Abstract
Study Design
A single large family, in which adolescent idiopathic scoliosis (AIS) and pectus excavatum (PE) segregate as an autosomal dominant condition, was evaluated. Genome-wide linkage analysis and candidate gene sequencing were performed.
Objective
To map the disease-causing locus in a large Caucasian family in which AIS and PE co-segregate.
Summary of Background Data
AIS and PE are common musculoskeletal conditions known to have a genetic component, though few genes have been identified for either. Genetic studies have been confounded by a lack of large families in which the disorders segregate.
Methods
Clinical examinations were performed on the proband, who underwent posterior spinal fusion, and twelve additional affected family members. To map a gene causing AIS and PE, a genome-wide linkage analysis was performed with the Affymetrix Mapping 10K XbaI array on thirteen affected and ten unaffected family members. Candidate genes were sequenced.
Results
AIS was present in thirteen female family members and PE was present in three males and one female. Genome-wide linkage analysis resulted in a linkage peak on chromosome 18q with a maximum parametric multipoint logarithm of the odds (LOD) score of 3.86. Recombinants delineated the critical genetic region to an interval of 6.4 cM between SNP_A-1519369 and SNP_A-1507702, corresponding to a 7.06 Mb region (hg18: chr18:26342508-34395660). The chromosome 18q linkage region contains more than 30 genes. Re-sequencing of the coding regions of 21 candidate genes in the region did not reveal any causative mutation.
Conclusion
Linkage analysis in this large family demonstrated a novel locus for AIS and PE on chromosome 18q. Because of the increased frequency of PE in family members of AIS patients, consideration of family members with PE as affected may increase the power of AIS genetic linkage studies.
Keywords: idiopathic scoliosis, pectus excavatum, genetics, linkage analysis
Key Points
- Adolescent idiopathic scolisis and pectus excavatum may have a similar genetic etiology.
- Linkage analysis demonstrated a novel locus for AIS and PE on chromosome 18q.
- Consideration of family members with PE as affected may increase the power of future AIS genetic linkage studies.
INTRODUCTION
Scoliosis refers to a structural lateral curvature of the spine measuring 10 degrees or more on an anteroposterior spine radiograph. Scoliosis is most often considered idiopathic and has a population prevalence of 0.2 to 3%, depending on the magnitude of the curve1. In contrast to neuromuscular and congenital scoliosis, for which both sexes are equally affected2, adolescent idiopathic scoliosis (AIS) predominantly affects girls. Males and females are nearly evenly affected at small curves (10 degrees to 25 degrees), but the ratio of females to males shifts markedly to 10:1 when the curvature is 40 degrees and greater3.
Genetic factors are thought to contribute to the development of scoliosis, as demonstrated by an increased incidence (6–11%) in first-degree relatives 4 5. Twin studies also suggest a high degree of heritability in scoliosis, as 73% of monozygous twins and 36% of dizygotic twins are concordant for scoliosis 6. Data regarding the inheritance pattern of AIS have been conflicting, with some studies suggesting that it is a complex trait, while others showing evidence for autosomal dominant transmission7. A recent study of AIS probands from the Salt Lake Valley, Utah region concluded that the majority of patients descended from common founders, and suggested a major autosomal dominant gene 8.
Previous analyses of several moderate-sized Caucasian families gave weak evidence for linkage to three loci on chromosomes 6p, distal 10q, and 18q 9. An additional locus on chromosome 17p 10 was identified in a large Italian family. However, linkage of scoliosis to these loci has not been replicated. Evidence for linkage to chromosome 19p was demonstrated in one large Chinese family with AIS11 and confirmed in a separate Caucasian family12, but no causative gene mutations have been found. More recently, linkage and association of AIS was demonstrated to single nucleotide polymorphisms on chromosome 8q within CHD7, the gene responsible for CHARGE syndrome, making it the first recognized AIS susceptibility gene 13.
The purpose of our study was to perform a genome-wide linkage scan in a nuclear family in which AIS and PE co-segregate. Identification of the chromosomal 18q locus in this family may hasten the discovery of a gene responsible for AIS in females and PE in males.
MATERIALS AND METHODS
Ascertainment and diagnosis of patients
The Washington University Human Subjects Committee approved this study and written informed consent was obtained for all individuals. DNA was obtained from twenty-one family members from a five generation North American Caucasian family in which AIS and PE co-segregate. Family members were examined in person when possible, and medical records, including spine radiographs were obtained in most cases (Table 1). This family consisted of thirteen females with adolescent idiopathic scoliosis. No males had scoliosis. One patient, the proband, had severe scoliosis requiring surgery. Four patients underwent bracing. Four patients reported worsening of the scoliosis with menopause, although this was not documented radiographically. Four individuals had PE, including three males, and one female (age 9 years). Individual IV:7 had surgical treatment of PE. Two individuals, the proband (IV:1) and IV:7, were examined by clinical geneticists and not found to have evidence of any connective tissue abnormality.
