Abstract
Immonium ions have been largely overlooked during the rapid expansion of mass spectrometry-based proteomics largely due to the dominance of ion trap instruments in the field. However, immonium ions are visible in hybrid quadrupole-time-of-flight (QTOF) mass spectrometers, which are now widely available. We have created the largest database to date of high-confidence sequence assignments to characterize the appearance of immonium ions in CID spectra using a QTOF instrument under “typical” operating conditions. With these data, we are able to demonstrate excellent correlation between immonium ion peak intensity and the likelihood of the appearance of the expected amino acid in the assigned sequence for phenylalanine, tyrosine, tryptophan, proline, histidine, valine, and the indistinguishable leucine and isoleucine residues. In addition, we have clearly demonstrated a positional effect whereby the proximity of the amino acid generating the immonium ion to the amino terminal of the peptide correlates with the strength of the immonium ion peak. This compositional information provided by the immonium ion peaks could substantially improve algorithms used for spectral assignment in mass spectrometry analysis using QTOF platforms.
Mass spectrometry has become an increasingly valuable tool in molecular biology. The most commonly employed strategy for identifying peptides employs low-energy collision-induced dissociation (CID) to produce characteristic fragments from which the sequence of the peptides can be determined. Tandem mass spectra generated by CID are typically dominated by cleavages along the peptide amide backbone, resulting in the formation of a series of overlapping b- and y-type ions.1-4 However, other fragmentation pathways can also occur during CID, some of which provide composition-specific information that can potentially improve the confidence of peptide assignments.5 One such pathway results in the formation of internal immonium ions. Immonium ions have the general structure RCH=NH2+ (where R is the amino acid side chain) and a mass of 27 Da less than the residue mass. These ions are formed following two cleavages surrounding a particular residue during CID, e.g., an a-type cleavage following either a b- or y-type cleavage,6 or can be formed directly by fragmentation of the N-terminal residue of the peptide ion.
Interest in obtaining residue-specific information from immonium ions began with the development of proteomics in the 1990s, though no effort has been large or exhaustive. The most thorough examination of immonium ions was performed by Falick et al. in 1993. They developed a model relating the presence of an immonium ion in high energy CID spectra to the presence of the corresponding amino acid.7 This model was tested against a database of 228 commercially available peptides with good results (67.5% correct predictions and a 4.8% incorrect rate). Since this work, few new insights have been gained.
Because the presence of immonium ions in CID spectra can provide compositional constraints on sequence assignment, some database search tools include this ion class in the search strategy. The original SEQUEST publication describes the use of immonium ions,8 but this function is not currently used. MASCOT has an option for utilizing the immonium ion peaks, though the specifics of how these peaks are included is not available.9 The search software ProbID10 also has an immonium ion scoring function. This function is not based on any predicted probability of residue appearance, however, and adjusts assignment scores only subtly. The ion accounting methodologies of the Waters Identity software also make use of the immonium ion intensity in peptide assignment (S. Geremanos, personal communication), but the precise methods used are proprietary. An alternative approach to utilizing immonium ions would be to use their presence as a tool for improving the probability scores of identified peptides using software such as PeptideProphet.11
In this study, we have created the largest database to date for characterizing the appearance of immonium ions in CID spectra. This data set is based upon high confidence peptide assignments from spectra obtained on a QTOF under typical operating conditions. With these data, we are able to identify those immonium ions that provide compositional information and relate immonium ion intensity to the likelihood of the appearance of the expected amino acid in the assigned sequence.
EXPERIMENTAL SECTION
Reagents
All reagents were purchased from Sigma (St. Louis, MO) unless otherwise noted. All experiments were performed using Saccharomyces cerevisiae (strain BY4741) prepared as described,12 alkylated using iodoacetamide, digested overnight with trypsin (Promega, Madison, WI), and dried in a low-pressure centrifuge. Peptides were separated into 10 fractions using strong cation exchange chromatography (SCX) as described5 using a polysulfethyl A 2.1 mm × 200 mm column (PolyLC, Columbia MD).
Mass Spectrometry and Database Searching
Each SCX fraction was run on an automated mass spectrometry system as described.13 Each sample (2 μL) was loaded onto a 75 μm internal diameter fused silica capillary column packed to a bed length of 10 cm with C18 spherical silica, 5 μm mean particle size resin (Magic C18Aq Michrom Bioresources). The loaded column was washed isocratically for 5 min with 0.1% formic acid (buffer A). Elution of peptides was performed using a linear gradient 10−35% buffer B (100% ACN) over 60 min at a flow rate of 200 nL/min using a split flow HP 1100 solvent delivery system (Agilent, Palo Alto, CA). Mass spectrometry including CID-MS/MS was performed in an automated fashion using the information dependent acquisition (IDA) option on a QSTAR Pulsar i (Applied Biosystems, Foster City, CA) equipped with an in-house built microspray device.14 The QSTAR was programmed to select the three most intense MS peaks for a single MS/MS scan of 2 s after which the selected precursor mass was excluded for 120 s. An IDA CE Parameter script was used for CID using the instrument-required slope intercept format with the form, CID voltage = slope(m/z) + constant. The slope and constants used in the script were (0.0575, 9), (0.0601, −5.42), (0.0539, −7), (0.0625, −6) for unknown charge states and 1+, 2+, 3+, and 4+, respectively. These values were determined in an earlier optimization study by repeatedly running a yeast cytoplasmic digest and optimizing the collision energy for each m/z range and charge state to achieve the maximal number of peptide identifications in that range. The ion transmission parameters used for MS were a scan range from m/z 300−1400 with ion transmission set to 33.3% at m/z 280, 450, and 900. For MS/MS, the scan range was from m/z 60−1800 with ion transmission set to 20% at m/z 40, 90, 180, 260, and 720.
