Abstract
Insulators define independent domains of gene function throughout the genome. The Drosophila gypsy insulator was isolated from the gypsy retrotransposon as a region that contains a cluster of binding sites for the Suppressor of Hairy-wing [Su(Hw)] protein. To study the effects of the gypsy insulator on gene expression within a single genomic domain, targeted gene replacement was used to exchange the endogenous yellow gene, located at cytological location 1A, with a set of gypsy-modified yellow genes. Replaced yellow genes carried a gypsy insulator positioned between the yellow promoter and either the upstream or the downstream tissue-specific enhancers. Whereas the gypsy insulator blocked the function of the upstream enhancers at the endogenous location, the downstream enhancers were not blocked. Investigation of the 1A region revealed two clustered Su(Hw)-binding sites downstream of the yellow gene, named 1A-2, that bind Su(Hw) in vivo and possess enhancer blocking function. We propose that interaction between 1A-2 and the gypsy insulator permits activation of yellow expression by enhancers in the neighboring achaete-scute complex, causing an apparent absence of the block of the downstream yellow enhancers. Based on these data, we suggest that 1A-2 is an endogenous Su(Hw) insulator that separates regulatory domains within the Drosophila genome.
Keywords: enhancers, chromatin, gypsy, gene replacement
Eukaryotic chromosomes are partitioned into distinct chromatin domains. These domains reflect the assembly of higher order structures that are proposed to fold in a manner that imparts functional independence (1, 2). Recent observations that up to 30% of genes within a eukaryotic genome are arranged in coexpressed clusters indicate that formation of independent structural domains may represent an important mechanism by which transcriptional fidelity is maintained within the genome (3-5).
Mechanisms used to delimit independent structural and functional domains are poorly understood. A specialized class of DNA elements, known as insulators, is implicated in these processes (6). Insulators are defined by two properties. First, insulators protect genes from chromosomal position effects that result from positive and negative influences of nearby chromatin (7-10). Second, insulators block enhancer and silencer action when positioned between these control elements and a promoter, but not when located upstream of the regulatory elements (11-14). The link between chromatin domains and insulators has been suggested by many studies, such as those of the chicken β-globin locus. This domain is defined by binding sites for the insulator protein, CCCTC-binding factor (CTCF), that separate the cluster of globin genes from the neighboring 5′-region of condensed chromatin and the independently regulated 3′-folate receptor gene (15, 16). CTCF sites mark the transitions of distinct nuclease susceptibility, histone hyperacetylation and methylation of lysine 4 of histone H3 (16-18). These observations imply that the CTCF insulator protein defines a structural domain that imparts independent regulatory function.
One well characterized Drosophila insulator is the gypsy insulator, which was isolated from the gypsy retrotransposon. The function of this insulator depends on two proteins, the zinc finger DNA-binding protein, Suppressor of Hairy-wing [Su(Hw)], and the BTB/POZ domain protein Modifier of mdg4 [Mod- (mdg4)]67.2 that is recruited to chromosomes through protein-protein interactions with the Su(Hw) protein (19-22). The gypsy insulator proteins colocalize at hundreds of genomic sites, with <1% of these sites representing positions of the gypsy retrotransposon (23). Although the function of the non-gypsy sites is unclear, it is proposed that these regions represent sites of endogenous insulators.
Recent studies demonstrate that gypsy insulators interact, altering gene expression in complex ways. A single gypsy insulator placed between an enhancer and promoter blocks enhancer-activated transcription, whereas two gypsy insulators placed in this position do not block the enhancer, an effect termed insulator neutralization or bypass (24-26). Based on these observations, it was proposed that the outcome of interactions between a pair of gypsy insulators depends on the location of the insulators (27). Interactions between gypsy insulators that flank an enhancer may prevent enhancer action by defining a loop domain of higher order chromatin structure that prevents enhancer-promoter communication, whereas interactions between two insulators located between the enhancer and promoter may facilitate enhancer action by decreasing the linear distance between the enhancer and promoter. These data imply that the gypsy insulator proteins may have versatile roles within the Drosophila genome.
To gain insights into gypsy insulator function, targeted gene replacement was used to insert the gypsy insulator at several positions within the endogenous yellow gene, located within cytological region 1A near the tip of the X chromosome. This gene encodes a protein responsible for dark pigmentation of larval and adult cuticle structures. Several independent tissue-specific enhancers, located upstream and downstream of the yellow promoter, are required for transcription (1). Studies of transgenic flies carrying gypsy insulator-modified yellow genes demonstrated that the gypsy insulator blocks every yellow enhancer when positioned between the enhancer and promoter (11). Surprisingly, enhancer blocking was not observed when a gypsy insulator was placed within the intron of the endogenous yellow gene, positioned between the bristle and tarsal claw enhancers and promoter. Investigation of the 1A region revealed an endogenous Su(Hw)-binding region, 1A-2, that has insulator properties. We propose that interactions between the intronic gypsy insulator and 1A-2 promote activation of yellow transcription by enhancers in the neighboring achaete-scute complex (AS-C). Our studies suggest that 1A-2 is an endogenous insulator that establishes the independence of two neighboring regulatory domains.
