Abstract
Pyrin, the familial Mediterranean fever protein, is found in association with the cytoskeleton in myeloid/monocytic cells and modulates IL-1β processing, NF-κB activation, and apoptosis. These effects are mediated in part through cognate interactions with the adaptor protein ASC, which shares an N-terminal motif with pyrin. We sought additional upstream regulators of inflammation by using pyrin as the bait in yeast two-hybrid assays. We now show that proline serine threonine phosphatase-interacting protein [PSTPIP1, or CD2-binding protein 1 (CD2BP1)], a tyrosine-phosphorylated protein involved in cytoskeletal organization, also interacts with pyrin. Recently, PSTPIP1/CD2BP1 mutations were shown to cause the syndrome of pyogenic arthritis, pyoderma gangrenosum, and acne (PAPA), a dominantly inherited autoinflammatory disorder mediated predominantly by granulocytes. Endogenous PSTPIP1/CD2BP1 and pyrin are coexpressed in monocytes and granulocytes and can be coimmunoprecipitated from THP-1 cells. The B box segment of pyrin was necessary and the B box/coiled-coil segment sufficient for this interaction, whereas the SH3 and coiled-coil domains of PSTPIP1/CD2BP1 were both necessary, but neither was sufficient, for pyrin binding. The Y344F PSTPIP1/CD2BP1 mutation, which blocks tyrosine phosphorylation, was associated with a marked reduction in pyrin binding in pervanadate-treated cells. PAPA-associated A230T and E250Q PSTPIP1/CD2BP1 mutations markedly increased pyrin binding as assayed by immunoprecipitation and, relative to WT, these mutants were hyperphosphorylated when coexpressed with c-Abl kinase. Consistent with the hypothesis that these mutations exert a dominant-negative effect on the previously reported activity of pyrin, we found increased IL-1β production by peripheral blood leukocytes from a clinically active PAPA patient with the A230T PSTPIP1/CD2BP1 mutation and in cell lines transfected with both PAPA-associated mutants.
The systemic autoinflammatory diseases are a group of heritable disorders characterized by seemingly unprovoked inflammation and notable for their relative lack of autoantibodies or antigen-specific T cells (1). One of the prototypic illnesses in this category is familial Mediterranean fever (FMF; MIM249100), a recessive disorder manifested by episodes of fever, serositis, arthritis, and skin rash, with marked accumulations of polymorphonuclear leukocytes in affected areas during attacks. Several of the autoinflammatory disease genes have been identified by positional cloning, with the broader goal of discovering novel pathways controlling inflammation and innate immunity (reviewed in ref. 2).
The FMF gene, MEFV, encodes a 781-aa protein denoted pyrin (or marenostrin) (3, 4), which is expressed primarily in neutrophils, eosinophils, and cytokine-activated monocytes (5, 6). Epitope-tagged full-length pyrin colocalizes with microtubules and the actin cytoskeleton (7). The N-terminal ≈90 amino acids of pyrin define a motif, variously called the PYRIN domain (8), PYD (9, 10), PAAD (11), or DAPIN (12), which is similar in structure to death domains, death effector domains, and caspase recruitment domains (CARDs). Like the latter three motifs, PYRIN domains participate in homotypic protein-protein interactions and are found in a number of proteins involved in the regulation of inflammation and apoptosis (8-12).
Through this N-terminal domain, pyrin interacts with apoptosis-associated speck-like protein with a CARD (ASC) (13), a bipartite adaptor molecule comprised of an N-terminal PYRIN domain and a C-terminal CARD (14). Either alone or in macromolecular complexes, ASC induces oligomerization and proinflammatory autocatalysis of caspase-1 (IL-1-converting enzyme) through cognate CARD-CARD interactions (10, 15, 16). In a mouse macrophage cell line, pyrin inhibits IL-1β activation, and full-length pyrin inhibits the interaction of ASC and caspase-1 in vitro (6). Pyrin also disrupts ASC-dependent apoptosis and NF-κB activation in transfected cells (17, 18). Moreover, mice expressing a truncated form of pyrin exhibit heightened inflammatory responses characterized by caspase-1 and IL-1β activation and IL-1-independent impairment of macrophage apoptosis (6). Taken together, these data indicate that pyrin is an important modulator of innate immunity.
To identify other proteins that might impinge on these pathways, we have performed yeast two-hybrid assays with pyrin as the bait. In the present paper, we describe the interaction of pyrin with a protein designated as proline serine threonine phosphatase-interacting protein 1 (PSTPIP1) when first described in the mouse (19) or CD2-binding protein 1 (CD2BP1) when the human ortholog was cloned (20). For the sake of simplicity, this PSTPIP1/CD2BP1 gene and the encoded protein will be denoted as PSTPIP1 in this report. PSTPIP1 is expressed predominantly in hematopoietic tissues and the lung (19, 20) and is the mammalian homolog of the yeast Schizosaccharomyces pombe CDC15p, a tyrosine-phosphorylated protein involved in the organization of the cytoskeleton (21). PSTPIP1 is found in association with cortical actin and with lamellipodia during most of the cell cycle and migrates to the cleavage furrow during cytokinesis (19). Ectopic expression of PSTPIP1 induces formation of filopodial membrane extensions, suggesting a role in actin reorganization (19). In T lymphocytes, PSTPIP1 serves as an adaptor bringing the cell-surface CD2 molecule into complexes with the Wiskott-Aldrich syndrome protein (WASP), thereby coupling the T cell receptor complex with the actin cytoskeleton to promote the formation of the “immunological synapse” (22). PSTPIP1 also acts as a bridge between PEST-type protein tyrosine phosphatases (PTPs) and both WASP (23) and cAbl tyrosine kinase (24).
