Abstract
Diminished Sonic Hedgehog (Shh) signaling is associated with the most common forebrain defect in humans, holoprosencephaly (HPE), which includes cyclopia, a phenotype also seen in mice and other vertebrates with defective Shh signaling. The secreted protein Shh acts as a crucial factor that patterns the ventral forebrain and is required for the division of the primordial eye field and brain into two discrete halves. Gli2 is one of three vertebrate transcription factors implicated as obligatory mediators of Shh signal transduction. Here, we show that loss-of-function mutations in the human GLI2 gene are associated with a distinctive phenotype (within the HPE spectrum) whose primary features include defective anterior pituitary formation and pan-hypopituitarism, with or without overt forebrain cleavage abnormalities, and HPE-like midfacial hypoplasia. We also demonstrate that these mutations lack GLI2 activity. We report on a functional association between GLI2 and human disease and highlight the role of GLI2 in human head development.
Holoprosencephaly (HPE) is characterized by a failure of midline division of the forebrain and can be caused by genetic or environmental insults (1). Clinical manifestations of HPE are variable and extend from simply closely spaced eyes (hypotelorism), to a failure of separation of the eye field and forebrain associated with cyclopia. The best understood causes of HPE are associated with actions that directly, or indirectly, affect Sonic Hedgehog (SHH) signaling (2). This signaling pathway culminates in the activation of target genes under the control of members of the GLI family of transcription factors that can direct either target gene activation or repression depending on Hedgehog (Hh) activity (3-6).
Three Gli genes have been implicated in the mediation of Shh signals in vertebrates (7). Shh signaling may be mediated by different Gli proteins in various contexts, with Gli1 and Gli2 being most important (5-17), although Gli3 has also been proposed to mediate Shh signals (8, 9). In addition, an antagonistic relationship between Gli3 and Shh is critical for early ventral neural tube patterning (5, 18). During development, Gli1 is strongly expressed along the midline in response to Shh signaling, and it is a faithful marker of a cell's response to Shh, whereas Gli2 and Gli3 are strongly expressed in more lateral regions, suggesting that they can be regulated by other factors. Knockout studies of mouse Gli1 have indicated its apparent redundancy and suggest that its function may be compensated by other Gli proteins (13-15). In contrast, in different organisms Gli2 and Gli3 have partially redundant functions (5, 7, 8, 10, 19-22), and each appears to exist either as a full-length activator or a C-terminally truncated repressor form (4-6, 8, 23, 24). Removal of the mouse Gli2 gene by targeted disruption leads to an embryonic lethal phenotype with defects in early brain and spinal cord development, which include absence of the floor plate (10-12), minor craniofacial defects (13), and a brain phenotype with expanded but thinner telencephalic vesicles and overtly reduced dorsal brain including the tectum and cerebellum (V. Palma and A.R.A., unpublished work). Mutation of Gli3 in (extra-toes J) mice results in an embryonic lethal phenotype affecting multiple organs including the brain, with a drastic reduction in cortical size (25, 26). In humans, alterations in SHH signaling are associated with a number of pathological states, but mutations in GLI1 or GLI2 have not yet been linked to any human disease. In contrast, Gli3 null mice partially recapitulate the defects seen in patients with GLI3 mutations in Greig syndrome, Pallister-Hall syndrome, or several distinct polydactyly disorders (23, 27). In the context of HPE, it was therefore important to elucidate which GLI protein(s) mediates SHH signaling in humans.
Materials and Methods
Isolation of the Human GLI2 Gene. Primer pairs were designed within a partial cDNA 3′ UTR sequence for human GLI2 and then tested for its ability to amplify from commercial genomic DNA (Clontech). Using this assay, the Physical Mapping Core, the National Human Genome Research Institute, identified a bacterial artificial chromosome (BAC) clone 433k6 (Research Genetics, Huntsville, AL) containing the 3′ UTR of human GLI2. Further analysis by direct sequencing of the BAC template established that this clone contains the initiator methionine (28) and thus extended through the entire coding region. Primers were designed based on direct comparison between the experimentally derived genomic sequence and that of the known cDNA isoforms. A total of eight coding exons were identified, and the immediate flanking sequences were determined.
Mutational Analysis. Mutations were identified by single-strand conformational polymorphism analysis and confirmed by DNA sequencing. Samples of genomic DNA from 390 unrelated patients that met the clinical criteria of HPE were obtained under informed consent according to the guidelines of the institutional review boards of the Children's Hospital of Philadelphia and the Division of Intramural Research, National Human Genome Research Institute, National Institutes of Health. The collection is representative of the entire HPE spectrum of clinical severity. Primers were designed to flank each experimentally determined exon sequence. The primer sets used to amplify exons 1-8 from genomic DNA templates are available on request. PCRs and screening were performed essentially as described (29).