Materials and Methods
Targeted Gene Replacement. Template plasmids used for gene replacement had an ≈7.7-kb yellow fragment (from a SalI to a BglII) that was modified by insertion of a gypsy insulator cassette at positions -900, -700, -300, or +660 relative to the transcription start site. The gypsy insulator cassette had the 350-bp insulator cloned between direct repeats of lox P sites, target sites for the Cre recombinase (28). Targeted gene replacement was carried out as described (29). The structure of the yellow gene in each putative replacement strain was assessed by Southern analysis. If this analysis showed evidence of an appropriately sized insertion, the promoter and insulator regions were PCR amplified and the resulting fragments sequenced. In all cases, only those lines that had the predicted sequence were studied further.
Phenotypic analysis involved crossing transgenic males to y1 w67c23 females and analysis of wing and body pigmentation in females, as described (26). Pigmentation was scored in 3- to 4-day-old female progeny by comparison to five parallel controls, where 1 represents null or nearly null, 5 represents WT, and scores of 2, 3, and 4 represent phenotypes of y2/y1, y82f29/y1#8, and y2/y1#8 flies, respectively. For each cross, 20-30 females were scored independently by at least two people. Flies of a given genotype, scored at different times, may show small differences in pigmentation score. For this reason, we only consider differences between scores of a unit or more to be significant (26).
Su(Hw) Ab and Western Analysis. The Su(Hw) Ab was an affinity purified, polyclonal rabbit Ab raised against a carboxyl-terminal peptide (RENKKKPVGEQEKA, Bethyl Laboratories, Montgomery, TX). Western analysis was done on protein extracts isolated from Canton S (CS, WT) and su(Hw) mutant third instar larvae. The su(Hw) genotypes studied were su(Hw)v/su(Hw)f and su(Hw)v/su(Hw)v, where the su(Hw)f allele encodes a protein mutated in finger 10 and su(Hw)v is a protein null, carrying a deletion of the su(Hw) and the neighboring RpII15 promoter (30, 31). Both su(Hw) mutant backgrounds completely reverse gypsy insulator effects. Protein from one larval equivalent was loaded on an 8% SDS polyacrylamide gel. Proteins were detected on Western blots by using the Su(Hw) Ab (1:3,300 dilution) followed by an anti-rabbit secondary Ab-conjugated to horseradish peroxidase. These analyses showed an ≈130-kDa protein that was present in CS larvae but reduced or absent in su(Hw) mutant larvae (see Fig. 3A), with two additional cross-reacting bands demonstrating equal loading between lanes. Importantly, the Su(Hw) Ab recognized only one protein in nuclear extracts isolated from CS embryos (see Fig. 3, N.E., 10 μg loaded).
Fig. 3.
ChIP of pSI-binding sites. (A) Shown is a Western blot of third-instar larval proteins isolated from three different su(Hw) genotypes probed with a Su(Hw) Ab. The v/f allele represents su(Hw)v/su(Hw)f heterozygotes, and v/v represents su(Hw)v/su(Hw)v flies. N.E. corresponds to embryonic nuclear extract from CS. Sizes in kilodaltons are shown on the left. (B) Representative examples of PCR products obtained from chromatin material that was directly purified (Input, IN) or immunoprecipitated with Su(Hw) or nonspecific (N.S.) Ab. (C) The average of at least three ChIP experiments is displayed. Results are expressed as percentage of input DNA. Gray bars, Su(Hw); black bars, nonspecific Ab.
Chromatin Immunoprecipitation (ChIP). Chromatin was prepared from nuclei isolated from third instar larvae. This stage was chosen because large quantities of larvae were available, and procedures for nuclear isolation were established (32). The Su(Hw) protein is ubiquitously expressed, suggesting that this stage accurately reflects genomic binding of protein. Nuclei were isolated as described (32), except that buffer A was prepared with Hepes-KOH (pH 7.6) and supplemented with 1× protease inhibitor mixture (Roche) and 0.1 mg/ml PMSF. Purified nuclei were resuspended in buffer A plus 0.1% Nonidet P-40 (≈108 nuclei/ml) and cross-linked with 1% formaldehyde for 5 min at room temperature. Nuclei were lysed and the chromatin sheared to an average length of ≈700 bp by sonication and immunoprecipitated as described (Upstate Biotechnology, Lake Placid, NY). In each ChIP experiment, a chromatin solution containing ≈20 μg of genomic DNA was incubated with either 1 μg of Su(Hw) Ab or normal rabbit IgG (Sigma). The precipitated products were analyzed by PCR, by using 10% of the precipitated DNA or 0.1% of the input DNA for all primers except the gypsy primers where 1% and 0.01% were used, respectively. PCR conditions were tested for each primer set to ensure amplification was in the linear range. PCR products were analyzed on a 6% polyacrylamide gel and digital images of ethidium bromide stained gels were quantitated by using IMAGEJ software (National Institutes of Health). ChIP was performed on three separate chromatin preparations.