Spectra were searched against a yeast subset of a nonredundant protein database acquired from the NCI's Advanced Biomedical Computing Center using SEQUEST (version 27) with a semitryptic option. Identifications were filtered based on a PeptideProphet11 probability of 0.9 using the INTERACT program.15 This corresponds to a false discovery rate of 1.6%. Identifications were sorted based on peptide sequence, and the first spectrum for each peptide in the sorted list was arbitrarily chosen as representative for that peptide. All data files in the mzXML format are available upon request.
Data Analysis
We adapted a program written to analyze neutral losses5 that extracted immonium ion intensity relative to base peak intensity from each spectrum along with the number of each residue present, the assigned peptide sequence, and the charge state of the precursor ion. The reference immonium ion masses used were per www.abrf.org/ResearchGroups/Mass-Spectrometry/EPosters/ms97quiz/residueMasses.html. Each reported immonium ion intensity corresponds to the maximum intensity within a m/z ±0.5 tolerance of each immonium ion mass. All peak intensities were rounded to two decimal places, thus peaks less than 0.5% of the base peak were assigned a zero value.
Statistical Analysis
All analysis and figures were made using the R statistical package. A boxplot of intensities versus number of residues in a peptide was calculated and fitted with a fitted linear regression including 99% confidence intervals for the mean intensity for each number of residues. After applying the Bonferroni correction for multiple comparisons, the familywise confidence level of the intervals over all residue occurrence counts simultaneously is approximately 95%.16
To relate immonium ion intensity to the probability of occurrence in the assigned sequence, the data for each immonium ion were grouped into six sets corresponding to base-peak-normalized immonium ion intensities of zero, 1−19%, 20−39%, 40−59%, 60−79%, and 80−100% for each charge state. For each set of normalized immonium ion intensities, the percentage of the spectral assignments containing the residue of interest was determined, as was a 99% individual confidence interval, which corresponds to the Bonferroni-corrected 95% confidence of all comparisons taken simultaneously for each amino acid. Calculations were made independently for doubly- and triply-charged data.
For the determination of the frequency of amino acid presence in yeast, a script queried the above yeast database for all unique tryptic peptides in the mass range of 800−1900 Da for doubly-charged and 1300−2900 Da for triply-charged (Supporting Information Table 2). These data were then sorted for the presence or absence of each of the studied amino acid residues. The above ranges include 95% of all the peptides identified for the respective charge states.
RESULTS
Visualizing Immonium Ions with a Modified Spectral Viewer
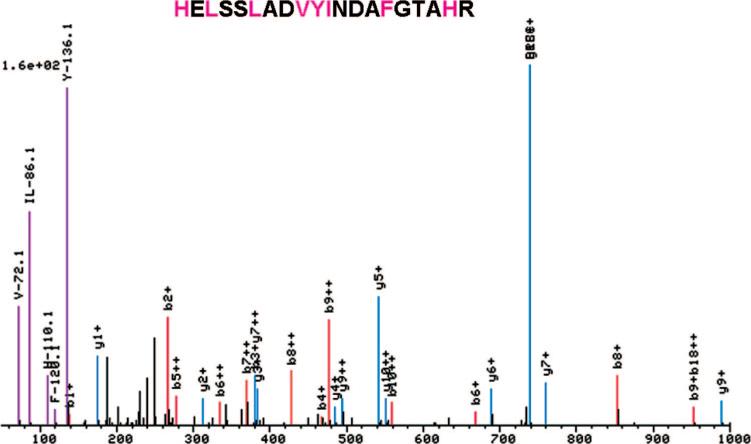
While observation of the individual spectra acquired during standard operation of our ABI QSTAR Pulsar i mass spectrometer, a correlation was observed between the presence of an immonium ion in the spectra and the presence of the corresponding amino acid in the peptide assignment. Our spectral viewer, a component of the Trans-Proteomic Pipeline package17 was adapted to permit easy viewing of all immonium ions, thus allowing comparison with the assigned peptide sequence. Several immonium ions were observed frequently in the CID spectra. For example, Figure 1 shows a spectrum where ions at m/z values corresponding to the immonium ions of valine (m/z 72.08), leucine/isoleucine (m/z 86.09), histidine (m/z 110.07), phenylalanine (m/z 120.08), and tryptophan (m/z 159.02) were observed and where these amino acids were also present in the assigned sequence. Using the modified spectral viewer, we were able to rapidly survey the frequency and intensity of all immonium ions. In addition to those mentioned above, the immonium ions of tyrosine (m/z 136.08) and proline (m/z 70.06) were also frequently observed, while for all other amino acids, immonium ions were not observed or observed at low frequency. We also noted that for the above amino acids, it was uncommon that a strong immonium ion was observed in the absence of the associated amino acid in the assigned peptide sequence.
Figure 1.
A single spectrum viewed using a modified spectral viewer, a component of the Trans-Proteomic Pipeline package. The viewer uniquely colors and labels peaks at the expected m/z of an immonium ion.
