Abstract
Arrestins ensure the timely termination of receptor signaling. The role of rhodopsin phosphorylation in visual arrestin binding was established more than 20 years ago, but the effects of the number of receptor-attached phosphates on this interaction remain controversial. Here we use purified rhodopsin fractions with carefully quantified content of individual phosphorylated rhodopsin species to elucidate the impact of phosphorylation level on arrestin interaction with three biologically relevant functional forms of rhodopsin: light-activated and dark phosphorhodopsin and phospho-opsin. We found that a single receptor-attached phosphate does not facilitate arrestin binding, two are necessary to induce high affinity interaction, and three phosphates fully activate arrestin. Higher phosphorylation levels do not increase the stability of arrestin complex with light-activated rhodopsin but enhance its binding to the dark phosphorhodopsin and phospho-opsin. The complex of arrestin with hyperphosphorylated light-activated rhodopsin is less sensitive to high salt and appears to release retinal faster. These data suggest that arrestin likely quenches rhodopsin signaling after the third phosphate is added by rhodopsin kinase. The complex of arrestin with heavily phosphorylated rhodopsin, which appears to form in certain disease states, has distinct characteristics that may contribute to the phenotype of these visual disorders.
Arrestins play a key role in the regulation of G protein-coupled receptor signaling. Arrestins bind to active phosphorylated forms of their cognate receptors precluding further G protein activation, directing receptors to the coated pits for internalization, and switching the signaling to alternative G protein-independent pathways (reviewed in Refs. 1–3). Rhodopsin is now widely considered a prototypical G protein-coupled receptor, although its activation-dependent phosphorylation was discovered long before the existence of G protein-coupled receptor family was even suspected (4, 5). Visual (rod) arrestin was the first member of the family discovered, and the fact that its binding to light-activated rhodopsin is greatly enhanced by rhodopsin phosphorylation was established more than 20 years ago (6). Remarkable selectivity of rod arrestin for P-Rh*3 is ensured by a sequential multi-site interaction mechanism. The first step involves low affinity “prebinding” of arrestin to rhodopsin-attached phosphates or to parts of rhodopsin that change conformation upon light-activation via phosphate or activation sensor sites, respectively. Following that, mobilization of additional hydrophobic binding sites occurs when both sensors are simultaneously engaged by an encounter of arrestin with P-Rh* (recently reviewed in Ref. 7–9). The role of receptor-attached phosphates in arrestin activation that “primes” it for high affinity binding and the mechanism of function of the “phosphate sensor” in arrestin molecule have been elucidated in great detail (reviewed in Ref. 3, 7). Rhodopsin has many phosphorylation sites and can be progressively phosphorylated to incorporate multiple phosphates (10, 11). Surprisingly, the effect of an increasing level of rhodopsin phosphorylation on arrestin binding still remains controversial. Every conceivable model has been proposed: that arrestin affinity gradually increases with the level of rhodopsin phosphorylation (12, 13) or that tight arrestin binding is an all-or-nothing event requiring a certain number of receptor-attached phosphates. This “magic number” was reported to be one (14, 15), two (16–18), or three (19, 20), and in some cases conflicting reports came from the same group. Here we prepared rhodopsin fractions with different levels of phosphorylation thoroughly characterized by mass spectroscopy. We used these preparations to unambiguously establish the number of phosphates necessary for arrestin binding to light-activated rhodopsin. Our data indicate that arrestin activation is essentially a threshold mechanism: unphosphorylated and monophosphorylated rhodopsin demonstrate the same low binding levels, so that at least two receptor-attached phosphates are necessary to promote arrestin binding above that observed with unphosphorylated light-activated rhodopsin (Rh*). Three phosphates promote higher arrestin binding, whereas arrestin complexes with rhodopsin carrying 4–7 phosphates have distinct functional characteristics. Interestingly, heavy phosphorylation (5–7 receptor-attached phosphates) also greatly enhances arrestin binding to the two unpreferred forms of rhodopsin: dark (inactive) phosphorhodopsin and phospho-opsin.
EXPERIMENTAL PROCEDURES
Materials
Monoclonal rhodopsin 4D2 antibody (21) was a generous gift of Dr. Robert Molday. Rabbit polyclonal antibody raised against F4C1 epitope (peptide VDPVDGVVLVDPDYL) was custom-made by Sigma.
Preparation of Rod Outer Segments and Phosphorylated Rhodopsin
All of the procedures were performed under dim red light illumination unless otherwise indicated. Rod outer segments were prepared from 50 frozen bovine retinas (W. L. Lawson Co., Lincoln, NE) exactly as described (22). The rod outer segments pellet was resuspended in 100 mM phosphate buffer, pH 7.4, to a final rhodopsin concentration 0.5 mg/ml, and rhodopsin was phosphorylated in the presence of 3 mM ATP, 3 mM GTP, and 5 mM MgCl2 in bright light according to Ref. 11. Different illumination times (5, 30, 60, and 180 min) were used to obtain rhodopsin with diverse phosphorylation levels. Rhodopsin was fully regenerated by 11-cis-retinal (two additions at 1:3 molar ratio) in 100 mM potassium phosphate buffer, pH 7.4, supplemented with 2% bovine serum albumin, 1 mM MgCl2, and 0.5 mM EDTA for 1 h at room temperature (22). After regeneration the rod outer segments were pelleted (20,000 rpm, 30 min in Sorvall SS34 rotor) and resuspended in 6.3 ml of 50 mM Tris acetate buffer, pH 6.9, containing 1 mM CaCl2 and 1 mM MnCl2. Rhodopsin was solubilized by the addition of octyl glucoside (0.7 ml of 15% solution) and incubation for 30 min at 0 °C. The sample was centrifuged at 100,000 rpm for 45 min to remove insoluble material. Solubilized rhodopsin was purified by affinity chromatography on a concanavalin A-Sepharose 4B (6 ml, 2 mg Rh/ml of bed volume) as described (23). Rhodopsin species with different levels of phosphorylation were separated by chromatofocusing on a Mono-P column (4 ml, HR 5/200) as described (23). The fractions were pooled based on the elution profile, and rhodopsin was reconstituted into liposomes made from mixed phospholipids (Sigma, P5638) by dialysis against 10 mM Hepes buffer, pH 7.5, containing 100 mM NaCl, as described (24). Rhodopsin content in each fraction was than measured by quantitative Western blotting, where known amounts of purified bovine rhodopsin were used to construct calibration curves on every blot, as described (25).
The orientation of reconstituted rhodopsin in all of the fractions used for the binding assay was compared using 4D2 monoclonal antibody against N-terminal rhodopsin residues 2–39 (21). To this end, proteoliposomes containing 0.1 µg of rhodopsin were incubated for 60 min at room temperature in 100 µl of 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 1 mg/ml bovine serum albumin with 4D2 monoclonal antibody (1:100). The samples were diluted with 150 µl of the same buffer and proteoliposomes along with bound 4D2 antibody were pelleted by centrifugation for 30 min at 20 °C at 90,000 rpm in a TLA 120.1 rotor in a Beckman TL100 ultracentrifuge. Parallel samples with the same amount of liposomes served as controls. The pellets were dissolved in SDS sample buffer, and the amount of bound 4D2 antibody (reflecting the amount of rhodopsin in inside-out orientation) was quantified by Western blot with anti-mouse horseradish peroxidase-conjugated antibody. We found no appreciable differences in the orientation of rhodopsin with different phosphorylation levels. Judging by similar binding of 4D2 antibody/0.1 µg of rhodopsin to repeatedly sonicated original phosphorylated disc membranes, equal amounts of rhodopsin in inside-out and right-side-out orientation were present in all fractions.