Primers for ChIP PCR, listed 5′-3′, and their product sizes were as follows: gypsy (311 bp) TCAAAAAATAAGTGCTGCATACTTTTT and GAGCACAATTGATCGGCTA; hsp26 (306 bp) TTCGCTTGTGGATGAACTC and TCCACCACCTTCACGTTGA; 1A-1 (276 bp) TTGCCTTGAAGAGATTGGTCG and CCTCAGACATAATTTGCCTGC; 1A-2 (269 bp) CTGCAAGCTAGATCCACCTG and CTTCGTCTACCGTTGTGC; and 1A-6 (371 bp) TCTACCTGTTGCATTATTCTCC and GCCTTTTAAGGTTACCTATTACAG.
Analysis of 1A-2. A 520-bp fragment, termed 1A-2, encompassed two gypsy consensus sites and was PCR amplified from CS DNA by using the primers 5′-TTTGAAGGTTAAGAGTTTACG-3′ and 5′-CTTCGTCTACCGTTGTGC-3′. The PCR product was cloned in the Topo II vector (Invitrogen) and sequenced. An intact 1A-2 was cloned between direct repeats of lox P sites and inserted into the yellow gene at position -900 relative to the transcription start site in the P element transformation vector that had the yellow and mini-white genes (26). Seven transgenic lines with a single transposon insertion were established in the host strain y1w67c23. Flies from five lines were crossed into the su(Hw)v/su(Hw)f genetic background to determine the contribution of the Su(Hw) protein to the observed yellow phenotype.
Results
P element-mediated germ line transformation has shown that the gypsy insulator blocks all of the yellow enhancers in a position-dependent manner (11). To test whether these effects were recapitulated at a single genomic location, we used targeted gene replacement to exchange the WT yellow gene with a gene carrying a gypsy insulator inserted 900-, 700-, or 300-bp upstream or 660-bp downstream of the promoter (Fig. 1). Successful gene replacement stocks were called y-900gin, y-700gin, y-300gin, and y+660gin, named for a gypsy insulator located at the indicated position.
Fig. 1.
Gypsy insulators converted into the endogenous yellow gene. (A) A map of the yellow gene. The coding region is represented by black rectangles, untranslated regions are represented by open rectangles, and enhancers are represented by shaded ovals. Enhancers are wing (W), body (B), bristle (Br), and tarsal claw (TC). The gypsy insulators (filled triangles) were inserted at positions -900, -700, -300, and +660 relative to the start site of transcription (bent arrow). Restriction sites are SalI (S), HinD III (H), BglII (G), and KpnI (K). (B) The level of yellow expression in the ygin lines is indicated for the wing, body, bristle, and tarsal claw tissues and was inferred by analyses of cuticle phenotype. (C) Southern blot analysis of the y+660gin genomic DNA digested with KpnI and probed with a SalIto BglII fragment of yellow that includes the entire gene. The first lane is CS, and the remaining lanes are three independent y+660gin lines. Sizes in kilobases are indicated on the left, with arrows on the right showing the sizes of hybridizing bands.
In the y-900gin, y-700gin, and y-300gin lines, the gypsy insulator was between the wing and body enhancers and yellow promoter. Phenotypic analysis showed that flies from each line had low levels of wing and body pigmentation (average pigmentation scores of 2, 2) and WT levels of bristle and tarsal claw pigmentation; a phenotype that matched that of y2 flies that carry a gypsy retrotransposon inserted at -700 bp (Fig. 1). These data demonstrate that the gypsy insulator produces a consistent block of the yellow wing and body enhancers when positioned between these enhancers and the promoter, irrespective of the precise location within the upstream regulatory region (11). Previous studies of gypsy insulator effects in the Drosophila affinidisjuncta alcohol dehydrogenase gene showed that insertion of the gypsy insulator within 500 bp of the promoter activated transcription in a manner that depended on the Su(Hw) protein (33). Such activating effects are not a general property of this insulator because we find that the gypsy insulator impedes the upstream enhancers in y-300gin flies.
In the y+660gin line, the gypsy insulator was inserted between the downstream bristle and tarsal claw enhancers and yellow promoter. Flies isolated from three independent y+660gin replacement lines showed WT pigmentation in all tissues, including the bristles and tarsal claws (average pigmentation scores of 5, 5 in the wing and body, Fig. 1). These results indicate that the gypsy insulator did not block the downstream enhancers, even though previous transgene studies demonstrated that a +660 gypsy insulator was functional in this yellow position (11).