Creation of a Database of High-Confidence Sequence Assignments for Analysis of Immonium Ion Peaks
We next sought to quantify the correlation between the intensity of the immonium ion peak and the appearance of the residues in each peptide assignment using a large database derived from spectra with high-confidence sequence assignments. An extract of baker's yeast was digested and fractionated by cation exchange chromatography. These fractions were then analyzed by LC–MS/MS on a QSTAR Pulsar i mass spectrometer using the standard operation parameters and searched using SEQUEST. Peptide assignments with a PeptideProphet11 probability of greater than 0.9 were incorporated into a data set, consisting of 2241 doubly-charged spectral assignments and 1542 triply-charged assignments. For each of these high confidence assignments, the intensity of the signal within a range of m/z ±0.5 of each immonium ion peak was extracted from the recorded spectrum and normalized to the intensity of the base peak (the most intense peak in the spectrum). In parallel, the number of occurrences of each corresponding amino acid residue was extracted to the same database (Supporting Information Table 1).
Correlation of Immonium Ion Strength with Number of Residues Present
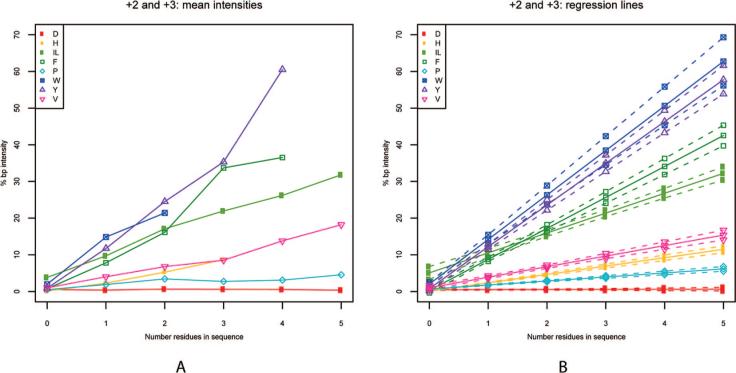
Because the secondary fragmentation events that produce specific immonium ions should occur at a rate proportional to the number of the associated residues present in a peptide, we hypothesized that the intensity of the immonium ion peak in a spectrum would correlate with the number of specific residues present. To test this hypothesis, we plotted the average base-peak-normalized immonium peak intensity against the number of associated residues present in the assigned peptide sequence. The resulting graph demonstrates a linear relationship between the average immonium ion intensity and the number of residues in the peptide assignment (Figure 2) for all of the amino acids in this study except for aspartic acid, which was included as a negative control. There was appreciable average immonium ion intensity even in cases where only a single residue was present. The slopes of the curves vary between immonium ions. Increasing slope (i.e., average ion intensity in a spectrum) represents the probability of immonium ion formation compared to the other b–y fragmentation events occurring during CID.
Figure 2.
(A) Average intensity of the immonium ion peak is plotted against the number of associated residues in the assigned sequence (including spectra without any immonium ion present) for all data. (B) Regression lines for each residue are plotted with 99% confidence intervals.
Relationship between Immonium Ion Intensity and Amino Acid Presence
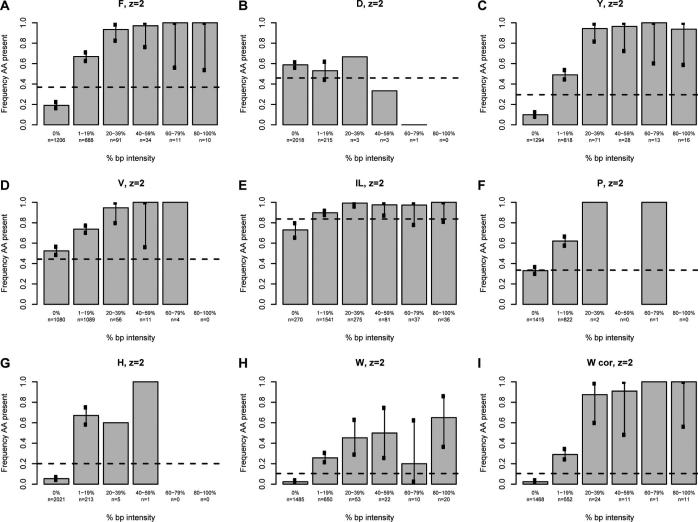
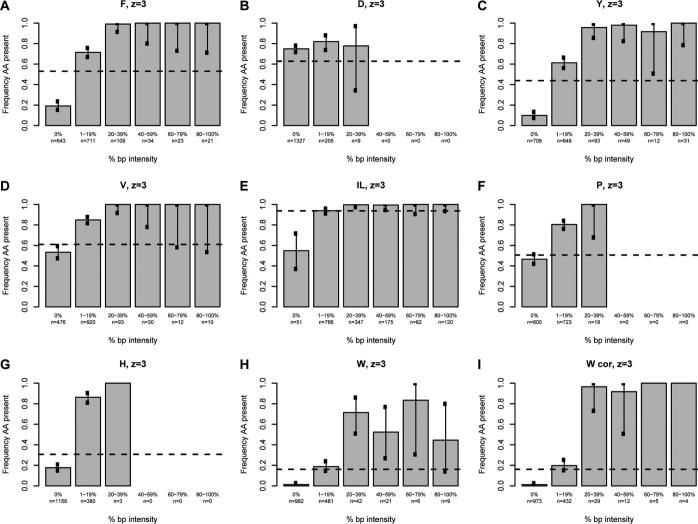
We next sought to determine whether a correlation exists between the intensity of the immonium ion and the probability of observing the associated amino acid in the assigned sequence using our high confidence spectra. For each immonium ion of interest, all spectra were divided into six groups based on normalized immonium intensity. Spectra with a normalized immonium ion intensity of zero were grouped together, while nonzero data were sorted into quintiles based on the base-peak-normalized immonium ion intensity (Figures 3 and 4). Because the analysis for all immonium ions is derived from the entire data set, each contains the same number of entries (2241 and 1542 for doubly- and triply-charged, respectively). A tabular version of these data is available in Supporting Information Table 1. The fraction of the assigned sequences in each of the six groups containing the corresponding amino acid was calculated for all intensity categories and plotted on the vertical axis. The error bars indicate the 99% individual confidence intervals, which correspond to the Bonferroni-corrected 95% confidence of all comparisons simultaneously for each amino acid. For the amino acids isoleucine, leucine, histidine, tyrosine, phenylalanine, proline, and tryptophan, there was a definite correlation between the intensity and the frequency of appearance in the sequence; details are discussed below.