Quantification of the Distribution of Rhodopsin with Different Levels of Phosphorylation
The methods we used for quantifying the extent of phosphorylation of bovine rhodopsin have been described in detail earlier (26). Briefly, purified bovine rhodopsin fractions from chromatofocusing were treated with AspN protease to release the C-terminal peptide of rhodopsin containing all of the known phosphorylation sites. The membranes were removed, and the peptides were analyzed by liquid chromatography-mass spectrometry (26). Quantification was based on the total number of ions detected at the masses corresponding to peptides containing between 0 and 7 phosphates. Signals from each species of peptide were corrected using factors determined previously (26) that account for the decrease in detection efficiency that occurs upon phosphorylation. The presence of phosphates on serines 334 and 343 in monophos-phorylated rhodopsin were determined from the abundance of unique b5 and b13 ions as described (26).
In Vitro Transcription and Translation
pGEM2-based plasmid containing bovine rod arrestin coding sequence with an “idealized” 5′-untranslated region (27) under the control of the SP6 promoter was linearized, transcribed, and translated in vitro in the presence of [3H]leucine and [14C]leucine (PerkinElmer Life Sciences) as previously described (16). The specific activity of the radiolabeled arrestin was 210–230 dpm/fmol.
Arrestin Purification
Untagged wild type bovine arrestin was expressed in Escherichia coli and purified to apparent homogeneity by sequential chromatography on heparin- and Q-Sepharose, as described (28).
Arrestin Binding to Rhodopsin
Arrestin binding to rhodopsin was performed as described (16, 18). Briefly, 75 fmol of in vitro translated radiolabeled arrestin were incubated in 50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 2 mM EDTA, and 1 mM EGTA with 0.15 µg of rhodopsin in a final volume of 75 µl for 5 min at 37 °C in room light (P-Rh*) or in the dark (dark P-Rh). After incubation, the samples were cooled and loaded onto 2-ml Sepharose 2B columns at 4 °C (under dim red light in case of dark P-Rh). Bound arrestin was eluted with rhodopsin-containing membranes in the void volume directly into scintillation vials. Nonspecific binding was determined by incubating arrestin with the same amount of liposomes as present in individual rhodopsin fractions and subtracted. The binding of purified arrestin to P-Rh* was performed using the same method, except that 1 µM pure arrestin was used instead of radiolabeled arrestin. To measure the kinetics of arrestin dissociation, the binding was performed at double concentrations of arrestin and rhodopsin, whereupon the samples were cooled on ice and diluted with ice-cold binding buffer. At indicated times (starting with t = 0) the aliquots were withdrawn, and the binding was determined, as described above. The data were analyzed using exponential decay equations in Graphpad Prizm to calculate the half-life of the complex.
Opsin was generated by treatment of the fractions with 10 mM NH2OH at room temperature in the light (2,000 lux) for 60 min. NH2OH was removed by dialyzing the samples three times for 1 h each against 1 liter of 10 mM Hepes-K, pH 7.4, 100 mM NaCl, 1 mM dithiothreitol, 0.1 mg/ml bovine serum albumin. Phospho-opsin was then quantified by Western blot and used in the direct binding assay as described for rhodopsin.
Statistical Analysis
Direct binding data were analyzed using one-way analysis of variance (SAS Institute, Cary, NC) with the phosphorylation level as a main factor, followed by a Bonferroni/Dunn post-hoc test with correction for multiple comparisons.
RESULTS
Purified Rhodopsin Preparations with Different Levels of Phosphorylation
One of the most likely sources of controversy in the published studies on rhodopsin phosphorylation levels required for arrestin binding is the use of phosphorhodopsin preparations that are not thoroughly characterized. These fall into two categories: “crude” phosphorhodopsin with wide range of phosphorylation levels (13, 16) or fractionated phosphorhodopsin that was not unambiguously characterized (12). In crude preparations the average phosphorylation level is usually known (16, 17), but the distribution of differentially phosphorylated species is not (this remains true even when this distribution is “calculated” based on assumptions that were not tested experimentally (13)). The most quantitative characterization of the distribution of different phosphorylated forms of rhodopsin requires proteolysis of rhodopsin C terminus followed by high pressure liquid chromatography and mass spectrometric quantification of peptides in various phosphorylation states (26). Therefore we phosphorylated bovine rhodopsin in isolated rod outer segments with endogenous rhodopsin kinase using a standard protocol (11, 12), fully regenerated phosphorhodopsin with 11-cis-retinal (22), purified and fractionated it by chromatofocusing (23), and then characterized the distribution of phosphorylated species in each fraction by mass spectrometry (26, 29). The results of this analysis clearly demonstrate the importance of thorough characterization of “purified” phosphorhodopsin. Very few fractions are actually homogeneous; most contain a mixture of different species, with the prevalent one or two representing 30–95% of the total (Table 1). However, 13 fractions of 18 contain more than 50% of a single form (Table 1). For binding experiments rhodopsin was reconstituted in phosphatidylcholine liposomes (24). Using the binding of the 4D2 monoclonal antibody directed against the rhodopsin N terminus (21), we ascertained that the orientation of reconstituted rhodopsin in all of the fractions was not significantly different (data not shown).
Table 1.