Site-Directed Mutagenesis. Multiple attempts to isolate the human GLI2 cDNA by RT-PCR from either the tumor cell line HUT102 or lymphoblastoid cell lines were unsuccessful for any sequences 5′ of exon 7 (data not shown). Therefore, standard recombinant techniques were used to synthetically create a functional version of human GLI2 using the fact that most of the coding region is contained within exon 8. Of the four isoforms described for human GLI2, the WT sequence that was generated was based on the GLI2 α form (28). Because the cDNA was created synthetically by PCR from a genomic DNA (BAC) template, sequence variations present in the BAC clone 433k6 are also present in the cDNA construct as the haplotype [207insT; 248delC; (A595P) 1783G>C; (G597R) 1789G>C; (T638A) 1912A>G; (S828A) 2482T>G]. All six of these sequence variations were subsequently also identified in the completely sequenced BAC clone ACO16764, deposited in the GenBank database, which contains the entire human GLI2 coding region. This finding suggests that the variations at these positions are unique to the HUT102 cell line from which the reference cDNA sequence was determined (positions are depicted in green in Fig. 1a; see constructs 12-14). Similar variations were not seen in the mutational screening of HPE patients. Furthermore, constructs designed to test the significance of these missense changes, namely P595A and R597G, or A638T, or A828S all had normal activity in functional assays. Similarly, several variations seen in both patients and controls are likely to represent polymorphisms: (K410R, construct 6) 1229A>G; (P987S) 2959C>T; and (N978D) 2932A>G (data not shown). The human GLI2 α coding region was subcloned in-frame with the N-myc tag of pCS2MT as described (5). Sequence variants corresponding to the alleles identified in human HPE patients were generated from this WT construct by primer-mediated mutagenesis (Transformer site-directed mutagenesis kit, Clontech), according to the manufacturer's instructions. Each construct was verified to contain only the intended sequence changes by bidirectional sequencing using an ABI 3100 (Applied Biosystems).
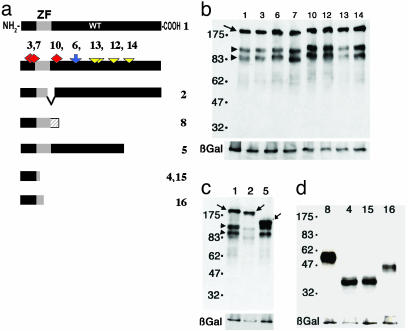
Fig. 1.
Summary of the predicted structure of the missense and loss-of-function alleles in GLI2. (a) The WT architecture of GLI2 is depicted in the top bar (WT construct 1; ZF = Zn finger). Shown below are the apparently unique missense variants seen in human HPE patients (red diamonds), putative polymorphisms (blue arrow), and HUT102 sequence variants (yellow triangles). Three of these variations predict missense changes (red diamonds). These three changes [V104M, 310G>A (construct 3), D88N, 262G>A (construct 7), and N273S, 818 A>G (construct 10)] behaved identically with the normal gene and are likely rare polymorphisms (Fig. 3c and data not shown). Similarly, apparent polymorphisms (found in normal controls), such as K410R, 1229 A>G (blue arrow, construct 6), behaved identically to the WT cDNA (construct 1) in all assays performed, as did the putative HUT102 reference cDNA variants (yellow triangles, constructs 12-14). The predicted architecture of the disease-related variants is shown below. Construct 2 represents the predicted outcome of a hypothetical RNA splicing event removing exon 5. Confirmation of this form was not attempted, and the related construct 8 represents the more likely form. Failure to execute alternative splicing predicts premature termination within intron 5 (hatched bar; construct 8). Constructs 5 (2274del1), 4 and 15 (W113X, 339G>A), and 16 (R168X, 502C>T) represent the predicted protein structures that are prematurely truncated. Note that constructs 4 and 15 are identical truncation mutations except that construct 4 includes also the V104M missense mutation (as does construct 3). (b) Western analysis of COS-7 cells transfected with N-myc-tagged GLI2 alleles and probed with anti-myc antibody to verify expression. Untransfected cells show no bands (data not shown). Cotransfected lacZ mRNA encoded the β-galactosidase (β-Gal) control protein. The arrow marks the predicted full-length protein, and the arrowheads identify two consistently seen smaller proteins that could represent processed forms or stable degradation products. Although these smaller bands resemble those seen with frog Gli2 (6), their significance is unknown and will require further study. (c) The deletions caused by variants 2 and 5 produce the expected truncated proteins that are smaller than the WT full-length product encoded by construct 1. Note that the smaller processed bands are still formed in both, but in allele 2 they migrate faster, indicating that an apparently specific cleavage occurs between the zinc fingers and the site of truncation of construct 5. (d) The predicted truncations of the rest of the alleles cause smaller proteins of the expected sizes (constructs 4, 8, 15, and 16).
Functional Studies. Injections into frog embryos were performed at the two-cell stage, injecting 2 or 0.5 ng of mRNA in 10 nl into one cell. 5-Bromo-4-chloro-3-indolyl β-d-galactoside reaction, anti-myc staining, and mounting were done as described at the tadpole stage (around stage 34) (5). Transfection in COS-7 cells, anti-myc staining, and Western blots were done as described (6). Early-passage C3H10T1/2 cells (American Type Culture Collection) were transfected with the appropriate pCS2 plasmids and assayed for alkaline phosphatase (AP) production 48 h after transfection through the nitroblue tetrazolium-5-bromo-4-chloro-3-indolyl phosphate reaction. Cells were doubly labeled with anti-myc antibodies and AP to estimate the efficiency of AP induction.
