Abstract
Immunization with vaccinia virus (VACV) resulted in long-lasting protection against smallpox and successful global eradication of the disease. VACV elicits strong cellular as well as humoral immune responses. Although neutralizing antibody is essential for protection, cellular immunity seems to be more important for recovery from infection in humans. We analyzed the immunodominance hierarchy of 73 previously identified VACV human CD8+ T cell epitopes restricted by HLA-A1, A2, A3, A24, B7 or B44 alleles or the alleles belonging to one of these supertypes in 56 donors after primary VACV immunization. Except for the responses to HLA-A24 supertype-restricted epitopes, there were no consistent patterns of epitope immunodominance among donors sharing the same HLA alleles or supertypes, which is in sharp contrast with the mouse studies. We, however, identified 12 epitopes that were recognized by ≥20% of donors sharing the same HLA allele; six of these contributed ≥20% of the total VACV-specific T cell response in at least one individual. VACV-specific CD8+ T cell responses targeted a group of epitopes, “relatively dominant” epitopes, without a strong immunodominance hierarchy in humans, which may be advantageous to humans to prevent the emergence of T cell escape mutants.
Keywords: vaccinia virus, poxvirus, CD8+ T cell epitope, immunodominance, HLA supertype
Introduction
Immunization with vaccinia virus (VACV) resulted in long-lasting protection against smallpox and was the successful approach used to eliminate natural smallpox infections worldwide. As a vaccine against smallpox, VACV elicits strong cellular as well as humoral immune responses. Although neutralizing antibody is essential for protection, cellular immunity seems to be more important for recovery from infection in humans [1, 2]. In children with immunological defects in cell-mediated immunity, VACV replicated without restriction, resulting in a continually progressive primary lesion, persistent viremia and widespread secondary viral infection of many organs. This response was particularly severe in patients with thymic aplasia, while those with defects in antibody production but satisfactory capacity to mount a cell-mediated immune response, e.g. congenital agammaglobulinemia, usually recovered normally after vaccination [3, 4]. In some cases of Bruton's syndrome a partially deficient cell-mediated immune mechanism might have been overwhelmed, but could be restored to effectiveness by administration of vaccinia immune globulin [5]. However, the presence of neutralizing antibody alone did not prevent the development of progressive vaccinia if cell-mediated immunity was defective [4, 6].
Today VACV has reemerged as a very important vaccine due to the concern about the potential use of smallpox virus as a bioweapon [7, 8], and also as an expression vector for immunization against other infectious diseases, such as HIV and cancer [9, 10]. Strong and long-lasting cellular immune responses, especially CTL responses are needed against such diseases.
In recent years nearly 100 VACV-specific CD8+ T cell epitopes have been identified in humans by us and several other groups [11-21](reviewed in [22]). Immunodominance is a phenomenon by which CD8+ T cell responses are predominantly directed to a few peptides (sometimes a single peptide), despite the presence of many potentially immunogenic peptides [23]. In VACV-infected C57BL/6 mice an H-2 Kb-restricted CD8+ T cell epitope B8R20-27 encoded by the B8R gene is clearly the immunodominant epitope [24]. In humans, however, it is not known if there is immunodominance or if epitope hierarchy is consistent among individuals sharing the same HLA alleles. The experiments in the papers that reported the identification of VACV-specific CD8+ T cell epitopes in humans were screening experiments and were not designed to determine immunodominance, although Oseroff et al. suggested that human anti-VACV CD8+ T cell responses were broad and did not target a single or a few epitopes [14], which was later called “immunodemocratic” [25]. They also suggested that the epitope G5R18–26 may be immunodominant in HLA-A*0201-positive individuals. However, their experiments used PBMC collected at various times within one year of vaccinia immunization from 21 donors who received primary immunization and 37 donors who received booster immunization. Therefore, it is likely that many factors, such as primary versus secondary immunization and the acute phase versus the memory phase of the immunization, influenced the epitope hierarchy in these donors, and could explain the lack of a consistent pattern of epitope hierarchy among donors sharing the same HLA alleles. In this report we quantitated VACV-specific CD8+ T cell responses in IFN-γ ELISPOT assays using 73 epitope peptides restricted by HLA-A1, A2, A3, A24, B7 or B44 alleles or the alleles belonging to one of these supertypes with PBMC of 56 HLA-typed individuals, who had received primary immunization with Dryvax® or its derivatives.
Materials and Methods
PBMC Donors
PBMC Donors in this study were healthy VACV-naïve volunteers who received smallpox vaccine, standard Dryvax®, ACAM 1000 or ACAM2000 [26, 27], by multiple cutaneous punctures made with a bifurcated needle in a clinical trial performed by Acambis (Cambridge, MA). The protocol was approved by the institutional review board and all experiments conformed to the relevant regulatory standards. Blood was drawn at day 0 (before vaccination) and day 45 after vaccination, and PBMC were separated and stored in liquid nitrogen. 56 donors were selected based on the HLA-A and B typing and the availability of the PBMC for our study.
HLA typing
MHC class I genotyping was performed by using Olerup SSP Combi-Kits for low resolution typing and Olerup SSP Kits for HLA-A and HLA-B for high resolution typing (Qiagen Inc., Valencia, CA).
Viruses
VACV New York City Board of Health (NYCBH) strain, the same strain used to produce Dryvax®, was provided by Gail Mazzara and Dennis Panicali of Applied Biotechnology, Inc, and was propagated and titrated in CV-1 cells (ATCC # CCL-70) as previously described [28].
Peptides
Peptides used in this study (listed in Table 1) were purchased from Anaspec Inc (San Jose, CA). They were HPLC-purified (>80% purity) and were confirmed by mass spectrometry.
Table 1. CD8+ T cell epitopes analyzed in this study and their conservation in Dryvax®.
| peptide name | gene name and (a.a. position)a | a.a. sequence | restricting allele | conservation in Dryvax® | temporal expressionb | reference |
|---|---|---|---|---|---|---|
| A1H1 | A24R (278–286) | ITDFNIDTY | A*0101 | Yes | E | [16] |
| A1H2 | B8R (139–147) | DMCDIYLLY | A1 supertype | Yes | E | [14] |
| A1H3 | B8R (153–162) | FGDSKEPVPY | A1 supertype | Yes | E | [14] |
| A1H4 | B8R (262–271) | FLSMLNLTKY | A1 supertype | Yes | E | [14] |
| A1H5 | C10L (297–305) | SQSDTVFDY | A1 supertype | Yes | E | [14] |
| A1H6 | C10L (298–306) | QSDTVFDYY | A1 supertype | Yes | E | [14] |
| A1H7 | C12L (326–334) | VYINHPFMY | A*2902 | Yes | E | [16] |
| A1H8 | D1R (156–164) | FTIDFKLKY | A1 supertype | Yes | E | [14] |
| A1H9 | D12L (11–19) | GTHVLLPFY | A1 supertype | Yes | E | [14] |
| A1H10 | F11L (259–267) | CMLTEFLHY | A1 supertype | Yes | E | [14] |
| A1H11 | VACWR008 (29–38) | VSVNNVCHMY | A1 supertype | Yes | E | [14] |
| A1H12 | VACWR008 (104–112) | QSITRSLIY | A1 supertype | Yes | E | [14] |
| A1H13 | VACWR013 (97–106) | VTDTNKFDNY | A1 supertype | VTDTNKFAHY | E | [14] |
| A1H14 | A27L (89-103)c | LRAAMISLAKKIDVQ | A1 | Yes | L | [18] |
| A2H1 | A6L (172–180) | ILSDENYLL | A2 supertype | Yes | L | [14] |
| A2H2 | A7L (342–350) | FLVIAINAM | A2 supertype | Yes | L | [14] |
| A2H3 | A36R (1–9) | MMLVPLITV | A2 supertype | Yes | E | [14] |
| A2H4 | A47L (169-177) | LLYAHINAL | A*0201 | Yes | IE | [12] |
| A2H5 | A55R (78–86) | YIYGIPLSL | A2 supertype | Yes | E | [14] |
| A2H6 | B22R/C16L (60-68) | CLTEYILWV | A*0201 | Yes | ?d | [12] |
| A2H7 | C7L (74–82) | KVDDTFYYV | A*0201 | Yes | E | [12] |
| A2H8 | E2L (249–257) | KIDYYIPYV | A2 supertype | Yes | E | [14] |
| A2H9 | E9L (107–115) | FLNISWFYI | A2 supertype | Yes | E | [14] |
| A2H10 | F12L (286–295) | NLFDIPLLTV | A2 supertype | Yes | E | [14] |
| A2H11 | F12L (404–412) | FLTSVINRV | A2 supertype | Yes | E | [14] |
| A2H12 | G5R (18–26) | ILDDNLYKV | A2 supertype | Yes | IE | [14] |
| A2H13 | H3L (184-192) | SLSAYIIRV | A*0201 | Yes | L | [11] |
| A2H14 | I4L (720–728) | SMHFYGWSL | A2 supertype | Yes | E | [14] |
| A2H15 | N2L (93–101) | YVNAILYQI | A2 supertype | Yes | IE | [14] |
| A2H16 | O1L (247–255) | GLNDYLHSV | A2 supertype | Yes | IE | [14] |