PSTPIP1 was first discovered in the mouse as a two-hybrid interactor of a PEST-type [originally so denoted because of regions rich in proline (P), glutamic acid (E), serine (S) and threonine (T)] PTP (19), and it is likely that PTP-induced variation in the phosphorylation of PSTPIP1 modulates its function. Dephosphorylation of PSTPIP1 occurs through the interaction of the tryptophan at residue 232 of its coiled-coil domain with the C-terminal proline-rich homology domain of PEST-type PTPs (25). cAbl appears to be the major kinase responsible for PSTPIP1 phosphorylation, and tryptic phosphopeptide mapping indicates that Y344 is the main phosphorylation site of PSTPIP1 (23). The phosphorylation status of PSTPIP1 does not affect its interaction with WASP in vivo (23).
Two missense substitutions in the coiled-coil domain of PSTPIP1 have recently been shown to cause the dominantly inherited autoinflammatory syndrome of pyogenic arthritis, pyoderma gangrenosum, and acne (PAPA; MIM604416) (26), which bears some clinical similarities to FMF. As in FMF, patients with PAPA syndrome develop neutrophil-rich sterile effusions of the joints (27). Cutaneous manifestations of PAPA syndrome include cystic acne, sterile abscesses at the sites of injections (pathergy), and expanding purulent ulcerating lesions known as pyoderma gangrenosum. Although these findings are more dramatic than usually seen in FMF, the skin manifestations of both are considered to be neutrophilic dermatoses (reviewed in ref. 28). By quantitative yeast two-hybrid assays, PAPA-associated PSTPIP1 mutant proteins show markedly diminished binding to PTP-PEST relative to WT (26), and thus one would predict that these mutant proteins should be hyperphosphorylated. Quantitative two-hybrid experiments revealed no detectable differences in the interaction of CD2 or WASP with mutant PSTPIP1, relative to WT (26).
We were therefore intrigued by the possibility that the clinical similarities between FMF and PAPA syndrome might be related to the fact that these disorders are caused by respective mutations in pyrin and PSTPIP1, proteins with a similar cellular and subcellular distribution that we have now found to interact with one another. We have assessed the functional significance of this interaction by performing coimmunoprecipitations and immunostaining, by defining the interacting domains, and by examining the effects of both PSTPIP1 phosphorylation and PAPA-associated mutations on the interaction with pyrin. Moreover, on the basis of the emerging understanding of the role of pyrin in inflammation, we have examined IL-1β processing in cell lines transfected with WT or mutant PSTPIP1 and in a clinically active PAPA patient.
Materials and Methods
Yeast Two-Hybrid Screen. Screening for pyrin-interacting proteins was performed by using a commercially available system, with pyrin as the bait and a cDNA library prepared from human neutrophils and activated monocytes as the prey. Further details are described in Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org. Deletion mutants were constructed by using PCR, and fragments were ligated into the original expression vector (pDBLeu for PSTPIP1 mutants and pPC86 for pyrin mutants). Point mutations were made by using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). Authenticity of all PCR fragments was confirmed by DNA sequencing. Quantitative assay for β-galactosidase was performed in liquid culture by using chlorophenol red-β-d-galactopyranoside (Roche Molecular Biochemicals) as the substrate according to the manufacturer's instructions (Invitrogen Life Technologies).
Plasmids. Full-length PSTPIP1 and deletion constructs were subcloned into pcDNA3.1 myc/His A (Invitrogen). Full-length pyrin was cloned in pCMV-SPORT (Invitrogen). To construct GST clones, PSTPIP1, pyrin, or their fragments were amplified by PCR by using oligonucleotides containing SpeI and NotI restriction sites, and the PCR product was ligated into a mammalian GST expression vector (pEGST). To construct the GFP-B box/coiled-coil (BBCC) clone, a fragment encoding amino acids 375-576 of pyrin was amplified by PCR by using oligonucleotides containing HindIII and BamHI restriction sites, and the PCR product was ligated to a pEGFP-C1 vector (Clontech). The cAbl plasmid was a kind gift from Silvio Gutkind (National Institute of Dental and Craniofacial Research, Bethesda, MD).
Antibodies. A GST fusion of the N-terminal 374 amino acids of human pyrin was used to generate a polyclonal anti-pyrin antiserum. Pyrin-specific antibodies were purified from immune serum by affinity chromatography by using full-length pyrin coupled to an Affi-Gel 10 support (Bio-Rad). Polyclonal antibodies against PSTPIP1 were generated by using full length PSTPIP1 fused to GST. Antibodies were purified from immune serum by the ImmunoPure purification kit (Pierce). Anti-myc monoclonal antibodies were obtained from Invitrogen and an antiphosphotyrosine monoclonal (4G10) from Upstate Biotechnology (Lake Placid, NY). The anti-PSTPIP1 and antiphosphotyrosine were crosslinked with Immunolinker (CytoSignal Research Products, Irvine, CA) in immunoprecipitation experiments.
Cell Culture and Transfection. HeLa and human embryonic kidney 293T cells were grown in DMEM supplemented with 10% (vol/vol) FBS and a mixture of penicillin and streptomycin. Cells were transfected by using FuGENE 6 (Roche Molecular Biochemicals). For phosphorylation assays, cells were treated with pervanadate solution containing 0.1 mM NaVO3 and 10 mM H2O2.