Figure 3.
The intensity of the base-peak-normalized immonium ion peaks plotted against the percentage of the population that contains the residue of interest for doubly-charged ions. The base-peak-normalized intensity of the immonium ion is grouped in five quintiles plus a zero value on the x-axis. The percentage of the population having the residue of interest is given on the y-axis. The horizontal line represents the proportion of sequences in the database (800−1900 Da) containing the residue. Each chart represents a single amino acid as indicated at the top; W cor indicates a plot corrected for confounding peaks.
Figure 4.
The intensity of base-peak-normalized immonium ion peaks plotted against the percentage of the population that contains the residue of interest for triply-charged ions. The intensity of the normalized immonium ion peak is grouped in five quintiles plus a zero value on the x-axis. The percentage of the population having the residue of interest is given on the y-axis. The horizontal line represents the proportion of sequences in the database (1300−2900 Da) containing the residue. Each chart represents a single amino acid as indicated at the top; W cor indicates a plot corrected for confounding peaks.
The plot for phenylalanine (Figure 3A) for doubly-charged peptide assignments illustrates the general findings for the informative immonium ions and is contrasted with the plot for aspartic acid (Figure 3B). For spectra lacking a phenylalanine immonium ion at m/z 120.08 (0% bp intensity column), 19% of the 1206 sequence assignments contain a phenylalanine while 81% do not. Phenylalanine occurrence in the assigned spectra is ∼67% when the immonium ion peak is between 1 and 19% of the base peak intensity. For normalized immonium ion intensity of 20% or greater, the frequency of phenylalanine occurrence in the assigned sequence is very close to 100% (94−100%). In contrast, Figure 3B shows that the intensity of the immonium ion associated with aspartic acid (the negative control) does not correlate with the likelihood of finding an aspartic acid in the assigned sequence. The frequency of the appearance of aspartic acid is roughly 55% for all intensity classes of the aspartic acid immonium ion at m/z 88.04. It is known from previous work that aspartic acid has a significantly higher propensity for cleavage at its C-terminus than other residues.18,19 This implies that this residue is more likely to be incorporated into a cyclic b-type oxazolone or an anhydride structure formed from this cleavage. Formation of the immonium ion from these structures would require two additional cleavages (e.g., loss of CO to form an a-type ion, then cleavage of the amide bond to form the immonium ion). Thus, it is clear from these two plots that the intensity of the phenylalanine immonium ion provides compositional information while that associated with aspartic acid does not.
The utility of the classification according to immonium ion intensity for suggesting peptide composition is apparent when compared with the distribution of all tryptic candidate peptides in the yeast proteome, ∼37% of which contain a phenylalanine. The percentage of all unique tryptic peptides in the yeast database between 800 and 1900 Da (a range including 95% of all identified doubly-charged peptides) containing each amino acid is indicated with the dotted lines in Figures 3 and 4 (and in tabular format in Supporting Information Table 2). When Figure 3A is looked at again, a spectrum lacking a phenylalanine immonium ion peak (zero intensity) is much less likely to match a sequence containing a phenylalanine than would be expected by chance; ∼19% of this class contained phenylalanine compared to ∼37% in the database as a whole. In the case that a phenylalanine immonium ion is seen at a normalized 1−19% of the base peak, a spectrum is a more likely to match to a sequence containing phenylalanine than would be expected by chance (∼67% vs ∼37%). The 6.5% of recorded spectra with normalized immonium peaks greater than or equal to 20% are much more likely to be assigned to peptides containing phenylalanine (∼95% vs 37%). The difference between the observed and expected frequencies of assignment to sequences containing phenylalanine gives utility to the phenylalanine immonium ion in predicting composition. Probabilistic algorithms for sequence assignment, de novo sequencing, or comparing similarly scored spectral assignments could incorporate such a likelihood ratio for each distinct range of base-peak-normalized immonium intensity. It is notable in Figure 3A that spectra with normalized phenylalanine ion intensity equal to or above 20% or at exactly zero contain highly informative information for peptide sequence assignment; these groups constitute approximately 60% of all high confidence spectra. It is anticipated that with larger training sets, the 1−20% group could be subdivided to improve prediction in this range. In the current data set, subdivision of the 1−19% data shows that the 11−19% range (n = 108) better predicts the presence of phenylalanine, 89% of sequence assignments contain the residue, than does the 1−10% range (n = 788), 64% of which contain phenylalanine (Supporting Information Table 3). This finding is most likely due to the fact that low-intensity spectral noise appearing at m/z ∼120 can give rise to false positive signal.
