Abstract
B-1a cells, an anatomically, phenotypically, and functionally distinct subset of B cells that produce the bulk of natural serum IgM and much of gut-associated IgA, are an important component of the early response to pathogens. Because the induced expression of CD5, a hallmark of B-1a cells, requires a nuclear factor of activated T cells (NFAT)-dependent enhancer, we examined the role of NFAT transcription factors in B-1a development. Here we show that the B-1a compartment is normal in mice lacking NFATc2 but essentially absent in mice lacking NFATc1. Loss of NFATc1 affects both peritoneal and splenic B-1a cells. Because there is a loss of B-1 cells defined by markers other than CD5, NFATc1 is not required simply for CD5 expression on B-1a cells. Using mixed-allotype chimeras and retroviral-mediated gene transduction we show that the requirement for NFATc1 is B cell-intrinsic. We also demonstrate that NFATc1 protein expression is elevated ≈5-fold in B-1a cells compared with B-2 cells. This is the first definitive demonstration of a B cell-intrinsic function for an NFAT family transcription factor.
The signaling events that lead to the differentiation of B-1a cells are not well understood. B-1a cells are phenotypically and functionally distinct from the bulk of B cells (e.g., B-2, B-0, and follicular) (reviewed in refs. 1 and 2); they are long-lived cells that express CD5, CD43, and high IgM together with little or no IgD and low CD45(B220). In mice, B-1a cells are known to play an important role in the response to T-cell-independent type-2 antigens (3) and produce the bulk of natural serum IgM (4-7). These antibodies help clear bacterial toxins (8) and are critically important for effective resistance to some pathogens (9). B-1a cells are also the source of many of the gut-associated plasma cells that produce IgA (7). These are important in maintaining homeostasis with intestinal flora (10). B-1a cells have a restricted repertoire, reactive with many common bacterial and self-antigens. Much evidence indicates that this is a consequence of the positive selection by self-antigens of cells into the B-1a subset (reviewed in ref. 1). We and others (11, 12) previously showed that crosslinking of the B cell receptor (BCR) on splenic B cells, to partially mimic positive selection by self-antigen, induces several aspects of the B-1a phenotype, including the expression of CD5. We subsequently demonstrated that this induction of CD5 is cyclosporin-sensitive and requires an enhancer located ≈2 kb upstream of the CD5 coding sequences (13, 14). This enhancer contains two functional nuclear factor of activated T cells (NFAT) sites (14) and is highly conserved in humans (15).
NFAT was originally identified as a Ca2+-inducible transcription factor necessary for the expression of IL-2 in T cells (16). It was subsequently determined that there is a family of four related proteins, encoded by four genes, that have similar properties and function similarly in vivo when ectopically expressed and in vitro in binding assays (17-21). Individual NFAT proteins are expressed in a variety of cell types, within and outside of the immune system, and they can bind to regulatory sequences of many different genes (reviewed in refs. 22-24).
Three Ca2+-sensitive members of the NFAT transcription factor family, NFATc1 (NFAT2, NFATc), NFATc2 (NFAT1, NFATp), and NFATc3 (NFAT4, NFATx), are expressed in B cells (21, 25). B cell NFAT is activated by BCR crosslinking or CD40 ligation and can transactivate NFAT-dependent reporter constructs (26-28). The physiological functions of NFAT in B cells remain unclear. Targeted disruption of NFATc2 was reported to result in a slightly increased B cell proliferative response to BCR or CD40 signaling (29). NFATc1-/- mice exhibit reduced levels of serum IgG1 (30, 31) and IgE (31), but this is apparently the result of impaired IL-4 production by T cells (30, 31). B cells in these mice were hyporesponsive to BCR or CD40 ligation ex vivo, but any in vivo consequences of this are not known (30, 31). These decreased responses may reflect a B cell-intrinsic function of NFATc1 or an effect on B cell development of the absence of NFATc1 in another cell type. In mice containing targeted disruptions of both NFATc2 and NFATc1 there was overproduction of IgG1 and IgE and a polyclonal plasma cell infiltration of end organs (32). The overproduction of IgG1 and IgE was independent of IL-4 and was observed in vitro with isolated B cells.
Because NFAT appears to be necessary for induction of CD5, the hallmark of B-1a cells, we initiated a study of B-1 development in NFAT-deficient mice.
Materials and Methods
Mice. NFATc2-/-, NFATc1-/+ double knockout mice on a BALB/c background were obtained from Laurie Glimcher (Harvard University School of Public Health, Boston). NFATc2-/- and NFATc1-/+ single knockout mice were obtained by breeding these mice with BALB/cByJ mice obtained from The Jackson Laboratory. Some NFATc2-/- mice were from crosses of NFATc2-/-, NFATc3-/+ double knockout mice, also obtained from Laurie Glimcher. Mice were genotyped by PCR by using protocols provided by Ann Ranger (Harvard Medical School, Boston) and Laurie Glimcher. All PCRs contained three primers. NeoPCR1 (AGCGTTGGCTACCCGTGATATTGCTGAAGA) was used in all reactions to prime sequences within the NEOR gene. To distinguish WT from disrupted NFATc2 alleles, two additional primers, NpIn1 (GCA AGCCTCAGTGACA A AGTATCCACT TCA) and NpEx1 (CCACGAGCTGCCCATGGTGGAGAGACAAGA), were included. For the NFATc1 allele, the additional primers were NcIn1 (CTTCCCTGATGTGTGTTGTGGCAGACAAGAT) and NcEx1 (GTTCCAGGTCAGCCAGAAATCAAGGGTCAT). For the NFATc3 allele, the additional primers were NFATc3 knockout (CCCAAGAAGATCAGAAGACTGTG) and NFATc3 WT (GCTCTAAAGATGGCTCCGTGCTTA). All PCRs used Invitrogen PCR buffer and Taq polymerase with 2.1 mM MgCl2 and other components at concentrations specified by Invitrogen. PCRs were at 94°C for 5 min followed by 40 cycles of 1 min each at 94°C, 60°C (NFATc2 and NFATc1) or 57°C (NFATc3), and 72°C followed by 10 min at 72°C. Products were resolved on a 1.8% agarose gel.