Immunoprecipitation and GST Pull-Down Assay. Cells were incubated in lysis buffer [0.05 M Tris·HCl, pH 7.5/0.3 M NaCl/0.1% BRIJ 97/2 mM EDTA/complete protease inhibitor mixture (Roche Molecular Biochemicals)] for 15 min on ice. For detection of tyrosine-phosphorylated proteins, 0.4 mM Na3VO4 was added to lysis buffer. The lysates were centrifuged at 16,000 × g at 4°C. Supernatants were incubated with anti-c-myc agarose beads (Clontech) for 2 h at 4°C, washed in lysis buffer and twice with PBS, and resuspended in sample buffer. Cell lysates or eluted samples were subjected to 10% SDS/PAGE, and proteins were transferred to polyvinylidene fluoride membranes and detected with appropriate antibodies. For GST pull-down, cells were lysed as described and incubated with glutathione Sepharose 4B beads (Amersham Pharmacia) for 16 h at 4°C. Beads were washed three times with lysis buffer, resuspended in sample buffer, and boiled 5 min at 95°C. Supernatants were subjected to 10% SDS/PAGE and Western blotting.
Immunofluorescence. After transfection, cells were fixed in 4% formaldehyde, permeabilized by using 0.5% Triton X-100 in PBS, and blocked in 10% goat serum, 1% BSA in PBS for 30 min. After antibody treatment, cells were visualized with a Zeiss LSM 410 confocal microscope.
In Vitro IL-1β Secretion. Cos-7L cells (Invitrogen) were cotransfected with expression vectors for human ASC (10 ng), procaspase-1 (30 ng), pyrin (100 ng), PSTPIP1 (500 ng, WT or mutant), and mouse pro-IL-1β (500 ng), ± c-Abl (300 ng), by using LipofectAMINE 2000 (Invitrogen). Supernatants from 24-well cultures were collected 24 h after transfection and assayed by ELISA for mouse IL-1β (R & D Systems).
Experiments with Human Blood Cells. Peripheral blood mononuclear cells, granulocytes, and monocytes were isolated as described in Supporting Text. Mononuclear cells (2 × 106) were seeded in six-well plates in 2 ml of serum-free RPMI medium 1640 for 1 h. Nonadherent cells were removed by aspiration, and wells were washed with PBS. After washing, 2 ml of complete media was added to each well for 1 h. Supernatants were analyzed by ELISA with the human inflammation cytometric bead array (BD Biosciences, San Jose, CA).
Results
Pyrin Binds to PSTPIP1. To identify pyrin-interacting proteins, we used a stringent yeast two-hybrid assay with low copy-number bait and prey vectors and three separate reporter genes to minimize false positives. Full-length human pyrin cDNA was cloned into the bait vector, and a size-selected cDNA library derived from human peripheral blood granulocytes and activated monocytes was used as the prey. From a total of ≈106 yeast transformants screened, five cDNA clones were identified as positive interactors. The sequence of three matched that of full-length human ASC. One of the other two was identical to full-length cDNAs encoding the 417-aa human PSTPIP1/CD2-binding protein 1 L (long splice variant), and the second differed only by a 9-nt deletion.
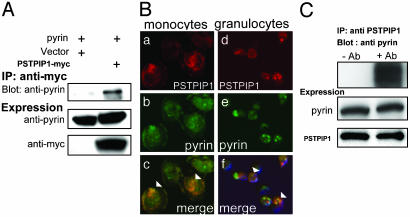
To confirm the interaction between PSTPIP1 and pyrin, we used both immunoprecipitation and fluorescence microscopy. Fig. 1A demonstrates that although similar amounts of pyrin were present in lysates of cells cotransfected with MEFV (encoding pyrin) and PSTPIP1-myc or empty vector, pyrin could be immunoprecipitated with anti-myc only in lysates of PSTPIP1-myc-transfected cells. Immunofluoresence microscopy showed diffuse patchy colocalization of transfected pyrin and PSTPIP1 (Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 1.
Pyrin interacts with PSTPIP1. HeLa cells were transiently cotransfected with PSTPIP1-myc (or vector) and pyrin-expressing plasmids. (A) Lysates were incubated with anti-myc agarose beads and washed, and eluates were separated on SDS/PAGE. Blots were probed with rabbit polyclonal antipyrin and with mouse monoclonal anti-myc. (B) Adherent human monocytes (a-c) and human granulocytes (d-f) were fixed and immunostained with polyclonal rabbit anti-PSTPIP1 followed by Alexa Fluor-568-conjugated goat anti-rabbit IgG to detect endogenous PSTPIP1 (a and d, red) and with Alexa Fluor-488-conjugated rabbit anti-pyrin (b and e, green). (c and f) Merge of PSTPIP1 and pyrin cellular localization images) demonstrating colocalization. Arrowheads indicate regions where staining overlap was most significant. (C) To assess endogenous protein interactions, lysates of THP-1 cells were subjected to immunoprecipitation with crosslinked rabbit polyclonal anti-PSTPIP1 (+Ab) or control serum (-Ab). Lysates were washed and separated on SDS/PAGE. Blots were probed with rabbit polyclonal anti-pyrin or anti-PSTPIP1.
We next examined the cellular distribution of endogenous pyrin and PSTPIP1. By immunofluorescence microscopy, these proteins were present and colocalized in human peripheral blood granulocytes and monocytes (Fig. 1B and Fig. 8, which is published as supporting information on the PNAS web site). Pyrin was not demonstrable in peripheral blood lymphocytes (not shown). To assess whether endogenous pyrin and PSTPIP1 bind one another, lysates of the THP-1 monocytic cell line were immunoprecipitated with crosslinked anti-PSTPIP1. As shown in Fig. 1C, pyrin was coimmunoprecipitated with PSTPIP1.