| A2H17 | VACWR148 (177–186) | YLYTEYFLFI | A2 supertype | YLYTEYFLFLe | L | [14] |
| A2H18 | A55R (391-399) | AMLNGLIYV | A*0201 | Yes | E | [55] |
| A2H19 | B5R (5-19)c | SVVTLLCVLPAVVYS | A2 | Yes | E/L | [18] |
| A2H19′ | B5R (8-16)f | TLLCVLPAV | ||||
| A3H1 | A8R (79–88) | AVKDVTITKK | A3 supertype | Yes | IE | [14] |
| A3H2 | A31R (86–94) | VTSSGAIYK | A3 supertype | Yes | E | [14] |
| A3H3 | B6R (154–163) | GTIAGGVCYY | A3 supertype | Yes | E | [14] |
| A3H4 | C5L (158–166) | KVMFVIRFK | A3 supertype | Yes | E/L | [14] |
| A3H5 | C7L (31–40) | KLKIISNDYK | A3 supertype | Yes | E | [14] |
| A3H6 | C9L (193–201) | ATSLDVINY | A3 supertype | Yes | E/L | [14] |
| A3H7 | D1R (152–161) | KTKNFTIDFK | A3 supertype | Yes | E | [14] |
| A3H8 | D5R (670–678) | YLLVKWYRK | A3 supertype | Yes | E | [14] |
| A3H9 | G8R (65–73) | IVFNLPVSK | A3 supertype | Yes | ?g | [14] |
| A3H10 | I3L (116–124) | AVYGNIKHK | A3 supertype | Yes | IE | [14] |
| A3H11 | J6R (332–340) | NQVKFYFNK | A3 supertype | Yes | E | [14] |
| A3H12 | VACWR013 (93–102) | KVLHVTDTNK | A3 supertype | Yes | E | [14] |
| A3H13 | VACWR195 (74–83) | AVFKDSFLRK | A3 supertype | Yes | IE | [14] |
| A24H1 | A48R (58–66) | TYNDHIVNL | A*2301 | Yes | IE | [16] |
| A24H2 | C6L (54–63) | RYYDGNIYE | A24 supertype | RYYDGNIYD | IE | [14] |
| A24H4 | D5R (349–357) | VWINNSWKF | A24 supertype | Yes | E | [14] |
| A24H5 | D5R (663–672) | RYRFAFLYLL | A24 supertype | Yes | E | [14] |
| B7H1 | B22R/C16L (53-61) | TVADVRHCL | B7 | Yes | ?d | [17] |
| B7H2 | C1L (102–111) | KPKPAVRFAI | B7 supertype | Yes | E | [14] |
| B7H3 | D1R (686–694) | HPRHYATVM | B7 supertype | Yes | E | [14] |
| B7H4 | F4L (6–14) | APNPNRFVI | B7 supertype | Yes | E | [14] |
| B7H5 | J2R (62-70) | EATKLCDVL | B*3502 | Yes | E | [17] |
| B7H6 | J6R (303-311) | MPAYIRNTL | B7 supertype | Yes | E | [14] |
| B7 | [17] | |||||
| B7H7 | O1L (335–344) | RPMSLRSTII | B7 supertype | Yes | IE | [14] |
| B44H1 | A3L (90–98) | DEVASTHDW | B*4403 | Yes | L | [16] |
| B44H2 | A3L (264–272) | YEFRKVKSY | B*4403 | Yes | L | [16] |
| B44H3 | A23R (287–295) | HDVYGVSNF | B*4403 | Yes | E | [16] |
| B44H4 | B8R (110–118) | TEYDDHINL | B44 supertype | Yes | E | [14] |
| B44H5 | C3L (120–128) | GESKSYCEL | B44 supertype | Yes | L | [14] |
| B44H6 | C7L(56-64) | DEVKGLTVF | B*1801 | Yes | E | [19] |
| B44H7 | D1R (126–134) | EERHIFLDY | B*4403 | Yes | E | [16] |
| B44H8 | D5R (298–306) | LENGAIRIY | B*4403 | Yes | E | [16] |
| B44H9 | D5R (691–699) | EEIPDFAFY | B*4403 | Yes | E | [16] |
| B44H10 | E3L (86–94) | DDVSREKSM | B*4403 | Yes | IE | [16] |
| B44H11 | G2R (181–189) | DELVDPINY | B44 supertype | Yes | E | [14] |
| B44H12 | I3L (173–181) | IEGELESLS | B*4403 | Yes | IE | [16] |
| B44H13 | M2L (38–46) | AELTIGVNY | B*4403 | Yes | IE | [16] |
| B44H14 | F14.5L (41–49)h | EEQELLLLY | B*4403 | Yesi | E | [16] |
| B44H15 | VACWR013 (21–29) | DEIKCPNLN | B*4403 | VETKCSNLDj | E | [16] |
| B44H16 | B5R (105-119)c | TKYFRCEEKNGNTSW | B44 | Yes | E/L | [18] |
: Gene names are based on nomenclature for Copenhagen strain. When not conserved in Copenhagen strain, nomenclature for WR strain is used. Misidentification of the genes in the original publications are corrected.
: Based on the reference [48].
: Minimal epitope was not determined
: B22R/C16L was not analyzed
: When compared with the WR sequence, the C-termial “I” in the original paper should be “L”
: Predicted HLA-A2 binding motif in B5R (5-19) peptide by HLA Peptide Binding Predictions (http://www.bimas.cit.nih.gov/molbio/hla_bind/) [56] and SYFPEITHI (http://www.syfpeithi.de/Scripts/MHCServer.dll/EpitopePrediction.htm)[57]
: G8R gene did not fit any class
: Also known as F3
: ORF was identified by BLAST search on the genomic DNA sequences
: Not conserved only in Acambis 3 strain
IFN-γ-ELISPOT assay
IFN-γ-ELISPOT assays were performed as previously described [26, 29]. Briefly, 96-well filtration plates (MSIPS4W; Millipore) were coated with 15 μg of mouse anti-human IFN-γ monoclonal antibody (clone D1K; Mabtech, Cincinnati, OH) per ml at 4°C overnight. Plates were blocked with RPMI-10% FBS for at least 2 hours. 2×105 PBMCs cells were added in RPMI-10% FBS. For stimulation peptides were added at 4 μg/ml or VACV (NYCBH strain) at an moi of 1, and plates were incubated for 16 hours at 37°C. The cells were removed by washing, and biotinylated mouse anti-human IFN-γ monoclonal antibody (clone 7-B6-1; Mabtech) was then added and left for 2 hours at room temperature, followed by a 1:400 dilution of streptavidin horse radish peroxidase for 30 min. Substrate (NovaRED™, Vector Laboratories, Burlingame, CA) was added and left for 10 min at room temperature. The plates were read by ImmunoSpot® S4 pro Analyzer and analyzed using ImmunoSpot® 4.0 software (CTL Analyzers LLC, Cleveland, OH). The frequency of IFN-γ-producing cells was calculated as a number of spots per 106 cells after subtracting the number of spots in negative control wells (cells incubated with medium alone). If the number of spots became negative after subtraction, it was treated as 0 in statistical analyses. Experiments were performed in triplicate.
Statistical analyses
Statistical analyses were performed using Microsoft Excel 2003.
Results
CD8+ T cell epitopes analyzed in this study and their conservation in Dryvax®
At the beginning of this study approximately 100 VACV-specific CD8+ T cell epitopes restricted by various human MHC class I alleles had been identified. Many of the HLA-A*0201-, A*1101- and B*0702-restricted epitopes were identified using HLA-transgenic mice. Some of them were, however, only recognized in the HLA-transgenic mice and not in humans [14, 15, 30]. Because of the limitation of available PBMCs for this analysis, we decided to include only epitope peptides to which specific T cells had been detected in humans. Although most of the epitopes were identified based on the sequence information of modified vaccinia Ankara (MVA) strain (GenBank accession number U94848) [31] or the Western Reserve (WR) strain (NC_006998), the donors of the PBMCs in this study were vaccinated with Dryvax®, Acambis 1000 or Acambis 2000 [26, 27]. Dryvax® has never been cloned and is expected to be a quasispecies. To assess the possible effects of sequence variation at T cell epitopes, we analyzed two genomic sequences of the strains isolated from a pox lesion from individuals who received Dryvax®, strain 3737 (DQ377945) and strain DUKE (DQ439815) [32], and two genomic sequences of the clones derived from the NYCBH strain, which was the parental strain of Dryvax®, Acambis clone 3 (AY313848) and Acambis 2000 (AY313847). Table 1 shows the conservation of all 73 epitope peptides used in this study grouped by the restricting HLA alleles. Only four of the 73 epitope peptides, VACWR01397–106 (A1H13), C6L54–63 (A24H2), VACWR01321–29 (B44H15) and VACWR148177–186 (A2H17) had amino acid changes in Dryvax®. We used these published epitope peptides in our assays rather than the peptides based on the NYCBH strains; however, these peptides induced IFN-γ production in the PBMC from individuals who were vaccinated with Dryvax® [14]. One amino acid change at the C-terminus of the VACWR148177–186 (A2H17) may be a typographical error, since the WR strain, on which the peptide sequence was based, does not have the same amino acid change. In Table 1 epitopes are grouped by HLA supertypes, by which the epitope is restricted. Sette et al. categorized HLA-A and HLA-B alleles into HLA supertypes based on the similarity of binding peptides, which are called “supermotifs” [33-35]. Since one paper reported that the peptide binding repertoire of HLA-A*2902, which was categorized as a member of the HLA-A1 supertype, overlapped with that of A24 supertype [35], peptide C12L326–334 (A1H7) was tested in both HLA-A1 supertype-positive and HLA-A*2402-positive donors. The data were analyzed based on the latest classification of HLA class I supertypes [36], which is slightly different from previous classifications.
T cell responses of donors' PBMC to live VACV
We quantitated the number of VACV-specific IFN-γ-producing cells by ELISPOT assays in donors' PBMC after live VACV (NYCBH strain) stimulation. One donor's day 0 PBMC had 55.0 IFN-γ-producing cells per 106 PBMC, suggesting that this donor may not have been naïve, and, therefore, the data from this donor were not included in further analyses. In the remaining 55 donors the T cell responses to live VACV were (average ± SD): day 0 (per-vaccination) 1.2 ± 3.5 per 106 PBMC and day 45 post-vaccination 317.1 ± 416.6 per 106 PBMC. These are comparable to our previous reports [12, 17].