RAG-2-/- mice on a BALB/c background were obtained from Taconic Farms and bred in the Tufts University School of Medicine animal facility. Ighb congenic BALB/c mice for making mixed-allotype chimeras were strain C.BKa-Ighb/IcrSmn (obtained from The Jackson Laboratory and bred in our facility).
Fetal Liver Reconstitutions. Single-allotype fetal liver reconstitutions of RAG-2-/- mice were performed as follows. Timed matings were set up of NFATc1-/+ or NFATc2-/-, NFATc1-/+ mice. On day 13.5, fetal livers were harvested and a cell suspension was made by pipetting them up and down 25 times in complete medium (14) supplemented with 10% heat-inactivated FCS (10% complete medium). One-tenth volumes of the suspensions were used for genotyping by PCR as described above. The remaining cells were washed once in complete medium with 1% FCS (1% complete medium) and resuspended in 0.4 ml of 1% complete medium. Each resuspended liver was injected into one RAG-2-/- mouse that had received 300 rad of γ radiation 4-24 h earlier and 10 units of heparin (i.p.) ≈1 h earlier. In some experiments disaggregated fetal livers were frozen in complete medium with 20% heat-inactivated FCS and 10% DMSO after removal of 1/10th volume for genotyping and the 1% complete medium wash. For injection, cells were thawed, diluted in 10% complete medium, pelleted at 250 × g, washed twice in 1% complete medium, resuspended in 0.4 ml of 1% complete medium, and injected as for the nonfrozen cells.
For the first mixed-allotype reconstitution experiment, two mice were injected i.v. with pooled, thawed frozen fetal liver cells such that each mouse received one fetal liver equivalent of NFATc1-/- cells and 0.75 fetal liver equivalents of C.BKa-Ighb/IcrSmn cells. One mouse was injected with pooled, thawed frozen fetal liver cells containing one fetal liver equivalent of WT IgHa cells (obtained from a mating of NFATc1-/+ mice) and 0.75 fetal liver equivalents of WT cells derived from C.BKa-Ighb/IcrSmn cells. Each IgHa pool was derived from four fetal livers, and the IgHb pool was derived from three fetal livers. For the second mixed-allotype reconstitution experiment, five mice were each injected i.v. with one fetal liver equivalent of pooled NFATc1-/- cells and one fetal liver equivalent of pooled C.BKa-Ighb/IcrSmn cells. Five additional mice were each injected i.v. with one fetal liver equivalent of pooled WT, IgHa cells (obtained from a mating of NFATc1-/+ mice) and one fetal liver equivalent of pooled C.BKa-Ighb/IcrSmn cells. In this experiment, each IgHa pool was derived from 5 fetal livers and the IgHb pool was derived from 10 fetal livers.
Flow Cytometry. Cells were stained with the indicated antibodies in Dulbecco's PBS including Ca2+ and Mg2+ (catalog no. 21300-058, Invitrogen) plus 1% rabbit serum, 0.1% NaAzide (staining buffer), and a supernatant of the monoclonal anti-FcγR antibody 2.4G2. All antibodies and fluorochrome-coupled streptavidin were obtained from Pharmingen. Phosphatidylcholine (PtC)-binding cells were detected by staining with liposomes encapsulating carboxyfluorescein as described (33). The liposomes were a gift of Steve Clarke (Duke University, Durham, NC). Standard procedures were used for cytometry (34), which was performed on a FACSCalibur (Becton Dickinson).
Immunoblotting. To isolate splenic B-2 cells, spleens were disaggregated and red blood cells were lysed by treatment with ammonium chloride as described (13). Cells were stained for fluorescence-activated cell sorting (FACS) with biotinylated anti-CD3ε, biotinylated anti-Ter119, phycoerythrin (PE)conjugated anti-CD5, FITC-conjugated anti-CD4, and PE-conjugated anti-CD8 antibodies followed by secondary staining with PE-conjugated streptavidin. Unstained viable cells [propidium iodide (PI)-excluding] in the lymphocyte gate were purified. To isolate peritoneal B-1 cells, cells were obtained by peritoneal lavage with staining buffer and stained with biotinylated anti-CD23, biotinylated anti-CD3ε, biotinylated anti-Ter119, FITC-conjugated anti-CD4, and PE-conjugated anti-CD8 antibodies followed by secondary staining with PE-conjugated streptavidin. Unstained, viable (PI-excluding) cells in the lymphocyte gate were isolated. Cells were pelleted by centrifugation for 10 min at 250 × g and lysed in 50 μl of SDS/PAGE sample loading buffer per 2 × 106 cells. Samples were boiled for 5 min and frozen. Subsequently, 50 μl of each lysate was run on a SDS/7.5% PAGE gel, and immunoblotting was performed by standard methods. The blots were probed by using mouse monoclonal 7A6 anti-NFATc1 antibody (Santa Cruz Biotechnology), rabbit polyclonal anti-NFATc2 antibody 67.1 (gift from Anjana Rao, Center for Blood Research, Harvard University, Boston), and mouse monoclonal anti-HSP-60 (H-1) antibody (Santa Cruz Biotechnology).