Southern analysis was undertaken to analyze the structure of the yellow gene in y+660gin flies. Genomic DNA was isolated, digested with KpnI, transferred to Nytran, and hybridized with a fragment including the complete yellow gene (Fig. 1). Hybridization in WT flies (CS) showed two bands, an ≈5-kb band that contained 5′-yellow sequences and a second 1.1-kb band that contained the yellow promoter and extended ≈0.15 kb downstream of the +660 insertion point. A third fragment, corresponding to sequences downstream of the 1.1-kb KpnI fragment, was not detected, possibly because the size was >39 kb. Analysis of genomic DNA isolated from each of the y+660gin convertant lines showed the ≈5-kb fragment, suggesting that a gross rearrangement of the gene during replacement did not occur. The y+660gin flies had a second band of 1.4 kb that differed from the band in CS flies and was the expected size for insertion of the gypsy insulator. PCR amplification and sequence analysis of the promoter and insulator regions in the y+660gin lines confirmed that the y+660gin alleles had a WT promoter and gypsy insulator inserted at +660. These data suggest that the lack of enhancer blocking in flies may be caused by sequences other than the +660 gypsy insulator.
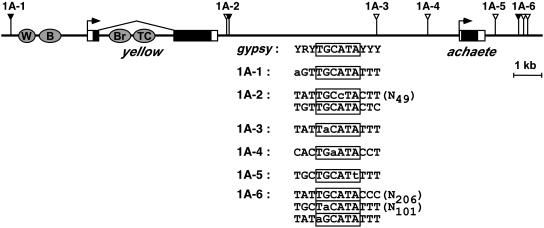
Su(Hw)-Binding Sites Downstream of the yellow Gene. We postulated that the loss of enhancer blocking by the +660 gypsy insulator might reflect insulator neutralization caused by interactions between the +660 insulator and a nearby endogenous Su(Hw) insulator. To identify a putative Su(Hw) insulator (pSI), several features of the gypsy insulator were considered. The gypsy insulator contains 12, closely spaced, degenerate Su(Hw)-binding sites that share a TGCATA core and are embedded in AT-rich sequences (19, 34). Insertion into, or deletion of, binding sites within the region disrupts the insulator function (35-37), with an apparent threshold of four binding sites required for activity (38). Based on these data, we predicted that a pSI would have two features. First, the pSI would have sites that match the gypsy consensus site (YRYTGCATAYYY), with an intact TGCATA core (Fig. 2). Second, the pSI would contain at least four closely spaced Su(Hw) sites. Analysis of a 20-kb region encompassing the yellow gene revealed six regions that matched the consensus sequence, none of which met all of our predictions (Fig. 2). Three regions (1A-1, 1A-2, and 1A-6) had gypsy consensus sites with an intact TGCATA core, whereas the other sites had mutated core sequences. Only 1A-2 and 1A-6 contained two or more sites, causing us to consider these as pSIs.
Fig. 2.
Location of predicted Su(Hw)-binding sites surrounding the yellow locus. The positions of predicted Su(Hw)-binding sites in the yellow-achaete region are indicated, with intact (▾) or polymorphic (▿) TGCATA motifs as raised triangles. The sequence of each binding site compared to the gypsy consensus is shown. The TGCATA core is boxed, polymorphic nucleotides are in lowercase, and the number of nucleotides between core sites in a cluster is listed at the right.
ChIP experiments were undertaken to determine whether the Su(Hw) protein bound any of the Su(Hw) consensus sites in vivo (Fig. 3). As a positive control, primers in the gypsy retrotransposon that surrounded the gypsy insulator were used. These primers produced a robust PCR signal from material precipitated by the Su(Hw) Ab (2.0 ± 0.7% of the input DNA), whereas PCR amplification of the gypsy insulator in ChIP material from the nonspecific Ab produced a low signal (0.2 ± 0.1% of the input DNA; Fig. 3). As a negative control, primers corresponding to the heat shock protein (hsp) 26 coding region were used, because this gene is devoid of consensus Su(Hw)-binding sites. Only a low level of PCR amplification occurred from the ChIP material from either Ab (<0.1% of the input DNA; Fig. 3), confirming that the Su(Hw) protein does not associate with this gene.
