-1 could be accelerated by DHA through 26S proteosomes in an Erk-dependant manner. In Elovl5−/− livers, the amounts of total Erk 1/2 and phosphorylated forms of Erk 1/2 were not changed (data not shown); however, further studies are needed to determine whether the half-life of nuclear SREBP-1 is altered in livers of Elovl5−/− mice. These studies will require the development of antibodies capable of immunoprecipitating nuclear SREBP-1 so that pulse-chase experiments can be performed.
Recently, Wang et al. (54) reported the metabolic effects of adenoviral-mediated overexpression of ELOVL5. In vitro elongation assays using microsomes prepared from the livers of mice infected with an adenovirus overexpressing ELOVL5 confirmed that C18 and C20 PUFAs were substrates of ELOVL5. In agreement with the current study, liver triglyceride contents were decreased with the overexpression of ELOVL5. However, the relative levels of arachidonic acid and DHA in livers of the mice that overexpressed ELOVL5 were decreased despite a ∼2-fold increase in dihomo-γ-linolenic (C20:3, n-6). Thus, it is possible that the Δ5-desaturase becomes rate-limiting in the setting of ELOVL5 overexpression. In contrast to our results, the level of nSREBP-1 and its regulated genes were largely unchanged, with the exception of SCD-1, which was reduced by 67%. These discrepancies raise the possibility that metabolites of arachidonic acid or DHA generated in Elovl5−/− livers mediate the regulation of nSREBP-1 or that the increase in dihomo-γ-linolenic (C20:3, n-6) was sufficient to maintain normal levels of nSREBP-1.
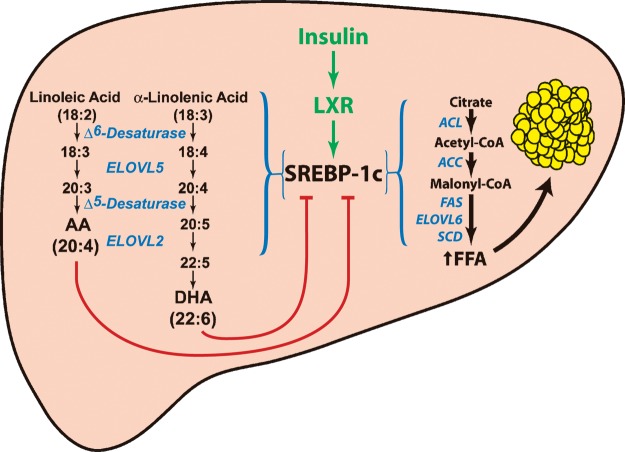
End-product feedback regulation of the cholesterol synthesis pathway was first described in 1933 (55). Recent work from the Brown and Goldstein laboratory has established that cholesterol binds to SCAP and alters the protein's conformation to allow binding to Insig, which in turn prevents the movement of the SCAP:SREBP-2 complex from the ER to the Golgi (48, 53). The regulation of SREBP-1 appears to be more complex. Insulin stimulates SREBP-1c transcription through the nuclear receptor LXR (Fig. 7) (22). When SREBP-1 is activated, the protein stimulates the transcription of genes involved in monounsaturated fatty acid synthesis enzymes as well as PUFA elongation and desaturation (29, 56, 57). Arachidonic acid and DHA are the end products of PUFA synthesis, while oleic acid (C18:1, n-9) is the predominant end product in the monounsaturated fatty acid synthesis pathway. The current studies suggest that the end products of endogenously synthesized PUFAs, but not monounsaturated fatty acid synthesis, feedback to tonically suppress SREBP-1 activity in the mouse liver (Fig. 7). Future studies are required to determine whether this end-product regulation is mediated through the PUFA inhibition of SREBP-1c cleavage as occurs with cholesterol-mediated regulation of SREBP-2, or by regulating the degradation of cleaved nuclear SREBP-1 protein.
Fig. 7.
End-product feedback regulation of SREBP-1 by endogenously synthesized PUFAs. Insulin activation of SREBP-1c transcription requires intact LXR binding sites in the promoter of the SREBP-1c. SREBP-1c transcriptionally activates genes required for the synthesis of monounsaturated fatty acids and PUFAs. PUFAs feedback and reduce the amount of nuclear SREBP-1 in the cell. ACC, acetyl-CoA carboxylase; ACL, ATP citrate lyase; FAS, fatty acid synthase; SCD, stearoyl-CoA desaturase.
Supplementary Material
Acknowledgments
The authors wish to thank David W. Russell for critical reading of the manuscript and Joseph L. Goldstein and Michael S. Brown for helpful suggestions. Norma Anderson, Scott Clark, Daniel Smith, Lauren Koob, Amy Smith, Richard Gibson, Anh Pho, Tuyet Dang, and Judy Sanchez provided excellent technical assistance.