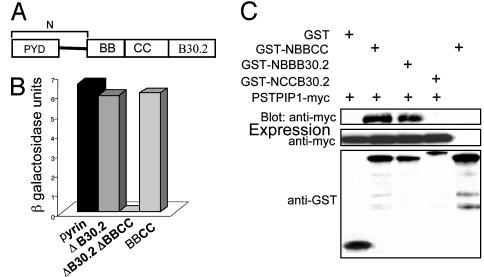
Delineation of the PSTPIP1-Binding Domain of Pyrin. Pyrin is comprised of at least four major domains (Fig. 2A): a 92-aa N-terminal PYRIN domain, a B box zinc finger (amino acids 375-407), an α-helical or potential coiled-coil (amino acids 408-594), and a B30.2 (rfp) domain (amino acids 598-774). To identify the domains in pyrin that interact with PSTPIP1, we performed quantitative yeast two-hybrid analysis, with the degree of interaction between pyrin variants and PSTPIP1 determined by measuring β-galactosidase activity (Fig. 2B). When the B30.2 domain was deleted, no significant reduction in binding was detected, relative to full-length pyrin. Deletion of both the B30.2 domain and the BBCC segment completely abolished binding. The BBCC segment alone exhibited levels of PSTPIP1 binding comparable to the WT control, indicating that this segment is also sufficient for the interaction with PSTPIP1. Moreover, in lysates of transfected HeLa cells, GFP-BBCC coprecipitated with PSTPIP1-myc (Fig. 9, which is published as supporting information on the PNAS web site), thus confirming that the BBCC segment can interact with PSTPIP1 in mammalian cells.
Fig. 2.
The B box domain of pyrin is necessary for the interaction with PSTPIP1. (A) Schematic diagram of the structural domains of pyrin. PYD, PYRIN domain; BB, B box; CC, coiled-coil. “N” denotes amino acids 1-374 (not to scale). (B) Yeast cells were cotransformed with WT pyrin or mutant constructs in pDBLeu and with PSTPIP1 in pPC86. Cells were subjected to a quantitative liquid culture assay for β-galactosidase activity. β-Galactosidase units indicate absorbance at 574 nm, normalized to culture density and incubation time, and represent the degree of interaction between PSTPIP1 and various pyrin mutants. (C) Cos 7L cells were cotransfected with PSTPIP1-myc and GST, GSTNBBCC, GST-NBBB30.2, or GST-NCCB30.2. GST-NBBCC was transfected without PSTPIP1-myc, as another control. Lysates were precipitated with glutathioneagarose beads, washed, eluted, and immunoblotted with anti-myc. Protein expression levels were confirmed with anti-GST or anti-myc.
To map the interacting domain of pyrin more precisely, we performed pull-down experiments with GST-tagged pyrin deletion mutants (Fig. 2C) that lacked either the B30.2 domain (lane 2), the coiled-coil domain (lane 3), or the B box domain (lane 4). Only the construct lacking the B box did not bind PSTPIP1-myc, indicating that this domain is necessary for the interaction.
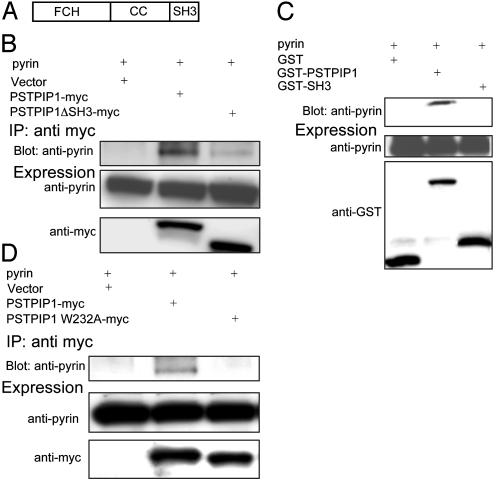
The SH3 and Coiled-Coil Domains of PSTPIP1 Are Both Necessary, but Neither Is Sufficient for the Interaction with Pyrin. The domain structure of PSTPIP1 (Fig. 3A) includes an N-terminal Fer-CIP4 homology domain (amino acids 12-93) (29), a coiled-coil (amino acids 123-288), and an SH3 domain (amino acids 360-416). Although the function of the N-terminal domain is unknown, the homologous domain of CIP4 interacts with microtubules (30). The coiled-coil domain of PSTPIP1 is the site of interaction with PEST-type PTPs (19, 23). The C-terminal SH3 domain of PSTPIP1 is necessary for the interaction of PSTPIP1 with c-Abl (24) and is both necessary and sufficient for the interaction with WASP (31).
Fig. 3.
The SH3 and coiled-coil domains of PSTPIP1 are necessary, but neither is sufficient for pyrin binding. (A) Schematic diagram of the structural domains of PSTPIP1. FCH, Fer-CIP4 homology domain; CC, coiled-coil (not to scale). (B) HeLa cells were transiently cotransfected with pyrin and with PSTPIP1-myc, PSTPIP1ΔSH3-myc, or vector. Lysates were immunoprecipitated with anti-myc and immunoblotted with anti-pyrin and with anti-myc. (C) HeLa cells were cotransfected with pyrin and GST, GST-PSTPIP1, or GST-SH3. Lysates were precipitated with glutathione-agarose beads, washed, eluted, and immunoblotted with anti-pyrin. Protein expression levels were confirmed with anti-GST or anti-pyrin. (D) HeLa cells were cotransfected with pyrin and myc-tagged PSTPIP1, PSTPIP1 W232A, or vector. Lysates were immunoprecipitated with anti-myc and immunoblotted with anti-pyrin or anti-myc.
Quantitative yeast two-hybrid analysis suggested that the SH3 domain of PSTPIP1 is necessary but not sufficient for the interaction with pyrin (Fig. 10, which is published as supporting information on the PNAS web site). To confirm this in mammalian cells, we performed coimmunoprecipitation and GST pull-down assays. HeLa cells were cotransfected with WT or mutant PSTPIP1-myc and pyrin. Deletion of the SH3 domain significantly reduced PSTPIP1 binding to pyrin (Fig. 3B). We also made GST fusion constructs of full-length PSTPIP1 or the SH3 domain alone, and then cotransfected these constructs into HeLa cells with the pyrin expression vector. Fig. 3C shows that whereas WT GST-PSTPIP1 efficiently precipitated pyrin, the GST-SH3 construct did not bind appreciable amounts of pyrin. Expression levels of a Fer-CIP4 fusion protein were insufficient to measure the interaction of this domain with pyrin.