Table 1.
Clinical characteristics of affected individuals
| Pt no. | Age | Sex | Pectus | Scoliosis | Cobb Angle | Treatment |
|---|---|---|---|---|---|---|
| II:2 | 88 | F | − | + | NA | None |
| III:1 | 45 | M | + | − | NA | None |
| III:4 | 50 | F | − | + | 45 | None |
| III:6 | 56 | F | − | + | NA | None |
| III:9 | 52 | F | − | + | 40 | None |
| III:13 | Deceased | F | − | + | 30 | Braced |
| IV:1 | 12 | F | − | + | 75 | Spinal fusion |
| IV:2 | 11 | F | + | − | NA | None |
| IV:5 | 18 | M | + | − | NA | None |
| IV:7 | 19 | M | + | − | NA | Surgery |
| IV:13 | 18 | F | − | + | 20 | Braced |
| IV:15 | 19 | F | − | + | 24 | Braced |
| IV:16 | 20 | F | − | + | 38 | Braced |
| IV:23 | 20 | F | − | + | 19 | None |
| V:1 | 20 | F | − | + | 20 | Braced |
Determination of the frequency of PE in families and individuals with AIS
Sixty-five AIS patients (51 male, 14 female) (58 Caucasian, 5 African American, 2 unknown) evaluated in a pediatric orthopaedic clinic were contacted by telephone. A three-generation family history was obtained from each family, with particular questions asked regard to the presence of pectus excavatum or pectus carinatum and presence of other signs or symptoms of connective tissue disease.
10K SNP Genotyping