Retroviral Constructions and Infections. The murine NFATc1 isoform a (GenBank accession no. GI7208617) complete coding sequence was amplified by PCR and a 5′ EcoR1 site and 3′ NotI site introduced by using the forward primer GTTACAGTGGAATTCAAACAGACACCATGCCAAATACCAGCTTTCCA and the reverse primer GTTACAGTCGCGGCCGCTCAGTAAAAACCTCCTCTCA. The forward primer also introduced the Kozak sequence from murine CD22 (35). The PCR template was cDNA, prepared by using the Invitrogen SuperScript 1 First-Strand Synthesis kit according to the manufacturer's instructions. The template for cDNA synthesis was total RNA prepared by using a Stratagene Absolutely RNA RT-PCR MiniPrep kit. The source of the RNA was purified splenic B cells that had been stimulated for 53 h with 10 μg/ml F(ab′)2 anti-IgM (Jackson ImmunoResearch). The PCR was performed by using KOD Hot Start DNA polymerase (Novagen) according to the manufacturer's instructions. The reaction times were 2 min at 94°C followed by 40 cycles of 30 seconds at 94°C, 30 seconds at 65°C, and 2.5 min at 68°C. The product was cleaned by using a QIAquick PCR kit (Qiagen, Valencia, CA), digested with EcoR1 and NotI, and cloned into EcoR1- and NotI-digested, gel-purified pMIG retroviral vector (36). The resulting construct was checked by automated sequencing at the Tufts University Sequencing Facility. Virus was prepared by transiently cotransfecting 293T cells with retroviral DNA and the ψEco packaging plasmid. Supernatants were collected every 8-16 h starting 24 h posttransfection and continuing to 56 h posttransfection. Supernatants were stored at -80°C, thawed, and pooled before infection of fetal liver cells.
Fetal liver cells, prepared and frozen as described above, were thawed and cultured on monolayers of IL-7-producing T220 fibroblasts (37, 38) that had received 2,800 rad of γ radiation. After 2 weeks in culture, cells were harvested, pelleted for 10 min at 250 × g, and resuspended at 1 × 106 cells per ml in viral supernatants plus 50 mM 2-mercaptoethanol and 8 μg/ml polybrene. After 3 h in a 37°C CO2 incubator, cells were pelleted as before, resuspended at ≈1 × 106 cells per ml in medium conditioned by T220 cells, and cultured overnight on a T220 monolayer. GFP+ cells were isolated by sorting on a MoFlo cell sorter (DakoCytomation, Fort Collins, CO) and introduced by tail vein injection into sublethally irradiated, heparin-treated RAG-2-/- mice.
Results
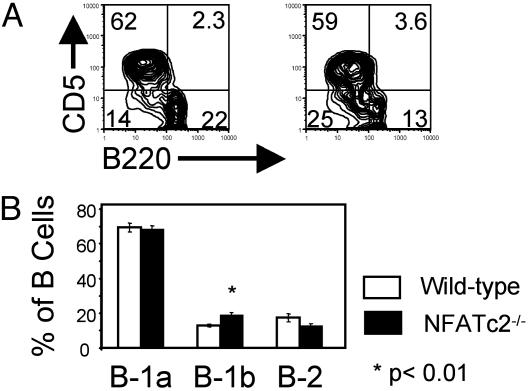
NFATc2 Is Dispensable for B-1a Development. To see whether loss of NFATc2 expression affects B-1 cell development, we examined B cell subsets in peritoneal washouts of NFATc2-/- and WT mice. The fraction of B-1a cells (B220+, CD5+) was comparable in NFATc2-/- and normal mice, whereas the fraction of B-1b cells (B220lo, CD5-) was increased ≈1.5-fold (P < 0.004) in NFATc2-/- compared with WT mice (Fig. 1). This increase may be accounted for by a decrease in the fraction of B-2 cells in NFATc2-/- mice (P < 0.09) (Fig. 1). We did not examine these mice further.
Fig. 1.
NFATc2 is dispensable for B-1a development and survival. (A) Representative FACS analysis of peritoneal cells from a BALB/cByJ mouse (Left) and an NFATc2-/- mouse (Right). Peritoneal washout cells were stained with FITC-conjugated anti-CD45 (B220), PE-conjugated anti-CD5 antibodies, and PI and then analyzed by flow cytometry. Populations shown are gated on B220+, viable (PI-excluding) lymphocytes. Numbers in quadrants are the percentages of B cells. (B) Summary of analysis of seven WT and seven NFATc2-/- mice. Error bars are the SEM. The P value is comparing the NFATc2-/- and WT cells from the same subset (B-1b) and was calculated by using a two-tailed t test. B-1a cells are B220+, CD5+; B-1b cells are B220lo, CD5-; and B-2 cells are B220hi, CD5-.
NFATc1 Is Essential for Normal B-1a Development and/or Survival. We next looked at the effect of targeted mutation of NFATc1 or of both NFATc2 and NFATc1 on B-1 development. Mice lacking NFATc1 die in utero at embryonic day 14.5 (30, 31). Therefore, to study NFATc1-deficient lymphocytes, sublethally irradiated RAG-2-/- mice were reconstituted with embryonic day 13.5 fetal liver cells from WT, NFATc1-/-, or NFATc2-/-, NFATc1-/- embryos.