Rhodopsin fractions with different phosphorylation level used for binding experiments
| N | Fraction name | Phosphate composition | |
|---|---|---|---|
| 1 | C4FT | 0 P | 0P-100% |
| 2 | C4W | 0/1 P | 0P-97.7%; 1P-2.3% |
| 3 | C4P4 | 0/1 Pa | 0P-50.6%; 1P-49.4% |
| 4 | C4P5 | 0/1 Pb | 0P-54.8%; 1P-45.2% |
| 5 | C4P6 | 0/1/2 P | 0P-5.4%; 1P-22.7%; 2P-71.9% |
| 6 | C4P7 | 0/1/2/3 P | 0P-3.7%; 1P-19.8%; 2P-42.4%; 3P-34.1% |
| 7 | C1P2 | 2/3/4 P | 2P-3%; 3P-95%; 4P-3% |
| 8 | C3P5 | 1/2/3/4/5 P | 1P-4.2%; 2P-17%; 3P-43.7%; 4P-32.6%; 5P-2.4% |
| 9 | C1P1 | 2/3/4 Pa | 2P-2%; 3P-22%; 4P-74% |
| 10 | C4P8 | 2/3/4 Pb | 2P-2.9%; 3P-17.1%; 4P-79.9% |
| 11 | C4P9 | 2/3/4/5 Pa | 2P-1.9%; 3P-15.2%; 4P-31.7%; 5P-37.2% |
| 12 | C3P6 | 2/3/4/5 Pb | 2P-3.4%; 3P-3.8%; 4P-39.7%; 5P-53.2% |
| 13 | C1P3 | 3/4/5 P | 3P-2%; 4P-9%; 5P-90% |
| 14 | C2P7 | 4/5/6 P | 4P-8.3%; 5P-70.1%; 6P-21.6% |
| 15 | C3P7 | 3/4/5/6/7 P | 3P-4.2%; 4P-18.6%; 5P-42.3%; 6P- 31.8%; 7P- 3.1% |
| 16 | C4P10 | 4/5/6 Pa | 4P-17.9%; 5P-31.6%; 6P-50.5% |
| 17 | C3P9 | 5/6/7 Pa | 5P-14.1%; 6P-41%; 7P-44.8% |
| 18 | C3P8 | 5/6/7 Pb | 5P-4.3%; 6P-30%; 7P-65.5% |
Phosphorylation Dependence of Arrestin Binding to Phosphorhodopsin
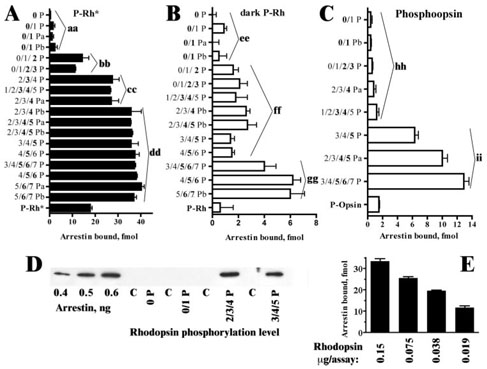
Next we tested rhodopsin fractions of defined composition for their ability to interact with arrestin using our standard direct binding assay with radiolabeled arrestin produced in cell-free translation (16, 30). The use of arrestin with specific activity of >200 dpm/fmol makes this assay sensitive enough to detect reliably even relatively low specific binding of arrestin to Rh* and modest changes in the binding to P-Rh* (31, 32). To increase the reliability of the data, we tested all 18 fractions, even though some of them have very similar composition (Table 1). We found that arrestin binding to fractions containing up to 50% monophosphorylated rhodopsin was not different from the binding to unphosphorylated rhodopsin, suggesting that a single receptor-attached phosphate is not sufficient to enhance arrestin binding (Fig. 1A). The addition of the second phosphate increases arrestin binding ~7-fold, even in the fraction where only 72% of rhodopsin is doubly phosphorylated, with the rest being monophosphorylated (Fig. 1A). The fractions containing >80% of rhodopsin with three phosphates demonstrate even higher binding, whereas rhodopsin with four phosphates shows a small but statistically significant further increase. Higher levels of phosphorylation (up to seven phosphates) do not increase arrestin binding beyond the levels observed with four phosphates (Fig. 1A).
Figure 1. The effect of rhodopsin phosphorylation level on arrestin binding.
Purified reconstituted light-activated (A) or dark (B) phosphorhodopsin or phospho-opsin (C) (0.15 µg) with the indicated levels of phosphorylation (Table 1) was incubated with 75 fmol of radiolabeled arrestin in 75 µl for 5 min at 37 °C in the light (A and C) or in the dark (B), and then bound arrestin was separated from free and quantified, as described under “Experimental Procedures.” The binding data were analyzed using one-way analysis of variance with phosphorylation level as a main factor, followed by a Bonferroni/Dunn post-hoc test with correction for multiple comparisons. The binding levels were grouped (aa–ii) so that they are not significantly different within the group and significantly different from other groups on the same panel (p < 0.0001 in all cases). D, purified arrestin (final concentration, 1 µM) was incubated with 0.15 µg of indicated rhodopsin preparation or equal amount of empty liposomes (C lones indicate control), as described under “Experimental Procedures.” Bound arrestin was separated from free by gel filtration, and ⅙ of the sample was subjected to SDS-PAGE and visualized by Western blot using rabbit polyclonal antibody raised against a peptide spanning residues 38–53 of bovine rod arrestin. E, the effect of the amount of binding-competent rhodopsin (fraction C3P5) on arrestin binding. Indicated amounts of phosphorylated rhodopsin were used in a standard binding assay with 1 nM radiolabeled arrestin. Note that a single rhodopsin-attached phosphate does not affect arrestin binding to any of the functional forms of rhodopsin tested.
Based on the available data (20, 33), we cannot rule out the possibility that different phosphorylation sites differentially affect arrestin interaction, so that only rhodopsin with the phosphate at certain positions binds arrestin effectively. Therefore, to test whether the absence of an increase of arrestin binding to monophosphorylated rhodopsin could be attributable to the prevalence of rhodopsin with a single phosphate attached in the “wrong” position, we analyzed the phosphorylated sites in C4P5 fraction (Table 1) by mass spectrometry and found that rhodopsin species phosphorylated at Ser343 and Ser334 were virtually equally represented. Because only half of rhodopsin in this fraction was phosphorylated (Table 1), each species of monophosphorhodopsin represents only a small fraction of the total. To determine whether we did not observe an increase of arrestin binding in the fractions containing monophosphorylated rhodopsin because of the presence of an insufficient amount of binding-competent phosphorhodopsin, we tested the effect of 2-, 4-, and 8-fold reduction of the phosphorhodopsin concentration in the assay on arrestin binding (Fig. 1E). We found that even with as little as 0.019 µg (0.49 pmol) of a binding-competent rhodopsin in the assay (fraction C3P5 with >2 phosphates per rhodopsin), more than 11 fmol of arrestin is bound (Fig. 1E), i.e. >3 times more than the binding to the standard amount (0.15 µg or 3.9 pmol) of unphosphorylated and monophosphorylated rhodopsin (Fig. 1A). Thus, the absence of detectable enhancement of arrestin binding by a single rhodopsin-attached phosphate is unlikely to be attributable to the small content of an individual binding-competent rhodopsin species. Previous in vivo analysis of rhodopsin phosphorylation showed that Ser343 is phosphorylated first, whereas Ser334 is dephosphorylated last (29). In the context of these findings, these data indicate that neither rhodopsin monophos-phorylated at Ser343 immediately after the bleach nor long-lived species with a phosphate attached to Ser334 binds arrestin better than unphosphorylated light-activated rhodopsin. Thus, at least two phosphates are necessary to increase arrestin binding above the level characteristic for the unphosphorylated Rh*, three are required for high affinity interaction, and four result in maximum binding.
One of the limitations of the direct binding assay is that achievable concentrations of radiolabeled arrestin are in the nanomolar range (16, 17). Therefore we tested whether rhodopsin phosphorylation levels affect arrestin binding in the same way at more physiological micromolar concentrations, using unphosphorylated rhodopsin and selected fractions with phosphorylation levels ranging from one to five rhodopsin-attached phosphates. To this end, 1 µM of purified bovine arrestin was incubated with 0.15 µg of rhodopsin, and then bound arrestin was separated from free by gel filtration, and equal aliquots of the eluate were subjected to Western blotting (Fig. 1D). These experiments confirmed that light-activated monophospho-rylated rhodopsin does not bind more arrestin than unphosphorylated rhodopsin and that rhodopsin carrying two to three phosphates or four to five phosphates shows virtually the same binding (Fig. 1D).