DNA was obtained from saliva samples (OraGene Self-Collection kit, Genotek, Ottawa, Canada) from 22 family members consisting of thirteen affected (nine with AIS, and four with PE) and nine unaffected family members. A genome-wide search was undertaken using an Affymetrix GeneChip Mapping 10K XbaI Array, containing 10,055 SNPs. These SNPs have a mean intermarker distance of 210kb and an average heterozygosity of 0.38. Call rates were >93%.
Linkage analysis
Genotyping was performed by the NIH Neuroscience Microarray Consortium. Microarray data were analyzed with GDAS v2 software. Because of the large size of the pedigree the family was separated into data two separate branches for linkage analysis. All linkage programs were accessed through the EasyLinkage package 14 15. PedCheck was used for detection of Mendelian errors16 and non-Mendelian errors were identified with Merlin17. All genotype inconsistencies were zeroed out from the database. Simulated two-point parametric analysis was performed with SLINK18 using a total of 1000 replicates for a biallelic marker (ie SNP). Two-point parametric linkage analysis was performed with SuperLink v1.519. Multipoint analysis was performed with GENEHUNTER20 using sets of 100 markers, and repeated with sets of 60 markers for verification. Individuals with AIS or PE were considered affected for the linkage analyses. Haplotypes were created with GENEHUNTER and viewed on Haplopainter21. LOD score graphs are displayed using EasyLinkage software.