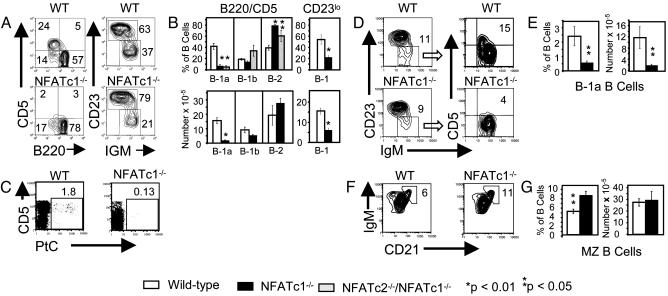
Analysis of B cell subsets in peritoneal washouts, which are greatly enriched for B-1 cells, revealed that the fraction of B-1a cells [defined as CD5+, CD45(B220)+] was reduced ≈6-fold (P < 0.00014) and the number of B-1a cells was reduced ≈9-fold (P < 0.0002) in NFATc1-/- compared with WT chimeras (Fig. 2 A and B). This decrease could be due simply to an inability to express CD5, consistent with our identification of an NFAT-dependent enhancer upstream of the murine CD5 gene (14). If this is the case, there should be no decrease in the fraction or number of B-1 cells (B-1a plus B-1b) identified by markers other than CD5. This is what is observed in CD5 knockout mice (39), but it is not what we observed in NFATc1-/- chimeras. Instead, we see a 2- to 3-fold decrease in the fraction and number of B-1 cells defined as B220lo, as IgMhi and CD23lo, or as B220lo and CD23lo (Fig. 2 A and B). The number of B-1 cells in the mutant chimeras is similar to the number of B-1b cells in WT chimeras, consistent with the selective loss of B-1a cells (Fig. 2B). To further document the loss of B-1a cells in NFATc1-/- chimeras, we determined the fraction of peritoneal B cells that were PtC-specific. PtC-specific B cells are restricted almost entirely to the B-1a subset in WT mice (40), and a normal fraction of these cells is found in the peritonea of CD5 knockout mice despite their inability to express CD5 (39). Again, in contrast to CD5 knockout mice, NFATc1-deficient chimeras had a severe reduction in the fraction of PtC-specific B cells (Fig. 2C). These results show that peritoneal B-1a cells fail to develop, expand, or survive in NFATc1-/- chimeras.
Fig. 2.
NFATc1 is essential for B-1a development or survival. RAG-2-/- mice were reconstituted with WT or NFATc1-/- fetal liver cells and analyzed after 8 or more weeks. (A-C) Analysis of peritoneal cells. (A) Representative FACS analysis of peritoneal B cells from a WT and an NFATc1-/- chimera. Staining was with anti-B220 and anti-IgM as described in Fig. 1 or with FITC-conjugated anti-IgM, biotinylated anti-CD23 detected with PE-conjugated streptavidin, and PI. Populations shown are gated on IgM+, viable lymphocytes. The B220/CD5 results are representative of nine WT and six NFATc1-/- chimeric mice, and the IgM/CD23 results are representative of three WT and four NFATc1-/- chimeras. (B) Percent and absolute number of B cells in each subset. B-1a, B-1b, and B-2 are defined in Fig. 1. B-1 cells are CD23lo and, depending on the experiment, either IgMhi or B220lo. For the B220/CD5 staining, percentages are given for nine WT, six NFATc1-/-, and three NFATc2-/-NFATc1-/- double knockout chimeras. Absolute numbers are given for five WT and five NFATc1-/- chimeras. The CD23lo results summarize percentages of six WT and six NFATc1-/- chimeras and the absolute numbers of five WT and five NFATc1-/- chimeras. (C) Binding of B cells to PtC liposomes. Cells were stained with PtC liposomes encapsulating carboxyfluorescein, PE-conjugated anti-CD5, biotinylated anti-CD23 detected with allophycocyanin (APC)-conjugated streptavidin, and PI. Viable B cells are shown. B cells were defined as either CD23-, CD5lo or CD23+, CD5-. Results shown are representative of three WT and two NFATc1-/- chimeras. (D-G) Analysis of splenic cells. (D) FACS analysis of splenic B cells. Cells were stained with FITC-conjugated anti-IgM, PE-conjugated anti-CD5, biotinylated anti-CD23 (detected with APC-conjugated streptavidin), and PI and analyzed by flow cytometry. Populations shown are viable B cells. To more readily identify B-1a cells, CD23lo B cells were gated as shown and analyzed for CD5 expression. In the IgM/CD23 plots, numbers are the percentages of B cells in the CD23lo gate. In the IgM/CD5 plots, numbers are the percent of CD23lo cells that are CD5+. Results are representative of four WT and five NFATc1-/- chimeras. (E) Percent and absolute number of B cells that are CD5+. Four WT and five NFATc1-/- chimeras were analyzed. (F) Cells were stained with FITC-conjugated anti-CD21, biotinylated anti-IgM detected with APC-conjugated streptavidin, and PI. Viable IgM+ cells are shown. MZ B cells are identified as CD21hi, IgMhi.(G) Percent and absolute number of MZ B cells. Three WT and two NFATc1-/- chimeras were analyzed as in F.In B, E, and G, P values compare mutant to WT cells of the same subset and were calculated by a two-tailed t test. Error bars are ± SEM.
We also examined splenic B-1a cells in WT and NFATc1-/- mice. This population makes up only a small fraction of B cells in the spleen. To see splenic B-1a cells, splenocytes were stained with anti-IgM, anti-CD23, and anti-CD5. IgM and CD5 levels were then examined on CD23lo gated cells. As shown in Fig. 2D, this reveals a distinct CD5+ population which, in the WT chimera shown, constituted 14.6% of the CD23lo B cells and 1.7% of total B cells. This population was significantly reduced in NFATc1-/- chimeras, both in total number (6-fold, P < 0.025) and as a fraction of B cells (4-fold, P < 0.02)(Fig. 2 D and E).
Splenic marginal zone (MZ) B cells share some phenotypic and functional properties with B-1a cells (reviewed in ref. 41); therefore, we examined the fraction and absolute numbers of these cells (defined as IgMhi, CD21hi) in WT and NFATc1-/- chimeric mice. Unlike B-1a cells, the fraction and number of MZ B cells was not diminished in the NFATc1-deficient chimeras (Fig. 2 F and G). In fact, the frequency (but not absolute number) of MZ B cells was somewhat increased in NFATc1-deficient chimeras.