Results and Discussion
Among 390 patients with HPE, we identified seven heterozygous sequence variations in GLI2 (Fig. 1a) that were apparently unique, because they were not found in >200 chromosomes from normal individuals. Four pedigrees segregating GLI2 loss-of-function mutations are shown in Fig. 2. Clinical findings of these four families are summarized in Table 1. Although phenotypic penetrance was variable, the principal feature in common among patients was abnormal pituitary gland formation and/or function, accompanied by variable craniofacial abnormalities. Similarly, brain findings including HPE were highly variable and could be caused by additional environmental or genetic influences superimposed on the GLI2 haploinsufficent state. None of the four probands is known to be a mutation carrier in either SHH, GLI1, PATCHED1, SMOH, ZIC2, or any previously described HPE gene (data not shown).
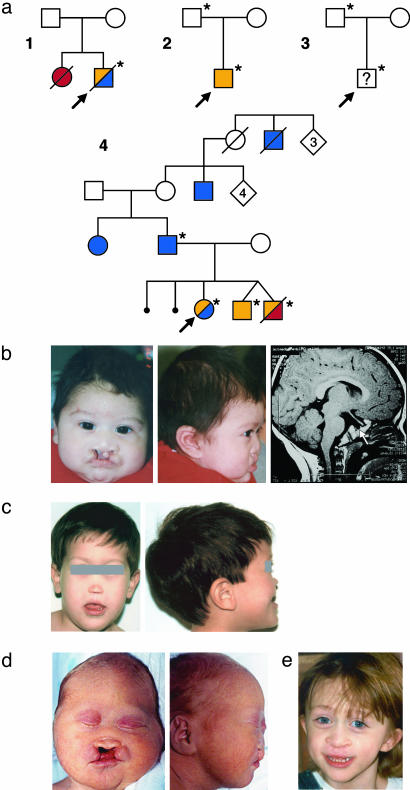
Fig. 2.
Clinical findings in families with mutations in GLI2. (a) Pedigrees of four families (nos. 1-4) who have individuals with GLI2 loss-of-function mutations (*). Probands are indicated by an arrow, HPE is indicated by red symbols, hypopituitarism is indicated by orange symbols, and polydactyly is indicated by blue symbols. (b) Proband in family 1 with severe midfacial hypoplasia, bilateral cleft lip and palate, postaxial polydactyly, and pituitary hypoplasia (arrow). (c) Proband in family 2 with extreme midfacial hypoplasia, pseudomedian cleft lip, and severe growth retardation. (d) Deceased sibling in family 4 with midface hypoplasia bilateral cleft lip and palate, and absent pituitary and brain findings consistent with HPE (data not shown). (e) Proband in family 4 with midface hypoplasia, repaired cleft lip and palate, postaxial polydactyly, and absent pituitary on MRI (data not shown). The deceased male in the first generation of family 4 is reported to have had cleft lip and cleft palate in addition to polydactyly. These pedigrees are consistent with autosomal dominant transmission of a null GLI2 disease susceptibility gene.
Table 1. Clinical findings in GLI2 mutation carriers.
| Family | Mutation | Constructs | Phenotype |
|---|---|---|---|
| 1 (proband) | W113X (339G>A) | 4, 15 | Bilateral cleft lip and palate (Fig. 2b), microcephaly, hypotelorism, single central incisor (removed), postaxial hexadactyly, growth hormone deficiency associated with pituitary hypoplasia, without other obvious forebrain anomalies. |
| 1 (sister) | Deceased | Autopsy findings included hypotelorism, single nostril, hypoplastic palate and maxilla, normal digits, absent anterior lobe of the pituitary, alobar HPE, and hydrocephalus. DNA was not available for testing. Neither parent is a mutation carrier consistent with gonadal mosaicism. | |
| 2 (proband) | IVS5 + 1G>A | 2, 8 | Hypotelorism, single nares, extreme midface hypoplasia (Fig. 2c), microcephaly, developmental delay, pseudomedian cleft lip, severe growth retardation, growth hormone deficiency, no obvious forebrain anomalies on computerized tomography. |
| 2 (father) | IVS5 + 1G>A | 2, 8 | Apparently normal. Clinical reevaluation was not possible. |
| 3 (proband) | R168X (502C>T) | 16 | Male patient referred with HPE findings; however, detailed findings were not available. |
| 3 (father) | R168X (502C>T) | 16 | Apparently normal. |
| 4 (proband) | 2274del1 | 5 | Repaired cleft lip and palate (Fig. 2e), pan-hypopituitarism, optic nerve hypoplasia, absent pituitary on MRI, bilateral postaxial polydactyly. |
| 4 (twin brother) | 2274del1 | 5 | Pan-hypopituitarism of both male twins. One sibling died at 5 months of age with midline cleft lip and palate (Fig. 2d), hypotelorism, flat midface, absent pituitary, an abnormal configuration of lower midline structures, and partial agenesis of the corpus callosum by head ultrasound. |
| 4 (twin brother) | Deceased | ||
| 4 (father) | 2274del1 | 5 | Father and paternal aunt with normal intelligence and postaxial polydactyly that may represent a microform. Note that other relatives who were unavailable for testing have postaxial polydactyly and cleft lip and palate (Fig. 1a).