The association of the Su(Hw) protein was tested by using primers that flanked the pSIs. The 1A-2 primers amplified a robust product from chromatin precipitated by the Su(Hw) Ab, whereas only a low level of product was produced from the ChIP material produced by incubation with the nonspecific Ab (1.3 ± 0.3% and <0.1%, respectively; Fig. 3). The level of the 1A-2 PCR product from the Su(Hw) ChIP material implies that the Su(Hw) protein associates with this region in vivo. In contrast, a low level of PCR product was produced by primers surrounding 1A-6 in chromatin that was precipitated by either the Su(Hw) or nonspecific Ab (<0.1% of input, Fig. 3). Similarly, a low level of PCR amplification of ChIP material was obtained by using primers surrounding the four pSIs with a single Su(Hw)-binding site (Fig. 3 and data not shown). Based on these in vivo studies, we conclude that 1A-2 is the only pSI that binds the Su(Hw) protein.
Enhancer Blocking by the pSI 1A-2. To test whether 1A-2 had properties of an insulator, an enhancer-blocking assay was used. This assay provides a direct assessment of the ability of a sequence to prevent regulatory interactions. To this end, the P[y1A-2] transposon was constructed that carried 1A-2 inserted 900 bp upstream of the yellow transcription start, between the wing and body enhancers and promoter. Seven P[y1A-2] lines were established and phenotypic analyses were conducted. We found that P[y1A-2] flies had low levels of wing and body pigmentation (Fig. 4, an average pigmentation score of 3, 2+), which is similar to the level found in transgenic P[En G] flies that carry a gypsy insulator inserted at the same position in the yellow gene (pigmentation scores of 3, 2; 26). Importantly, the bristle and tarsal claw phenotypes of the P[y1A-2] flies were WT, demonstrating that the low level of expression in the wing and body was not caused by insertion of a silencer. Although the average phenotype among the P[y1A-2] lines was similar to that of P[En G] flies, none of these lines had pigmentation levels as low as that found in y2 flies or the flies from the upstream ygin convertant lines (pigmentation scores of 2, 2), whereas several P[En G] lines did. These data suggest that 1A-2 establishes a weaker enhancer block than the gypsy insulator, consistent with the fewer number of Su(Hw)-binding sites associated with this insulator. Insertion of 1A-2 upstream of the yellow wing and body enhancers did not interfere with enhancer function (data not shown), demonstrating that 1A-2 effects on gene expression are position-dependent. The 1A-2 region was independently identified due to disruption of enhancer activated transcription in the achaete-scute gene cluster and subsequently shown to block the white eye enhancer (39). Taken together these data suggest that 1A-2 has general enhancer blocking effects.
Fig. 4.
1A-2 acts as an enhancer blocker. (Top) Structure of the P[y1A-2] transposon. A DNA fragment containing 1A-2 (▿) was inserted between the upstream enhancers and promoter of a yellow gene in a P transposon (black boxes with P show P element ends). Symbols are described in Fig. 1. (Bottom) Phenotype of P[y1A-2] flies. The average wing and body (W, B) pigmentation score in a WT [su(Hw)+] and mutant [su(Hw)-] background is listed. A “+” indicates average level of pigmentation was slightly greater than that of the corresponding control.
To test the contribution of the Su(Hw) protein to enhancer blocking by 1A-2, we crossed flies from five P[y1A-2] lines into a su(Hw)v/su(Hw)f mutant background. In four of five cases, the level of pigmentation in su(Hw) WT and mutant P[y1A-2] flies was not substantially different, whereas the fifth line showed an increase in body pigmentation only (Fig. 4). Our findings suggest that enhancer blocking by 1A-2 is less dependent on the Su(Hw) protein than previously reported (39) and imply that the Su(Hw) protein cooperates with other protein(s) bound to 1A-2 to prevent enhancer-promoter communication.
Discussion
The Su(Hw) insulator protein binds hundreds of sites within the Drosophila genome. The vast majority of these regions have not been characterized because few correspond to the gypsy insulator. Isolation of endogenous Su(Hw) sites has been hampered by the distinct sequence organization of these sites relative to the gypsy insulator. Here we report the identification of the first endogenous Su(Hw)-binding region, 1A-2, which was discovered through effects on enhancer blocking by the gypsy insulator. Several features of 1A-2 differ from the gypsy insulator. The 1A-2 contains two Su(Hw)-binding sites with nonidentical core sequences, spaced by 49 bp. In contrast, the gypsy insulator has 12 Su(Hw) sites that contain an invariant TGCATA core and are distanced an average of 26-28 bp. The mutated core site in 1A-2 binds the Su(Hw) protein in vitro (B. L. Gilmore, M. S. Wold, and P. K. G., unpublished data), demonstrating that the TGCATA core is not essential for DNA association. These data support the proposition that the Su(Hw) protein binds a broad spectrum of endogenous sequences.