Multiple sites on arrestin participate in high affinity binding to P-Rh*, some of which do not directly interact with receptor-attached phosphates (16, 31, 32, 34). According to the current model of arrestin-rhodopsin interaction, the most important function of the receptor-attached phosphates is to “activate” arrestin by stabilizing the high affinity receptor-binding state (18, 35, 36). Transition between the inactive and active state involves a global conformational rearrangement of the arrestin molecule (Refs. 32, 35, and 37–43; reviewed in Refs. 7–9). In contrast, specific arrestin binding to dark phosphorhodopsin that has been detected by several independent methods (16, 31, 44, 45) is largely mediated by the phosphates. Therefore, to gauge the direct contribution of rhodopsin-attached phosphates, we measured arrestin binding to dark P-Rh (Fig. 1B). In this case we also found no appreciable difference between unphosphorylated and monophosphorylated rhodopsin. The presence of two to five phosphates increases the binding ~2–3-fold, whereas the fractions containing >30% of rhodopsin with six and seven phosphates demonstrated 4–6-fold higher binding then dark Rh (Fig. 1B and Table 1). Thus, at least two phosphates are necessary to promote arrestin binding via purely electrostatic interactions. Although the receptor-binding surface of arrestin has multiple exposed positive charges (31), even as many as six or seven phosphates on the inactive rhodopsin support the binding levels that are only ⅙ of the binding to the corresponding light-activated forms (Fig. 1, A and B).
In the retina light-activated rhodopsin decays to phospho-opsin as it releases all-trans-retinal. This process occurs much more slowly than the rate at which the photoresponse is inactivated by rhodopsin phosphorylation, arrestin binding, and transducin inactivation. Thus, inactivation of rhodopsin by retinal release likely occurs while arrestin is bound. It is generally believed that the decay of active rhodopsin into opsin facilitates the release of bound arrestin (16). Therefore the effect of phos-phorylation level on arrestin-opsin interaction is biologically important. In our earlier studies with heterogeneous preparations of phospho-opsin, we found that arrestin binds this functional form of rhodopsin surprisingly well, better than dark P-Rh (16). We prepared phospho-opsin from selected fractions and tested its ability to bind arrestin (Fig. 1C). We found that phospho-opsin with up to four phosphates binds arrestin at about the level of dark P-Rh phosphorylated to the same extent, whereas opsin with five or more phosphates demonstrates about three times higher binding than corresponding dark P-Rh (Fig. 1C). Thus, heavy phosphorylation enhances arrestin binding to phospho-opsin to a much greater extent than to other functional forms of rhodopsin.
Rhodopsin Phosphorylation Level and the Stability of the Arrestin-Receptor Complex
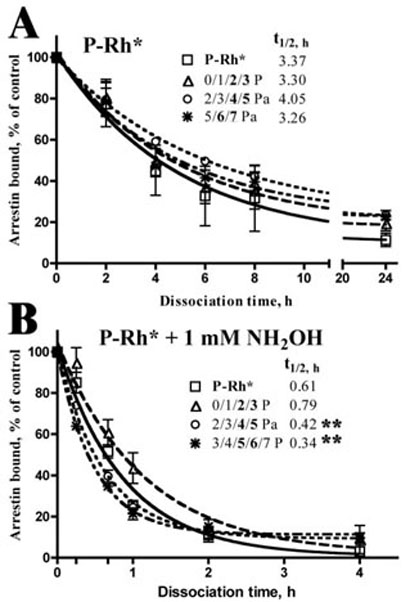
To ensure reliable signal shut-off, arrestin has to stay bound at least until P-Rh* decays to phospho-opsin. The loss of the active receptor conformation is generally believed to serve as the signal for arrestin release (2, 7–9). Ultimately arrestin must dissociate to allow receptor dephos-phorylation (46) and recycling back to the active pool. Thus, for the proper function of rod photoreceptors, the timing of arrestin release is as important as its timely binding. Therefore we compared the effect of the phosphorylation level of P-Rh* and phospho-opsin on the stability of the arrestin-receptor complex. To this end, after the standard binding in concentrated samples, we cooled them on ice, diluted to the final concentration of 1 nM radiolabeled arrestin and 0.15 µg of rhodopsin/75 µl, and measured arrestin that remains bound in aliquots withdrawn at different time points from 0 to 24 h (Fig. 2). The half-life of the complex with P-Rh* was essentially the same (3.3–4 h) regardless of rhodopsin phosphorylation, which varied from two to three up to six to seven phosphates/molecule (Fig. 2A). As could be expected, the presence of hydroxylamine that facilitates the removal of retinal from light-activated rhodopsin by chemically reacting with it to form retinal oxime (47) significantly accelerated arrestin dissociation (Fig. 2B). Interestingly, under these conditions arrestin release from rhodopsin with high levels of phosphorylation (four to five and five to six phosphates/mol) was significantly faster (half-life, 0.3–0.4 h) than from rhodopsin containing two to three phosphates (half-life, 0.6–0.8 h). These data suggest that in heavily phosphorylated rhodopsin (with >4 attached phosphates), the release of arrestin and retinal are interdependent. Conceivably, arrestin binding to highly phosphorylated rhodopsin increases retinal accessibility to hydroxylamine, and retinal removal in its turn accelerates arrestin dissociation.
Figure 2. The effect of phosphorylation level on the stability of arrestin-rhodopsin complex.
After the standard 5-min incubation at 37 °C of radiolabeled arrestin with P-Rh* in a concentrated sample, the reaction mix was diluted with binding buffer without (A) or with (B) 1 mM hydroxylamine. Aliquots were taken at indicated times to determine the amount of remaining bound arrestin, which was expressed as a percentage of that bound at time 0. Phosphorhodopsin fractions used are fully described in Table 1. Half-life of the complex (t½) was calculated using exponential decay model in GraphPad Prizm. The only phosphorylation-dependent difference in arrestin dissociation rate was found in the presence of hydroxylamine, where the dissociation was faster in case of heavily phosphorylated 2/3/4/5 and 3/4/5/6/7 fractions (*, p < 0.05, as compared with the 0/1/2/3 fraction).
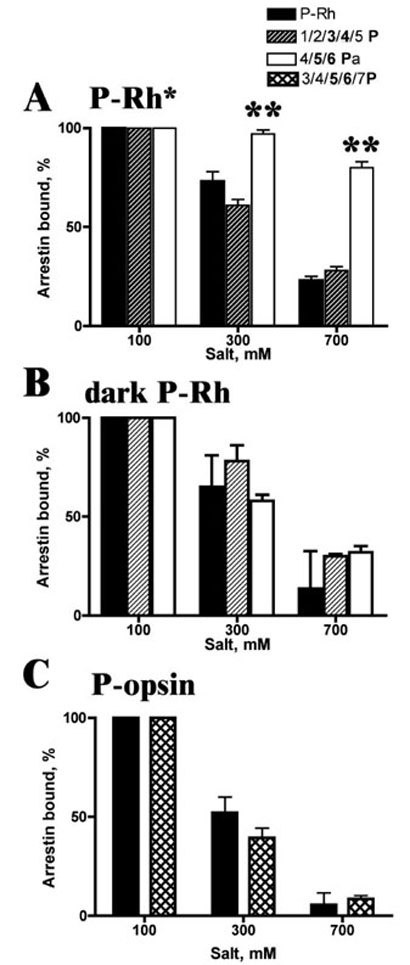
Arrestin binding to dark phosphorhodopsin is mostly mediated by ionic interactions and therefore is very sensitive to high salt inhibition (16). In contrast, the binding to P-Rh* is fairly resistant to high salt because it involves additional hydrophobic interactions (16, 43). The engagement of these additional interactions by multi-phosphorylated P-Rh and opsin could explain enhanced arrestin binding to dark P-Rh and phospho-opsin carrying five or more phosphates (Fig. 1, B and C). To test this hypothesis, we compared salt sensitivity of arrestin binding to P-Rh*, dark P-Rh, and phospho-opsin preparations with three to four and five to six phosphates (Fig. 3). We found that the salt sensitivity of arrestin binding to dark P-Rh and phospho-opsin does not depend on the number of rhodopsin-attached phosphates, suggesting that additional phosphates do not change the chemical nature of arrestin interactions with nonpreferred functional forms of rhodopsin. Unexpectedly, we found that arrestin binding to its preferred target, P-Rh*, becomes even more resistant to salt inhibition when rhodopsin is heavily phosphorylated (Fig. 3), suggesting that arrestin transition into the active high affinity receptor-binding state involving the engagement of additional binding sites may be a graded rather than “all-or-nothing” process. This hypothesis is consistent with the stepwise increase in arrestin binding to rhodopsin with two, three, and four or more phosphates (Fig. 1A).