Peptide-specific ELISPOT responses in day 0 PBMC and the determination of cutoff value
In the 55 donors, the average (± SD) number of peptide-specific IFN-γ-producing cells in the day 0 PBMC was 2.8 ± 22.8 per 106 PBMC. There were, however, three donors whose day 0 PBMC had very high number of IFN-γ-producing cells against one of the VACV peptides (71.7, 91.7 and 950.0 IFN-γ-producing cells per 106 PBMC); however, responses against live VACV were not detected, and the number of IFN-γ-producing cells responding to these peptides decreased after vaccination. Based on the possibility that these responses represented “heterologous immunity”, we removed these donors from the calculation of threshold values for the peptide ELISPOT assay. Without these donors the average (± SD) number of IFN-γ-producing cells against each peptide in day 0 PBMC was 2.1 ± 4.0 per 106 PBMC. In the subsequent analyses we used average + 3SD (14.1 per 106 PBMC) as a cutoff value for positive peptide-specific responses, after subtraction of the day 0 value.
Epitope-specific T cell responses after vaccination
Epitope-specific T cell responses in donors, all of whose HLA-A and HLA-B alleles belonged to one of these six HLA supertypes (HLA-A1, A2, A3, A24, B7 or B44)
As shown in Table 1, most VACV-specific T cell epitopes are restricted by common HLA-A1, A2, A3, A24, B7 or B44 alleles, and the rest are restricted by alleles belonging to the supertypes of these alleles. There were 17 donors, all of whose HLA-A and HLA-B alleles belonged to one of these six HLA supertypes (HLA-A1, A2, A3, A24, B7 or B44). In these donors, if the sum of the VACV peptide-specific CD8+ T cell responses restricted by a given allele represents all the VACV-specific CD8+ T cell responses restricted by the allele, and if most VACV-specific CD8+ T cell responses are restricted by HLA-A or HLA-B alleles, the sum of the peptide-specific CD8+ T cell responses should be comparable to the CD8+ T cell responses to live VACV. Table 2 summarizes the responses to live VACV and the responses to peptides grouped by HLA supertype in 14 of the 17 donors. Day 45 PBMCs from the other three donors had very low responses to VACV (fewer than 10.0 IFN-γ-producing cells per 106 PBMC) probably due to low viability of cells, and we did not include the data from these donors in the analysis. On average the sum of the responses to all peptides tested was 39% of the response to the live VACV, but this varied considerably from donor to donor (range 0 to 101%). Considering that VACV-specific CD4+ T cells also contribute to the IFN-γ response to live virus, the peptide sets had good coverage of total VACV-specific CD8+ T cell responses, although IFN-γ-ELISPOT assays using live VACV sometimes underestimate the responses to the whole virus [14, 17, 37]. In some donors the responses to the peptides were close to zero, suggesting there are additional VACV epitopes yet to be identified. In two donors T cell responses to a single peptide constituted >25% of the response to the live VACV- peptide A48R58–66 (A24H1) in donor 2051 (30%) (Figure 1A) and peptide I3L116–124 (A3H10) in donor 2035 (26%) (Figure 1B). In the other donors the highest single peptide responses constituted less than 12% of the ELISPOT response to live VACV.
Table 2. Epitope-specific T cell responses in donors, all of whose HLA-A and HLA-B alleles belonged to one of these six HLA supertypes (HLA-A1, A2, A3, A24, B7 or B44).
| IFN-γ-producing cells / million PBMC |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| donors | live VACV | all peptides | all peptides/VACV | A1 peptides (14 peptides) |
A2a (19) |
A3 (13) |
A24 (5) |
B7 (7) |
B44 (16) |
| 2081 | 161.7 | 138.3 | 85.5% | 17% | 33% | ntb | nt | 42% | 8% |
| 2026 | 76.7 | 8.3 | 10.8% | 40% | nt | 40% | nt | 20% | 0% |
| 2036 | 298.3 | 63.3 | 21.2% | 21% | nt | 24% | nt | 34% | 21% |
| 2012 | 381.7 | 141.7 | 37.1% | nt | 29% | nt | 19% | 2% | 49% |
| 2022 | 118.3 | 28.3 | 23.9% | nt | nt | 0% | 41% | 59% | nt |
| 2057 | 33.3 | 0.0 | 0.0% | ||||||
| 2059 | 413.3 | 80.0 | 19.4% | nt | nt | 98% | 2% | nt | 0% |
| 2035 | 250.0 | 136.7 | 54.7% | nt | nt | 71% | nt | 5% | 24% |
| 2090 | 68.3 | 3.3 | 4.9% | nt | nt | 0% | nt | 100% | 0% |
| 2020 | 113.3 | 15.0 | 13.2% | nt | nt | 56% | nt | 44% | nt |
| 2042 | 738.3 | 392.7 | 53.2% | nt | nt | 77% | nt | 23% | nt |
| 2051 | 301.7 | 303.3 | 100.6% | nt | nt | nt | 43% | 26% | 31% |
| 2055 | 90.0 | 86.7 | 96.3% | 44% | nt | nt | nt | nt | 56% |
| 2084 | 268.3 | 66.7 | 24.2% | nt | 97% | nt | nt | nt | 3% |
: Response to peptide A2H19′, not A2H19, was used for calculation
: Not tested
Figure 1. Epitope-specific T cell responses to VACV in donor 2051 (A) and donor 2035 (B).
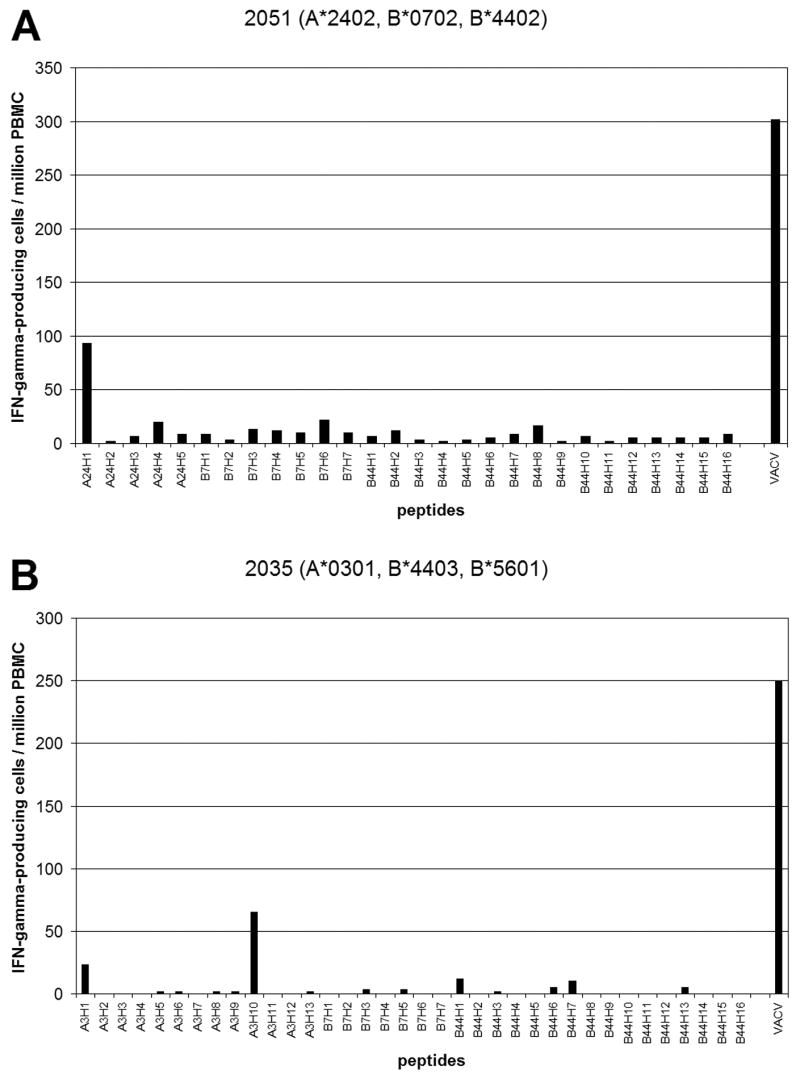
The number of IFN-γ-producing cells specific to each epitope (per 106 PBMC) is shown. Day 0 values were subtracted from the day 45 values. The amino acid sequence of each peptide is shown in Table 1. VACV: The number of IFN-γ-producing cells responding to live VACV.
Epitope-specific T cell responses in donors sharing the same HLA allele or allele belonging to the same HLA supertypes
We tested donors who had at least one HLA allele belonging to the HLA-A1, A2, A3, A24, B7 or B44 supertype (Figures 2, 3, 4, 5, 6A, 6B and 7). The number of IFN-γ-producing cells specific to each epitope was compared among the donors sharing the same HLA alleles or HLA supertypes. When analyzing ELISPOT data, donors whose day 45 PBMC had very low responses to VACV (fewer than 10 per 106 PBMC) were not included. In the analyses below the percentage of the response to a given peptide was calculated using the response to VACV as a denominator, not the sum of the responses to all peptides tested to avoid an overestimation of the “dominant” peptide in donors whose peptide-specific responses were low in general.
Figure 2. Epitope-specific T cell responses in donors sharing HLA-A*0101 allele or an allele belonging to the HLA-A1 supertype.

The number of IFN-γ-producing cells specific to each HLA-A*0101 or HLA-A1 supertype-restricted epitope (per 106 PBMC) is shown. Day 0 values were subtracted from the day 45 values. More detailed data are in supplemental Table S1.
Figure 3. Epitope-specific T cell responses in donors sharing HLA-A*0201 allele or an allele similar to the HLA-A2.