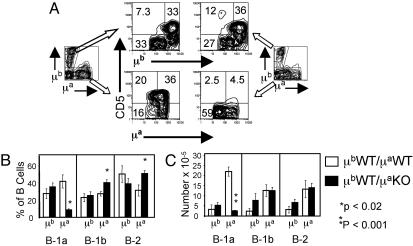
The Effect of NFATc1 Disruption on B-1a Development Is B Cell-Intrinsic. To determine whether the effect of disruption of NFATc1 on B-1a development was B cell-intrinsic, we made mixed-allotype chimeras in which WT fetal liver cells of the IgHb allotype were cotransferred with NFATc1-/- fetal liver cells of the IgHa allotype. This allowed us to determine whether the loss of B-1a cells was restricted to cells actually carrying the targeted alleles. As controls, we made mixed-allotype chimeras in which fetal liver cells of both allotypes were WT. Fig. 3 shows the analysis of B cell subsets in peritonea of mixed-allotype chimeric mice. The fraction of IgHa B-1a cells in chimeras made with mutant IgHa fetal liver cells was ≈5-fold lower than the fraction in chimeras made with WT IgHa cells (P < 0.0007, Fig. 3 A and B). The fraction of IgHb B-1a cells was unaffected by the genotype of the cotransferred IgHa cells (Fig. 3 A and B). The absolute number of IgHa B-1a cells in mutant chimeras was also reduced (average, 7.6-fold; P < 0.006) compared with the number in WT chimeras (Fig. 3C) whereas there was no statistically significant difference in the absolute number of IgHb B-1a cells in mutant versus WT chimeras. Thus, the effect of NFATc1 mutation on peritoneal B-1a development and/or survival is B cell-intrinsic. Similar results were obtained in the spleens of the mixed-allotype chimeric mice (data not shown).
Fig. 3.
The requirement for NFATc1 in peritoneal B-1a development/survival is B cell-intrinsic. (A) RAG-2-/- mice were reconstituted with WT Ighb fetal liver cells mixed with Igha fetal liver cells that were either WT (Left) or NFATc1-/- (Right). Peritoneal cells were stained with FITC-conjugated anti-μa, PE-conjugated anti-μb, biotinylated anti-CD5 detected with APC-conjugated streptavidin, and PI and analyzed by flow cytometry. Viable lymphocytes were gated on μa+ or μb+ cells as shown and CD5 expression on the gated populations analyzed. Results are representative of four WT and six NFATc1-/- chimeras. (B) The fraction of peritoneal B cells in different subsets as a percentage of total peritoneal B cells. B-1a cells are defined as μ+, CD5+; B-1b cells as μhi, CD5-; and B-2 cells as μlo, CD5-. Results shown are for four WT and six NFATc1-/- chimeras. P values compare mutant to WT cells of the same subset and allotype and were calculated by a two-tailed t test. Error bars are ± SEM. (C) The absolute number of peritoneal B cells in different subsets defined as in B. Results are for three WT and four NFATc1-/- chimeras. Statistics are as in B.
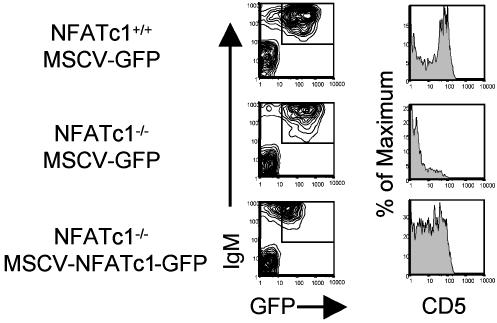
Rescue of B-1a Cells by Ectopic Expression of NFATc1 in NFATc1-/- Fetal Liver Progenitors. We next attempted to determine whether the B-1 compartment in NFATc1-/- chimeras could be rescued by ectopic expression of NFATc1. To do this, we made use of the fact that B cell progenitors maintained in vitro by culture with IL-7 give rise to mature B cells upon injection into lymphocyte-deficient mice (42, 43). We isolated fetal liver cells from day-13.5 embryos of NFATc1-/- or NFATc1+/+ littermates and cultured them for 2 weeks with IL-7-producing T220 fibroblasts (37, 38). The cells were infected with a retroviral vector encoding either an internal ribosome entry site-containing bicistronic NFATc1/GFP RNA (NFATGFP) or the same vector encoding only GFP (empty vector). The following day, GFP+ cells were sorted and injected into sublethally irradiated RAG-2-/- mice. Peritoneal cells were harvested and analyzed 1-2 months after reconstitution. As shown in Fig. 4, WT cells infected with empty vector gave rise to peritoneal CD5+ B cells in the recipient mice. NFATc1-/- cells, in contrast, were ineffective in generating CD5+ cells when infected with empty vector. Infection of NFATc1-/- cells with NFATGFP significantly, but not completely, corrected the defect in generation of CD5+ B cells.
Fig. 4.
Ectopic expression of NFATc1 enables NFATc1-/- pre-B cells to generate B-1a cells. WT or NFATc1-/- fetal liver cells were cultured on Il-7-producing T220 fibroblasts and infected with the indicated retroviruses, and on the following day GFP+ cells were injected i.v. into sublethally irradiated Rag-2-/- mice. Thirty-one days later, peritoneal cells were harvested and stained with PE-conjugated anti-IgM antibody, biotinylated anti-CD5 antibody visualized with APC-conjugated streptavidin, and PI. GFP+, IgM+ viable lymphocytes, gated as shown, were analyzed for CD5 expression. This result is representative of three independent experiments.