We next asked whether the coiled-coil domain of PSTPIP1 might also be necessary for the interaction with pyrin. Studies of alanine mutants have previously demonstrated a unique motif around W232 in the coiled-coil domain that is involved in PSTPIP1 binding to PTP HSCF, a PEST-type PTP (25). The W232A PSTPIP1 mutation abolished binding to PTP HSCF, although the mutant protein retained the ability to interact with the actin cytoskeleton and to undergo tyrosine phosphorylation. To examine the role of the W232 residue in the interaction of PSTPIP1 with pyrin, we coexpressed pyrin with WT PSTPIP1-myc or PSTPIP1 W232A-myc in HeLa cells, and cell lysates were immunoprecipitated with anti-myc antibody. As shown in Fig. 3D, the W232A mutation completely abolished binding to pyrin.
Pyrin Binding Is Markedly Increased by Tyrosine Phosphorylation of PSTPIP1. The disruption of pyrin binding in the W232A PSTPIP1 mutant could be due to a direct interaction with pyrin at this site or could be an indirect consequence of this mutation's known ability to interfere with PSTPIP1 binding to PEST-type phosphatases. If the latter mechanism were true, one might predict that the degree of PSTPIP1-pyrin interaction would vary inversely with the phosphorylation status of PSTPIP1, because low PTP binding, and thus increased phosphorylation, would be associated with W232A's decreased binding to pyrin.
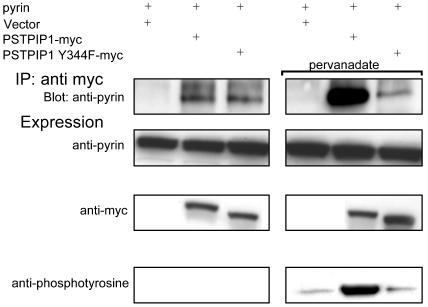
To examine this issue, we studied PSTPIP1 binding to pyrin in the presence or absence of pervanadate, a pan-PTP inhibitor. Because Y344 has been identified as the major tyrosine phosphorylation site of PSTPIP1 (23), we examined the phenylalanine mutant at this residue, which would be predicted to show impaired phosphorylation in the presence of pervanadate. As shown in Fig. 4 Left, in the absence of pervanadate, there was minimal phosphorylation of either WT or Y344F mutant PSTPIP1, and pyrin was equally precipitated by WT or mutant protein. Preincubation of the cells with pervanadate (Fig. 4 Right) caused a marked increase in phosphorylation of the WT PSTPIP1, and a relatively greater increase in the binding of WT PSTPIP1 to pyrin, compared with the Y344F mutant. Pyrin binding is therefore positively correlated with PSTPIP1 phosphorylation status, and the W232A mutation probably does not block pyrin binding by an indirect effect of reduced PTP PEST binding.
Fig. 4.
Tyrosine phosphorylation of PSTPIP1 increases PSTPIP1-pyrin binding. HeLa cells were transiently cotransfected with pyrin and with PSTPIP1-myc, PSTPIP1 Y344F-myc, or vector. Cells were either untreated (Left) or were incubated for 10 min with the phosphatase inhibitor pervanadate before immunoprecipitation (Right). Lysates were immunoprecipitated with anti-myc and immunoblotted with anti-pyrin, anti-myc, or antiphosphotyrosine. Phosphorylation of PSTPIP1 was examined with antiphosphotyrosine (Bottom).
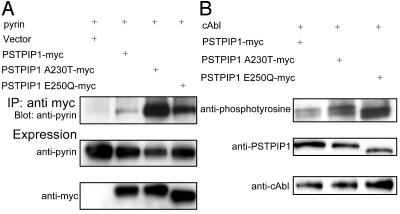
The PAPA Syndrome-Associated A230T and E250Q PSTPIP1 Mutations Cause Increased Pyrin Binding and Are Hyperphosphorylated When Coexpressed with cAbl Kinase. PAPA syndrome is a severe auto-inflammatory disorder caused by two dominantly inherited mutations, A230T and E250Q, in the coiled-coil domain of PSTPIP1 (26). Moreover, these two mutations have previously been shown to impair PTP PEST binding to PSTPIP1 in the yeast two-hybrid system. The effect of these mutations on pyrin binding is unknown. Because the two mutations are very near to the W232 residue, one could hypothesize either decreased pyrin binding through a direct effect on PSTPIP1's coiled-coil binding site, or increased pyrin binding because of overphosphorylation of the PSTPIP1 protein.
Fig. 5A directly addresses this question in lysates from cells transfected with pyrin and WT or mutant PSTPIP1-myc. Relative to WT PSTPIP1, the two disease-associated mutants exhibited markedly increased binding to pyrin. To probe the mechanism of increased pyrin binding, we studied the phosphorylation status of WT and mutant PSTPIP1 in the presence of cAbl kinase. Consistent with our previous data relating phosphorylation status to pyrin binding, we found that the two mutant proteins were hyperphosphorylated, relative to WT (Fig. 5B).
Fig. 5.