|
| 4 (paternal aunt) | 2274del1 | 5 |
To functionally characterize the consequences of the different mutations detected in GLI2, we used the frog embryo as an assay system. Previous analyses have shown that misexpressed Gli proteins, through microinjection of synthetic mRNAs, induce distinct phenotypes in developing tadpoles (30). Injection of WT GLI2 mRNA in frog embryos induced the formation of epidermal tumors or hyperplasias (Fig. 3a). This phenotype resembles that induced by GLI1 (30) but differs from the phenotype of frog Gli2, which induces secondary tails (ventroposterior mesoderm). Human GLI2 and frog Gli2 thus appear to have divergent functions in this assay. In contrast, human GLI1 and GLI2 behave similarly, inducing tumors that resemble basal cell carcinomas (BCCs) molecularly (30). This finding is in agreement with the consistent expression of GLI1 and GLI2 in sporadic human BCCs, with the ability of both frog Gli1 and human GLI1 to induce tumors in tadpoles (30), and with the ability of human GLI1 and mouse Gli2 to induce BCC-like tumors in mice (31, 32).
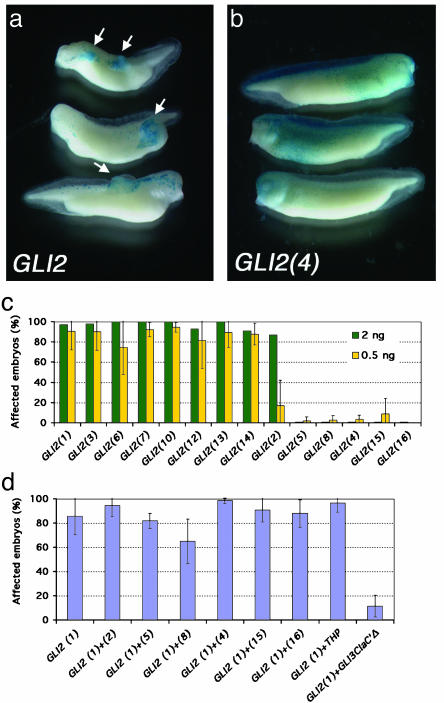
Fig. 3.
Targeting GLI2 to the ectoderm leads to the development of skin tumors. (a) Either 2 or 0.5 ng of GLI2 synthetic mRNA was injected along with tracer amounts of lacZ mRNA to mark injected cells. (b) An example of a loss-of-function allele (construct 4) where 5-bromo-4-chloro-3-indolyl β-d-galactoside-stained cells do not form tumors and show a pattern indistinguishable from that of lacZ mRNA-only injections (data not shown). (c) Histograms showing the percentage of affected embryos with overgrowths after misexpression of the different GLI2 alleles. Experiments were performed at two different concentrations, 0.5 ng (yellow) and 2.0 ng (green) per embryo. The 2-ng concentration was done once, and the 0.5-ng concentration was repeated three or four times per sample. The mean ± SEM is represented in the histogram. Each sample in each experiment had 20-70 embryos. All of the missense variants behaved similarly to WT, suggesting that they do not affect GLI2 function. On the other hand, allele 2 showed a less penetrant phenotype at low concentrations, indicating that it could be a hypomorph. This lower induction was significantly different from that of allele 6 (P < 0.05). Allele 8, which represents the more likely form, and allele 15 showed traces of activity, but the mean values were not significantly different from that of allele 4 (P > 0.5 for both), which had no activity. These alleles (nos. 8, 4, 15) plus alleles 5 and 16 are scored as loss-of-function in this assay. (d) Coinjection of WT GLI2 mRNA with each of the mutant constructs (in amounts of 0.5 ng of each) was assessed for synergy or antagonism. No evidence of dominant negative activity was demonstrable. Each allele was tested between 2 and 10 different times, and each sample had between 20 and 80 embryos. The bars represent the means ± SEM.
As a second functional assay, we tested for the induction of AP in mouse C3H10T1/2 cells transfected with the GLI2 allelic constructs. Activating Gli function in these cells triggers Hh-induced osteogenic differentiation that induces AP expression (6). AP induction was observed with GLI1, GLI2, and frog Gli1, but not with frog Gli2 (6) (Fig. 4 a and b and data not shown), thus allowing for the functional analyses of GLI2 variants. The mutant GLI2 alleles failed to show any activity in the in vivo tadpole or in vitro osteogenic assays (Figs. 3 and 4), with the exception of construct 2 (Fig. 1), which showed varying activity at different levels in tadpoles and may thus be a hypomorph (Fig. 3). Construct 2 was tested based on a hypothetical splicing event and is not proven to exist experimentally, with construct 8 representing a more plausible form. Our result highlights the sensitivity of the tadpole in vivo assay, but it remains unclear whether such mutation may behave as a null or a hypomorph in humans. In contrast, all polymorphisms showed WT activity in both assays (Figs. 3 and 4).