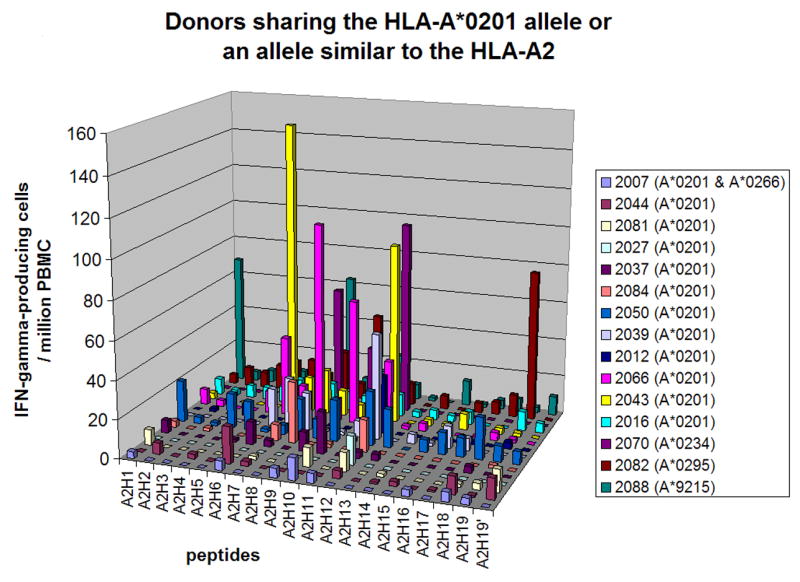
The number of IFN-γ-producing cells specific to each HLA-A*0201- or HLA-A2-restricted epitope (per 106 PBMC) is shown. Day 0 values were subtracted from the day 45 values. More detailed data are in supplemental Table S2.
Figure 4. Epitope-specific T cell responses in donors sharing HLA-A3 allele or an allele belonging to the HLA-A3 supertype.
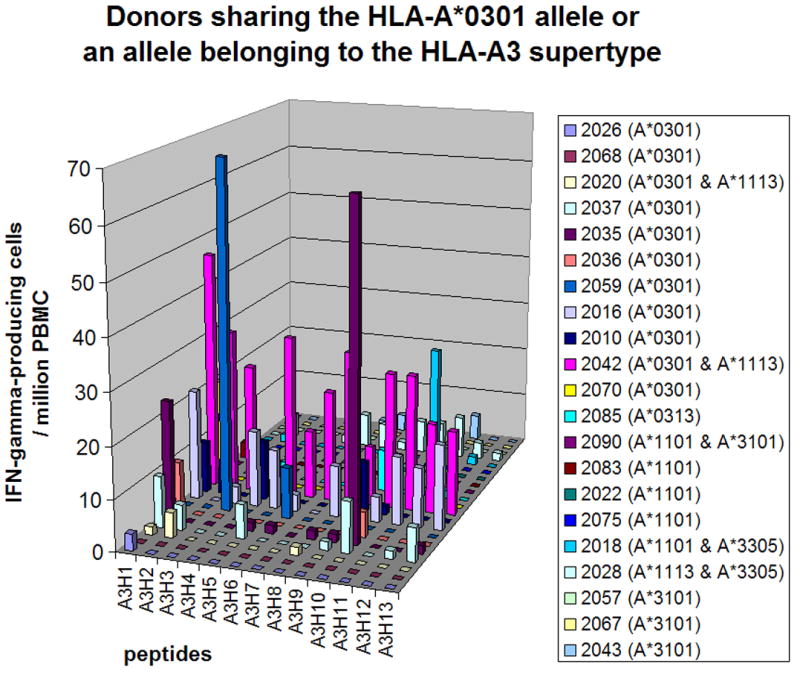
The number of IFN-γ-producing cells specific to each HLA-A3- or HLA-A3 supertype-restricted epitope (per 106 PBMC) is shown. Day 0 values were subtracted from the day 45 values. More detailed data are in supplemental Table S3.
Figure 5. Epitope-specific T cell responses in HLA-A*2402-positive donors.
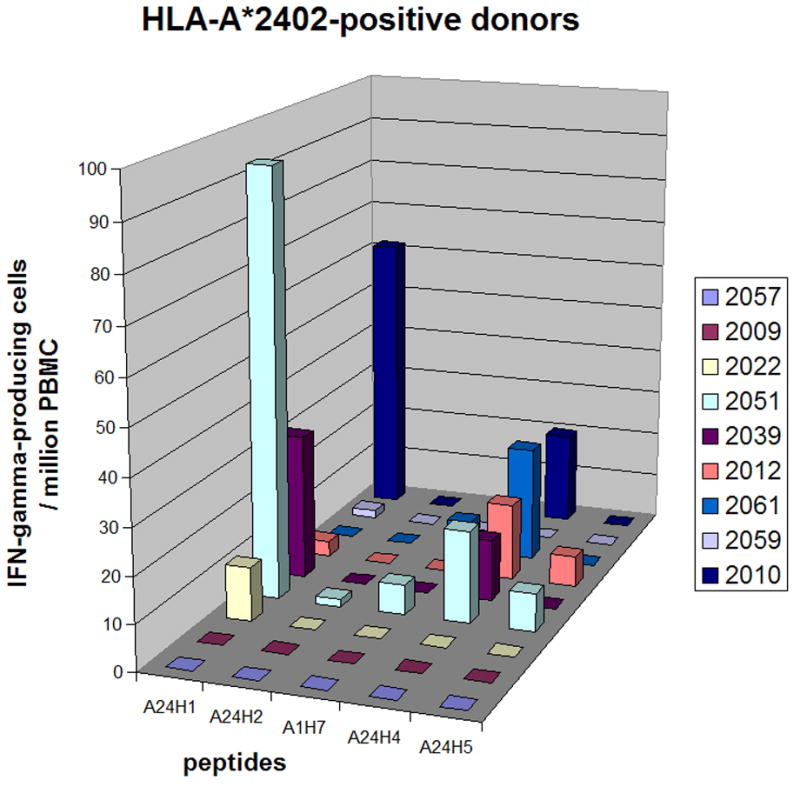
The number of IFN-γ-producing cells specific to each HLA-A*2402- or HLA-A24 supertype-restricted epitope (per 106 PBMC) is shown. All donors were HLA-A*2402-positive. Day 0 values were subtracted from the day 45 values. More detailed data are in supplemental Table S4.
Figure 6. Epitope-specific T cell responses in donors sharing HLA-B*0702 allele (A) or an allele belonging to the HLA-B7 supertype (B).
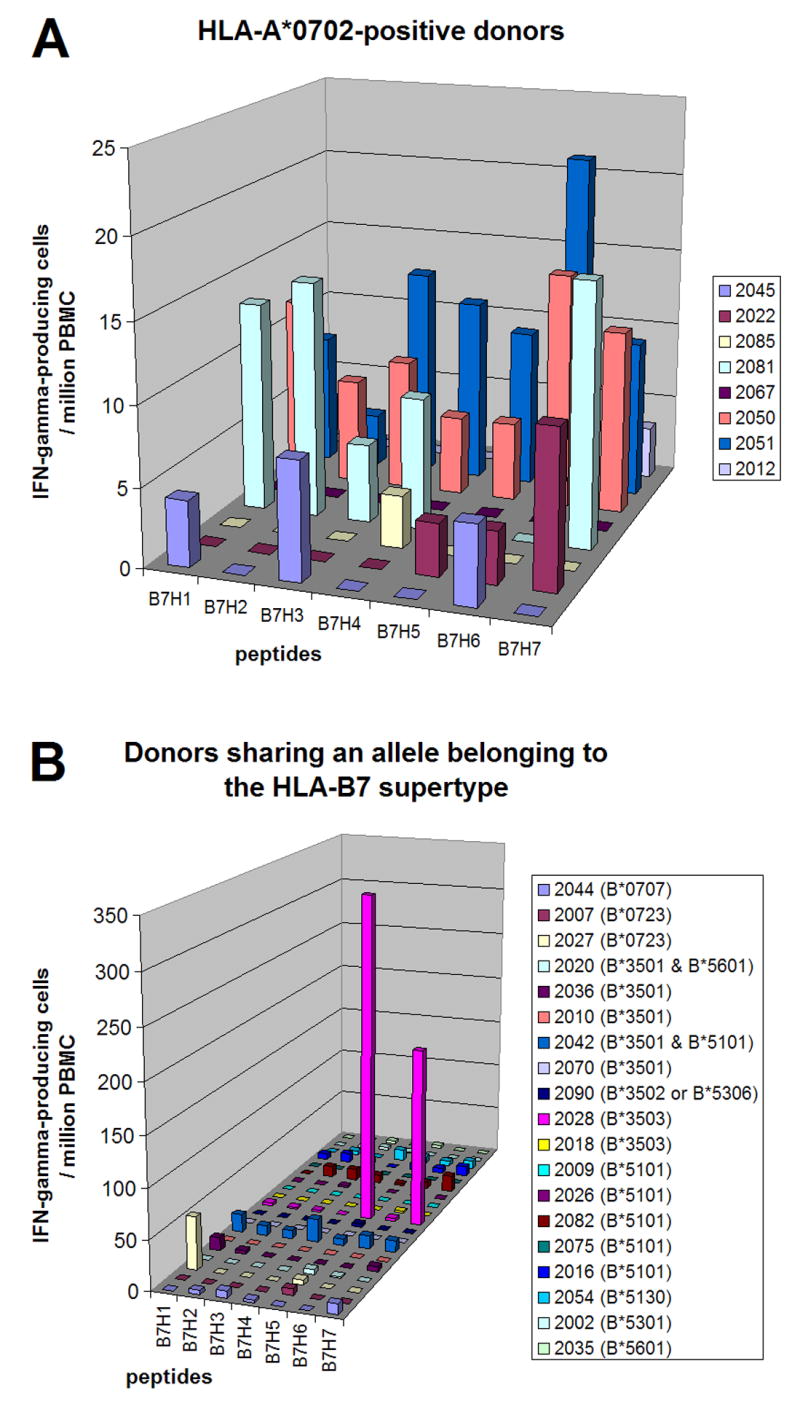
The number of IFN-γ-producing cells specific to each HLA-B7- or HLA-B7 supertype- restricted epitope (per 106 PBMC) is shown. Day 0 values were subtracted from the day 45 values. More detailed data are in supplemental Table S5.
Figure 7. Epitope-specific T cell responses in donors sharing an allele belonging to the HLA-B44 supertype.