Abbreviations
ACC, acetyl-CoA carboxylase
AGPAT, 1-acylglycerol-3-phosphate-O-acyltransferase
ChREBP, carbohydrate regulatory element-binding protein
CREB, cAMP response element binding protein
DAG, diacylglycerol
DHA, docosahexaenoic acid
DGAT, diacylglycerol acyltransferase
ELOVL, elongation of very long chain fatty acids
EPA, eicosapentaenoic acid
ER, endoplasmic reticulum
GLC, gas-liquid chromatography
G6PD, glucose-6-phosphate dehydrogenase
GPAT, glycerol-3-phosphate acyl transferase
MGAT, monoacylglycerol acyltransferase
SCAP, SREBP cleavage-activating protein
SREBP, sterol regulatory element-binding protein
SCD-1, stearoyl-CoA desaturase 1
TFR, transferrin receptor
Published, JLR Papers in Press, October 6, 2008.
Footnotes
This work was supported by grants from the Perot Family Foundation and National Institutes of Health HL-38049, DK-081182. Y-A Moon was supported by an American Heart Association Texas Affiliate, Beginning Grant-in-Aid Award.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one table.
References
- 1.Cinti D. L., L. Cook, M. N. Nagi, and S. K. Suneja. 1992. The fatty acid chain elongation system of mammalian endoplasmic reticulum. Prog. Lipid Res. 31 1–51. [DOI] [PubMed] [Google Scholar]
- 2.Prasad M. R., M. N. Nagi, D. Ghesquier, L. Cook, and D. L. Cinti. 1986. Evidence for multiple condensing enzymes in rat hepatic microsomes catalyzing the condensation of saturated, monounsaturated, and polyunsaturated acyl coenzyme A. J. Biol. Chem. 261 8213–8217. [PubMed] [Google Scholar]
- 3.Nugteren D. H. 1965. The enzymatic chain elongation of fatty acids by rat-liver microsomes. Biochim. Biophys. Acta. 106 280–290. [DOI] [PubMed] [Google Scholar]
- 4.Moon Y-A., N. A. Shah, S. Mohapatra, J. A. Warrington, and J. D. Horton. 2001. Identification of a mammalian long chain fatty acyl elongase regulated by sterol regulatory element-binding proteins. J. Biol. Chem. 276 45358–45366. [DOI] [PubMed] [Google Scholar]
- 5.Tvrdik P., R. Westerberg, S. Silve, A. Asadi, A. Jakobsson, B. Cannon, G. Loison, and A. Jacobsson. 2000. Role of a new mammalian gene family in the biosynthesis of very long chain fatty acids and sphingolipids. J. Cell Biol. 149 707–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tvrdik P., A. Asadi, L. P. Kozak, J. Nedergaard, B. Cannon, and A. Jacobsson. 1997. Cig30, a mouse member of a novel membrane protein gene family, is involved in the recruitment of brown adipose tissue. J. Biol. Chem. 272 31738–31746. [DOI] [PubMed] [Google Scholar]
- 7.Zhang K., M. Kniazeva, M. Han, W. Li, Z. Yu, Z. Yang, Y. Li, M. L. Metzker, R. Allikmets, D. J. Zack, et al. 2001. A 5-bp deletion in ELOVL4 is associated with two related forms of autosomal dominant macular dystrophy. Nat. Genet. 27 89–93. [DOI] [PubMed] [Google Scholar]
- 8.Leonard A. E., E. G. Bobik, J. Dorado, P. E. Kroeger, L. T. Chuang, J. M. Thurmond, J. M. Parker-Barnes, T. Das, Y. S. Huang, and P. Mukerji. 2000. Cloning of a human cDNA encoding a novel enzyme involved in the elongation of long-chain polyunsaturated fatty acids. Biochem. J. 350 765–770. [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuzaka T., H. Shimano, N. Yahagi, T. Yoshikawa, M. Amemiya-Kudo, A. H. Hasty, H. Okazaki, Y. Tamura, Y. Iizuka, K. Ohashi, et al. 2002. Cloning and characterization of a mammalian fatty acyl-CoA elongase as a lipogenic enzyme regulated by SREBPs. J. Lipid Res. 43 911–920. [PubMed] [Google Scholar]
- 10.Jakobsson A., R. Westerberg, and A. Jacobsson. 2006. Fatty acid elongases in mammals: their regulation and roles in metabolism. Prog. Lipid Res. 45 237–249. [DOI] [PubMed] [Google Scholar]
- 11.Leonard A. E., B. Kelder, E. G. Bobik, L. T. Chuang, C. J. Lewis, J. J. Kopchick, P. Mukerji, and Y. S. Huang. 2002. Identification and expression of mammalian long-chain PUFA elongation enzymes. Lipids. 37 733–740. [DOI] [PubMed] [Google Scholar]