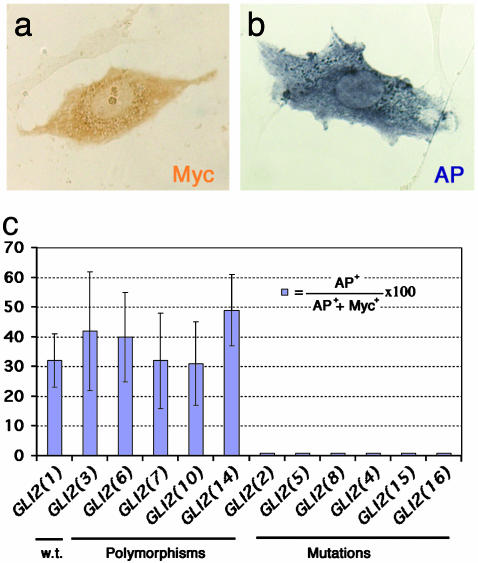
Fig. 4.
Osteogenic lineage induction by GLI2 alleles in C3H10T1/2 cells. (a) WT and mutant alleles were transfected into cells, and the encoded proteins were detected by using an antibody against the N-terminal myc tag. (b) AP activity is detected when C3H10T1/2 differentiates into an osteogenic lineage. (c) Plot of the mean ± SEM of ratios of AP-positive cells per total transfected cells in 10 independent fields of cells, with one representative experiment shown. The apparent polymorphisms behave similarly to the normal control. The mutant alleles (nos. 2, 5, 8, 4, 15 and 16) lose the ability to induce AP activity.
We next examined whether any of the GLI2 mutants could influence the activity of the normal GLI2 protein. Equivalent amounts of WT and mutant mRNAs were coinjected, and the injected tadpoles were measured for any change in tumor formation (Fig. 3d). None of the mutant alleles manifested dominant negative activity in these studies. Similarly, a naturally occurring short isoform of GLI2, called THP (amino acids 1-521) (28), also failed to display measurable dominant negative activity. As a positive control for repressor function we used GLI3Cla′Δ that lacks the C-terminal activator domain, but retains its N-terminal repression domain (6). GLI3Cla′Δ inhibited the function of WT GLI2 (Fig. 3d). These findings are consistent with the possible absence of repressor function in the shorter N-terminal domain of GLI2 as compared with that of its mouse or frog homologs, which contain dominant repressor function in their N termini (3, 4, 6). However, we have not directly investigated the putative processing of the human GLI2 protein to a repressor form. Nevertheless, these results raise the possibility that if GLI2 were cleaved to yield C-terminally deleted forms, such truncation may lead to the loss of activating function without the concurrent gain of repressor activity.
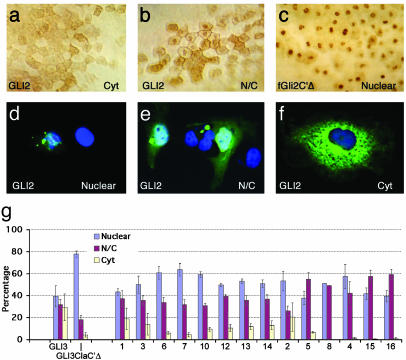
We used N-terminal Myc tags to assess the subcellular localization of the normal and altered GLI2 proteins in frog embryos injected with synthetic mRNA (Fig. 5 a-c) and in transfected mammalian COS-7 cells (Fig. 5 d-f). Gli2 proteins in other species encode C-terminally truncated nuclear repressors or full-length activators that are both cytoplasmic and nuclear (4-6). In injected frog embryos, human GLI2 protein, like frog Gli2, can localize both in the nucleus and the cytoplasm or just in the cytoplasm, probably depending on the state of the expressing cells (Fig. 5 a and b), and shows a pattern resembling that of an N-terminal deletion form of frog Gli1 (6). The different alleles had similar subcellular distribution patterns (data not shown), and none showed nuclear-only localization (compare with Fig. 5c). In contrast, constructs 4, 8, 15, and 16 rarely presented a cytoplasmic-only distribution. This finding is in accordance with the existence of a cytoplasmic localization signal in the C-terminal region in the Gli proteins (3). In COS cells the distributions of transfected WT and variant GLI2 proteins were similar, all showing variable nucleo-cytoplasmic distribution (Fig. 5 d-g).
Fig. 5.
Subcellular localization of GLI2 protein in animal cap cells of injected gastrula (stage 11) frog embryos. The normal and variant proteins were detected by using an antibody against the N-terminal myc tag. Representative examples of blastomere staining patterns are shown and quantified. Background levels were essentially none. (a) Prominent cytoplasmic staining, compared with combined nuclear and cytoplasmic staining (b). As a control for primarily nuclear staining (c) we used a frog Gli2 construct deleted for its C terminus (fGli2C′Δ) and previously shown to exhibit nuclear accumulation (6). A similar subcellular localization in transfected COS-7 cells was scored as primarily nuclear (d), both nuclear and cytoplasmic (e), or exclusively cytoplasmic (f). (g) Histograms showing the distribution of the different alleles in transfected COS-7 cells. Each allele was transfected three or four times, and >100 cells were counted in each experiment.