The analysis of tyrosine is very similar to that of phenylalanine (Figure 3C). In spectra without any detectable immonium ion peak (∼58% of all spectra recorded), only ∼10% of the assigned sequences contain tyrosine, while 29% of all tryptic sequences between 800 and 1900 Da in the database contain this residue. Those spectra with 1−19% base-peak-normalized immonium ion intensity are somewhat more likely (∼49%) than the population as a whole to be assigned to a sequence containing tyrosine. Spectra with a base-peak-normalized immonium ion intensity of 20% or greater (∼6% of the acquired spectra) are much more likely to include a tyrosine in the assigned sequence than would be expected by chance (∼95% vs 29%). Like the phenylalanine data, the most informative ranges (the zero intensity and 20−100% intensity) constitute 63% of all measured spectra. Also like phenylalanine, spectra with tyrosine immonium intensity from 11−19% of the base peak predicted tyrosine content better than those with intensities from 1−10% of the base peak (80 vs 46%) (Supporting Information Table 3).
A study of the characteristics of immonium ions associated with valine, isoleucine, and leucine make it clear that other factors, including the frequency of the secondary fragmentation events which produce the immonium ion peak, the frequency of the amino acid in the database, and the presence of confounding peaks at the expected m/z for the immonium ion, can all play important roles in the utility of immonium ion peaks. The plot for valine (Figure 3D) is different from phenylalanine and tyrosine in three ways and illustrates these points. First, valine is a common amino acid and appears in peptides more frequently (44%) than phenylalanine and tyrosine. Second, unlike phenylalanine and tyrosine, 52% of high confidence spectra lacking an immonium ion peak actually contain a valine in the assigned sequence. This false negative rate is much higher than that of phenylalanine and tyrosine (52% vs 19% and 10%, respectively). More importantly, this rate is quite close to the frequency of valine containing peptides in the database, such that spectra lacking a valine immonium ion are assigned a valine at a frequency close to that expected by chance. Thus, the absence of the immonium ion has little power to predict the composition of the assigned spectrum. This high “false negative” rate results from the gas-phase properties of the valine. Similar to that for aspartic acid discussed above, it is known from previous work that valine, isoleucine, and leucine all have a higher propensity for cleavage at their C-terminus than other residues.19 Again, this implies that they are more likely to be incorporated into a cyclic b-type oxazolone formed from this cleavage and therefore unlikely to undergo further dissociation to yield an immonium ion. Thus, it is consistent with the gas phase properties of the residue that the immonium ion for valine is observed less often.
Despite the above differences, when present, the valine immonium ion peak is a good predictor of the presence of the amino acid. In Figure 3D, for all immonium ion peaks of 1−19% normalized intensity, 73% contain a valine, compared to ∼44% of theoretical peptide candidates. Spectra with very intense immonium peaks are again highly likely to contain a valine; however, this population is quite small, constituting only ∼3% of all measured spectra. Subdivision of the 1−19% group (Supporting Information Table 3) shows that an immonium ion of 11−20% of base peak intensity is an excellent predictor of valine presence in the assigned spectra (∼95%), though it constitutes only a small fraction of the 1−19% group (96 of nearly 1100). In conclusion, the valine immonium ion provides compositional information. However, the failure to indicate composition when absent, as in approximately 50% of the recorded spectra, makes this immonium ion peak less useful than those discussed earlier.
Leucine and isoleucine (Figure 3E) are abundant amino acids and must be considered together because they produce indistinguishable immonium ions. Like the case of valine, the absence of an I/L immonium ion does not predict very well whether the assigned spectra will contain an I/L. In the absence of any immonium ion, ∼70% of the spectra are still assigned to a sequence containing an isoleucine or leucine. This is less than the expected frequency of peptides containing either of these residues (84%), though not markedly so. This is again likely due to the gas phase properties of the leucine and isoleucine which, like valine, are more likely to be incorporated into the cyclic oxazolone structure of a b-type ion. The 1−19% group is indistinguishable from the population as a whole for I/L and is uninformative. Like the other informative peaks described above, intense base-peak-normalized I/L immonium ions are excellent predictors of the presence of an isoleucine or leucine residue. However, the utility of this information is somewhat limited because the difference between the frequency of assignment to sequences including isoleucine or leucine, ∼97%, is only slightly above that expected by chance alone (84%). This is an example of how the amino acid frequency can limit the utility of the immonium ion peak.
Proline shows similar behavior to valine. A large percentage of the recorded spectra (63%) have no immonium ion detectable, and this population contains a proline residue at about the same frequency as would be expected by chance (33%). For spectra that have an immonium ion of 1−19% of the base peak, the assigned sequences include proline residues at a substantially greater frequency than is expected by chance (62% vs 33%). Subdivision of the 1−19% data (Supporting Information Table 3) shows that of 86% of spectra containing proline immonium ions are in the 0−5% range. Despite the low intensity, these spectra predict the presence of an immonium ion (60% vs 33% expected by chance). Thus, the absence of an immonium ion is uninformative while the presence of an immonium ion, even at low intensity, is quite informative.
Histidine shows behavior with characteristics that could have substantial impact in predicting the composition of the assigned sequence for a large portion of the measured spectra. Spectra lacking an immonium ion (constituting 90% of all spectra) are much less likely (∼5%) to have a histidine in the assigned sequence than would be expected by chance (∼20%). The presence of any normalized immonium ion intensity is also highly informative; with 70% of spectra having a sequence assigned containing a histidine (vs ∼20% by chance). Like proline, the histidine immonium ion is low intensity. In nearly 75% of all occurrences, the immonium ion has an intensity of less than 5% of the base peak (Supporting Information Table 3). It is clear from these data that in nearly all cases, the immonium ion state for histidine provides information regarding the presence or absence of this amino acid.