Figure 3. Salt sensitivity of arrestin interactions with different functional forms of rhodopsin.
Arrestin binding to the P-Rh* (A), dark P-Rh (B), and phospho-opsin (C) fractions with the indicated distribution of phosphorylated species (Table 1) was measured under standard (100 mM) ionic strength or in the presence of additional potassium acetate, bringing total ionic strength to 300 and 700 mM.
DISCUSSION
G protein-coupled receptors are the largest known family of proteins encoded by ~4% of genes in animals from Caenorhabditis elegans to mammals (48). The signaling of most members of this tremendously diverse family is regulated by a surprisingly uniform mechanism: active receptor is phosphorylated by a specialized kinase, whereupon arrestin binds to active phosphoreceptor, precluding further G protein activation and often redirecting the signaling to alternative pathways (2, 3, 9, 49). Phototransduction was the signaling pathway in which receptor phosphorylation and its role in arrestin binding were discovered (4–6). The molecular mechanism whereby receptor-attached phosphates activate arrestin and facilitate its transition into a high affinity receptor-binding state has been described (16, 32, 38, 43), and the structural basis of arrestin activation is well understood (18, 35–37, 50, 51). However, it is still unclear how the number of rhodopsin-attached phosphates regulates arrestin binding and how many phosphates are required for arrestin activation. Different answers to this question have distinct biological implications. Clarification of this point would improve the mechanistic understanding of arrestin binding to different G protein-coupled receptors, which is arguably one of the most ubiquitous regulatory protein-protein interactions occurring in virtually every eukaryotic cell (9). An additional issue depending on this answer in the visual system is the mechanism ensuring a remarkable reproducibility of single photon response in rods. One of the explanations of this reproducibility is based on the assumption that rhodopsin is deactivated through a large number of steps (52), the great majority of which is believed to be sequential phosphorylation by rhodopsin kinase (20, 33, 53, 54), whereas in other models multiplicity of inactivation steps is not required (55).4 Direct binding studies with numerous reconstituted fractions containing different phosphorhodopsin species suggest that as soon as light-activated rhodopsin has two to three phosphates, arrestin can bind it with high affinity (Fig. 1), forming complexes with sufficient stability to ensure high fidelity of signal shut-off (Fig. 2). Additional phosphates only marginally enhance arrestin binding to P-Rh* (Fig. 1). Arrestin activation appears to involve three distinct thresholds that require two, three, and four rhodopsin-attached phosphates, respectively (Fig. 1).
The limitation of the direct binding assay is that only nanomolar concentrations of radiolabeled arrestin can be achieved (16, 17). In rod photoreceptors arrestin is expressed at about 0.8:1 ratio to rhodopsin (25, 57, 58), which, considering that rhodopsin concentration in the outer segment is 2–3 mM (59), translates into extremely high intracellular concentrations. However, there are two mechanisms that limit the concentration of active arrestin monomer, which is the only species that can bind rhodopsin (60). In low light conditions where rods operate, the great majority of arrestin is kept away from the compartment where rhodopsin resides (57), with the proportion present in the outer segments estimated at 1–7% (25, 58, 61). Moreover, rod arrestin self-associates, forming tetramers in a cooperative fashion (60, 62, 63). The content of rod arrestin in the outer segment in the dark along with recently measured self-association constants (60) yield an estimate of the concentration of free arrestin monomer at ~10 µM. Our experiments show that relative arrestin binding to rhodopsin preparations with different phosphorylation levels is essentially the same at 1 nM and 1 µM arrestin (Fig. 1, A and D), validating the physiological relevance of our data.
Because we did not have purified monophosphorylated rhodopsin preparations carrying the phosphate at each of the seven possible sites, our data do not formally rule out a theoretical possibility that a single phosphate in certain places could be sufficient for tight arrestin binding. However, this seems unlikely for several reasons. First, we observed no phosphorylation-induced enhancement of arrestin binding by phosphates at Ser343 and Ser334 in fraction C4P5 (Fig. 1 and Table 1). Second, the salt bridge between Arg175 and Asp296 was shown to function as the main phosphate sensor in arrestin (32, 35, 43) in such a way that its disruption by charge reversal mutations from either side (R175E or D296R) “preactivates” arrestin, enabling its high affinity binding to Rh*, whereas its reconstruction even in the opposite configuration (by a combination of R175E and D296R mutations) restores normal arrestin selectivity for P-Rh* (35, 36). These results imply a purely electrostatic mechanism of phosphate-induced arrestin activation, making it unlikely that a single rhodopsin-attached phosphate in one position cannot turn it on, but the same phosphate attached to a different serine or threonine can. It is worth noting that both the structure (36, 50, 51) and function (39, 41) of the phosphate sensor are remarkably conserved in the arrestin family, and its ability to respond to the phosphates regardless of the sequence context of the phosphorylated residues is necessary to enable four mammalian arrestin subtypes to “serve” hundreds of structurally diverse G protein-coupled receptors.
Considering that arrestin is expressed in photoreceptors at a much higher level than rhodopsin kinase (25, 58, 59), it seems very unlikely that rhodopsin phosphorylation proceeds much further than the minimum required for arrestin binding in vivo. Indeed, mono-, di-, and tri-phosphorylated rhodopsin was found to predominate in wild type mouse rods with negligible proportion of highly phosphorylated species (29, 56, 61), and arrestin-dependent acceleration of photoresponse recovery is clearly detectable in mice expressing mutant rhodopsin that has only three phosphorylation sites (20). These data in conjunction with our results (Figs. 1 and 2) indicate that rhodopsin inactivation is likely achieved in three or four steps (two to three phosphorylation events followed by arrestin binding), favoring the models that explain limited variability of the single photon response by a small number of steps of rhodopsin inactivation (55).4 However, the rhodopsin C terminus in different mammalian species has six to seven fairly well conserved phosphorylation sites, suggesting that their abundance is biologically important. Moreover, the elimination of even one or two of six sites in mouse rhodopsin has detectable effects on photoresponse kinetics, and the elimination of three substantially slows down rhodopsin inactivation (20, 33). It is tempting to speculate that these “supernumerary” sites are necessary to increase the probability that transient relatively low affinity interaction of rhodopsin kinase with light-activated rhodopsin will be productive, i.e. will result in the phosphorylation of at least one of the sites. This would become increasingly important with the progression of phosphorylation, e.g. after two sites are already phosphorylated, wild type rhodopsin would retain four accessible targets, whereas the mutant carrying three sites would have only one left. This consideration appears to be particularly important because the inactivation rate at the rhodopsin level was shown to be extremely fast, with a half-life of active rhodopsin of <80 ms (64).