- 12.Vasireddy V., Y. Uchida, N. Salem, Jr., S. Y. Kim, M. N. A. Mandal, G. B. Reddy, R. Bodepudi, N. L. Alderson, J. C. Brown, H. Hama, et al. 2007. Loss of functional ELOVL4 depletes very long-chain fatty acids (>=C28) and the unique {omega}-O-acylceramides in skin leading to neonatal death. Hum. Mol. Genet. 16 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W., R. Sandhoff, M. Kono, P. Zerfas, V. Hoffmann, B. C. Ding, R. L. Proia, and C. X. Deng. 2007. Depletion of ceramides with very long chain fatty acids causes defective skin permeability barrier function, and neonatal lethality in ELOVL4 deficient mice. Int. J. Biol. Sci. 3 120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jump D. B., and S. D. Clarke. 1999. Regulation of gene expression by dietary fat. Annu. Rev. Nutr. 19 63–90. [DOI] [PubMed] [Google Scholar]
- 15.Jump D. B., S. D. Clarke, A. Thelen, M. Liimatta, B. Ren, and M. Badin. 1996. Dietary polyunsaturated fatty acid regulation of gene transcription. Prog. Lipid Res. 35 227–241. [DOI] [PubMed] [Google Scholar]
- 16.Sampath H., and J. M. Ntambi. 2005. Polyunsaturated fatty acid regulation of genes of lipid metabolism. Annu. Rev. Nutr. 25 317–340. [DOI] [PubMed] [Google Scholar]
- 17.Hannah V. C., J. Ou, A. Luong, J. L. Goldstein, and M. S. Brown. 2001. Unsaturated fatty acids down-regulate SREBP isoforms 1a and 1c by two mechanisms in HEK-293 cells. J. Biol. Chem. 276 4365–4372. [DOI] [PubMed] [Google Scholar]
- 18.Xu J., M. Teran-Garcia, J. H. Y. Park, M. T. Nakamura, and S. D. Clarke. 2001. Polyunsaturated fatty acids suppress hepatic sterol regulatory element-binding protein-1 expression by accelerating transcript decay. J. Biol. Chem. 276 9800–9807. [DOI] [PubMed] [Google Scholar]
- 19.Yahagi N., H. Shimano, A. H. Hasty, M. Amemiya-Kudo, H. Okazaki, Y. Tamura, Y. Iizuka, F. Shionoiri, K. Ohashi, J-i. Osuga, et al. 1999. A crucial role of sterol regulatory element-binding protein-1 in the regulation of lipogenic gene expression by polyunsaturated fatty acids. J. Biol. Chem. 274 35840–35844. [DOI] [PubMed] [Google Scholar]
- 20.Kim H. J., M. Takahashi, and O. Ezaki. 1999. Fish oil feeding decreases mature sterol regulatory element-binding protein 1 (SREBP-1) by down-regulation of SREBP-1c mRNA in mouse liver. A possible mechanism for down-regulation of lipogenic enzyme mrnas. J. Biol. Chem. 274 25892–25898. [DOI] [PubMed] [Google Scholar]
- 21.Ou J., H. Tu, B. Shan, A. Luk, R. A. DeBose-Boyd, Y. Bashmakov, J. L. Goldstein, and M. S. Brown. 2001. Unsaturated fatty acids inhibit transcription of the sterol regulatory element-binding protein-1c (SREBP-1c) gene by antagonizing ligand-dependent activation of the LXR. Proc. Natl. Acad. Sci. USA. 98 6027–6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen G., G. Liang, J. Ou, J. L. Goldstein, and M. S. Brown. 2004. Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver. Proc. Natl. Acad. Sci. USA. 101 11245–11250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dentin R., P. D. Denechaud, F. Benhamed, J. Girard, and C. Postic. 2006. Hepatic gene regulation by glucose and polyunsaturated fatty acids: a role for ChREBP. J. Nutr. 136 1145–1149. [DOI] [PubMed] [Google Scholar]
- 24.Dentin R., F. Benhamed, J. P. Pegorier, F. Foufelle, B. Viollet, S. Vaulont, J. Girard, and C. Postic. 2005. Polyunsaturated fatty acids suppress glycolytic and lipogenic genes through the inhibition of ChREBP nuclear protein translocation. J. Clin. Invest. 115 2843–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren B., A. P. Thelen, J. M. Peters, F. J. Gonzalez, and D. B. Jump. 1997. Polyunsaturated fatty acid suppression of hepatic fatty acid synthase and S14 gene expression does not require peroxisome proliferator-activated receptor alpha. J. Biol. Chem. 272 26827–26832. [DOI] [PubMed] [Google Scholar]
- 26.Goldstein J. L., R. B. Rawson, and M. S. Brown. 2002. Mutant mammalian cells as tools to delineate the sterol regulatory element-binding protein pathway for feedback regulation of lipid synthesis. Arch. Biochem. Biophys. 397 139–148. [DOI] [PubMed] [Google Scholar]