The results for tryptophan (Figure 3H) show unique and informative behavior, with different tendencies than immonium ions discussed earlier. Spectra lacking tryptophan immonium ions are very unlikely (∼2%) to be assigned a sequence containing a tryptophan. This is much lower than the appearance of tryptophan-containing tryptic peptides in the proteome as a whole (10.4%). This informative absence of any tryptophan immonium ion peak is quite common; 66% of all spectra recorded were in this group. The remaining 34% of spectra with 1% or more base-peak-normalized tryptophan immonium ion peaks are also informative regarding the presence of tryptophan. However, unlike phenylalanine and tyrosine, the correlation never rises much higher than 50%. This indicates that there are other sources of ions at the same m/z creating false signals. Possible sources for such confounding signals are internal fragments and dipeptides.
Upon analyzing the data for tryptophan, we carefully searched through the spectra and identified the source of the confounding signals as a-type and b-type dipeptide fragments. A complete listing of dipeptide fragments that can interfere with the measurement of immonium ions is given in (Supporting Information Table 4) The most common cause is an a2 peak indistinguishable from the tryptophan immonium ion at m/z 159.09. The amino termini combinations (in any order) of the a2 ions of serine and valine (m/z 159.11), alanine and aspartic acid (m/z 159.08), and glycine and glutamic acid (m/z 159.08) occurred in the data set 82 times; 74 of those occurrences were associated with a measurable signal recorded for the tryptophan immonium ion and therefore resulted in false positive categorizations. Because the data extraction was performed at m/z ±0.5, distinction of the immonium ion from these a2 ions was not possible. The mass analyzer resolving power of the QSTAR generally will not permit resolution of the tryptophan immonium ion at m/z 159.09 from the combinations at m/z 159.08 (required resolution ∼16 000), though resolution is possible between m/z 159.09 and 159.11 (required resolution ∼8000).
Singly charged b2 fragments from the amino termini dipeptide sequences (again, in any order) threonine and glycine or serine and alanine produce ions at m/z 159.08 and 159.07, respectively, and were originally recorded as tryptophan immonium ions. A total of 60 of the 62 occurrences produced a measurable signal indistinguishable from the tryptophan immonium ion. The instrument resolution required to distinguish these peaks is ∼16 000 and ∼8000, respectively. Finally, the amino terminal pairing of alanine and asparagine (m/z 158.09) can generate an a2 ion, whose M + 1 isotope could generate a false positive ion. A total of 23 of the 29 occurrences of peptides containing this dipeptide generated measurable signal initially assigned to the tryptophan immonium ion (with lower intensity signal than those described above).
When the data for tryptophan is replotted following removal of 173 spectra containing the amino terminal sequences that cause confounding peaks, the predictive power of the presence of a tryptophan immonium ion is dramatically improved (Figure 3I, labeled W cor). Hence, the immonium ion at m/z 159.09 indicates the presence of either an immonium ion or one of the possible dipeptides present at the amino termini; these findings could be integrated into a software algorithm to predict composition. Further refinement of peak picking would permit discrimination of confounding peaks by the potential presence of these dipeptide sequences, depending on the operating resolution of the TOF.
Given the confounding peaks observed for the tryptophan immonium ion peaks, each of the data sets was reviewed for potential confounding peaks. None of the a-type and b-type dipeptide fragments interfered with the measurement of the other immonium ions discussed above. While the b1 ion of any peptide with an amino terminal alanine produces a fragment at m/z 72.03, indistinguishable from the valine immonium ion peak at m/z 72.08, analysis of these spectra indicated that b1 alanine ions were no more likely to generate a signal incorrectly assigned to the valine immonium ion than the population of sequence assignments without valine (data not shown). This is consistent with published reports that b1 ions of alanine are not stable and that the only b1 ions observed for the 20 common amino acids are of lysine20 and methionine.21
Additional confounding peaks were considered that corresponded to the loss of a single water or ammonia from a-type, b-type, and y-type dipeptides, in addition to the possibility of multiple losses of water or ammonia, or combinations of water and ammonia (all potential confounders are listed in Supporting Information Table 4). In contrast to the findings for tryptophan above, none of the potentially interfering dipeptides appeared to be enriched in spectra containing an immonium ion without the associated residue in the assigned sequence (data not shown).
The analysis of triply-charged peptides yielded plots very similar to those of the doubly-charged ions (Figure 4). The analysis is complicated by the increased number of potentially confounding peaks that result from the presence of doubly-charged di- and tripeptide fragment ions, in addition to the singly-charged dipeptides discussed above. These confounding peaks are listed in Supporting Information Table 4. Careful examination of all combinations of interfering peaks showed no evidence of confounding peaks for the spectra of triply charged peptides, with the exception of the singly charged a-type dipeptides that confound the tryptophan plot (data not shown).
There are a few subtle differences between the doubly- and triply-charged data. Most significantly, because sequences matching triply-charged spectra are larger (1300−2900 Da vs 800−1900 Da), the frequency of peptides containing each amino acid is higher. This has some effect on the magnitude of the difference between the observed and expected frequencies for each immonium ion. However, for each individual immonium ion, there are few major changes. For tyrosine, immonium ion intensity between 1 and 19% of the base peak is a slightly better predictor of the presence of tyrosine in the assigned sequence (61% vs 49%). For isoleucine/leucine, the absence of an immonium ion (0%) is more suggestive of the absence of the residue in the triply-charged state compared to the doubly-charged state (54% vs 73%). Given the increased proportion of the population containing isoleucine or leucine, 94%, the absence of the isoleucine/leucine ammonium ion peak is informative regarding the composition; note, however, that this class of spectra constituted only 3% of the database. For triply-charged spectra, tryptophan is again extremely unlikely to be in a sequence assigned to spectra lacking the characteristic immonium ion. Only 11 of the 962 triply-charged spectra lacking a tryptophan immonium ion were assigned to sequences that included this residue. Again, this is highly informative as it is far less than the expected frequency of tryptophan-containing peptides in the database (16%).