The human SHH gene was the first HPE gene to be identified (29) and suggested that components of its signaling machinery might also contribute to HPE-like disorders. We initiated this study to determine whether GLI2 is important for the division of the eye field and forebrain in humans as analysis of Gli function in frogs, mice, and zebrafish have given dissimilar results. In frog embryos, gain and interference with function analyses indicate that Gli2 and Gli3 act both as activators and repressors (6) and are first involved in ventroposterior mesodermal development (19, 20), whereas later they function in neurogenesis (33) and ventral neural tube patterning (5, 34). Here, Gli1 mimics Shh signaling and appears to act in floor plate induction downstream of Shh signaling, whereas Gli2 and Gli3 antagonize this function (5, 34). Gli1 and Gli2, in contrast, can induce motor neuron development and Gli3 inhibits this activity (5). These analyses in frog embryos have led to the idea that Gli proteins act in a context-dependent combinatorial fashion: a varying Gli code (5-7).
Analyses in mice indicate that Gli1 and Gli2 function mainly as activators (8, 10-16), Gli1 function is redundant (13, 14), Gli2 is essential for floor pate development (11, 12), and Gli3 partly antagonizes Shh signaling (14, 18, 35). Moreover, the single or combined loss of Gli1 and/or Gli2 does not lead to cyclopia, as seen in mice lacking Shh (36). Other factors, possibly Gli3 or Zic proteins, could prevent the development of cyclopia in the absence of these Shh mediators. Interestingly, the cyclopia and severe microcephaly seen in Shh-/- mice is partially compensated by a simultaneous reduction in the function of Gli3 (18, 35). Provided that the function of Gli3 is held in check, possibly its repressor function, a remarkable degree of ventral growth and patterning of the brain and spinal cord can proceed in the absence of Shh ligand. In contrast, Shh-/-;Gli2-/- mice phenotypically resemble Shh nulls (14). The functional divergence of Gli proteins in different organisms is further confirmed by recent results in zebrafish. In this species, Gli2 acts mainly as a repressor, whereas Gli1 is required for floor plate development (24, 37). Taken together, the results in mice and zebrafish support the context-dependent varying Gli code hypothesis (5-7). Indeed, the different functions of Gli proteins in model systems make it difficult to predict a priori which GLI protein(s) might be important in the ventral forebrain in humans, even though limited gain-of-function analyses of human GLI1 and GLI3 in frogs and mice have suggested a conserved molecular function of these GLIs across species (5, 16, 19, 20, 30, 31, 33, 34).
Our results suggest that human GLI2 is an autosomal dominant disease susceptibility gene and that its activator role is critical in the developing human ventral forebrain and face. It is the pituitary and facial structures that are the most sensitive to a reduction in GLI2 activity. Furthermore, the division of the eye field and ventral forebrain in humans likely occurs because of the concerted action of both GLI2 and GLI3 similar to the combined roles suggested for the spinal cord (14). In addition to early midline defects, Gli2 mutant mice show defects in dorsal brain growth (V. Palma and A.R.A., unpublished work), a phenotype that is consistent with the action of Shh in the mouse dorsal brain (38) but that we have not observed in our patients. This discrepancy could be caused by the functional divergence of Gli proteins in various species or the possibility that the requirement of GLI2 for brain growth in humans, as in mice, is not haploinsufficient. Interestingly, the occasional pituitary hypoplasia and/or HPE seen in patients with Pallister-Hall syndrome carrying mutations in GLI3 could be consistent with the possibility that GLI3 also partakes in the mediation of the SHH signal (8, 9) in the ventral forebrain (35, 39) and that this cannot always be compensated by the activity of GLI2 and related factors.
Recently, the ZIC2 gene, which also encodes a Zn-finger transcription factor that can bind the same target sequence as Gli proteins, was identified as a common cause of HPE (40). Unlike the patients identified in this study, who show prominent ventral craniofacial and pituitary findings associated with highly variable brain findings, the described cases with loss-of-function mutations in ZIC2 typically have few facial findings despite often quite dramatic brain defects. This observation suggests that both GLI2 and ZIC2 may act in parallel and that diminished function of both might result in the reconstitution of a complete spectrum of HPE findings. It is interesting that Zic and Gli factors have been described to physically and functionally interact in certain contexts (33, 41).
Our data support a growing body of evidence that Shh signaling is necessary for an early step in pituitary formation (42). In the zebrafish, mutations in Gli2 result in ventral forebrain abnormalities and transdifferentiation of the adenohypophysis into a lens (24). Pharmacological inhibition of cholesterol synthesis in rats, which inhibits Hh signaling, can cause HPE-like findings including alterations in pituitary formation (43). Tissue-targeted inhibition of Shh function in mice is also a cause of pituitary organogenesis failure (44). Finally, mice with simultaneous inactivation of Gli1 and Gli2 completely lack a pituitary gland (13). We are interested to see whether further examples of GLI2 mutations will highlight additional functions of GLI2 in human development and whether these can be ascertained on the basis of clinical and endocrinological findings similar to those seen in our present patients.