RESULTS
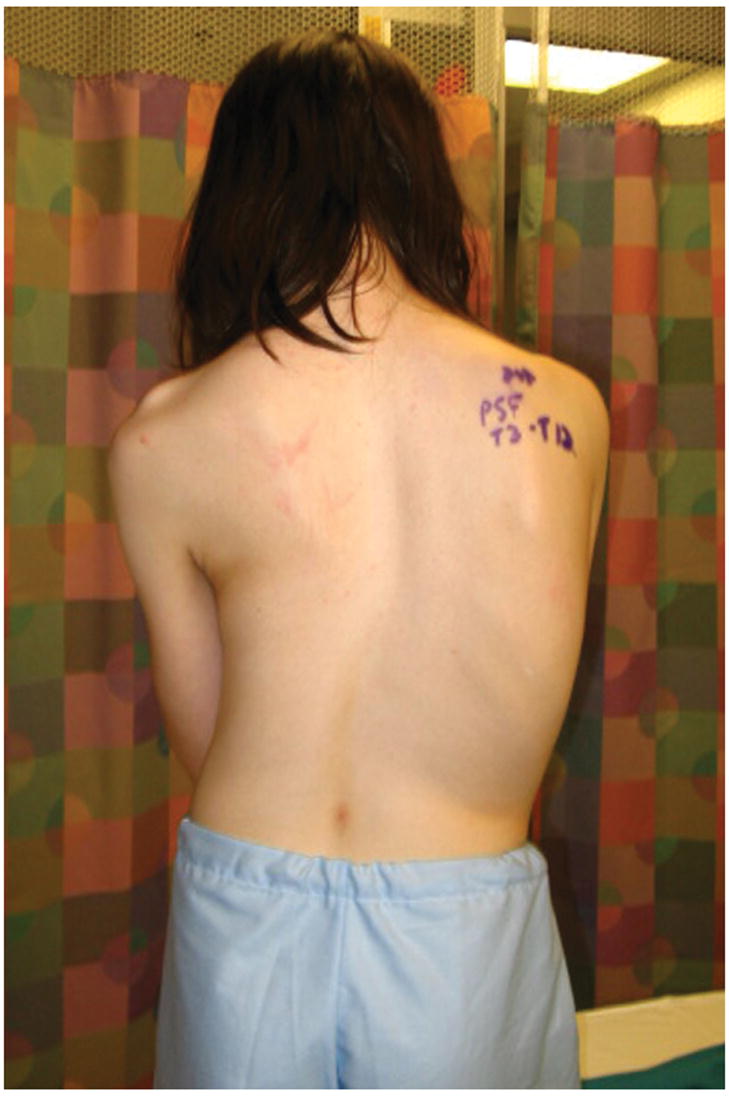
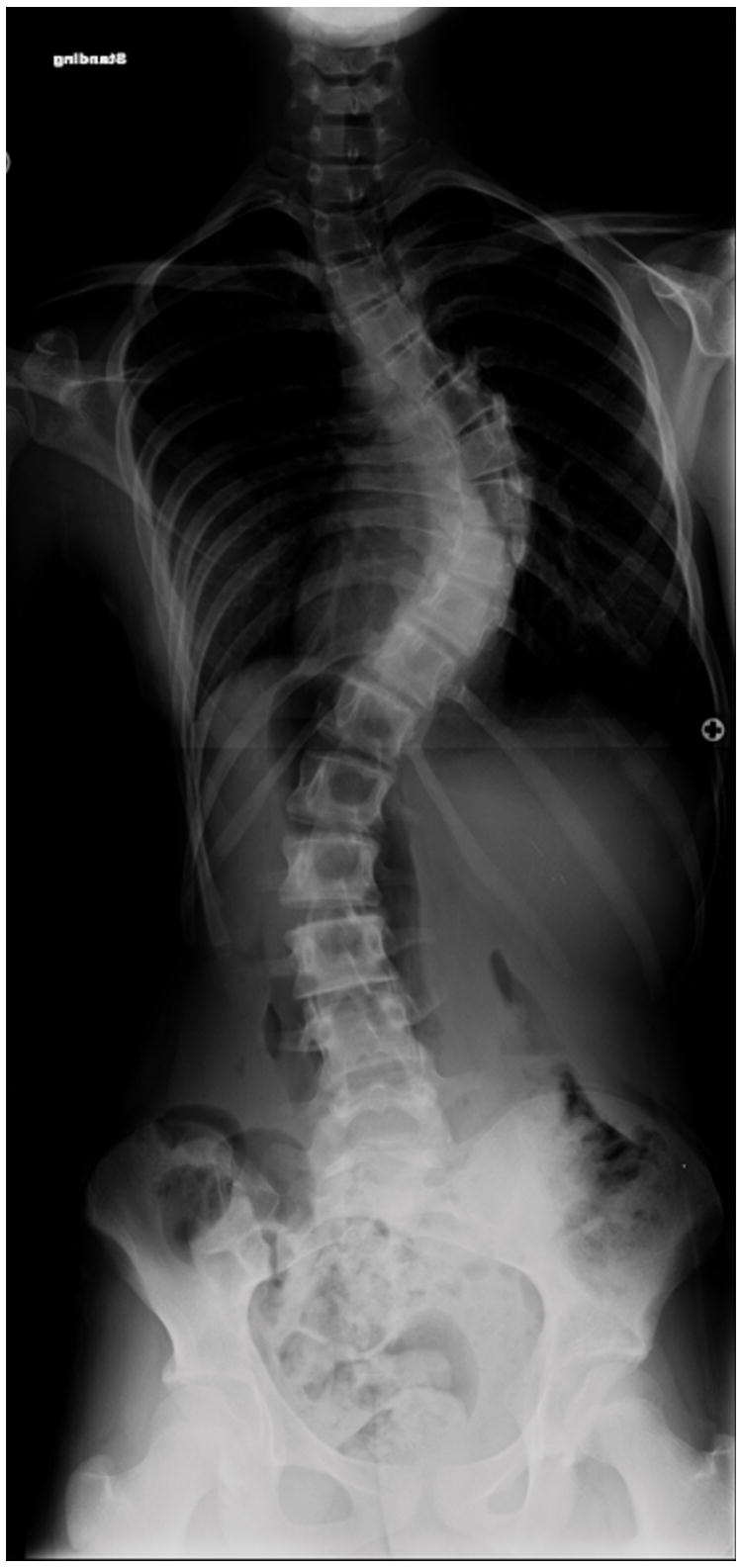
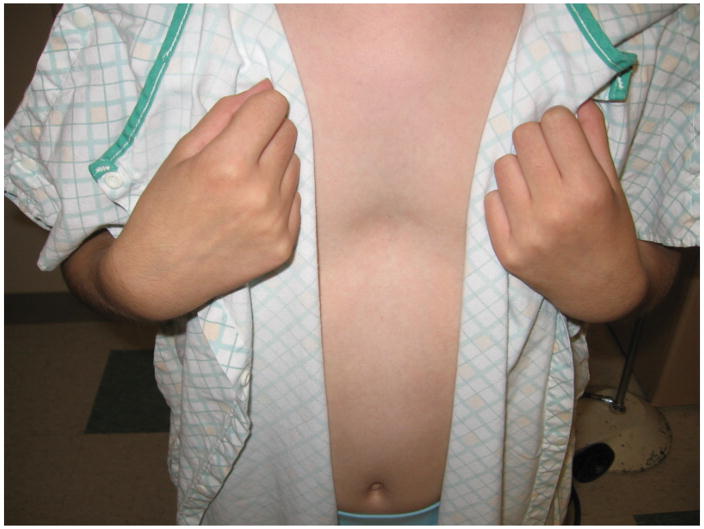
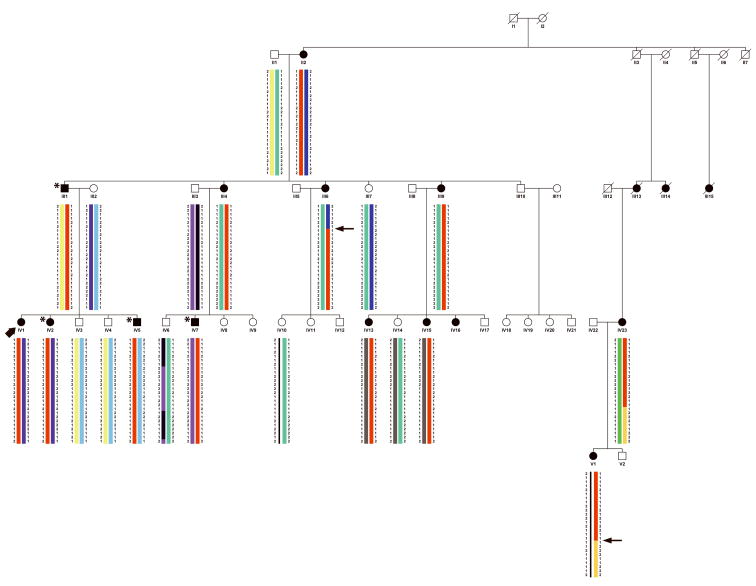
We identified a five-generation Caucasian family in which AIS and PE segregate. Of thirteen females affected with scoliosis, clinical data and DNA is available for nine (Table 1). There is significant phenotypic variability in scoliosis severity, ranging from >70 degrees in the proband (Figure 1A and 1B), to 15 degrees in several individuals. Four individuals have PE, including three males and one female (age 11 years) (Figure 1C). No individuals have both scoliosis and PE. Several individuals, including the grandmother of the proband, were never evaluated for scoliosis, but had significant scoliosis on exam. Significant variability in severity was also seen with pectus excavatum, including one individual that underwent surgical correction of the condition.
Figure 1.
Clinical photos of family members. A, Photograph of proband (age 12 years) before undergoing posterior spinal fusion for progressive idiopathic scoliosis. B. Anteroposterior upright spinal radiograph of proband before spinal fusion demonstrating a 75 degree main right thoracic curve. C, Photograph of proband’s sister (age 9 years) demonstrating pectus deformity.