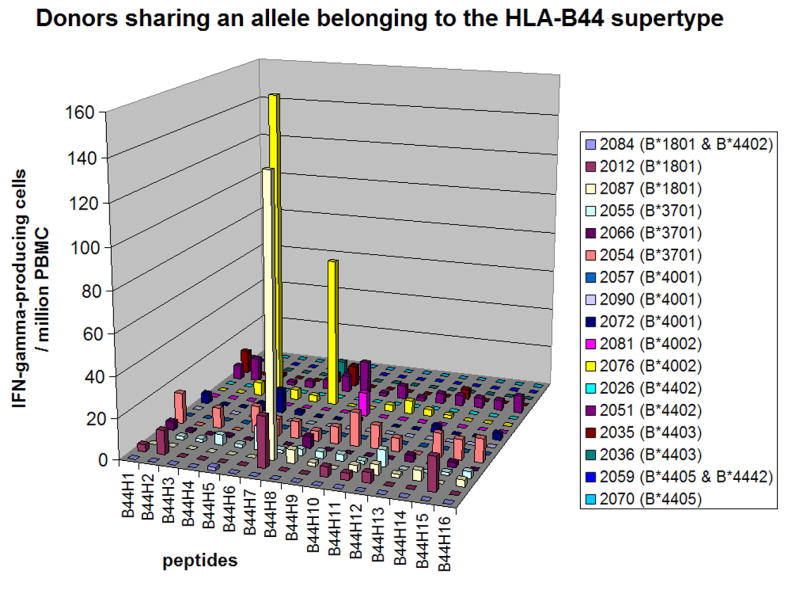
The number of IFN-γ-producing cells specific to each HLA-B44 supertype-restricted epitope (per 106 PBMC) is shown. Day 0 values were subtracted from the day 45 values. More detailed data are in supplemental Table S6.
Donors sharing the HLA-A *0101 allele or an allele belonging to the HLA-A1 supertype
Figure 2 shows the responses to HLA-A1 supertype-restricted epitopes among HLA-A*0101-positive donors and the donors who did not have HLA-A*0101 but had other A1 supertype alleles, such as A*2501, A*2601, A*2902, A*3001, A*3002 and A*3004 (more detailed data are in supplemental Table S1). Although there was no consistent pattern of epitope hierarchy among these donors, some epitopes were recognized more frequently than others. For example, C10L298–306 (A1H6) was recognized by 42% (5/12) of the HLA-A*0101-positive donors and 30% (6/20) of all HLA-A1 supertype-positive donors. All of the other epitopes were recognized by less than 20% of the donors. Among those the HLA-A*2902-restricted C12L326–334 (A1H7) epitope was recognized by both of the donors who were HLA-A*2902 positive as compared to only one out of 12 HLA-A*0101-positive donors and none of the 6 HLA-A1 supertype-positive donors or 9 HLA-A*2402-positive donors (one of whom was also HLA-A*0101-positive) (Figure 5 and Table S4), suggesting that the C12L326–334 epitope is highly specific to HLA-A*2902 and is not presented by other alleles. Except for donor 2081, donors who had HLA-A1 supertype alleles other than A*0101 or A*2902 did not recognize any HLA-A1 supertype-restricted epitopes. The dominant peptide in each donor contributed to an average of 8% (range, 0% -16%) of the responses against VACV among 12 HLA-A*0101-positive donors and 10% (range, 0% - 46%) among the other 8 HLA-A1 supertype-positive donors. The B8R139–147 (A1H2) epitope contributed 46% of the responses against VACV in donor 2003, who was HLA-A*2902- and A*3001-positive.
Donors sharing the HLA-A *0201 allele
Figure 3 shows the responses to HLA-A2 supertype-restricted epitopes among 12 HLA-A*0201-positive donors and three donors who were HLA-A2-positive by low resolution HLA typing but were found to have alleles whose supertype have not yet been assigned (HLA-A*0234, A*0295 and A*9215 [36]) (more detailed data are in supplemental Table S2). Peptide G5R18–26 (A2H12 in Figure 3) was recognized by 58% (7/12) of HLA-A*0201-positive donors, E2L249–257 (A2H8) by 42% (5/12), B22R/C16L60-68 (A2H6) by 33% (4/12), C7L74-86 by 25% (3/12) and F12L286–295 (A2H10) by 25% (3/12). The most dominant peptide in each donor contributed to an average of 14% (range, 3% - 36%) of the responses against VACV. Peptides E2L249–257 (A2H8), G5R18–26 (A2H12), and B22R/C16L60-68 (A2H6) also contributed more than 20% of the responses to VACV at least in one donor. Donors 2070 (HLA-A*0234-positive), 2082 (HLA-A*0295-positive) and 2088 (HLA-A*9215-positive) recognized three to four HLA-A2-restricted epitopes. Peptide B5R5-19 (A2H19), SVVTLLCVLPAVVYS, whose minimal epitope has not been determined [18], was also recognized by donor 2082. The predicted HLA-A2 binding motif in this 15mer peptide, TLLCVLPAV (A2H19′), was not recognized, however.
Donors sharing the HLA-A*0301 allele or an allele belonging to the HLA-A3 supertype
Figure 4 shows the responses to HLA-A3 supertype-restricted epitopes among HLA-A*0301-positive donors and the donors who did not have HLA-A*0301 but had other A3 supertype alleles, such as A*0313, A*1101, A*1113, A*3101 and A*3305 (more detailed data are in supplemental Table S3). Peptide A8R79–88 (A3H1) was recognized by 36% (4/11) of the HLA-A3-positive donors. All of the other epitopes were recognized by less than 20% of the donors. Peptide I3L116–124 (A3H10), which had been identified using PBMC from HLA-A3-positive donors [14] and using splenocytes from immunized HLA-A*1101-transgenic mice [15], was recognized by two HLA-A*0301-positive donors and one HLA-A*1101-positive donor. Donors who had HLA-A3 supertype alleles other than A*0301 or A*1101 did not recognize any HLA-A3 supertype-restricted epitopes. The most dominant peptide in each donor contributed to an average of 7% (range, 0% - 26%) of the responses against VACV among 11 HLA-A*0301-positive donors and 2% (range, 0% - 7%) among the other 10 HLA-A3 supertype-positive donors. Peptide I3L116–124 (A3H10) contributed 26% of the responses to VACV in donor 2035 (described in the previous section).
Donors sharing the HLA-A*2402 allele
Figure 5 shows the responses to HLA-A24 supertype-restricted epitopes among HLA-A*2402-positive donors (more detailed data are in supplemental Table S4). In these donors HLA-A24-restricted T cell responses were directed against either A48R58–66 (A24H1) or D5R349–357 (A24H4), or both. None of the other three epitopes was recognized. Peptide A48R58–66 was HLA-A*2301-restricted and three donors who recognized this epitope were HLA-A*2402-positive, not A*2301-positive, suggesting that HLA-A*2301 does belong to the HLA-A24 supertype. The most dominant peptide in each donor contributed to an average of 8% (range, 0% - 31%) of the responses against VACV. Peptide A48R58–66 (A24H1) contributed 31% of the responses to VACV in donor 2051 (described in the previous section).
Donors sharing the HLA-B*0702 allele or an allele belonging to the HLA-B7 supertype
We analyzed 8 HLA-B*0702-positive donors and 19 donors who did not have HLA-B*0702 but had other B7 supertype alleles, such as B*0707, B*0723, B*3501, B*3502, B*3503, B*5101, B*5130, B*5301 and B*5601. Figure 6A and 6B shows the responses to HLA-B7 supertype-restricted epitopes (more detailed data are in supplemental Table S5). Peptide J6R303-311 (B7H6) was recognized by 25% (2/8) of the HLA-B*0702-positive donors. Donors positive for HLA-B*0723, B*3501, B*3503 and B*5101 recognized one to two HLA-B7 supertype-restricted epitopes. The most dominant peptide in each donor contributed to an average of 14% (range, 0% - 236%) of the responses against VACV among all 27 donors. One donor (2028) was an outlier in this analysis, with T cell responses to J2R62-70 (B7H5) and O1L335–344 (B7H7) far higher than the responses against VACV (236% and 129%, respectively). Excluding this donor the average was 5% (range, 0% - 32%). Peptide B22R/C16L53-61 (B7H1) contributed 32% of the responses to VACV in donor 2027.
Donors sharing alleles belonging to the HLA-B44 supertype
We analyzed 17 donors sharing an allele belonging to the HLA-B44 supertype, such as B*1801, B*3701, B*4001, B*4002, B*4402, B*4403, B*4405 and B*4442. Figure 7 shows the responses to HLA-B44 supertype-restricted epitopes (more detailed data are in supplemental Table S6). Peptide D1R126-134 (B44H7) was recognized by 67% (2/3) of donors positive for HLA-B*1801. This epitope had been identified with HLA-B*4403-positive donor's PBMC [16], however, it was not recognized by PBMC from HLA-B*4403-positive donors in this study. There were no other epitope peptides recognized by more than 20% of these donors. This may reflect the fact that most HLA-B44-restricted epitopes tested (Table 1) were identified by using HLA-B*4403-positive donor's PBMC and there were only two HLA-B*4403-positive donors in our study. Donors positive for HLA-B*3701, B*4002 and B*4402 recognized one to two HLA-B44 supertype-restricted epitopes. The most dominant peptide in each donor contributed to an average of 5% (range, 0% - 27%) of the response against VACV. Peptide D1R126–134 (B44H7) also contributed 27% of the responses to VACV in donor 2087.
Discussion
In contrast to genetically homogeneous laboratory mice, immunodominance of T cell epitopes in humans is less clear [25], with some exceptions. Almost all HLA-B8-positive subjects responded to all three HLA-B8-restricted EBV epitopes when they were infected with EBV [38]. All HLA-A2-positive subjects responded to the HLA-A2-restricted epitope encoded by influenza A matrix protein 1 (M158-66) [39]. Boon et al. also reported that the epitope specificity of the CTL response is influenced by the phenotype of the other HLA molecules [39]. In VACV infection of HLA-transgenic mice epitope-specific CTL responses restricted by the human MHC class I molecule were also influenced by the presence of mouse MHC class I alleles [15].