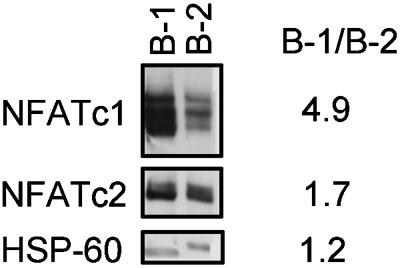
B-1a Cells Exhibit Elevated Expression of NFATc1. NFATc1 could be necessary for the generation of B-1a cells, their maintenance, or both. To determine whether NFATc1 might play an ongoing role in mature peritoneal B-1a cells, we examined NFATc1 expression in purified peritoneal B-1a cells and compared it to that in purified splenic B cells. The results are shown in Fig. 5. Whole-cell lysates of B-1a cells contained 5-fold more NFATc1 than did lysates from an equivalent number of splenic B cells. NFATc2 levels were ≈2-fold higher in B-1a cells compared with B-2 cells. As a loading control we looked at the level of HSP-60, which, as expected, was similar in B-1 and B-2 cells.
Fig. 5.
B-1a cells express high levels of NFATc1 compared with B-2 cells. Whole-cell extracts of FACS-purified peritoneal B-1 cells and splenic B-2 cells were analyzed by Western blot. A total of 2 × 106 cell equivalents were loaded per lane. The blot was probed sequentially with anti-NFATc1, anti-NFATc2, and anti-HSP-60 antibodies, and staining was visualized in each case by enhanced chemiluminescence. Peritoneal B-1a cells were 96% pure, and splenic B-2 cells were 89% pure.
Discussion
In the present study we examined the role of NFATc1 in the development and/or survival of B-1a cells. Mice reconstituted with NFATc1-/- fetal liver essentially lacked peritoneal and splenic B-1a cells. Unlike some mutations that lead to the loss of B-1a cells, such as xid, the loss of NFATc1 does not result in the accumulation of B cells with an immature phenotype (ref. 31 and our unpublished results). By making mixed-allotype chimeras in which fetal liver cells of the IgHa allotype were NFATc1-/- and those of the IgHb allotype were WT, we showed that the inability to generate B-1a cells was restricted to cells lacking NFATc1. Thus, the requirement for NFATc1 in B-1a cell development is B cell-intrinsic. This result is bolstered by experiments in which in vitro cultured B cell progenitors from WT but not NFATc1-/- embryos gave rise to CD5+ peritoneal B cells after transfer to RAG-2-/- recipients. Retroviral transduction of a WT NFATc1 cDNA partially corrected the defect in NFATc1-/- pre-B cells.
We believe that a B cell-intrinsic function for an NFAT factor has not previously been demonstrated. Peng et al. (32) showed that there were elevated levels of serum IgG1 and IgE that were IL-4-independent in NFATc2-/-, NFATc1-/- double knockout mice. Isolated splenic B cells from these mice spontaneously produced high levels of IgG1 and IgE in vitro, leading the authors to suggest that there was an intrinsic B cell defect in these mice. However, the data did not rule out the alternative of B cell-extrinsic effects acting before the isolation of the splenic B cells. The same caveat applies to the alterations in proliferative responses seen in B cells isolated from NFATc2 (29) and NFATc1 (30, 31) single knockout mice.
The missing B-1a cells in NFATc1-/- mice do not appear in the CD5- B-1b population. Furthermore, B cells specific for PtC, which are almost entirely B-1a in WT chimeras, are lost in the NFATc1-/- chimeras (see Fig. 2C). Thus, NFATc1 is not required simply for CD5 expression. The loss of B-1a cells is not secondary to the loss of CD5, because there is a normal fraction of peritoneal B-1 cells as well as PtC binding cells in CD5 knockout mice (39).
Splenic MZ B cells share some phenotypic characteristics with B-1 cells and, like them, provide a first line of thymus-independent antibody response against pathogens (3, 44). Studies with transgenic mice show that expression of certain antigen receptor specificities drives B cells into the B-1 subset whereas expression of others drives cells into the MZ subset (45). The fact that NFATc1 is not required for MZ B cell development or survival (Fig. 2 F and G) underscores the selective requirement for NFATc1 in the B-1a as compared with other B cell subsets.
Targeted disruptions of any of a number of genes whose products function within the BCR signaling pathway, or act to modulate it, result in a loss or decrease of B-1a cells (reviewed in ref. 1). Many of these proteins, including phospholipase Cγ, PKC, phosphatidylinositol 3-kinase, CD19, CD21, Bruton's tyrosine kinase (BTK), B cell linker protein and vav-1, act upstream of Ca2+ influx and therefore presumably function upstream of NFAT. Our study indicates that this is sufficient to explain their effect on B-1a cells. Of course, these proteins may provide other functions also necessary for B-1a development.
In addition to NFATc1, the transcription factors Oct-2 and aiolos have been shown to be important for B-1a development/survival, because mice carrying targeted disruptions of these genes have a severe reduction in B-1a cells (46, 47). In both cases the effects on B cell development are broad. In Oct-2-deficient chimeric mice, in addition to the loss of B-1a cells, there is a large reduction in the fraction of mature (IgMlo, IgDhi) peripheral B cells, impaired BCR-mediated activation in vitro, and impaired T cell-independent and -dependent responses in vivo (46, 48). Aiolos-deficient mice have a severe reduction in recirculating B cells and evidence of spontaneous B cell activation in vivo as well as hyperresponsiveness to BCR-mediated activation in vitro (47). In contrast, the phenotype of NFATc1-deficient B cells, apart from the loss of B-1a cells reported here, is mild. B cell development appears normal, although delayed. In vitro, BCR-mediated activation is impaired (30, 31). Thus, NFATc1 appears to be particularly important for B-1a development/survival.