Linkage analysis and locus identification
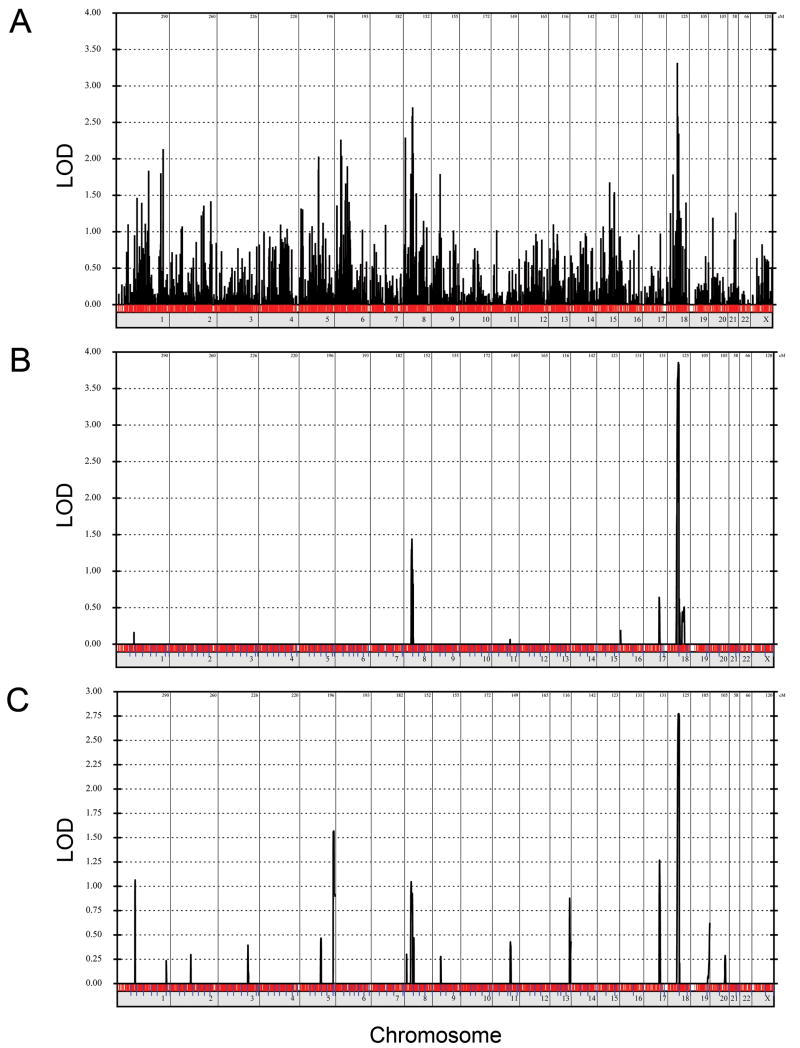
To map the gene causing AIS and PE, a genome-wide linkage scan was performed with 10K SNP genotypes. All parametric analyses assumed autosomal dominant inheritance with 80% penetrance, disease allele frequency of 0.1%, and 1% phenocopies. For the initial evaluations, individuals with AIS and PE were considered affected. Two point LOD score analysis resulted in a LODmax of 3.31 on chromosome 18 at marker SNP_A1515867, with a secondary peak of 2.70 on chromosome 8 at marker SNP_A-1510387 (Figure 2A). For comparison, the maximal simulated two-point LOD score calculated by SLINK under the same conditions was 3.8. Parametric multipoint linkage analysis for AIS and PE revealed a maximum multipoint LODmax of 3.86 at chromosome 18q12.1–12.2 between 50 and 70 cM (Figure 2B). Only one other linkage peak with a LOD>1 was present and was located on chromosome 8p12 at 60cM (LOD 1.48).
Figure 2.
Genome-wide linkage analysis of family 6061. (A) Two-point linkage analysis considering individuals with AIS and PE as affected was performed assuming autosomal dominant inheritance with penetrance of 80%, phenocopy rate of 1%, and disease allele frequency of 0.1%. (B) Multipoint linkage analysis considering individuals with AIS and PE as affected using the same parameters as above. Analysis was performed with marker sets of 100. (C) Multipoint linkage analysis considering individuals with AIS as affected using the same parameters.
Linkage analysis was repeated under the same conditions, except that individuals with PE were considered unaffected, and linkage peaks were produced over the same regions (Figure 2C). Because individual IV2 was prepubertal, her affected status was considered unknown for this analysis. The LODmax for the AIS only analysis was reduced at 2.77 (Figure 2C) and corresponded to the same chromosome 18q region identified in the AIS and PE analysis(Figure 2B).
Haplotype analysis demonstrated 38 contiguous SNPs that were common to all individuals with AIS or PE (Figure 3). A critical centromeric recombination event occurred in individual III:6 and a critical telomeric recombination occurred in individual IV:23 (or in a non-genotyped ancestor). These recombination events narrow the conserved haplotype to a 6.6cM (7.3 Mb) region of chromosome 18q12.1–q12.2. The critical recombinants that delineated the candidate region occurred in family members with AIS.
Figure 3.