In our study there were no consistent patterns of epitope hierarchy among donors sharing the same HLA allele or HLA supertypes, except for HLA-A*2402. It is interesting that CD8+ T cell responses to VACV and EBV, double-stranded DNA viruses with similar genome sizes [40, 41], differ in terms of epitope immunodominance. However, some epitopes were recognized more often than others. Twelve epitopes, C10L298–306, C12L326–334, G5R18–26, E2L249–257, B22R/C16L60-68, C7L74-86, F12L286–295, A8R79–88, D5R349–357, A48R58–66, J6R303-311 and D1R126-134, were recognized by more than 20% of the donors sharing the same HLA allele (at least by two donors), and six of them (shown in bold) contributed to more than 20 % of the total VACV responses of at least one donor. Five other epitopes, B8R139–147, I3L116-124, J2R62-70, O1L335–344, and B22R/C16L53-61, also contributed to more than 20 % of the total VACV responses of at least one donor (summarized in Table 3). These results suggest that there are no strong immunodominant epitopes in VACV, as previously proposed [14], but there are groups of “relatively dominant” epitopes restricted by the same HLA allele. Alternatively, authentic immunodominant epitopes may be still unidentified. Considering that there were only two epitopes restricted by HLA-B7 or B44 recognized by more than 20% of the donors, many more epitopes need to be identified to perform this type of analysis for these HLA-B alleles. Another caveat of this study is that these PBMC donors received one of three smallpox vaccines, Dryvax®, ACAM 1000 and ACAM2000, which are very similar, but not identical. Although almost all CD8+ T cell epitopes analyzed in this study are identical among the four genomic sequences of the Dryvax® and the derivatives from the Dryvax®, there is a possibility that amino acid differences outside of the epitope peptides affect the processing of the epitopes [42-44].
Table 3. “Relatively dominant” epitopes.
| peptide name in the figures | gene name and (a.a. position) | restricting allele | recognized by | Temporal Expression |
|---|---|---|---|---|
| A1H6 | C10L (298–306) | A1 supertype | 42% (5/12) of A*0101 (+) donors | early |
| A1H7 | C12L (326–334) | A*2902 | 100% (2/2) of A*2902 (+) donors | early |
| A2H12 | G5R (18–26) | A2 supertype | 58% (7/12) of A*0201 (+) donors | immediate-early |
| A2H8 | E2L (249–257) | A2 supertype | 42% (5/12) of A*0201 (+) donors | early |
| A2H6 | B22R/C16L (60-68) | A*0201 | 33% (4/12) of A*0201 (+) donors | ?a |
| A2H7 | C7L (74–82) | A*0201 | 25% (3/12) of A*0201 (+) donors | early |
| A2H10 | F12L (286–295) | A2 supertype | 25% (3/12) of A*0201 (+) donors | early |
| A3H1 | A8R (79–88) | A3 supertype | 36% (4/11) of A*0301 (+) donors | immediate-early |
| A24H4 | D5R (349–357) | A24 supertype | 44% (4/9) of A*2402 (+) donors | early |
| A24H1 | A48R (58–66) | A*2301 | 33% (3/9) of A*2402 (+) donors | immediate-early |
| B7H6 | J6R (303-311) | B7 or B7 supertype | 25% (2/8) of B*0702 (+) donors | early |
| B44H7 | D1R (126–134) | B*4403 | 67% (2/3) of B*1801 (+) donors | early |
| A1H2 | B8R (139–147) | A1 supertype | 46% of VACV-specific cells of A*2902 & A*3001 (+) donor | early |
| A2H10 | F12L (286–295) | A2 supertype | 38% of VACV-specific cells of A*0295 (+) donorb | |
| A2H6 | B22R/C16L (60-68) | A*0201 | 36% of VACV-specific cells of A*0201 (+) donor | |
| A2H12 | E2L (249–257) | A2 supertype | 25% of VACV-specific cells of A*0201 (+) donor | |
| 21% of VACV-specific cells of A*0295 (+) donorb | ||||
| A2H8 | G5R (18–26) | A2 supertype | 22% of VACV-specific cells of A*0201 (+) donor | |
| A3H1 | I3L (116–124) | A3 & A*1101 | 26% of VACV-specific cells of A*0301 (+) donor | immediate-early |
| A24H1 | A48R (58–66) | A*2301 | 31% of VACV-specific cells of A*2402 (+) donor | |
| B7H5 | J2R (62-70) | B*3502 | 236% of VACV-specific cells of B*3503 (+) donor | early |
| B7H7 | O1L (335–344) | B7 supertype | 129% of VACV-specific cells of B*3503 (+) donor | immediate-early |
| B7H1 | B22R/C16L (53-61) | B7 | 32% of VACV-specific cells of B*0723 (+) donor | ?a |
| B44H7 | D1R (126–134) | B*4403 | 27% of VACV-specific cells of B*1801 (+) donor |
: B22R/C16L was not analyzed
: HLA supertype asignment of A*0295 has not yet been determined.
In this study we used PBMC drawn at day 45 after vaccination, which was the only postvaccination time point available. VACV-specific CD8+ T cell responses peak at about two weeks after vaccination, and then start to decline [12, 45]. If earlier PBMCs samples had been analyzed, we might have detected more epitope peptides positive in our assays. However, epitopes that are positive only at earlier time points and become negative by day 45 are not likely to be important for immunological memory against VACV.
In previous reports [12, 17] we hypothesized that early gene products may be more likely to have CD8+ T cell epitopes, since in both humans and mice all of the then-known CD8+ T cell epitopes to cytomegalovirus were encoded by immediate-early phase proteins [46, 47], and we biased our screening toward the early gene products. Other groups took less biased and more comprehensive approaches to identify CD8+ T cell epitopes to VACV [14, 16]. The majority of identified epitopes are, however, encoded by early genes. Recently Assarsson et al. analyzed the complete VACV transcriptome and proposed to separate immediate-early genes from early genes, categorizing the VACV open reading frames into four classes of genes, immediate-early, early, early/late and late genes [48]. Of the 73 epitopes analyzed in this study, epitopes encoded by genes which belong to the immediate-early and the early class were overrepresented (p<0.0000005 by chi-square analysis). Three epitopes, G8R65–73, B22R/C16L53-61 and B22R/C16L60-68, were not included in the statistical analysis, because the G8R gene did not fit any class and the temporal expression pattern of B22R/C16L was not experimentally determined in the report due to the lack of conservation of the B22R/C16L gene in the WR strain (Table 4). Among the 17 “relatively dominant” epitopes, five are encoded by immediate-early genes and ten are encoded by early genes; the other two are encoded by the B22R/C16L gene. Immediate-early and early gene products were overrepresented in the “relatively dominant” epitopes in comparison to all 73 epitopes (p<0.05 by chi-square analysis) (Table 4). This may mean that the proteins expressed earlier can compete against the other proteins for MHC class I binding.
Table 4. Epitopes and temporal expression of the proteins encoding the epitopes.
| immediate-early | early | early/late | late | ?a | total | |
|---|---|---|---|---|---|---|
| all ORFs (WR) | 35 | 73 | 26 | 60 | - | 194 |
| all epitopes | 13 | 45 | 4 | 8 | 3 | 73 |
| relatively dominant epitopes | 5 | 10 | 0 | 0 | 2 | 17 |
| other epitopes | 8 | 35 | 4 | 8 | 1 | 58 |
: G8R gene did not fit any class and B22R/C16L was not analyzed [48]
HLA-transgenic mice lacking endogenous murine class I MHC protein expression have been used to model human MHC class I-restricted CTL responses and are useful to identify CD8+ T cell epitopes restricted by human MHC class I molecules (reviewed in [49]). In influenza A virus infection the “immunodominant” matrix protein 1 epitope restricted by HLA-A2.1 in humans was also dominant in these HLA-A2.1-transgenic mice [50, 51], which was also the case with a hepatitis B virus epitope [52]. In comparison, only four of 23 VACV CD8+ T cell epitopes detected in HLA-transgenic mice have also been detected by human PBMC, C7L74–82, G5R18–26 and H3L184-192 restricted by HLA-A*0201 and I3L116–124 restricted by HLA-A3 (human) or HLA-A*1101 (mouse) [11, 14, 15, 30, 53]. Three of these four epitopes, C7L74–82, G5R18–26 and I3L116–124, are among the 17 relatively dominant epitopes. This may mean that when an epitope is recognized by both human samples and HLA-transgenic mice, the epitope has a good chance to be a relatively dominant epitope. However, the overall utility of using HLA-transgenic mice as a guide for detecting human T cell responses to VACV appears to be low. In contrast to the epitopes identified using human PBMCs, especially “relatively dominant” epitopes, late gene products were well-represented in the epitopes identified by using HLA-transgenic mice [15]. As we suggested previously [30], when a virus is large, such as VACV, and encodes many high binding peptides to human MHC class I molecules, slight differences in antigen processing, antigen presentation, and TCR repertoire between human and transgenic mouse cells may influence epitope selection significantly.
Our analysis also reveals useful information regarding the practicality of the concept of HLA supertypes and supermotifs. T cells specific to the HLA-A*2301-restricted A48R58–66 epitope were detected in three of nine donors who were HLA-A*2402-positive and not A*2301-positive. Three donors, who were HLA-A*0234-, A*0295-, or A*9215-positive, respectively (HLA supertype assignment of these alleles have not yet been determined), also recognized several HLA-A2-restricted epitopes. Besides these HLA-A24 supertype- or A2 supertype-restricted epitopes, there were no epitopes recognized in the context of multiple HLA alleles in the supertype. Therefore, cross-reactivity among the alleles belonging to the same HLA supertype may be epitope-dependent, or the degree of similarity of the peptide binding motif among the alleles belonging to the same HLA supertype may vary, e.g. high for the HLA-A24 and A2 supertypes and low for the others. It was previously reported that three HLA-B*0702-restricted VACV-specific epitopes (identified using HLA-B*0702-transgenic mice) did not bind to other B7 supertype molecules, such as B*3501, B*5101, B*5301 and B*5401 [15].