Finally, our results raise the question of why other NFAT family members cannot substitute for the loss NFATc1 in B-1a cell development/maintenance. In addition to NFATc1, NFATc2 and NFATc3 are known to be present in peripheral B cells (21, 25), and here we show that at least NFATc2 is present in B-1a cells and at a level somewhat higher than that in B-2 cells. All NFATs (except NFAT5) are activated by the elevation of cytoplasmic Ca2+. Furthermore, they bind to similar sites and can transactivate a reporter construct driven by the IL-2 promoter or a synthetic 3x-NFAT element (17-21). Differences in binding, in particular in the requirement for cooperative interaction with AP-1, have been observed for some sites (19). Thus, NFATc1 may have a higher affinity for the regulatory element(s) of the target(s) critical for B-1a development. Alternatively, it may be the level of total NFAT protein that is important, and NFATc1 may represent the major NFAT protein in B-1a cells or their precursors. We are currently attempting to address this issue using the retroviral transduction system described in Fig. 4. This will allow us to see the effect on B-1a development of overexpression of different NFAT proteins.
The progression of B cells through multiple developmental and phenotypic stages is driven by alterations in programs of gene expression. These changes are, in turn, a consequence of changes in the activities and expression of transcription factors (reviewed in ref. 49). The pattern of transcription factor expression and activity underlying the induction and maintenance of the B-1a phenotype is not well characterized. Determining the targets of NFATc1 in B-1a cells and understanding how NFATc1 expression and activity are regulated during B-1a development should provide insight into the development and function of B-1a cells and the mechanisms of NFAT action.
Acknowledgments
We thank Paul McLean and E. Macario Herrera for useful discussions and technical assistance, Allen Parmalee for cell sorting, Jason Cyster for advice on making mixed-allotype chimeras, Laurie Glimcher and Ann Ranger for NFAT knockout mice and genotyping protocols, and Anjana Rao for anti-NFATc2 antibody. This work was funded by National Institutes of Health Grant AI15803 (to H.H.W.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: APC, allophycocyanin; BCR, B cell receptor; FACS, fluorescence-activated cell sorting; MZ, marginal zone; NFAT, nuclear factor of activated T cells; PE, phycoerythrin; PI, propidium iodide; PtC, phosphatidylcholine.
References
- 1.Berland, R. & Wortis, H. H. (2002) Annu. Rev. Immunol. 20, 253-300. [DOI] [PubMed] [Google Scholar]
- 2.Hardy, R. R. & Hayakawa, K. (2001) Annu. Rev. Immunol. 19, 595-621. [DOI] [PubMed] [Google Scholar]
- 3.Martin, F., Oliver, A. M. & Kearney, J. F. (2001) Immunity 14, 617-629. [DOI] [PubMed] [Google Scholar]
- 4.Herzenberg, L. A., Stall, A. M., Lalor, P. A., Sidman, C., Moore, W. A. & Parks, D. R. (1986) Immunol. Rev. 93, 81-102. [DOI] [PubMed] [Google Scholar]
- 5.Lalor, P. A., Herzenberg, L. A., Adams, S. & Stall, A. M. (1989) Eur. J. Immunol. 19, 507-513. [DOI] [PubMed] [Google Scholar]
- 6.Forster, I. & Rajewsky, K. (1987) Eur. J. Immunol. 17, 521-528. [DOI] [PubMed] [Google Scholar]
- 7.Kroese, F. G., Butcher, E. C., Stall, A. M., Lalor, P. A., Adams, S. & Herzenberg, L. A. (1989) Int. Immunol. 1, 75-84. [DOI] [PubMed] [Google Scholar]
- 8.Reid, R. R., Prodeus, A. P., Khan, W., Hsu, T., Rosen, F. S. & Carroll, M. C. (1997) J. Immunol. 159, 970-975. [PubMed] [Google Scholar]
- 9.Baumgarth, N., Herman, O. C., Jager, G. C., Brown, L. E., Herzenberg, L. A. & Chen, J. (2000) J. Exp. Med. 192, 271-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Macpherson, A. J., Gatto, D., Sainsbury, E., Harriman, G. R., Hengartner, H. & Zinkernagel, R. M. (2000) Science 288, 2222-2226. [DOI] [PubMed] [Google Scholar]
- 11.Cong, Y. Z., Rabin, E. & Wortis, H. H. (1991) Int. Immunol. 3, 467-476. [DOI] [PubMed] [Google Scholar]
- 12.Rothstein, T. L., Kolber, D. L., Murphy, T. P. & Cohen, D. P. (1991) J. Immunol. 147, 3728-3735. [PubMed] [Google Scholar]
- 13.Teutsch, M., Higer, M., Wang, D. & Wortis, H. W. (1995) Int. Immunol. 7, 381-392. [DOI] [PubMed] [Google Scholar]
- 14.Berland, R. & Wortis, H. H. (1998) J. Immunol. 161, 277-285. [PubMed] [Google Scholar]
- 15.Berland, R. & Wortis, H. H. (2000) Curr. Top. Microbiol. Immunol. 252, 131-140. [DOI] [PubMed] [Google Scholar]
- 16.Shaw, J. P., Utz, P. J., Durand, D. B., Toole, J. J., Emmel, E. A. & Crabtree, G. R. (1988) Science 241, 202-205. [DOI] [PubMed] [Google Scholar]