Dark phosphorhodopsin can be generated in vivo by phosphorylation of more rhodopsin molecules than are activated by light (65–67) or by regeneration of rhodopsin before dephosphorylation has been completed. Rhodopsin kinase was shown to be activated by direct binding to the light-activated rhodopsin (68). Apparently, because of a high concentration of rhodopsin in the disc, membrane-activated kinase can phosphorylate not only the rhodopsin molecule it interacts with, but also its neighbors. Phosphorylation of unbleached pigment by this mechanism might contribute to light adaptation of cone photoreceptors (69). Rod arrestin specifically binds dark phosphorhodopsin (Figs. 1 and 3) (16, 45). Our data suggest that the phosphorylation of inactive rhodopsin can play a role in light adaptation in rods via two mechanisms, which are not mutually exclusive. First, the signaling by prephosphorylated rhodopsin would be quenched faster because it would need fewer additional phosphorylation events to reach the threshold necessary for the high affinity arrestin binding (Fig. 1A). Even monophos-phorylation of dark rhodopsin can play this role. In addition, by virtue of its higher affinity for arrestin than that of microtubules which hold rod arrestin in the inner segment of dark-adapted photoreceptors (61), dark phosphorhodopsin would increase the concentration of arrestin in the outer segments, thereby facilitating rhodopsin inactivation. Our data (Fig. 1B) indicate that multi-phosphorylated inactive rhodopsin would be more effective in this regard. Relatively slow rhodopsin dephosphorylation in vivo (29) suggests that these mechanisms may well operate in rods.
Ultimately light-activated rhodopsin releases all-trans-retinal. Its return to the active pool requires the release of bound arrestin, dephosphorylation, and regeneration with 11-cis-retinal. Recent studies suggest that the release of retinal and bound arrestin are interdependent (47, 70). We found that complexes of arrestin with rhodopsin with two to three phosphates and five to six phosphates have similarly high stability (Fig. 2A), possibly because of relatively high arrestin affinity for phospho-opsin with bound all-trans-retinal (47). The addition of hydroxylamine, that reacts with retinal and forcibly removes it from light-activated rhodopsin, facilitates arrestin dissociation. Under these conditions, arrestin dissociates from heavily phosphorylated rhodopsin carrying four to six phosphates faster than from rhodopsin that only has two to three phosphates (Fig. 2B). These data suggest that in arrestin complexes with rhodopsin that has four to six phosphates, retinal is more accessible for hydroxylamine. Interestingly, at the same phosphorylation level opsin demonstrates substantially higher arrestin binding than dark phosphorhodopsin (Fig. 1, B and C), indicating that phospho-opsin, especially carrying five or more phosphates, has significant ability to retain arrestin in the outer segment. Indeed, arrestin return to the inner segment in the dark is slow, and its kinetics closely correlates with the disappearance of multi-phosphorylated rhodopsin species (61).
Recent findings show that depending on the position and number of receptor-attached phosphates, the same nonvisual arrestin can form functionally distinct complexes with N-formyl-peptide receptor (71), V2 vasopressin receptor (72), and angiotensin II receptor (73) (reviewed in Ref. 3). Importantly, these complexes link receptor to different arrestin-mediated signaling pathways (72, 73). We found that arrestin complexes with rhodopsin containing five or more phosphates differ from those with rhodopsin carrying two to four phosphates in terms of stability in the presence of hydroxylamine (Fig. 2B) and high salt sensitivity (Fig. 3). These data suggest that, similarly to nonvisual subtypes, rod arrestin can form structurally and functionally distinct complexes with rhodopsin. Several rhodopsin mutants associated with autosomal dominant retinitis pigmentosa that demonstrate constitutive activity in vitro were shown to be constitutively hyperphosphorylated and bound to arrestin (74, 75). Persistent association of mammalian rod arrestin with hyperphosphorylated rhodopsin apparently causes cellular dysfunction (76) and contributes to photoreceptor death (77), similar to earlier observations in Drosophila (78, 79). These complexes in mammalian photoreceptors were hypothesized to cause disease phenotypes by inducing rhodopsin mislocalization (76, 77). The recent discovery that rod arrestin directly interacts with protein kinase JNK3 and ubiquitin ligase Mdm2 (8), both of which participate in life-ordeath decisions in the cell, suggests that erroneous arrestin-dependent signaling may also play a role in the demise of photoreceptors. Our data show that rod arrestin complexes with hyperphosphorylated rhodopsin do indeed differ from “normal” ones containing rhodopsin with two to three phosphates and provide a model for further studies of the functional capabilities of these complexes to elucidate the molecular mechanism of retinal degeneration caused by activating rhodopsin mutations.
Acknowledgments
We are grateful to Drs. T. Shinohara, Rosalie K. Crouch, and Robert S. Molday for bovine rod arrestin cDNA, 11-cis-retinal, and anti-rhodopsin 4D2 antibody, respectively.
This work was supported by National Institutes of Health Grants EY11500 (to V. V. G.) and EY06641 (to J. B. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: P-Rh*, phosphorylated light-activated rhodopsin; P-Rh, phosphorylated rhodopsin; Rh*, unphosphorylated light-activated rhodopsin.