In a previous report [17] we suggested that it may be useful to quantitate VACV epitope-specific CD8+ T cell responses as a surrogate marker for a “take”, when a smallpox vaccine does not induce a pox lesion, such as with the MVA strain. At least in HLA-A*0201 or HLA-A*2402-positive individuals it is probably feasible to use groups of epitopes to monitor CD8+ T cell responses to MVA, or to compare CD8+ T cell responses to the vector and the transgene product after immunization with recombinant VACV.
In this study we analyzed the VACV-specific CD8+ T cell epitope hierarchy using PBMC from donors who received primary VACV immunization. VACV is not a natural human pathogen. But as a closely related poxvirus, infection by VACV induces solid protection against smallpox, and most CD8+ T cell epitopes and proteins encoding the CD8+ T cell epitopes are conserved in variola (smallpox) viruses (summarized in [22]). Smallpox is thought to have emerged as human disease about 5000 years ago [2]. The exposure of humans to smallpox is, therefore, relatively limited in time, and evolutionary pressures would support survival of some hosts for periods of time to allow virus replication to proceed and virus dissemination to susceptible contacts to occur. CD8+ T cells presumably help to reduce the viral load and clear infections allowing most smallpox victims to survive and produce offspring who would become susceptible to infection. It would be an advantage to the survival of infected humans if the CD8+ T cell responses were numerous and “redundant”. In the case of a mutation at a CD8+ T cell epitope multiple other CD8+ T cells responses remain and could eliminate infected cells. Similar redundancy was reported very recently for the neutralizing antibody responses against VACV in humans [54].
Supplementary Material
Acknowledgments
We thank Christine Turcotte and Denise Marengo for assistance with HLA typing and Gail Mazzara and Dennis Panicali of Applied Biotechnology, Inc. for VACV NYCBH strain. We thank Acambis, Inc for providing PBMC samples from vaccine recipients and Dr. Thomas Monath for supporting our efforts to define human T cell epitopes in VACV. We thank Dr. Jeffrey S. Kennedy for assisting us in obtaining these PBMC samples. This work was supported by the National Institute of Allergy and Infectious Diseases/National Institute of Health grant U19 AI-057319.
List of abbreviations used
- VACV
vaccinia virus
- NYCBH
New York City Board of Health strain
- MVA
modified vaccinia Ankara strain
- WR
Western Reserve strain
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fenner F, Henderson DA, Arita I, Jezek Z, Ladnyi ID. Smallpox and its Eradication. Geneza, Switzerland: The World Health Organization; 1988. [Google Scholar]
- 2.Esposito JJ, Fenner F. Poxviruses. In: Knipe DM, Howley PM, editors. Fields Virology. Vol. 2. Philadelphia: Lippincott Wiliams & Wilkins; 2001. p. 2885. [Google Scholar]
- 3.Fulginiti VA, Kempe CH, Hathaway WE, Pearlman DS, Serber OF, Jr, Eller JJ, Joyner JJ, Sr, Robinson A. Progressive vaccinia in immunologically deficient individuals. Birth defects original article series. 1968;4:129. [Google Scholar]
- 4.Kempe CH. Acceptance of the Howland Award. Pediatr Res. 1980;14(11):1155. doi: 10.1203/00006450-198011000-00002. [DOI] [PubMed] [Google Scholar]
- 5.Freed ER, Duma RJ, Escobar MR. Vaccinia necrosum and its relationship to impaired immunologic responsiveness. Am J Med. 1972;52(3):411. doi: 10.1016/0002-9343(72)90031-9. [DOI] [PubMed] [Google Scholar]
- 6.Hansson O, Johansson SGO, Vahlquist B. Vaccinia gangrenosa with normal humora antibodies. A case possibly due to deficient cellular immunity treated with N-methylisatin β-thiosemicarbazone (compound 33T57, Marboran) Acta paediatrica scandinavica. 1966;55:264. doi: 10.1111/j.1651-2227.1966.tb17653.x. [DOI] [PubMed] [Google Scholar]
- 7.Henderson DA, Inglesby TV, Bartlett JG, Ascher MS, Eitzen E, Jahrling PB, Hauer J, Layton M, McDade J, Osterholm MT, O'Toole T, Parker G, Perl T, Russell PK, Tonat K. Smallpox as a biological weapon: medical and public health management. Working Group on Civilian Biodefense. Jama. 1999;281(22):2127. doi: 10.1001/jama.281.22.2127. [DOI] [PubMed] [Google Scholar]
- 8.Rosenthal SR, Merchlinsky M, Kleppinger C, Goldenthal KL. Developing new smallpox vaccines. Emerg Infect Dis. 2001;7(6):920. doi: 10.3201/eid0706.010602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carroll MW, Moss B. Poxviruses as expression vectors. Curr Opin Biotechnol. 1997;8(5):573. doi: 10.1016/s0958-1669(97)80031-6. [DOI] [PubMed] [Google Scholar]
- 10.Shen Y, Nemunaitis J. Fighting cancer with vaccinia virus: teaching new tricks to an old dog. Mol Ther. 2005;11(2):180. doi: 10.1016/j.ymthe.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 11.Drexler I, Staib C, Kastenmuller W, Stevanovic S, Schmidt B, Lemonnier FA, Rammensee HG, Busch DH, Bernhard H, Erfle V, Sutter G. Identification of vaccinia virus epitope-specific HLA-A*0201-restricted T cells and comparative analysis of smallpox vaccines. Proc Natl Acad Sci U S A. 2003;100(1):217. doi: 10.1073/pnas.262668999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Terajima M, Cruz J, Raines G, Kilpatrick ED, Kennedy JS, Rothman AL, Ennis FA. Quantitation of CD8+ T cell responses to newly identified HLA-A*0201-restricted T cell epitopes conserved among vaccinia and variola (smallpox) viruses. J Exp Med. 2003;197(7):927. doi: 10.1084/jem.20022222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Snyder JT, Belyakov IM, Dzutsev A, Lemonnier F, Berzofsky JA. Protection against lethal vaccinia virus challenge in HLA-A2 transgenic mice by immunization with a single CD8+ T-cell peptide epitope of vaccinia and variola viruses. J Virol. 2004;78(13):7052. doi: 10.1128/JVI.78.13.7052-7060.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oseroff C, Kos F, Bui HH, Peters B, Pasquetto V, Glenn J, Palmore T, Sidney J, Tscharke DC, Bennink JR, Southwood S, Grey HM, Yewdell JW, Sette A. HLA class I-restricted responses to vaccinia recognize a broad array of proteins mainly involved in virulence and viral gene regulation. Proc Natl Acad Sci U S A. 2005;102(39):13980. doi: 10.1073/pnas.0506768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pasquetto V, Bui HH, Giannino R, Mirza F, Sidney J, Oseroff C, Tscharke DC, Irvine K, Bennink JR, Peters B, Southwood S, Cerundolo V, Grey H, Yewdell JW, Sette A. HLA-A*0201, HLA-A*1101, and HLA-B*0702 Transgenic Mice Recognize Numerous Poxvirus Determinants from a Wide Variety of Viral Gene Products. J Immunol. 2005;175(8):5504. doi: 10.4049/jimmunol.175.8.5504. [DOI] [PubMed] [Google Scholar]
- 16.Jing L, Chong TM, McClurkan CL, Huang J, Story BT, Koelle DM. Diversity in the acute CD8 T cell response to vaccinia virus in humans. J Immunol. 2005;175(11):7550. doi: 10.4049/jimmunol.175.11.7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terajima M, Cruz J, Leporati AM, Demkowicz WE, Jr, Kennedy JS, Ennis FA. Identification of vaccinia CD8+ T-cell epitopes conserved among vaccinia and variola viruses restricted by common MHC class I molecules, HLA-A2 or HLA-B7. Hum Immunol. 2006;67(7):512. doi: 10.1016/j.humimm.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 18.Tang J, Murtadha M, Schnell M, Eisenlohr LC, Hooper J, Flomenberg P. Human T-cell responses to vaccinia virus envelope proteins. J Virol. 2006;80(20):10010. doi: 10.1128/JVI.00601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sahin U, Tureci O, Graf C, Meyer RG, Lennerz V, Britten CM, Dumrese C, Scandella E, Wolfel T, Ludewig B. Rapid molecular dissection of viral and bacterial immunomes. Eur J Immunol. 2006;36(4):1049. doi: 10.1002/eji.200535538. [DOI] [PubMed] [Google Scholar]
- 20.Ostrout ND, McHugh MM, Tisch DJ, Moormann AM, Brusic V, Kazura JW. Long-term T cell memory to human leucocyte antigen-A2 supertype epitopes in humans vaccinated against smallpox. Clin Exp Immunol. 2007;149(2):265. doi: 10.1111/j.1365-2249.2007.03401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer RG, Graf C, Britten CM, Huber C, Wolfel T. Rapid identification of an HLA-B*1501-restricted vaccinia peptide antigen. Vaccine. 2007;25(24):4715. doi: 10.1016/j.vaccine.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 22.Kennedy R, Poland GA. T-Cell epitope discovery for variola and vaccinia viruses. Rev Med Virol. 2007;17(2):93. doi: 10.1002/rmv.527. [DOI] [PubMed] [Google Scholar]