The in vivo requirements of DNA binding by the Su(Hw) protein are unclear. We characterized a second cluster of three Su(Hw) sites, called pSI 1A-6, and found that these sequences do not bind the Su(Hw) protein in vivo. Previous studies suggest that one critical determinant of Su(Hw) association is the spacing between binding sites (38). In 1A-6, the Su(Hw) sites are spaced at ≈100 bp apart, a greater distance than found in the gypsy or 1A-2 insulators. These observations indicate that in vivo binding by the Su(Hw) protein may require the presence of at least two sites within 100 bp. Further studies are needed to more fully understand the determinants of Su(Hw) DNA binding.
Transgene studies demonstrated that 1A-2 prevented transcriptional activation by the yellow enhancers in a position-dependent manner, without inactivation of the promoter. Based on these findings, we conclude that 1A-2 has properties of a genomic insulator. It was unexpected that Su(Hw)-binding regions with fewer than four sites would establish a block of enhancers, as previous studies showed that synthetic insulators with fewer than four tandem copies of a gypsy insulator site did not (38). The distinct sequence composition of 1A-2 relative to the synthetic gypsy insulators may account for these differences. The Su(Hw) sites in 1A-2 may have a higher intrinsic affinity than the gypsy insulator sites, thereby increasing DNA occupancy by the Su(Hw) protein such that only two sites are needed for an enhancer block. Alternatively, 1A-2 may bind additional protein(s) that cooperate with the Su(Hw) protein to block enhancer action. This possibility is consistent with our observation that mutation of the Su(Hw) protein, as assessed by phenotypic analysis of su(Hw)v/su(Hw)f flies, had a minimal, if any, effect on enhancer blocking by 1A-2. Although it is possible that the distinct sequence of 1A-2 permits DNA association by the Su(Hw)f protein due to different zinc finger requirements for binding, we favor the alternative that other proteins are needed for the residual block. Interestingly, a 125-bp fragment of 1A-2 that includes only the Su(Hw)-binding sites does not confer enhancer blocking (39), demonstrating that the 1A-2 Su(Hw) sites are not sufficient for insulator activity. The identity of the putative insulator protein is unknown, as 1A-2 does not contain binding sites for the known Drosophila insulator proteins, Zw5, BEAF, or GAGA factor (40-42).
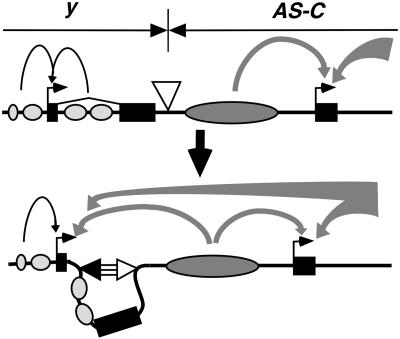
The 1A-2 element was identified because a gypsy insulator inserted at +660 in the yellow intron did not reduce pigmentation in the bristle and tarsal claws of y+660gin flies, although it was positioned between the bristle and tarsal claw enhancers and promoter. We propose that these effects result from interactions between the +660 gypsy insulator and the downstream 1A-2 element, located ≈4.4 kb away (Fig. 5). Previous studies have demonstrated that interactions between two gypsy insulators result in an apparent neutralization of insulator function because these associations bring distant enhancers to a target promoter, not because the insulators are inactivated (25). Based on these findings, we predict that transcription in the bristles and tarsal claws in y+660gin flies results from the action of enhancers downstream of the loop formed by 1A-2 and the gypsy insulator, not from the yellow bristle and tarsal claw enhancers that are present in the constrained loop that is presumed to prevent enhancer action. This supposition is supported by the properties of the neighboring genes. 1A-2 is 8.5 kb upstream of the achaete gene, one of a cluster of genes within AS-C that encode basic helix-loop-helix transcription factors required for the development of extra sensory organs in the fly, such as the bristles and hairs (reviewed in ref. 43). This complex contains multiple enhancer regions distributed over a ≈90-kb region (44). We postulate that enhancers from the AS-C drive transcription in the bristles and tarsal claws of the y+660gin flies because these elements are active at the same time and in the same tissues as the yellow intronic enhancers.
Fig. 5.
A model for yellow activation in the bristles of y+660gin flies. In a WT chromosome, the 1A-2 insulator (▿) separates the yellow and AS-C regulatory domains and prevents cross-regulation by enhancers (curved black and gray arrows, Top). In the y+660gin allele, the gypsy insulator (▾) interacts with the 1A-2 insulator, allowing bypass of enhancers from the AS-C to activate yellow transcription. The thick arrows indicate the possible participation of additional enhancers in AS-C other than those shown.