Acknowledgments
We thank the families for their participation in these studies. We also recognize the excellent support of the Physical Mapping Core Facility of the National Human Genome Research Institute. We thank the current members of the Muenke and Ruiz i Altaba laboratories for critical reading of the manuscript. This study was supported by National Institutes of Health Grants HD01218 and HD29862 (to J.E.M.), a Hirschl Award (to A.R.A.), National Institutes of Health grants from the National Institute of Neurological Disorders and Stroke and the National Cancer Institute (to A.R.A.), a Human Science Frontiers Program Postdoctoral Fellowship (to J.L.M.), and the Division of Intramural Research, National Human Genome Research Institute, National Institutes of Health (M.M.).
Abbreviations: HPE, holoprosencephaly; Hh, Hedgehog, SHH, Sonic Hh; AP, alkaline phosphatase; BAC, bacterial artificial chromosome.
References
- 1.Muenke, M. & Beachy, P. A. (2001) in The Metabolic and Molecular Bases of Inherited Disease, eds. Scriver, C. R., Beaudet, A. L., Sly, W. S. & Valle, D. (McGraw-Hill, New York), 8th ed., pp. 6203-6230.
- 2.Roessler, E. & Muenke, M. (2001) BioEssays 23, 888-900. [DOI] [PubMed] [Google Scholar]
- 3.Aza-Blanc, P., Ramirez-Weber, F.-A., Laget, M.-P., Schwartz, C. & Kornberg, T. B. (1997) Cell 89, 1043-1053. [DOI] [PubMed] [Google Scholar]
- 4.Aza-Blanc, P., Lin, H. Y., Ruiz i Altaba, A. & Kornberg, T. B. (2000) Development (Cambridge, U.K.) 127, 4293-4301. [DOI] [PubMed] [Google Scholar]
- 5.Ruiz i Altaba, A. (1998) Development (Cambridge, U.K.) 125, 2203-2212. [DOI] [PubMed] [Google Scholar]
- 6.Ruiz i Altaba, A. (1999) Development (Cambridge, U.K.) 126, 3205-3216. [DOI] [PubMed] [Google Scholar]
- 7.Ruiz i Altaba, A., Palma, V. & Dahmane, N. (2002) Nat. Rev. Neurosci. 3, 24-33. [DOI] [PubMed] [Google Scholar]
- 8.Sasaki, H., Nishizaki, Y., Hui, C., Nakafuku, M. & Kondoh, H. (1999) Development (Cambridge, U.K.) 126, 3915-3924. [DOI] [PubMed] [Google Scholar]
- 9.Dai, P., Akimaru, H., Tanaka, Y., Maekawa, T., Nakafuku, M. & Ishii, S. (1999) J. Biol. Chem. 19, 8143-8152. [DOI] [PubMed] [Google Scholar]
- 10.Mo, R., Freer, A. M., Zinyk, D. L., Crackower, M. A., Michaud, J., Heng, H. H., Chik, K. W., Shi, X. M., Tsui, L. C., Cheng, S. H., et al. (1997) Development (Cambridge, U.K.) 124, 113-123. [DOI] [PubMed] [Google Scholar]
- 11.Ding, Q., Motoyama, J., Gasca, S., Mo, R., Sasaki, H., Rossant, J. & Hui, C. C. (1998) Development (Cambridge, U.K.) 125, 2533-2543. [DOI] [PubMed] [Google Scholar]
- 12.Matise, M. P., Epstein, D. J., Park, H. L., Platt, K. A. & Joyner, A. L. (1998) Development (Cambridge, U.K.) 125, 2759-2770. [DOI] [PubMed] [Google Scholar]
- 13.Park, H. L., Bai, C., Platt, K. A., Matise, M. P., Beeghly, A., Hui, C. C., Nakashima, M. & Joyner, A. L. (2000) Development (Cambridge, U.K.) 127, 1593-1605. [DOI] [PubMed] [Google Scholar]
- 14.Bai, C. B. & Joyner, A. L. (2001) Development (Cambridge, U.K.) 128, 5161-5172. [DOI] [PubMed] [Google Scholar]
- 15.Bai, C. B., Auerbach, W., Lee, J. S., Stephen, D. & Joyner, A. L. (2002) Development (Cambridge, U.K.) 129, 4753-4761. [DOI] [PubMed] [Google Scholar]
- 16.Hynes, M., Stone, D. M., Dowd, M., Pitts-Meek, S., Goddard, A., Gurney, A. & Rosenthal, A. (1997) Neuron 19, 15-26. [DOI] [PubMed] [Google Scholar]
- 17.Sasaki, H., Hui, C., Nakafuku, M. & Kondoh, H. (1997) Development (Cambridge, U.K.) 124, 1313-1322. [DOI] [PubMed] [Google Scholar]
- 18.Littingtung, Y. & Chiang, C. (2000) Nat. Neurosci. 3, 979-985. [DOI] [PubMed] [Google Scholar]
- 19.Brewster, R., Mullor, J. L. & Ruiz i Altaba, A. (2000) Development (Cambridge, U.K.) 127, 4395-4405. [DOI] [PubMed] [Google Scholar]