- 27.Horton J. D., J. L. Goldstein, and M. S. Brown. 2002. SREBPs:activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109 1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimomura I., H. Shimano, J. D. Horton, J. L. Goldstein, and M. S. Brown. 1997. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J. Clin. Invest. 99 838–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horton J. D., N. A. Shah, J. A. Warrington, N. N. Anderson, S. W. Park, M. S. Brown, and J. L. Goldstein. 2003. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. USA. 100 12027–12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimomura I., Y. Bashmakov, S. Ikemoto, J. D. Horton, M. S. Brown, and J. L. Goldstein. 1999. Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proc. Natl. Acad. Sci. USA. 96 13656–13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foretz M., C. Guichard, P. Ferre, and F. Foufelle. 1999. Sterol regulatory element binding protein-1c is a major mediator of insulin action on the hepatic expression of glucokinase and lipogenesis-related genes. Proc. Natl. Acad. Sci. USA. 96 12737–12742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Repa J. J., G. Liang, J. Ou, Y. Bashmakov, J. M. Lobaccaro, I. Shimomura, B. Shan, M. S. Brown, J. L. Goldstein, and D. J. Mangelsdorf. 2000. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRα and LXRβ. Genes Dev. 14 2819–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Botolin D., Y. Wang, B. Christian, and D. B. Jump. 2006. Docosahexaneoic acid (22:6,n-3) regulates rat hepatocyte SREBP-1 nuclear abundance by Erk- and 26S proteasome-dependent pathways. J. Lipid Res. 47 181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimano H., J. D. Horton, R. E. Hammer, I. Shimomura, M. S. Brown, and J. L. Goldstein. 1996. Overproduction of cholesterol and fatty acids causes massive liver enlargement in transgenic mice expressing truncated SREBP-1a. J. Clin. Invest. 98 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimano H., I. Shimomura, R. E. Hammer, J. L. Goldstein, M. S. Brown, and J. D. Horton. 1997. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J. Clin. Invest. 100 2115–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamilton J. G., and K. Comai. 1988. Rapid separation of neutral lipids, free fatty acids and polar lipids using prepacked silica Sep-Pak columns. Lipids. 23 1146–1149. [DOI] [PubMed] [Google Scholar]
- 37.Shimomura I., H. Shimano, B. S. Korn, Y. Bashmakov, and J. D. Horton. 1998. Nuclear sterol regulatory element-binding proteins activate genes responsible for the entire program of unsaturated fatty acid biosynthesis in transgenic mouse liver. J. Biol. Chem. 273 35299–35306. [DOI] [PubMed] [Google Scholar]
- 38.Engelking L. J., H. Kuriyama, R. E. Hammer, J. D. Horton, M. S. Brown, J. L. Goldstein, and G. Liang. 2004. Overexpression of Insig-1 in the livers of transgenic mice inhibits SREBP processing and reduces insulin-stimulated lipogenesis. J. Clin. Invest. 113 1168–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakai J., A. Nohturfft, D. Cheng, Y. K. Ho, M. S. Brown, and J. L. Goldstein. 1997. Identification of complexes between the COOH-terminal domains of sterol regulatory element-binding proteins (SREBPs) and SREBP cleavage-activating protein. J. Biol. Chem. 272 20213–20221. [DOI] [PubMed] [Google Scholar]
- 40.Liang G., J. Yang, J. D. Horton, R. E. Hammer, J. L. Goldstein, and M. S. Brown. 2002. Diminished hepatic response to fasting/refeeding and LXR agonists in mice with selective deficiency of SREBP-1c. J. Biol. Chem. 277 9520–9528. [DOI] [PubMed] [Google Scholar]
- 41.Yang J., J. L. Goldstein, R. E. Hammer, Y-A. Moon, M. S. Brown, and J. D. Horton. 2001. Decreased lipid synthesis in livers of mice with disrupted site-1 protease gene. Proc. Natl. Acad. Sci. USA. 98 13607–13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moon Y-A., and J. D. Horton. 2003. Identification of two mammalian reductases involved in the two-carbon fatty acyl elongation cascade. J. Biol. Chem. 278 7335–7343. [DOI] [PubMed] [Google Scholar]