Several lines of evidence support the proposal that the endogenous role of 1A-2 is to prohibit regulatory interactions between the yellow gene and enhancers in the AS-C (Fig. 5). First, the 1A-2 element demonstrates insulator properties in an enhancer blocking assay. Second, data from genetic studies indicate that cross-regulatory interactions occur between the yellow and AS-C regulatory regions when a gypsy insulator is inserted. These data include our studies of y+660gin flies and previous studies of the “hairy-wing” class of achaete-scute alleles, a set of dominant, gain of function mutations that produce supernumerary chaetae and sensilla and were responsible for the original identification of the su(Hw) gene (45, 46). Our discovery of 1A-2 provides new insights into the changes in expression associated with the hairy-wing class of mutations. Two alleles, Hw1 and a spontaneous derivative of Hw1 called HwBS, carry an insertion of a gypsy retrotransposon within achaete that increases achaete mRNA levels, leading to an overproduction of a transcription factor that directs sensilla development (47). We propose that the increased achaete mRNA levels result from an interaction between 1A-2 and the gypsy insulator in the gypsy retrotransposon that brings yellow enhancers to the AS-C to activate transcription. This model is supported by two X-ray-induced revertants of Hw1 that appear to be associated with chromosomal rearrangements that have breakpoints in or downstream of the yellow regulatory region that contains the bristle enhancer (47), indicating that the yellow enhancers participate in the generation of the hairy-wing phenotype. Our y+660gin flies do not have a hairy-wing phenotype, suggesting that the yellow bristle or tarsal claw enhancers are the regulatory elements participating in the Hw1 phenotype but fail to do so in y+660gin flies because these enhancers are in a constrained loop (Fig. 5). We note that there are no discernable changes in AS-C gene expression in a su(Hw) mutant background. We propose that these effects may reflect functional redundancy in the 1A-2 insulator, a proposition that is supported by our experiments on enhancer blocking by 1A-2 in a su(Hw) mutant background. Further studies are needed to determine whether the natural role of 1A-2 is to establish an insulator that separates the yellow and AS-C regulatory domains.
Acknowledgments
We thank Stephanie Grade, Lori Wallrath, Ting Wu, and Oya Yazgan for critical reading of the manuscript and comments on the experimental design and interpretation. This work was supported by National Institute of Health Grants GM42539 (to P.K.G.) and GM61936 (to P.K.G. and C.-t. Wu, Harvard Medical School, Boston).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Su(Hw), Suppressor of Hairy-wing; pSI, putative Su(Hw) insulator; AS-C, achaete-scute complex; ChIP, chromatin immunoprecipitation; CS, Canton S.
References
- 1.Kuhn, E. J. & Geyer, P. K. (2003) Curr. Opin. Cell Biol. 15, 259-265. [DOI] [PubMed] [Google Scholar]
- 2.Labrador, M. & Corces, V. G. (2002) Cell 111, 151-154. [DOI] [PubMed] [Google Scholar]
- 3.Boutanaev, A. M., Kalmykova, A. I., Shevelyov, Y. Y. & Nurminsky, D. I. (2002) Nature 420, 666-669. [DOI] [PubMed] [Google Scholar]
- 4.De Gregorio, E., Spellman, P. T., Tzou, P., Rubin, G. M. & Lemaitre, B. (2002) EMBO J. 21, 2568-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spellman, P. T. & Rubin, G. M. (2002) J. Biol. 1, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.West, A. G., Gaszner, M. & Felsenfeld, G. (2002) Genes Dev. 16, 271-288. [DOI] [PubMed] [Google Scholar]
- 7.Roseman, R. R., Pirrotta, V. & Geyer, P. K. (1993) EMBO J. 12, 435-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roseman, R. R., Johnson, E. A., Rodesch, C. K., Bjerke, M., Nagoshi, R. N. & Geyer, P. K. (1995) Genetics 141, 1061-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kellum, R. & Schedl, P. (1991) Cell 64, 941-950. [DOI] [PubMed] [Google Scholar]
- 10.Chung, J. H., Whiteley, M. & Felsenfeld, G. (1993) Cell 74, 505-514. [DOI] [PubMed] [Google Scholar]
- 11.Geyer, P. K. & Corces, V. G. (1992) Genes Devel. 6, 1865-1873. [DOI] [PubMed] [Google Scholar]