Pedigree showing haplotype consisting of a 6.6cM (7.3 Mb) region of chromosome 18q12.1–q12.2 shared among all individuals affected with scoliosis or pectus excavatum (red bar). Individuals with scoliosis are shaded black. Individuals with pectus excavatum are shaded black and indicated with an asterisk. The proband is indicated with an arrowhead. Critical recombinants are indicated by arrows. Brackets enclose haplotypes that were inferred.
Candidate gene selection and mutation screening
The linkage region on chromosome 18q12.1–q12.2 contains approximately 36 predicted genes. All coding sequences of 21 genes were resequenced in two affected individuals(DTNA, B4GALT6, GALNT1, ZNF396, C18orf34, FAM59A, MAPRE2, DSC2, DSC3, MEP1B, FAM59A, KLHL14, C18orf34, KIAA1713, ZNF397, SNF24,ZNF396, C18Orf21,P15RS, SLC39A6, and ELP2). No coding mutations were identified.
Determination of the frequency of PE in AIS family members
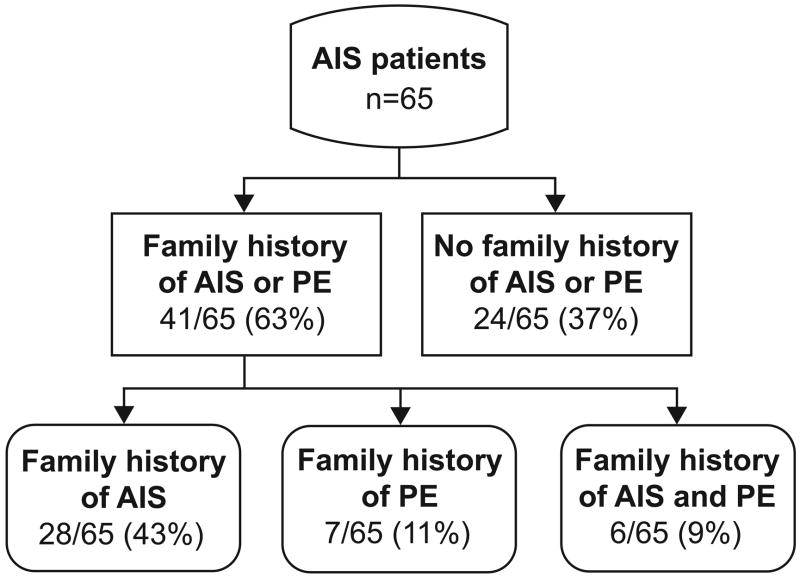
A family history was positive for AIS or PE in 63% (41/65) of patients with AIS (Figure 4). Although the majority of these cases were due to the presence additional family members with AIS, a family history of PE was present in 20% (13/65), and both AIS and PE was present in 9% (9/65).
Figure 4.
Frequency of pectus excavatum in AIS family members. AIS patients were selected randomly from a tertiary referral center for scoliosis and a detailed family history was obtained, specifically asking for evidence of connective tissue disorders, including pectus excavatum.
DISCUSSION
Linkage analysis of in this large family resulted in the identification of a novel locus for AIS and PE on chromosome 18q12.1–q12.2. The typical female predominance of AIS was present in this pedigree, and therefore this locus may represent a locus for common forms of AIS. A larger region of chromosome 18q, encompassing the current locus, was previously identified in a genome-wide linkage scan involving a large family with AIS9, although the linkage peak in that family was considerably more telomeric (70–120cM). However, it is possible that this represents the same locus that is described here in the current study of AIS and PE.
Although scoliosis is increased in frequency in patients with PE and their familiy members22, to our knowledge no study has previously determined the incidence of PE in AIS. An increased incidence of PE was noted both in family 6061 as well as in a clinic population of AIS patients, where a family history of PE was present in 20%, which is considerably higher than would be expected given the rate of PE in the general populations (1 out of every 400 children)23 24. PE and scoliosis are also common in connective tissue disorders including Ehlers-Danlos and Marfan syndrome. However, additional evidence of connective tissue disorders (long extremities, nearsightedness, aortic dilatation, mitral valve prolapse, easy bruising, lax joints, loose or excess skin, arthritis, and hernias) was absent in family 6061 and the 65 AIS patients surveyed. Furthermore, there was no evidence of linkage to chromosomal loci containing genes for Ehlers Danlos syndrome and Marfan syndrome in family 6061. Because our data show an increased incidence of PE in family members of AIS patients, it is likely that at least some of the genetic susceptibility genes for AIS may also contribute to the susceptibility to PE.