- 17.McCaffrey, P. G., Luo, C., Kerppola, T. K., Jain, J., Badalian, T. M., Ho, A. M., Burgeon, E., Lane, W. S., Lambert, J. N., Curran, T. et al. (1993) Science 262, 750-754. [DOI] [PubMed] [Google Scholar]
- 18.Northrop, J. P., Ho, S. N., Chen, L., Thomas, D. J., Timmerman, L. A., Nolan, G. P., Admon, A. & Crabtree, G. R. (1994) Nature 369, 497-502. [DOI] [PubMed] [Google Scholar]
- 19.Hoey, T., Sun, Y. L., Williamson, K. & Xu, X. (1995) Immunity 2, 461-472. [DOI] [PubMed] [Google Scholar]
- 20.Masuda, E. S., Naito, Y., Tokumitsu, H., Campbell, D., Saito, F., Hannum, C., Arai, K. & Arai, N. (1995) Mol. Cell. Biol. 15, 2697-2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho, S. N., Thomas, D. J., Timmerman, L. A., Li, X., Francke, U. & Crabtree, G. R. (1995) J. Biol. Chem. 270, 19898-19907. [DOI] [PubMed] [Google Scholar]
- 22.Rao, A., Luo, C. & Hogan, P. G. (1997) Annu. Rev. Immunol. 15, 707-747. [DOI] [PubMed] [Google Scholar]
- 23.Kiani, A., Rao, A. & Aramburu, J. (2000) Immunity 12, 359-372. [DOI] [PubMed] [Google Scholar]
- 24.Crabtree, G. R. & Olson, E. N. (2002) Cell 109, Suppl., S67-S79. [DOI] [PubMed] [Google Scholar]
- 25.Timmerman, L. A., Healy, J. I., Ho, S. N., Chen, L., Goodnow, C. C. & Crabtree, G. R. (1997) J. Immunol. 159, 2735-2740. [PubMed] [Google Scholar]
- 26.Verweij, C. L., Guidos, C. & Crabtree, G. R. (1990) J. Biol. Chem. 265, 15788-15795. [PubMed] [Google Scholar]
- 27.Choi, M. S., Brines, R. D., Holman, M. J. & Klaus, G. G. (1994) Immunity 1, 179-187. [DOI] [PubMed] [Google Scholar]
- 28.Venkataraman, L., Francis, D. A., Wang, Z., Liu, J., Rothstein, T. L. & Sen, R. (1994) Immunity 1, 189-196. [DOI] [PubMed] [Google Scholar]
- 29.Hodge, M. R., Ranger, A. M., Charles de la Brousse, F., Hoey, T., Grusby, M. J. & Glimcher, L. H. (1996) Immunity 4, 397-405. [DOI] [PubMed] [Google Scholar]
- 30.Yoshida, H., Nishina, H., Takimoto, H., Marengere, L. E., Wakeham, A. C., Bouchard, D., Kong, Y. Y., Ohteki, T., Shahinian, A., Bachmann, M., et al. (1998) Immunity 8, 115-124. [DOI] [PubMed] [Google Scholar]
- 31.Ranger, A. M., Hodge, M. R., Gravallese, E. M., Oukka, M., Davidson, L., Alt, F. W., de la Brousse, F. C., Hoey, T., Grusby, M. & Glimcher, L. H. (1998) Immunity 8, 125-134. [DOI] [PubMed] [Google Scholar]
- 32.Peng, S. L., Gerth, A. J., Ranger, A. M. & Glimcher, L. H. (2001) Immunity 14, 13-20. [DOI] [PubMed] [Google Scholar]
- 33.Arnold, L. W., Pennell, C. A., McCray, S. K. & Clarke, S. H. (1994) J. Exp. Med. 179, 1585-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coligan, J., Kruisbeek, A. M., Margulies, D. M., Shevach, E. M. & Strober, W., eds. (1999) Current Protocols in Immunology (Wiley, New York).
- 35.Nadler, M. J., McLean, P. A., Neel, B. G. & Wortis, H. H. (1997) J. Immunol. 159, 4233-4243. [PubMed] [Google Scholar]
- 36.Van Parijs, L., Refaeli, Y., Lord, J. D., Nelson, B. H., Abbas, A. K. & Baltimore, D. (1999) Immunity 11, 281-288. [DOI] [PubMed] [Google Scholar]
- 37.Borzillo, G. V., Endo, K. & Tsujimoto, Y. (1992) Oncogene 7, 869-876. [PubMed] [Google Scholar]
- 38.Lin, Q., Taniuchi, I., Kitamura, D., Wang, J., Kearney, J. F., Watanabe, T. & Cooper, M. D. (1998) J. Immunol. 160, 4681-4687. [PubMed] [Google Scholar]
- 39.Tarakhovsky, A., Muller, W. & Rajewsky, K. (1994) Eur. J. Immunol. 24, 1678-1684. [DOI] [PubMed] [Google Scholar]
- 40.Pennell, C. A., Mercolino, T. J., Grdina, T. A., Arnold, L. W., Haughton, G. & Clarke, S. H. (1989) Eur. J. Immunol. 19, 1289-1295. [DOI] [PubMed] [Google Scholar]
- 41.Martin, F. & Kearney, J. F. (2000) Immunol. Rev. 175, 70-79. [PubMed] [Google Scholar]
- 42.Rolink, A., Kudo, A., Karasuyama, H., Kikuchi, Y. & Melchers, F. (1991) EMBO J. 10, 327-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reininger, L., Winkler, T. H., Kalberer, C. P., Jourdan, M., Melchers, F. & Rolink, A. G. (1996) J. Exp. Med. 184, 853-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chumley, M. J., Dal Porto, J. M. & Cambier, J. C. (2002) J. Immunol. 169, 1735-1743. [DOI] [PubMed] [Google Scholar]
- 45.Martin, F. & Kearney, J. F. (2000) Immunity 12, 39-49. [DOI] [PubMed] [Google Scholar]
- 46.Humbert, P. O. & Corcoran, L. M. (1997) J. Immunol. 159, 5273-5284. [PubMed] [Google Scholar]
- 47.Wang, J. H., Avitahl, N., Cariappa, A., Friedrich, C., Ikeda, T., Renold, A., Andrikopoulos, K., Liang, L., Pillai, S., Morgan, B. A. & Georgopoulos, K. (1998) Immunity 9, 543-553. [DOI] [PubMed] [Google Scholar]
- 48.Corcoran, L. M. & Karvelas, M. (1994) Immunity 1, 635-645. [DOI] [PubMed] [Google Scholar]
- 49.Schebesta, M., Heavey, B. & Busslinger, M. (2002) Curr. Opin. Immunol. 14, 216-223. [DOI] [PubMed] [Google Scholar]