- 43.Matsuda M., B. S. Korn, R. E. Hammer, Y. A. Moon, R. Komuro, J. D. Horton, J. L. Goldstein, M. S. Brown, and I. Shimomura. 2001. SREBP cleavage-activating protein (SCAP) is required for increased lipid synthesis in liver induced by cholesterol deprivation and insulin elevation. Genes Dev. 15 1206–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitchell K. J., K. I. Pinson, O. G. Kelly, J. Brennan, J. Zupicich, P. Scherz, P. A. Leighton, L. V. Goodrich, X. Lu, B. J. Avery, et al. 2001. Functional analysis of secreted and transmembrane proteins critical to mouse development. Nat. Genet. 28 241–249. [DOI] [PubMed] [Google Scholar]
- 45.Stanford W. L., J. B. Cohn, and S. P. Cordes. 2001. Gene-trap mutagenesis: past, present and beyond. Nat. Rev. Genet. 2 756–768. [DOI] [PubMed] [Google Scholar]
- 46.Horton J. D. 2002. Sterol regulatory element-binding proteins: transcriptional activators of lipid synthesis. Biochem. Soc. Trans. 30 1091–1095. [DOI] [PubMed] [Google Scholar]
- 47.Ishii S., K. I. Izuka, B. C. Miller, and K. Uyeda. 2004. Carbohydrate response element binding protein directly promotes lipogenic enzyme gene transcription. Proc. Natl. Acad. Sci. USA. 101 15597–15602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goldstein J. L., R. A. DeBose-Boyd, and M. S. Brown. 2006. Protein sensors for membrane sterols. Cell. 124 35–46. [DOI] [PubMed] [Google Scholar]
- 49.Yang T., P. J. Espenshade, M. E. Wright, D. Yabe, Y. Gong, R. Aebersold, J. L. Goldstein, and M. S. Brown. 2002. Crucial step in cholesterol homeostasis:sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 110 489–500. [DOI] [PubMed] [Google Scholar]
- 50.Yabe D., M. S. Brown, and J. L. Goldstein. 2002. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc. Natl. Acad. Sci. USA. 99 12753–12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu J., M. T. Nakamura, H. P. Cho, and S. D. Clarke. 1999. Sterol regulatory element binding protein-1 expression is suppressed by dietary polyunsaturated fatty acids. A mechanism for the coordinate suppression of lipogenic genes by polyunsaturated fats. J. Biol. Chem. 274 23577–23583. [DOI] [PubMed] [Google Scholar]
- 52.Yabe D., R. Komuro, G. Liang, J. L. Goldstein, and M. S. Brown. 2003. Liver-specific mRNA for Insig-2 down-regulated by insulin: implications for fatty acid synthesis. Proc. Natl. Acad. Sci. USA. 100 3155–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Engelking L. J., G. Liang, R. E. Hammer, K. Takaishi, H. Kuriyama, B. M. Evers, W-P. Li, J. D. Horton, J. L. Goldstein, and M. S. Brown. 2005. Schoenheimer effect explained - feedback regulation of cholesterol synthesis in mice mediated by Insig proteins. J. Clin. Invest. 115 2489–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y., M. Torres-Gonzalez, S. Tripathy, D. Botolin, B. Christian, and D. B. Jump. 2008. Elevated hepatic fatty acid elongase-5 activity affects multiple pathways controlling hepatic lipid and carbohydrate composition. J. Lipid Res. 49 1538–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schoenheimer R., and F. Breusch. 1933. Synthesis and destruction of cholesterol in the organism. J. Biol. Chem. 103 439–448. [Google Scholar]
- 56.Matsuzaka T., H. Shimano, N. Yahagi, M. Amemiya-Kudo, T. Yoshikawa, A. H. Hasty, Y. Tamura, J-i. Osuga, H. Okazaki, Y. Iizuka, et al. 2002. Dual regulation of mouse {Delta}5- and {Delta}6-desaturase gene expression by SREBP-1 and PPAR{alpha}. J. Lipid Res. 43 107–114. [PubMed] [Google Scholar]
- 57.Wang Y., D. Botolin, J. Xu, B. Christian, E. Mitchell, B. Jayaprakasam, M. G. Nair, J. M. Peters, J. V. Busik, L. K. Olson, et al. 2006. Regulation of hepatic fatty acid elongase and desaturase expression in diabetes and obesity. J. Lipid Res. 47 2028–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.