There are well known, significant, and nearly opposite gender differences in the incidence of PE and AIS that were also seen in our study. PE is more common in males (4:1 male to female ratio)24 and this is consistent with inheritance of PE in family 6061 in which three out of four affected were male. In family 6061, all individuals affected with AIS were female and there were no individuals with both AIS and PE. However, the eleven year-old sister of the proband has PE and inherited the common haplotype on chromosome 18q, and based on her age, remains at risk for developing AIS. Identification of the genes responsible for AIS and PE may eventually allow us to better understand these gender specific effects on skeletal development.
Because of the large number of affected individuals in family 6061, we were able to narrow the candidate interval to a relatively small region [6.6cM (7.3 Mb)] of chromosome 18q12.1–q12.2 Mb, containing <40 genes. There are several interesting candidate genes on chromosome 18q, including DTNA and B4GALT7. DTNA encodes alpha-dystrobrevin and is a member of the dystrophin family. Duchenne muscular dystrophy is an X-linked neuromuscular condition that results from mutations in dystrophin, which is often associated with scoliosis25. Recessive mutations in B4GALT7 (xylosylprotein 4-beta-galactosyltrasferase I) results in the progeroid form of Ehlers-Danlos syndrome26 27 and is therefore a likely candidate for AIS and PE. Although the exons of these genes and 19 other candidate genes were sequenced and mutations not identified, we did not evaluate for mutations within noncoding, regulatory regions surrounding these genes. For many common diseases such as AIS, disease associated variants have been identified in the noncoding sequence. This was recently demonstrated in the association of AIS with a single nucleotide polymorphism (SNP) on chromosome 8q12.2 that is located in an intron of CHD713, the gene responsible for CHARGE syndrome. Despite finding an association of AIS with this intronic SNP, mutations in CHD7 were not identified, and the functional effects of this SNP remain unknown. Disease-causative noncoding mutations are much more difficult to identify given the large amount of conserved DNA sequence flanking most genes and it is currently not cost effective to sequence all of these noncoding regions.
To summarize, we have identified an increased incidence of PE in a large multigenerational family with AIS as well as in an independent cohort of AIS patients. Because the etiology of these two conditions are likely related, we considered both phenotypes as affected in our linkage analysis, and were therefore able to identify a novel genetic locus for AIS and PE on chromosome 18q. Although familial cases of PE have been reported22, linkage studies have not been published and it will be interesting to see if PE is linked to the same region on chromosome 18q. Confirmation of linkage in additional AIS families will be needed to determine if the chromosome 18 locus represents a common locus for AIS. In the near future, genome-wide association studies including hundreds or thousands of AIS patients may also identify SNPs within the chromosome 18q region that are associated with AIS, thus confirming our linkage findings, and identifying the genetic basis for these common musculoskeletal disorders.
Acknowledgments
C.A.G. is supported by NIH NINDS K12 Award (NS01690) and the Children’s Discovery Institute. M.B.D. is supported by The Cotrel Foundation, the Shriners Hospital for Children, and the Saint Louis Children’s Hospital Foundation.
References
- 1.Weinstein SL. The thoracolumbar spine. Philadelphia: Lippincot Company; 1994. [Google Scholar]
- 2.Tsirikos AI, Chang WN, Dabney KW, et al. Life expectancy in pediatric patients with cerebral palsy and neuromuscular scoliosis who underwent spinal fusion. Dev Med Child Neurol. 2003;45:677–82. doi: 10.1017/s0012162203001269. [DOI] [PubMed] [Google Scholar]
- 3.Lenke LDM. Idiopathic scoliosis. 3. Philadelphia: Lippincot, Williams & Wilkins; 2004. [Google Scholar]
- 4.Wynne-Davies R. Familial (idiopathic) scoliosis. A family survey. J Bone Joint Surg Br. 1968;50:24–30. [PubMed] [Google Scholar]
- 5.Riseborough EJ, Wynne-Davies R. A genetic survey of idiopathic scoliosis in Boston, Massachusetts. J Bone Joint Surg Am. 1973;55:974–82. [PubMed] [Google Scholar]