- 20.Mullor, J. L., Dahmane, N., Sun, T. & Ruiz i Altaba, A. (2001) Curr. Biol. 11, 769-773. [DOI] [PubMed] [Google Scholar]
- 21.Hardcastle, Z., Mo, R., Hui, C. C. & Sharpe, P. T. (1998) Development (Cambridge, U.K.) 125, 2803-2811. [DOI] [PubMed] [Google Scholar]
- 22.Motoyama, J., Liu, J., Mo, R., Ding, Q., Post, M. & Hui, C. C. (1998) Nat. Genet. 20, 54-57. [DOI] [PubMed] [Google Scholar]
- 23.Shin, S. H., Kogerman, P., Lindström, E., Toftgard, R. & Biesecker, L. G. (1999) Proc. Natl. Acad. Sci. USA 96, 2880-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlstrom, R. O., Talbot, W. S. & Schier, A. F. (1999) Genes Dev. 13, 388-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hui, C. C. & Joyner, A. L. (1993) Nat. Genet. 3, 241-246. [DOI] [PubMed] [Google Scholar]
- 26.Franz, T. (1994) Acta Anat. 150, 38-44. [DOI] [PubMed] [Google Scholar]
- 27.Radhakrishna, U., Bornholdt, D., Scott, H. S., Patel, U. C., Rossier, C., Engel, H., Bottani, A., Chandal, D., Blouin, J. J., Solanki, J. V., et al. (1999) Am. J. Hum. Genet. 65, 645-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanimura, A., Dan, S. & Yoshida, M. (1998) J. Virol. 72, 3958-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roessler, E., Belloni, E., Gaudenz, K., Jay, P., Berta, P., Scherer, S. W., Tsui, L.-C. & Muenke, M. (1996) Nat. Genet. 14, 357-360. [DOI] [PubMed] [Google Scholar]
- 30.Dahmane, N., Lee, J., Robins, P., Heller, P. & Ruiz i Altaba, A. (1997) Nature 389, 876-881. [DOI] [PubMed] [Google Scholar]
- 31.Nilsson, M., Unden, A. B., Krause, D., Malmqwist, U., Raza, K., Zaphiropoulos, P. G. & Toftgard, R. (2000) Proc. Natl. Acad. Sci. USA 97, 3438-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grachtchouk, M., Mo, R., Yu, S., Zhang, X., Sasaki, H., Hui, C. C. & Dlugosz, A. A. (2000) Nat. Genet. 24, 216-217. [DOI] [PubMed] [Google Scholar]
- 33.Brewster, R., Lee, J. & Ruiz i Altaba, A. (1998) Nature 393, 579-583. [DOI] [PubMed] [Google Scholar]
- 34.Lee, J., Platt, K. A., Censullo, P. & Ruiz i Altaba, A. (1997) Development (Cambridge, U.K.) 124, 2537-2552. [DOI] [PubMed] [Google Scholar]
- 35.Rallu, M., Machold, R., Gaiano, N., Corbin, J. G., McMahon, A. P. & Fishell, G. (2002) Development (Cambridge, U.K.) 129, 4963-4974. [DOI] [PubMed] [Google Scholar]
- 36.Chiang, C., Litingtung, Y., Lee, E., Young, K. E., Corden, J. L., Westphal, H. & Beachy, P. A. (1996) Nature 383, 407-413. [DOI] [PubMed] [Google Scholar]
- 37.Karlstrom, R. O., Tyurina, O. V., Kawakami, A., Nishioka, N., Talbot, W. S., Sasaki, H. & Schier, A. F. (2003) Development (Cambridge, U.K.) 130, 1549-1564. [DOI] [PubMed] [Google Scholar]
- 38.Dahmane, N., Sanchez, P., Gitton, Y., Palma, V., Sun, T., Beyna, M., Weiner, H. & Ruiz i Altaba, A. (2001) Development (Cambridge, U.K.) 128, 5201-5212. [DOI] [PubMed] [Google Scholar]
- 39.Roessler, E. & Muenke, M. (2003) Hum. Mol. Genet. 12, R15-R25. [DOI] [PubMed] [Google Scholar]
- 40.Brown, S. A., Warburton D., Brown, L. Y., Yu, C., Roeder, E. R., Stengel-Rutkowski, S., Hennekam, R. C. M. & Muenke, M. (1998) Nat. Genet. 20, 180-183. [DOI] [PubMed] [Google Scholar]
- 41.Koyabu, Y., Nakata, K., Mizugishi, K., Aruga, J. & Mikoshiba, K. (2001) J. Biol. Chem. 278, 6889-6892. [DOI] [PubMed] [Google Scholar]
- 42.Sheng, H. & Westphal, H. (1999) Trends Genet. 15, 236-240. [DOI] [PubMed] [Google Scholar]
- 43.Gofflot, F., Kolf-Clauw, M., Clotman, F., Roux, C. & Picard, J. J. (1999) Am. J. Med. Genet. 87, 207-216. [DOI] [PubMed] [Google Scholar]
- 44.Treier, M., O'Connell, S., Gleiberman, A., Price, J., Szeto, D. P., Burgess, R., Chuang, P.-T., McMahon, A. P. & Rosenfeld, M. G. (2001) Development (Cambridge, U.K.) 128, 377-386. [DOI] [PubMed] [Google Scholar]