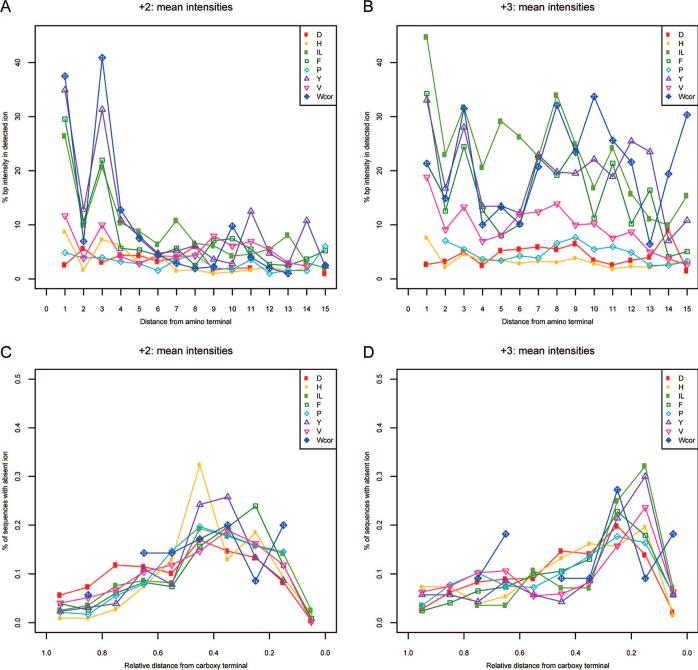
Location of the Residue in the Amino Acid Sequence Impacts Immonium Ion Intensity
In examination of the sequences that gave the strongest immonium ion peaks, it was noted that the proximity of the amino acid generating the immonium ion to the amino terminus of the peptide correlated with the strength of the immonium ion. This trend was studied more carefully by plotting the average of all base-peak-normalized immonium ion intensities for doubly-charged ions against the location of the associated residue from the amino terminus. These data are shown in parts A and B of Figure 5 for doubly- and triply-charged peptides, respectively. It is clear from Figure 5A that for all immonium ion peaks, with the exception of aspartic acid, that there is a correlation of intensity with distance from the amino terminus. It is also quite notable that immonium ion intensity is much lower when any residue is located in the second position from the amino terminus. This suggests that b2 ions are particularly stable against the formation of an immonium ion from the second amino acid in the peptide sequence (whereas the second amino acid of the peptide sequence is easily incorporated into the oxazolone ring). However, it is also possible that yn−1 ions (where the second amino acid of the peptide sequence is at the N-terminus) are not formed at high abundance. In addition, careful scrutiny of our database showed that even residues that are known to produce weak immonium ions, such as aspartic acid and glutamine, do produce immonium ions when they are situated at or near the amino terminus, though these peaks tend to be weak and occur less regularly (data not shown).
Figure 5.
Relationship of base-peak-normalized immonium ion intensity to the position of the associated amino acid in the assigned peptide sequence. Average normalized immonium ion intensity for each residue is plotted against the absolute position of the residue in the assigned sequence for doubly-charged (A) and triply-charged spectra (B). The fraction of the population of all spectra that contain the associated residue but fail to produce the expected immonium ion peak plotted against relative position in the assigned sequence for doubly-charged (C) and triply-charged spectra (D).
For triply-charged peptides shown in Figure 5B, the same phenomenon is evident. However, also notable in this figure is that residues situated well away from the amino terminus frequently generate intense immonium ions. The most plausible explanation for this phenomenon is a secondary fragmentation of the ion formed from the initial b–y-type fragmentation event. In such a scenario, a doubly-charged y-type ion would undergo formation of an immonium ion with a similar relationship to that seen in Figure 5A.
As a corollary to the findings above, we also noted that when a residue was present in a sequence assigned to a spectrum that lacked the expected immonium ion, the residue tended to be located near the carboxy terminus of the sequence. This observation was also confirmed graphically (parts C and D of Figure 5) by plotting the fraction of this population (residue present/immonium ion absent) against the relative position within the sequence for each amino acid. These plots demonstrate that for tryptophan, tyrosine, phenylalanine, and histidine, the closer the residue is to the carboxy terminus, the more likely it is to fail to produce an immonium ion. Note that the decrease in the probability of occupying the last position is due to the fact that the use of trypsin typically results in an arginine or lysine at the carboxy terminus, and thus this position is only infrequently occupied by any of the immonium ion-forming residues of interest. Lastly, we also observed that the aspartic acid control follows the same trend as the other residues. This is because, as noted above, aspartic acid can form an immonium ion when the residue is at or near the amino terminus. Thus, in aspartic acid-containing spectra that fail to include an immonium ion peak, the aspartic acid sequence position is mildly skewed toward the carboxy terminus, though less so than with other residues.
DISCUSSION
Immonium ions have been largely overlooked in the process of assigning a peptide sequence to an experimentally derived mass spectrum. This is presumably due to the success of ion trap platforms which impose a lower m/z limit on retained ions well above the m/z values of immonium ions.22 With the emergence of robust commercial hybrid-TOF platforms, immonium ions may be of increasing utility in search algorithms for these instrument types. In this work, we demonstrate that CID on an ABI QSTAR Pulsar i routinely produces immonium ions that are informative of the composition of the assigned peptide sequence. Because the voltage and ion transmission settings of the instrument were optimized for maximal peptide identification in shotgun proteomics experiments long before the presence of immonium ions was noted, we believe our data was collected under “typical” operation conditions. Also, as the presence of immonium ions has been stable over a number of years of operation, our results do not represent a peculiarity of instrument settings. Because original descriptions of this phenomenon utilized triple quadrupole mass spectrometers, and the same phenomenon has been seen on a Waters QTOF Premier system recently installed in our facility (data not shown), we believe that this phenomenon is generalizable to CID fragmentation reactions in a quadrupole.