PAPA-associated PSTPIP1 mutations increase PSTPIP1-pyrin binding and are hyperphosphorylated. (A) HeLa cells were transiently cotransfected with pyrin and PSTPIP1-myc, PSTPIP1 A230T-myc, PSTPIP1 E250Q-myc, or vector. Lysates were immunoprecipitated with anti-myc and immunoblotted with anti-pyrin and anti-myc. (B) 293T cells were transiently cotransfected with WT PSTPIP1-myc or myc-tagged mutants and cAbl. Proteins were immunoprecipitated with anti-myc agarose beads. Tyrosine phosphorylation of WT PSTPIP1 or PSTPIP1 A230T and PSTPIP1 E250Q was detected by blotting with antiphosphotyrosine (Top). Protein expression levels were confirmed by immunoblot with anti-PSTPIP1 (Middle) or anti-cAbl (Bottom).
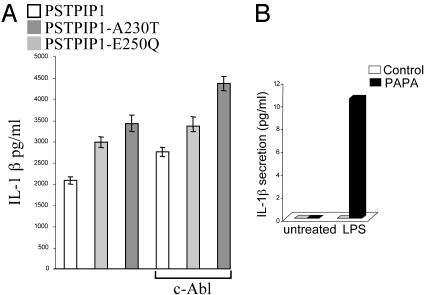
Effect of PAPA-Associated PSTPIP1 Mutations on IL-1β Production. We hypothesized that the accentuated interaction of PSTPIP1 with pyrin might contribute to the clinical phenotype in PAPA by sequestering pyrin, thus shifting the regulatory balance in favor of increased IL-1β processing, as has been demonstrated in pyrin-deficient mice (6). Fig. 6A explores the effect of WT or mutant PSTPIP1 in a recently described system for reconstituting IL-1β responses in vitro (15), in which COS-7L cells are transfected with human pyrin, ASC, caspase-1, and mouse IL-1β expression constructs. With or without cotransfected c-Abl, IL-1β secretion was accentuated in cells transfected with the PAPA-associated PSTPIP1 mutants. Fig. 6B examines IL-1β production in vitro from a PAPA patient with the A230T mutation and active pyoderma gangrenosum (Fig. 11, which is published as supporting information on the PNAS web site), compared with healthy controls. Lipopolysaccharide-induced IL-1 production was substantially increased in the patient relative to controls. Levels of other cytokines including IL-10, IL-4, IFNγ, IL-2, and IL-5 were undetectable, whereas IL-6 and IL-12p70, which are inducible by IL-1β, were also increased (not shown).
Fig. 6.
Effect of PAPA-associated PSTPIP1 mutations on IL-1 secretion. (A) COS-7L cells were cotransfected with expression plasmids for mouse IL-1β and human pyrin, ASC, caspase-1, and PSTPIP1, with or without c-Abl. Supernatants were assayed for IL-1β at 24 h (mean of four replicate experiments). Without c-Abl, WT vs. A230T, P < 0.01, WT vs. E250Q, P < 0.003; with c-Abl, WT vs. A230T, P < 0.004, WT vs. E250Q, P < 0.008. (B) Adherent monocytes from a patient with active PAPA syndrome or healthy controls either were not treated or were stimulated with 1 μg/ml lipopolysaccharide for 1 h. Supernatants were assayed for IL-1β.
Discussion
This paper defines the interaction between two leukocyte proteins, pyrin and PSTPIP1, and provides evidence linking PAPA syndrome, an inherited autoinflammatory disorder of the skin and joints, with abnormalities in pyrin-PSTPIP1 binding. Mutations in pyrin itself lead to FMF, a related but clinically distinct condition. The identification of MEFV, the FMF gene, has facilitated the elucidation of a new pathway regulating myeloid/monocytic inflammation and innate immunity. By placing PSTPIP1 in this schema, we have demonstrated a direct role for protein phosphorylation in the control of pyrin's actions and have added a fifth clinical disorder to the list of pyrin-related autoinf lammatory diseases. Our data suggest that other PSTPIP1-associated proteins may also indirectly influence the pyrin pathway, and that mutations in these proteins may define yet other inflammatory disorders.
The evidence that pyrin binds PSTPIP1 in a biologically meaningful way is strong. We initially discovered the interaction through a stringent yeast two-hybrid analysis of pooled granulocyte/monocyte cDNA that identified as its only other positive ASC, a protein that binds and functionally interacts with pyrin both in vitro and in vivo (6, 13, 17). In the present report, we have confirmed the PSTPIP1-pyrin interaction by coimmunoprecipitation and immunostaining of the respective transfected and endogenous proteins. Moreover, pyrin and PSTPIP1 are expressed in an intersecting and relatively circumscribed subset of hematopoietic cells, and both associate with the actin cytoskeleton (6, 7, 19).
Recent data assign to pyrin an important role in the regulation of inflammation. Although the net effect of pyrin in transfections may depend on the cellular context (32), several reports indicate that pyrin inhibits proinflammatory signaling pathways in vitro (17, 18). Moreover, in mouse RAW monocytic cells, pyrin inhibits the interaction of ASC with caspase-1, and pyrindeficient mice exhibit a phenotype of endotoxin sensitivity associated with excessive activation of caspase-1 and IL-1β and with a defect in macrophage apoptosis (6). A major implication of this latter study is that autoinflammatory disorders such as FMF may represent an exaggerated innate immune response to various signals, including microbial products.
The mutations in PSTPIP1 underlying PAPA syndrome lead to hyperphosphorylation and a marked increase in the strength of the interaction with pyrin. Increased pyrin-PSTPIP1 binding may, in turn, modulate pyrin's normal immunoregulatory function by preventing its interaction with other proteins such as ASC, thus explaining the clinical phenotype of PAPA patients and the excess in IL-1β production we observed in a PAPA patient with pyoderma gangrenosum. Consistent with the clinical picture, such increase-of-function mutations would be expected to be inherited in an autosomal dominant fashion and to predispose to inflammatory lesions predominated by granulocytes and monocytes. Increased levels of IL-6 and IL-12p70 observed in the monocytes of a patient may also contribute to the pathogenesis of PAPA syndrome. These cytokines function downstream of IL-1β and are likely to amplify the inflammatory cascade triggered by PSTPIP1 mutations.