P. Bisegna, G. Caruso, D. Andreucci, L. Shen, V. V. Gurevich, H. E. Hamm, and E. DiBenedetto, submitted for publication.
References
- 1.Carman CV, Benovic JL. Curr. Opin. Neurobiol. 1998;8:335–344. doi: 10.1016/s0959-4388(98)80058-5. [DOI] [PubMed] [Google Scholar]
- 2.Lefkowitz RJ, Shenoy SK. Science. 2005;308:512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- 3.Gurevich VV, Gurevich EV. Pharmacol. Ther. 2006;110:465–502. doi: 10.1016/j.pharmthera.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuhn H, Dreyer WJ. FEBS Lett. 1972;20:1–6. doi: 10.1016/0014-5793(72)80002-4. [DOI] [PubMed] [Google Scholar]
- 5.Bownds D, Dawes J, Miller J, Stahlman M. Nat. New. Biol. 1972;237:125–127. doi: 10.1038/newbio237125a0. [DOI] [PubMed] [Google Scholar]
- 6.Kuhn H, Hall SW, Wilden U. FEBS Lett. 1984;176:473–478. doi: 10.1016/0014-5793(84)81221-1. [DOI] [PubMed] [Google Scholar]
- 7.Gurevich VV, Gurevich EV. Trends Pharmacol. Sci. 2004;25:59–112. doi: 10.1016/j.tips.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 8.Song X, Raman D, Gurevich EV, Vishnivetskiy SA, Gurevich VV. J. Biol. Chem. 2006;281:21491–21499. doi: 10.1074/jbc.M603659200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gurevich EV, Gurevich VV. Genome Biol. 2006;7:236. doi: 10.1186/gb-2006-7-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilden U, Kuhn H. Biochemistry. 1982;21:3014–3022. doi: 10.1021/bi00541a032. [DOI] [PubMed] [Google Scholar]
- 11.Wilden U, Kuhn H. Biochemistry. 1982;21:3014–3022. doi: 10.1021/bi00541a032. [DOI] [PubMed] [Google Scholar]
- 12.Wilden U. Biochemistry. 1995;34:1446–1454. doi: 10.1021/bi00004a040. [DOI] [PubMed] [Google Scholar]
- 13.Gibson SK, Parkes JH, Liebman PA. Biochemistry. 2000;39:5738–5749. doi: 10.1021/bi991857f. [DOI] [PubMed] [Google Scholar]
- 14.Ohguro H, Van Hooser JP, Milam AH, Palczewski K. J. Biol. Chem. 1995;270:14259–14262. doi: 10.1074/jbc.270.24.14259. [DOI] [PubMed] [Google Scholar]
- 15.Krupnick JG, Gurevich VV, Benovic JL. J. Biol. Chem. 1997;272:18125–18131. doi: 10.1074/jbc.272.29.18125. [DOI] [PubMed] [Google Scholar]
- 16.Gurevich VV, Benovic JL. J. Biol. Chem. 1993;268:11628–11638. [PubMed] [Google Scholar]
- 17.Gurevich VV, Dion SB, Onorato JJ, Ptasienski J, Kim CM, Sterne-Marr R, Hosey MM, Benovic JL. J. Biol. Chem. 1995;270:720–731. doi: 10.1074/jbc.270.2.720. [DOI] [PubMed] [Google Scholar]
- 18.Vishnivetskiy SA, Schubert C, Climaco GC, Gurevich YV, Velez M-G, Gurevich VV. J. Biol. Chem. 2000;275:41049–41057. doi: 10.1074/jbc.M007159200. [DOI] [PubMed] [Google Scholar]
- 19.Ohguro H, Johnson RS, Ericsson LH, Walsh KA, Palczewski K. Biochemistry. 1994;33:1023–1028. doi: 10.1021/bi00170a022. [DOI] [PubMed] [Google Scholar]
- 20.Mendez A, Burns ME, Roca A, Lem J, Wu LW, Simon MI, Baylor DA, Chen J. Neuron. 2000;28:153–164. doi: 10.1016/s0896-6273(00)00093-3. [DOI] [PubMed] [Google Scholar]
- 21.Hicks D, Molday RS. Exp. Eye Res. 1986;42:55–71. doi: 10.1016/0014-4835(86)90017-5. [DOI] [PubMed] [Google Scholar]
- 22.McDowell JH. Methods Neurosci. 1993;15:123–130. [Google Scholar]
- 23.McDowell JH, Nawrocki JP, Hargrave PA. Methods Enzymol. 2000;315 doi: 10.1016/s0076-6879(00)15835-5. [DOI] [PubMed] [Google Scholar]
- 24.Niu L, Kim JM, Khorana HG. Proc. Natl. Acad. Sci. U. S. A. 2002;99:13409–13412. doi: 10.1073/pnas.212518899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanson SM, Gurevich EV, Vishnivetskiy SA, Ahmed MR, Song X, Gurevich VV. Proc. Natl. Acad. Sci. U.S.A. 2007;104:3125–3128. doi: 10.1073/pnas.0610886104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee KA, Craven KB, Niemi GA, Hurley JB. Protein Sci. 2002;11:862–874. doi: 10.1110/ps.3870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gurevich VV. Methods Enzymol. 1996;275:382–397. doi: 10.1016/s0076-6879(96)75023-1. [DOI] [PubMed] [Google Scholar]
- 28.Gurevich VV, Benovic JL. Methods Enzymol. 2000;315:422–437. doi: 10.1016/s0076-6879(00)15859-8. [DOI] [PubMed] [Google Scholar]
- 29.Kennedy MJ, Lee KA, Niemi GA, Craven KB, Garwin GG, Saari JC, Hurley JB. Neuron. 2001;31:87–101. doi: 10.1016/s0896-6273(01)00340-3. [DOI] [PubMed] [Google Scholar]
- 30.Gurevich VV, Benovic JL. J. Biol. Chem. 1992;267:21919–21923. [PubMed] [Google Scholar]
- 31.Hanson SM, Gurevich VV. J. Biol. Chem. 2006;281:3458–3462. doi: 10.1074/jbc.M512148200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gurevich VV, Benovic JL. Mol. Pharmacol. 1997;51:161–169. doi: 10.1124/mol.51.1.161. [DOI] [PubMed] [Google Scholar]
- 33.Doan T, Mendez A, Detwiler PB, Chen J, Rieke F. Science. 2006;313:530–533. doi: 10.1126/science.1126612. [DOI] [PubMed] [Google Scholar]
- 34.Vishnivetskiy SA, Hosey MM, Benovic JL, Gurevich VV. J. Biol. Chem. 2004;279:1262–1268. doi: 10.1074/jbc.M308834200. [DOI] [PubMed] [Google Scholar]
- 35.Vishnivetskiy SA, Paz CL, Schubert C, Hirsch JA, Sigler PB, Gurevich VV. J. Biol. Chem. 1999;274:11451–11454. doi: 10.1074/jbc.274.17.11451. [DOI] [PubMed] [Google Scholar]
- 36.Hirsch JA, Schubert C, Gurevich VV, Sigler PB. Cell. 1999;97:257–269. doi: 10.1016/s0092-8674(00)80735-7. [DOI] [PubMed] [Google Scholar]
- 37.Vishnivetskiy SA, Hirsch JA, Velez M-G, Gurevich YV, Gurevich VV. J. Biol. Chem. 2002;277:43961–43968. doi: 10.1074/jbc.M206951200. [DOI] [PubMed] [Google Scholar]
- 38.Gray-Keller MP, Detwiler PB, Benovic JL, Gurevich VV. Biochemistry. 1997;36:7058–7063. doi: 10.1021/bi963110k. [DOI] [PubMed] [Google Scholar]
- 39.Celver J, Vishnivetskiy SA, Chavkin C, Gurevich VV. J. Biol. Chem. 2002;277:9043–9048. doi: 10.1074/jbc.M107400200. [DOI] [PubMed] [Google Scholar]
- 40.Gurevich VV, Pals-Rylaarsdam R, Benovic JL, Hosey MM, Onorato JJ. J. Biol. Chem. 1997;272:28849–28852. doi: 10.1074/jbc.272.46.28849. [DOI] [PubMed] [Google Scholar]
- 41.Kovoor A, Celver J, Abdryashitov RI, Chavkin C, Gurevich VV. J. Biol. Chem. 1999;274:6831–6834. doi: 10.1074/jbc.274.11.6831. [DOI] [PubMed] [Google Scholar]