- 6.Kesling KL, Reinker KA. Scoliosis in twins. A meta-analysis of the literature and report of six cases. Spine. 1997;22:2009–14. doi: 10.1097/00007632-199709010-00014. discussion 15. [DOI] [PubMed] [Google Scholar]
- 7.Axenovich TI, Zaidman AM, Zorkoltseva IV, et al. Segregation analysis of idiopathic scoliosis: demonstration of a major gene effect. Am J Med Genet. 1999;86:389–94. [PubMed] [Google Scholar]
- 8.Ogilvie JW, Braun J, Argyle V, et al. The search for idiopathic scoliosis genes. Spine. 2006;31:679–81. doi: 10.1097/01.brs.0000202527.25356.90. [DOI] [PubMed] [Google Scholar]
- 9.Wise CA, Barnes R, Gillum J, et al. Localization of susceptibility to familial idiopathic scoliosis. Spine. 2000;25:2372–80. doi: 10.1097/00007632-200009150-00017. [DOI] [PubMed] [Google Scholar]
- 10.Salehi LB, Mangino M, De Serio S, et al. Assignment of a locus for autosomal dominant idiopathic scoliosis (IS) to human chromosome 17p11. Hum Genet. 2002;111:401–4. doi: 10.1007/s00439-002-0785-4. [DOI] [PubMed] [Google Scholar]
- 11.Chan V, Fong GC, Luk KD, et al. A genetic locus for adolescent idiopathic scoliosis linked to chromosome 19p13.3. Am J Hum Genet. 2002;71:401–6. doi: 10.1086/341607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alden KJ, Marosy B, Nzegwu N, et al. Idiopathic scoliosis: identification of candidate regions on chromosome 19p13. Spine. 2006;31:1815–9. doi: 10.1097/01.brs.0000227264.23603.dc. [DOI] [PubMed] [Google Scholar]
- 13.Gao X, Gordon D, Zhang D, et al. CHD7 gene polymorphisms are associated with susceptibility to idiopathic scoliosis. Am J Hum Genet. 2007;80:957–65. doi: 10.1086/513571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindner TH, Hoffmann K. easyLINKAGE: a PERL script for easy and automated two-/multi-point linkage analyses. Bioinformatics. 2005;21:405–7. doi: 10.1093/bioinformatics/bti009. [DOI] [PubMed] [Google Scholar]
- 15.Hoffmann K, Lindner TH. easyLINKAGE-Plus--automated linkage analyses using large-scale SNP data. Bioinformatics. 2005;21:3565–7. doi: 10.1093/bioinformatics/bti571. [DOI] [PubMed] [Google Scholar]
- 16.O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–66. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abecasis GR, Cherny SS, Cookson WO, et al. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 18.Cottingham RW, Jr, Idury RM, Schaffer AA. Faster sequential genetic linkage computations. Am J Hum Genet. 1993;53:252–63. [PMC free article] [PubMed] [Google Scholar]
- 19.Fishelson M, Geiger D. Exact genetic linkage computations for general pedigrees. Bioinformatics. 2002;18(Suppl 1):S189–98. doi: 10.1093/bioinformatics/18.suppl_1.s189. [DOI] [PubMed] [Google Scholar]
- 20.Kruglyak L, Daly MJ, Reeve-Daly MP, et al. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet. 1996;58:1347–63. [PMC free article] [PubMed] [Google Scholar]
- 21.Thiele H, Nurnberg P. HaploPainter: a tool for drawing pedigrees with complex haplotypes. Bioinformatics. 2005;21:1730–2. doi: 10.1093/bioinformatics/bth488. [DOI] [PubMed] [Google Scholar]
- 22.Creswick HA, Stacey MW, Kelly RE, Jr, et al. Family study of the inheritance of pectus excavatum. J Pediatr Surg. 2006;41:1699–703. doi: 10.1016/j.jpedsurg.2006.05.071. [DOI] [PubMed] [Google Scholar]
- 23.Molik KA, Engum SA, Rescorla FJ, et al. Pectus excavatum repair: experience with standard and minimal invasive techniques. J Pediatr Surg. 2001;36:324–8. doi: 10.1053/jpsu.2001.20707. [DOI] [PubMed] [Google Scholar]
- 24.Chung CS, Myrianthopoulos NC. Factors affecting risks of congenital malformations. II. Effect of maternal diabetes on congenital malformations. Birth Defects Orig Artic Ser. 1975;11:23–38. [PubMed] [Google Scholar]
- 25.Kinali M, Main M, Eliahoo J, et al. Predictive factors for the development of scoliosis in Duchenne muscular dystrophy. Eur J Paediatr Neurol. 2007;11:160–6. doi: 10.1016/j.ejpn.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 26.Okajima T, Fukumoto S, Furukawa K, et al. Molecular basis for the progeroid variant of Ehlers-Danlos syndrome. Identification and characterization of two mutations in galactosyltransferase I gene. J Biol Chem. 1999;274:28841–4. doi: 10.1074/jbc.274.41.28841. [DOI] [PubMed] [Google Scholar]
- 27.Almeida R, Levery SB, Mandel U, et al. Cloning and expression of a proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I. A seventh member of the human beta4-galactosyltransferase gene family. J Biol Chem. 1999;274:26165–71. doi: 10.1074/jbc.274.37.26165. [DOI] [PubMed] [Google Scholar]