This model does not exclude the possibility that PAPA-associated mutations might also act through other pathways in which PSTPIP1 participates. Nevertheless, against the possibility that the A230T and E250Q substitutions have a major effect through CD2, it should be noted that these mutants bind normally to CD2 in a yeast two-hybrid system (26), and that CD2 is not expressed in granulocytes, the major cell type involved in the inflammatory lesions of PAPA syndrome. The PAPA-associated PSTPIP1 mutants also bind normally to WASP in the yeast two-hybrid system (26), and the phosphorylation status of PSTPIP1 does not affect its interaction with WASP in vivo (23). Moreover, the experiments reported in Fig. 5 indicate that cAbl can interact and phosphorylate mutant PSTPIP1. The A230T and E250Q substitutions do abrogate PSTPIP1's interaction with PTP PEST (26), at least one consequence of which is the aforementioned increase in binding to pyrin. Further studies are in progress to examine PSTPIP1 phosphorylation in leukocyte subsets with disease flares and remissions.
We have mapped the PSTPIP1 interaction domain of pyrin to the B box zinc finger and have specifically ruled out a major PSTPIP1-binding site in the C-terminal B30.2 (rfp) domain of pyrin. This latter domain is a hot spot for disease-associated mutations in pyrin, and our data therefore indicate it is unlikely that these mutations act by altering the binding of pyrin to PSTPIP1. In fact, in the yeast two-hybrid assay, the three FMF-associated mutations in the B30.2 domain we tested had no effect on binding to PSTPIP1 (data not shown).
PAPA syndrome is now the fifth human autoinflammatory disease caused by mutations in pyrin or a related protein. The prototype illness, FMF, is caused by recessive mutations in pyrin itself and shares with PAPA the neutrophilic predominance of inflammatory lesions. Three other disorders, familial cold autoinflammatory syndrome (MIM120100), Muckle-Wells syndrome (MIM191900), and the neonatal onset multisystem inflammatory disease (MIM607115), are all caused by dominantly inherited mutations in CIAS1 (33-35). CIAS1 encodes cryopyrin, an inflammatory regulator with an N-terminal PYRIN domain that appears to oppose the action of pyrin on ASC. We have screened >1,000 patients with periodic inflammatory disease for mutations in these and other genes and have found a genetic basis in only ≈40% (unpublished data), raising the possibility that some of these undiagnosed patients may have mutations in other pyrin-related genes.
This report presents data extending a new pathway regulating inflammation and strongly suggests yet another connection between a protein in this pathway and human disease. At least equally exciting, though, are the therapeutic implications of these findings. Especially for those with severe pyoderma gangrenosum, the therapeutic options available to PAPA patients are frequently ineffective or are wrought with serious side effects. With the current availability of anakinra, a recombinant IL-1 receptor antagonist, it is possible that the inflammatory phenotype of PAPA syndrome may be susceptible to more effective control. Preliminary data on the response of two PAPA patients to this agent over several months are encouraging. Larger clinical trials with this agent hold the promise of establishing a treatment for this very serious condition, while at the same time subjecting our hypotheses to a more rigorous test.
Supplementary Material
Abbreviations: PSTPIP1, proline serine threonine phosphatase-interacting protein 1; PAPA, pyogenic arthritis with pyoderma gangrenosum and acne; FMF, familial Mediterranean fever; CARD, caspase recruitment domain; ASC, apoptosis-associated speck-like protein with a CARD; WASP, Wiskott-Aldrich syndrome protein; PTP, protein tyrosine phosphatase; BBCC, B box/coiled-coil.
References
- 1.Galon, J., Aksentijevich, I., McDermott, M. F., O'Shea, J. J. & Kastner, D. L. (2000) Curr. Opin. Immunol. 12, 479-486. [DOI] [PubMed] [Google Scholar]
- 2.Kastner, D. L. (2003) in Rheumatology, eds. Hochberg, M. C., Silman, A. J., Smolen J. S., Weinblatt, M. E. & Weisman, M. H. (Mosby, Edinburgh), 3rd Ed., pp. 1717-1734.