- 23.Yewdell JW, Del Val M. Immunodominance in TCD8+ responses to viruses: cell biology, cellular immunology, and mathematical models. Immunity. 2004;21(2):149. doi: 10.1016/j.immuni.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 24.Tscharke DC, Karupiah G, Zhou J, Palmore T, Irvine KR, Haeryfar SM, Williams S, Sidney J, Sette A, Bennink JR, Yewdell JW. Identification of poxvirus CD8+ T cell determinants to enable rational design and characterization of smallpox vaccines. J Exp Med. 2005;201(1):95. doi: 10.1084/jem.20041912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yewdell JW. Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity. 2006;25(4):533. doi: 10.1016/j.immuni.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 26.Weltzin R, Liu J, Pugachev KV, Myers GA, Coughlin B, Blum PS, Nichols R, Johnson C, Cruz J, Kennedy JS, Ennis FA, Monath TP. Clonal vaccinia virus grown in cell culture as a new smallpox vaccine. Nat Med. 2003;9(9):1125. doi: 10.1038/nm916. [DOI] [PubMed] [Google Scholar]
- 27.Monath TP, Caldwell JR, Mundt W, Fusco J, Johnson CS, Buller M, Liu J, Gardner B, Downing G, Blum PS, Kemp T, Nichols R, Weltzin R. ACAM2000 clonal Vero cell culture vaccinia virus (New York City Board of Health strain)--a second-generation smallpox vaccine for biological defense. Int J Infect Dis. 2004;8 2:S31. doi: 10.1016/j.ijid.2004.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terajima M, Van Epps HL, Li D, Leporati AM, Juhlin SE, Mustonen J, Vaheri A, Ennis FA. Generation of recombinant vaccinia viruses expressing Puumala virus proteins and use in isolating cytotoxic T cells specific for Puumala virus. Virus Res. 2002;84(12):67. doi: 10.1016/s0168-1702(01)00416-6. [DOI] [PubMed] [Google Scholar]
- 29.Ennis FA, Cruz J, Demkowicz WE, Jr, Rothman AL, McClain DJ. Primary induction of human CD8+ cytotoxic T lymphocytes and interferon-gamma-producing T cells after smallpox vaccination. J Infect Dis. 2002;185(11):1657. doi: 10.1086/340517. [DOI] [PubMed] [Google Scholar]
- 30.Terajima M, Ennis FA. Using HLA-transgenic mice to identify immunodominant human CD8+ T cell epitopes--does (genome) size matter? Immunol Lett. 2006;105(1):97. doi: 10.1016/j.imlet.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 31.Antoine G, Scheiflinger F, Dorner F, Falkner FG. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology. 1998;244(2):365. doi: 10.1006/viro.1998.9123. [DOI] [PubMed] [Google Scholar]
- 32.Li G, Chen N, Feng Z, Buller RM, Osborne J, Harms T, Damon I, Upton C, Esteban DJ. Genomic sequence and analysis of a vaccinia virus isolate from a patient with a smallpox vaccine-related complication. Virol J. 2006;3:88. doi: 10.1186/1743-422X-3-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sette A, Sidney J. HLA supertypes and supermotifs: a functional perspective on HLA polymorphism. Curr Opin Immunol. 1998;10(4):478. doi: 10.1016/s0952-7915(98)80124-6. [DOI] [PubMed] [Google Scholar]
- 34.Sette A, Sidney J. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics. 1999;50(34):201. doi: 10.1007/s002510050594. [DOI] [PubMed] [Google Scholar]
- 35.Sidney J, Southwood S, Sette A. Classification of A1- and A24-supertype molecules by analysis of their MHC-peptide binding repertoires. Immunogenetics. 2005;57(6):393. doi: 10.1007/s00251-005-0004-2. [DOI] [PubMed] [Google Scholar]
- 36.Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 2008;9:1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith CL, Mirza F, Pasquetto V, Tscharke DC, Palmowski MJ, Dunbar PR, Sette A, Harris AL, Cerundolo V. Immunodominance of poxviral-specific CTL in a human trial of recombinant-modified vaccinia Ankara. J Immunol. 2005;175(12):8431. doi: 10.4049/jimmunol.175.12.8431. [DOI] [PubMed] [Google Scholar]
- 38.Woodberry T, Suscovich TJ, Henry LM, Davis JK, Frahm N, Walker BD, Scadden DT, Wang F, Brander C. Differential targeting and shifts in the immunodominance of Epstein-Barr virus--specific CD8 and CD4 T cell responses during acute and persistent infection. J Infect Dis. 2005;192(9):1513. doi: 10.1086/491741. [DOI] [PubMed] [Google Scholar]
- 39.Boon AC, de Mutsert G, Graus YM, Fouchier RA, Sintnicolaas K, Osterhaus AD, Rimmelzwaan GF. The magnitude and specificity of influenza A virus-specific cytotoxic T-lymphocyte responses in humans is related to HLA-A and -B phenotype. J Virol. 2002;76(2):582. doi: 10.1128/JVI.76.2.582-590.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moss B. Poxviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Vol. 2. Philadelphia: Lippincott Wiliams & Wilkins; 2001. p. 2849. [Google Scholar]
- 41.Kieff E, Rickinson AB. Epstein-Barr Virus and Its Replication. In: Knipe DM, Howley PM, editors. Fields Virology. Vol. 2. Philadelphia: Lippincott Williams & Wilkins; 2001. p. 2511. [Google Scholar]
- 42.Del Val M, Schlicht HJ, Ruppert T, Reddehase MJ, Koszinowski UH. Efficient processing of an antigenic sequence for presentation by MHC class I molecules depends on its neighboring residues in the protein. Cell. 1991;66(6):1145. doi: 10.1016/0092-8674(91)90037-y. [DOI] [PubMed] [Google Scholar]
- 43.Yellen-Shaw AJ, Wherry EJ, Dubois GC, Eisenlohr LC. Point mutation flanking a CTL epitope ablates in vitro and in vivo recognition of a full-length viral protein. J Immunol. 1997;158(7):3227. [PubMed] [Google Scholar]
- 44.Yellen-Shaw AJ, Eisenlohr LC. Regulation of class I-restricted epitope processing by local or distal flanking sequence. J Immunol. 1997;158(4):1727. [PubMed] [Google Scholar]
- 45.Miller JD, van der Most RG, Akondy RS, Glidewell JT, Albott S, Masopust D, Murali-Krishna K, Mahar PL, Edupuganti S, Lalor S, Germon S, Del Rio C, Mulligan MJ, Staprans SI, Altman JD, Feinberg MB, Ahmed R. Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity. 2008;28(5):710. doi: 10.1016/j.immuni.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 46.Reddehase MJ, Koszinowski UH. Significance of herpesvirus immediate early gene expression in cellular immunity to cytomegalovirus infection. Nature. 1984;312(5992):369. doi: 10.1038/312369a0. [DOI] [PubMed] [Google Scholar]
- 47.Reddehase MJ. The immunogenicity of human and murine cytomegaloviruses. Curr Opin Immunol. 2000;12(4):390. doi: 10.1016/s0952-7915(00)00106-0. [DOI] [PubMed] [Google Scholar]
- 48.Assarsson E, Greenbaum JA, Sundstrom M, Schaffer L, Hammond JA, Pasquetto V, Oseroff C, Hendrickson RC, Lefkowitz EJ, Tscharke DC, Sidney J, Grey HM, Head SR, Peters B, Sette A. Kinetic analysis of a complete poxvirus transcriptome reveals an immediate-early class of genes. Proc Natl Acad Sci U S A. 2008;105(6):2140. doi: 10.1073/pnas.0711573105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pascolo S. HLA class I transgenic mice: development, utilisation and improvement. Expert Opin Biol Ther. 2005;5(7):919. doi: 10.1517/14712598.5.7.919. [DOI] [PubMed] [Google Scholar]
- 50.Vitiello A, Marchesini D, Furze J, Sherman LA, Chesnut RW. Analysis of the HLA-restricted influenza-specific cytotoxic T lymphocyte response in transgenic mice carrying a chimeric human-mouse class I major histocompatibility complex. J Exp Med. 1991;173(4):1007. doi: 10.1084/jem.173.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pascolo S, Bervas N, Ure JM, Smith AG, Lemonnier FA, Perarnau B. HLA-A2.1-restricted education and cytolytic activity of CD8(+) T lymphocytes from beta2 microglobulin (beta2m) HLA-A2.1 monochain transgenic H-2Db beta2m double knockout mice. J Exp Med. 1997;185(12):2043. doi: 10.1084/jem.185.12.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pajot A, Michel ML, Fazilleau N, Pancre V, Auriault C, Ojcius DM, Lemonnier FA, Lone YC. A mouse model of human adaptive immune functions: HLA-A2.1-/HLA-DR1-transgenic H-2 class I-/class II-knockout mice. Eur J Immunol. 2004;34(11):3060. doi: 10.1002/eji.200425463. [DOI] [PubMed] [Google Scholar]
- 53.Di Nicola M, Carlo-Stella C, Mortarini R, Baldassari P, Guidetti A, Gallino GF, Del Vecchio M, Ravagnani F, Magni M, Chaplin P, Cascinelli N, Parmiani G, Gianni AM, Anichini A. Boosting T cell-mediated immunity to tyrosinase by vaccinia virus-transduced, CD34(+)-derived dendritic cell vaccination: a phase I trial in metastatic melanoma. Clin Cancer Res. 2004;10(16):5381. doi: 10.1158/1078-0432.CCR-04-0602. [DOI] [PubMed] [Google Scholar]
- 54.Benhnia MR, McCausland MM, Su HP, Singh K, Hoffmann J, Davies DH, Felgner PL, Head S, Sette A, Garboczi DN, Crotty S. Redundancy and plasticity of neutralizing antibody responses are cornerstone attributes of the human immune response to the smallpox vaccine. J Virol. 2008;82(7):3751. doi: 10.1128/JVI.02244-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dong Y, Denny TN. HLA-A2-restricted human CD8(+) cytotoxic T lymphocyte responses to a novel epitope in vaccinia virus that is conserved among orthopox viruses. J Infect Dis. 2006;194(2):168. doi: 10.1086/505224. [DOI] [PubMed] [Google Scholar]
- 56.Parker KC, Bednarek MA, Coligan JE. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J Immunol. 1994;152(1):163. [PubMed] [Google Scholar]
- 57.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50(34):213. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