- 12.Kellum, R. & Schedl, P. (1992) Mol. Cell. Biol. 12, 2424-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holdridge, C. & Dorsett, D. (1991) Mol. Cell. Biol. 11, 1894-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mallin, D. R., Myung, J. S., Patton, J. S. & Geyer, P. K. (1998) Genetics 148, 331-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prioleau, M. N., Nony, P., Simpson, M. & Felsenfeld, G. (1999) EMBO J. 18, 4035-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saitoh, N., Bell, A. C., Recillas-Targa, F., West, A. G., Simpson, M., Pikaart, M. & Felsenfeld, G. (2000) EMBO J. 19, 2315-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Litt, M. D., Simpson, M., Gaszner, M., Allis, C. D. & Felsenfeld, G. (2001) Science 293, 2453-2455. [DOI] [PubMed] [Google Scholar]
- 18.Litt, M. D., Simpson, M., Recillas-Targa, F., Prioleau, M. N. & Felsenfeld, G. (2001) EMBO J. 20, 2224-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spana, C., Harrison, D. A. & Corces, V. G. (1988) Genes Devel. 2, 1414-1423. [DOI] [PubMed] [Google Scholar]
- 20.Georgiev, P. G. & Gerasimova, T. I. (1989) Mol. Gen. Genet. 220, 121-126. [DOI] [PubMed] [Google Scholar]
- 21.Ghosh, D., Gerasimova, T. I. & Corces, V. G. (2001) EMBO J. 20, 2518-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gause, M., Morcillo, P. & Dorsett, D. (2001) Mol. Cell. Biol. 21, 4807-4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerasimova, T. I., Gdula, D. A., Gerasimov, D. V., Simonova, O. & Corces, V. G. (1995) Cell 82, 587-597. [DOI] [PubMed] [Google Scholar]
- 24.Cai, H. N. & Shen, P. (2001) Science 291, 493-495. [DOI] [PubMed] [Google Scholar]
- 25.Muravyova, E., Golovnin, A., Gracheva, E., Parshikov, A., Belenkaya, T., Pirrotta, V. & Georgiev, P. (2001) Science 291, 495-498. [DOI] [PubMed] [Google Scholar]
- 26.Kuhn, E. J., Viering, M. M., Rhodes, K. M. & Geyer, P. K. (2003) EMBO J. 22, 2463-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mongelard, F. & Corces, V. G. (2001) Nat. Struct. Biol. 8, 192-194. [DOI] [PubMed] [Google Scholar]
- 28.Siegal, M. L. & Hartl, D. L. (1996) Genetics 144, 715-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morris, J. R., Geyer, P. K. & Wu, C. T. (1999) Genes Devel. 13, 253-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harrison, D. A., Gdula, D. A., Coyne, R. S. & Corces, V. G. (1993) Genes Devel. 7, 1966-1978. [DOI] [PubMed] [Google Scholar]
- 31.Harrison, D. A., Mortin, M. A. & Corces, V. G. (1992) Mol. Cell. Biol. 12, 928-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wallrath, L. L., Swede, M. J. & Elgin, S. C. R. (1998) in Chromatin: A Practical Approach, ed. Gould, H. (Oxford Univ. Press, Oxford), pp. 59-77.
- 33.Wei, W. & Brennan, M. D. (2001) Mol. Cell. Biol. 21, 7714-7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marlor, R. L., Parkhurst, S. M. & Corces, V. G. (1986) Mol. Cell. Biol. 6, 1129-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoover, K. K., Gerasimova, T. I., Chien, A. J. & Corces, V. G. (1992) Genetics 132, 691-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geyer, P. K., Green, M. M. & Corces, V. G. (1988) Proc. Natl. Acad. Sci. USA 85, 8593-8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flavell, A. J., Alphey, L. S., Ross, S. J. & Leigh-Brown, A. J. (1990) Mol. Gen. Genet. 220, 181-185. [DOI] [PubMed] [Google Scholar]
- 38.Scott, K. C., Taubman, A. D. & Geyer, P. K. (1999) Genetics 153, 787-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Golovnin, A., Birukova, I., Romanova, O., Silicheva, M., Parshikov, A., Savitskaya, E., Pirrotta, V. & Georgiev, P. (2003) Development (Cambridge, U.K.) 130, 3249-3258. [DOI] [PubMed] [Google Scholar]
- 40.Gaszner, M., Vazquez, J. & Schedl, P. (1999) Genes Devel. 13, 2098-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao, K., Hart, C. M. & Laemmli, U. K. (1995) Cell 81, 879-889. [DOI] [PubMed] [Google Scholar]
- 42.Ohtsuki, S. & Levine, M. (1998) Genes Devel. 12, 3325-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Campuzano, S. & Modolell, J. (1992) Trends Genet. 8, 202-208. [DOI] [PubMed] [Google Scholar]
- 44.Gomez-Skarmeta, J. L., Rodriguez, I., Martinez, C., Culi, J., Ferres-Marco, D., Beamonte, D. & Modolell, J. (1995) Genes Dev. 9, 1869-1882. [DOI] [PubMed] [Google Scholar]
- 45.Lindsley, D. L. & Grell, E. H. (1968) Genetic Variations of Drosophila melanogaster (Carnegie Institution of Washington, Baltimore), Pub. No. 627.
- 46.Modolell, J., Bender, W. & Meselson, M. (1983) Proc. Natl. Acad. Sci. USA 80, 1678-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Campuzano, S., Balcells, L., Villares, R., Carramolino, L., Garcia-Alonso, L. & Modolell, J. (1986) Cell 44, 303-312. [DOI] [PubMed] [Google Scholar]