- 3.International FMF Consortium (1997) Cell 90, 797-807. [DOI] [PubMed] [Google Scholar]
- 4.French FMF Consortium (1997) Nat. Genet. 17, 25-31. [DOI] [PubMed] [Google Scholar]
- 5.Centola, M., Wood, G., Frucht, D. M., Galon, J., Aringer, M., Farrell, C., Kingma, D. W., Horwitz, M. E., Mansfield, E., Holland, S. M., et al. (2000) Blood 95, 3223-3231. [PubMed] [Google Scholar]
- 6.Chae, J. J., Komarow, H. D., Cheng, J., Wood, G., Raben, N., Liu, P. P. & Kastner, D. L. (2003) Mol. Cell 11, 591-604. [DOI] [PubMed] [Google Scholar]
- 7.Mansfield, E., Chae, J. J., Komarow, H. D., Brotz, T. M., Frucht, D. M., Aksentijevich, I. & Kastner, D. L. (2001) Blood 98, 851-859. [DOI] [PubMed] [Google Scholar]
- 8.Bertin, J. & DiStefano, P. S. (2000) Cell Death Differ. 7, 1273-1274. [DOI] [PubMed] [Google Scholar]
- 9.Martinon, F., Hofmann, K. & Tschopp, J. (2001) Curr. Biol. 11, R118-R120. [DOI] [PubMed] [Google Scholar]
- 10.Srinivasula, S. M., Poyet, J. L., Razmara, M., Datta, P., Zhang, Z. & Alnemri, E. S. (2002) J. Biol. Chem. 277, 21119-21122. [DOI] [PubMed] [Google Scholar]
- 11.Pawlowski, K., Pio, F., Chu, Z., Reed, J. C. & Godzik, A. (2001) Trends Biochem. Sci. 26, 85-87. [DOI] [PubMed] [Google Scholar]
- 12.Staub, E., Dahl, E. & Rosenthal, A. (2001) Trends Biochem. Sci. 26, 83-85. [DOI] [PubMed] [Google Scholar]
- 13.Richards, N., Schaner, P., Diaz, A., Stuckey, J., Shelden, E., Wadhwa, A. & Gumucio, D. L. (2001) J. Biol. Chem. 276, 39320-39329. [DOI] [PubMed] [Google Scholar]
- 14.Masumoto, J., Taniguchi, S., Ayukawa, K., Sarvotham, H., Kishino, T., Niikawa, N., Hidaka, E., Katsuyama, T., Higuchi, T. & Sagara, J. (1999) J. Biol. Chem. 274, 33835-33838. [DOI] [PubMed] [Google Scholar]
- 15.Wang, L., Manji, G. A., Grenier, J. M., Al-Garawi, A., Merriam, S., Lora, J. M., Geddes, B. J., Briskin, M., DiStefano, P. S. & Bertin, J. (2002) J. Biol. Chem. 277, 29874-29880. [DOI] [PubMed] [Google Scholar]
- 16.Martinon, F., Burns, K. & Tschopp, J. (2002) Mol. Cell 10, 417-426. [DOI] [PubMed] [Google Scholar]
- 17.Dowds, T. A., Masumoto, J., Chen, F. F., Ogura, Y., Inohara, N. & Nunez, G. (2003) Biochem. Biophys. Res. Commun. 302, 575-580. [DOI] [PubMed] [Google Scholar]
- 18.Masumoto, J., Dowds, T. A., Schaner, P., Chen, F. F., Ogura, Y., Li, M., Zhu, L., Katsuyama, T., Sagara, J., Taniguchi, S., et al. (2003) Biochem. Biophys. Res. Commun. 303, 69-73. [DOI] [PubMed] [Google Scholar]
- 19.Spencer, S., Dowbenko, D., Cheng, J., Li, W., Brush, J., Utzig, S., Simanis, V. & Lasky, L. A. (1997) J. Cell. Biol. 138, 845-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, J., Nishizawa, K., An, W., Hussey, R. E., Lialios, F. E., Salgia, R., Sunder-Plassmann, R. & Reinherz, E. L. (1998) EMBO J. 17, 7320-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fankhauser, C., Reymond, A., Cerutti, L., Utzig, S., Hofmann, K. & Simanis, V. (1995) Cell 82, 435-444. [DOI] [PubMed] [Google Scholar]
- 22.Badour, K., Zhang, J., Shi, F., McGavin, M. K., Rampersad, V., Hardy, L. A., Field, D. & Siminovitch, K. A. (2003) Immunity 18, 141-154. [DOI] [PubMed] [Google Scholar]
- 23.Côté, J. F., Chung, P. L., Théberge, J. F., Hallé, M., Spencer, S., Lasky, L. A. & Tremblay, M. L. (2002) J. Biol. Chem. 277, 2973-2986. [DOI] [PubMed] [Google Scholar]
- 24.Cong, F., Spencer, S., Coôté, J. F., Wu, Y., Tremblay, M. L., Lasky, L. A. & Goff, S. P. (2000) Mol. Cell 6, 1413-1423. [DOI] [PubMed] [Google Scholar]
- 25.Dowbenko, D., Spencer, S., Quan, C. & Lasky, L. A. (1998) J. Biol. Chem. 273, 989-996. [DOI] [PubMed] [Google Scholar]
- 26.Wise, C. A., Gillum, J. D., Seidman, C. E., Lindor, N. M., Veile, R., Bashiardes, S. & Lovett, M. (2002) Hum. Mol. Genet. 11, 961-969. [DOI] [PubMed] [Google Scholar]
- 27.Lindor, N. M., Arsenault, T. M., Solomon, H., Seidman, C. E. & McEvoy, M. T. (1997) Mayo Clin. Proc. 72, 611-615. [DOI] [PubMed] [Google Scholar]
- 28.Callen, J. P. (2002) Dermatol. Clin. 20, 409-419. [DOI] [PubMed] [Google Scholar]
- 29.Aspenstrom, P. (1997) Curr. Biol. 7, 479-487. [DOI] [PubMed] [Google Scholar]
- 30.Tian, L., Nelson, D. L. & Stewart, D. M. (2000) J. Biol. Chem. 275, 7854-7861. [DOI] [PubMed] [Google Scholar]
- 31.Wu, Y., Spencer, S. D. & Lasky, L. A. (1998) J. Biol. Chem. 273, 5765-5770. [DOI] [PubMed] [Google Scholar]
- 32.Stehlik, C., Fiorentino, L., Dorfleutner, A., Bruey, J. M., Ariza, E. M., Sagara, J. & Reed, J. C. (2002) J. Exp. Med. 196, 1605-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffman, H. M., Mueller, J. L., Broide, D. H., Wanderer, A. A. & Kolodner, R. D. (2001) Nat. Genet. 29, 301-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feldmann, J., Prieur, A. M., Quartier, P., Berquin, P., Certain, S., Cortis, E., Teillac-Hamel, D., Fischer, A. & de Saint Basile, G. (2002) Am. J. Hum. Genet. 71, 198-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aksentijevich, I., Nowak, M., Mallah, M., Chae, J. J., Watford, W. T., Hofmann, S. R., Stein, L., Russo, R., Goldsmith, D., Dent, P., et al. (2002) Arthritis Rheum. 46, 3340-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.