- 42.Pan L, Gurevich EV, Gurevich VV. J. Biol. Chem. 2003;278:11623–11632. doi: 10.1074/jbc.M209532200. [DOI] [PubMed] [Google Scholar]
- 43.Gurevich VV, Benovic JL. J. Biol. Chem. 1995;270:6010–6016. doi: 10.1074/jbc.270.11.6010. [DOI] [PubMed] [Google Scholar]
- 44.Gurevich VV. J. Biol. Chem. 1998;273:15501–15506. doi: 10.1074/jbc.273.25.15501. [DOI] [PubMed] [Google Scholar]
- 45.Hanson SM, Francis DJ, Vishnivetskiy SA, Kolobova EA, Hubbell WL, Klug CS, Gurevich VV. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4900–4905. doi: 10.1073/pnas.0600733103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palczewski K, McDowell H, Jakes S, Ingebritsen TS, Hargrave PA. J. Biol. Chem. 1989;264:15770–15773. [PubMed] [Google Scholar]
- 47.Hofmann KP, Pulvermuller A, Buczylko J, Van Hooser P, Palczewski K. J. Biol. Chem. 1992;267:15701–15706. [PubMed] [Google Scholar]
- 48.Bockaert J, Pin JP. EMBO J. 1999;18:1723–1729. doi: 10.1093/emboj/18.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gurevich VV, Gurevich EV. Structure. 2003;11:1037–1042. doi: 10.1016/s0969-2126(03)00184-9. [DOI] [PubMed] [Google Scholar]
- 50.Han M, Gurevich VV, Vishnivetskiy SA, Sigler PB, Schubert C. Structure. 2001;9:869–880. doi: 10.1016/s0969-2126(01)00644-x. [DOI] [PubMed] [Google Scholar]
- 51.Sutton RB, Vishnivetskiy SA, Robert J, Hanson SM, Raman D, Knox BE, Kono M, Navarro J, Gurevich VV. J. Mol. Biol. 2005;354:1069–1080. doi: 10.1016/j.jmb.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 52.Kirkwood A, Lisman JE. J. Gen. Physiol. 1994;103:679–690. doi: 10.1085/jgp.103.4.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hamer RD, Nicholas SC, Tranchina D, Lamb TD, Jarvinen JL. Vis. Neurosci. 2005;22:417–436. doi: 10.1017/S0952523805224045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rieke F, Baylor DA. Biophys. J. 1998;75:1836–1857. doi: 10.1016/S0006-3495(98)77625-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whitlock GG, Lamb TD. Neuron. 1999;23:337–351. doi: 10.1016/s0896-6273(00)80784-9. [DOI] [PubMed] [Google Scholar]
- 56.Ablonczy Z, Darrow RM, Knapp DR, Organisciak DT, Crouch RK. Photochem. Photobiol. 2007;81:541–547. doi: 10.1562/2004-08-27-RA-294. [DOI] [PubMed] [Google Scholar]
- 57.Broekhuyse RM, Tolhuizen EF, Janssen AP, Winkens HJ. Curr. Eye Res. 1985;4:613–618. doi: 10.3109/02713688508999993. [DOI] [PubMed] [Google Scholar]
- 58.Strissel KJ, Sokolov M, Trieu LH, Arshavsky VY. J. Neurosci. 2006;26:1146–1153. doi: 10.1523/JNEUROSCI.4289-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pugh EN, Jr., Lamb TD. In: Handbook of Biological Physics. Molecular Mechanisms in Visual Transduction. Stavenga DG, DeGrip WJ, Pugh E. N. eds Jr., editors. Elsevier, Amsterdam: 2000. pp. 183–255. [Google Scholar]
- 60.Hanson SM, Van Eps N, Francis DJ, Altenbach C, Vishnivetskiy SA, Arshavsky VY, Klug CS, Hubbell WL, Gurevich VV. EMBO J. 2007;26:1726–1736. doi: 10.1038/sj.emboj.7601614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nair KS, Hanson SM, Mendez A, Gurevich EV, Kennedy MJ, Shestopalov VI, Vishnivetskiy SA, Chen J, Hurley JB, Gurevich VV, Slepak VZ. Neuron. 2005;46:555–567. doi: 10.1016/j.neuron.2005.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Imamoto Y, Tamura C, Kamikubo H, Kataoka M. Biophys. J. 2003;85:1186–1195. doi: 10.1016/S0006-3495(03)74554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schubert C, Hirsch JA, Gurevich VV, Engelman DM, Sigler PB, Fleming KG. J. Biol. Chem. 1999;274:21186–21190. doi: 10.1074/jbc.274.30.21186. [DOI] [PubMed] [Google Scholar]
- 64.Krispel CM, Chen D, Melling N, Chen YJ, Martemyanov KA, Quillinan N, Arshavsky VY, Wensel TG, Chen CK, Burns ME. Neuron. 2006;51:409–416. doi: 10.1016/j.neuron.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 65.Binder BM, Biernbaum MS, Bownds MD. J. Biol. Chem. 1990;265:15333–15340. [PubMed] [Google Scholar]
- 66.Shi GW, Chen J, Concepcion F, Motamedchaboki K, Marjoram P, Langen R, Chen J. J. Biol. Chem. 2005;280:41184–41191. doi: 10.1074/jbc.M506935200. [DOI] [PubMed] [Google Scholar]
- 67.Binder BM, O'Connor TM, Bownds MD, Arshavsky VY. J. Biol. Chem. 1996;271:19826–19830. doi: 10.1074/jbc.271.33.19826. [DOI] [PubMed] [Google Scholar]
- 68.Palczewski K, Buczylko J, Kaplan MW, Polans AS, Crabb JW. J. Biol. Chem. 1991;266:12949–12955. [PubMed] [Google Scholar]
- 69.Kennedy MJ, Dunn FA, Hurley JB. Neuron. 2004;41:915–928. doi: 10.1016/s0896-6273(04)00086-8. [DOI] [PubMed] [Google Scholar]
- 70.Sommer ME, Smith WC, Farrens DL. J. Biol. Chem. 2005;280:6861–6871. doi: 10.1074/jbc.M411341200. [DOI] [PubMed] [Google Scholar]
- 71.Key TA, Foutz TD, Gurevich VV, Sklar LA, Prossnitz ER. J. Biol. Chem. 2003;278:4041–4047. doi: 10.1074/jbc.M204687200. [DOI] [PubMed] [Google Scholar]
- 72.Ren XR, Reiter E, Ahn S, Kim J, Chen W, Lefkowitz RJ. Proc. Natl. Acad. Set U. S. A. 2005;102:1448–1453. doi: 10.1073/pnas.0409534102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim J, Ahn S, Ren XR, Whalen EJ, Reiter E, Wei H, Lefkowitz RJ. Proc. Natl. Acad. Sci. U. S. A. 2005;102:1442–1447. doi: 10.1073/pnas.0409532102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rim J, Oprian DD. Biochemistry. 1995;34:11938–11945. doi: 10.1021/bi00037a035. [DOI] [PubMed] [Google Scholar]
- 75.Li T, Franson WK, Gordon JW, Berson EL, Dryja TP. Proc. Natl. Acad. Sci. U. S. A. 1995;92:3551–3555. doi: 10.1073/pnas.92.8.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chuang JZ, Vega C, Jun W, Sung CH. J. Clin. Investig. 2004;114:131–140. doi: 10.1172/JCI21136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen J, Shi G, Concepcion FA, Xie G, Oprian D, Chen J. J. Neurosci. 2006;26:11929–11937. doi: 10.1523/JNEUROSCI.3212-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alloway PG, Howard L, Dolph PJ. Neuron. 2000;28:129–138. doi: 10.1016/s0896-6273(00)00091-x. [DOI] [PubMed] [Google Scholar]
- 79.Kiselev A, Socolich M, Vinos J, Hardy RW, Zuker CS, Ranganathan R. Neuron. 2000;28:139–152. doi: 10.1016/s0896-6273(00)00092-1. [DOI] [PubMed] [Google Scholar]