To help researchers interpret TOF-MS/MS spectra, we have modified the viewing software within our Trans Proteomic Pipeline software to indicate the presence of immonium ions. The marked ions now allow users to quickly see the presence of these immonium ions to assist in evaluating the quality of a spectral assignment. This viewer should serve the QTOF user well; in our case, it has allowed us to quickly determine which immonium ions were the most consistent and intense and therefore worthy of quantitative analysis.
Using a large database of high-confidence MS/MS spectra from the QSTAR, we have found that across the data set, a number of trends can be observed regarding immonium ions. First, the number of immonium ion-associated residues in the assigned sequence correlated quite nicely with immonium ion intensity. This is not surprising as the secondary fragmentation event that results in the formation of an immonium ion (with the exception of immonium ions formed from an N-terminal residue) is a gas-phase event dependent on the properties of the residue, as well as its position in the peptide and sequence. Thus, for the group as a whole, it is expected that each occurrence of an immonium ion-generating residue is an independent event with an identical likelihood of undergoing secondary fragmentation. It is observed that, on average, tryptophan and tyrosine produce immonium ions that are typically a more intense component of the spectra than leucine/isoleucine, phenylalanine, valine, and histidine. The measured intensity of each immonium ion is actually the product of the two events required, the first being a b–y-type fragmentation event and the second being the a-type fragmentation event. Thus, the slopes in Figure 2 represent the co-occurrence of these events; the relative contribution of each is difficult to estimate.
Our database of high-confidence MS/MS QSTAR spectra has also allowed us to demonstrate that for phenylalanine, tyrosine, tryptophan, proline, histidine, and to a lesser extent valine and the indistinguishable leucine and isoleucine, the likelihood of finding a particular residue in the assigned sequence can be predicted by the strength of its immonium ion peak. The power of this observation depends on the difference between the likelihood of the appearance predicted by the immonium ion strength and the likelihood of appearance in peptides derived from the yeast database (as determined by amino acid frequency). For a number of residues, there is a substantial difference between these two values. For phenylalanine, the absence of the immonium ion indicates that the spectrum is roughly half as likely to be assigned to a sequence containing phenylalanine as would be expected by chance. Likewise, for any immonium ion abundance, the assigned sequence is much more likely to include a phenylalanine than would be expected by chance. This behavior is seen also with tyrosine, although lower intensity immonium ions are less robust predictors. In the case of tryptophan and histidine, the absence of the immonium ion indicates that the assigned sequence is much less likely (4-fold) to include either residue. The immonium ion intensity of valine is not quite as robust a predictor of the presence of this residue in the assigned sequence. For low-intensity valine immonium ions, the likelihood of the assigned sequence including valine is roughly 1.7 times that expected by chance. For more intense valine immonium ions, this ratio is over 2. Isoleucine and leucine immonium ions are the least informative due to the fact that their absence is not a good predictor of the absence of this residue in the assigned sequence, and their presence predicts a likelihood of appearance, that while high, is only slightly above that expected by chance.
We have used a global estimate for the likelihood of the presence of an amino acid by determining the frequency of the amino acid in peptides in the yeast proteome of lengths appropriate for doubly- and triply-charged peptides (800−1900 and 1300−2900 Da, respectively). However, the expected frequency actually varies quite substantially depending on the mass of the peptide. For example, 24% of all peptides in the mass range of 800−900 Da contain a phenylalanine, compared to 49% in the 1700−1800 Da range (Supporting Information Table 5). A larger data set would permit data to be subdivided by mass range, allowing for a better estimation of the prognostic power of the immonium ion peak. A larger data set would also facilitate refinement of the model relating immonium ion intensity with the expected residue's location in the assigned sequence. Such a model could assign to sequences a likelihood of correct assignment based on this relationship.
It is clear that the gas-phase properties of amino acids under both types of fragmentation events play a role in the production of the immonium ion. In the case of the hydrophobic amino acids valine, leucine, and isoleucine, the gas phase properties clearly do not favor the formation of immonium ions, except under particular conditions. This is in sharp contrast to histidine where production of an immonium ion happens fairly reliably. As discussed above, this is likely due to the cyclic oxazolone structure of leucine, isoleucine, and valine b-type ions which requires two additional cleavage events to produce the immonium ion. In this case, a larger data set might allow for the study of the effects of other parameters such as proton mobility23 to immonium ion generation.
There is a low level of spectral noise in the m/z range of each immonium ion. This noise is likely a characteristic of that region of the spectrum and due to the presence of low abundance b-H2O, b-NH3, a-H2O, a-NH3, y-H2O, and y-NH3 ions. Thus, the predictive power of immonium ions with very low intensity is quite variable. Discerning this precise threshold at which peaks become “informative” may hinge on instrument settings and the methods used to process the immonium ion peaks. The data presented herein represent a pilot study where instrument parameters were optimized for maximal peptide identification without regard to the immonium ions. More sophisticated strategies for noise reduction, peak picking, and immonium ion peak normalization would likely improve upon the findings in this initial report.
One final and important aspect of our findings is that we have analyzed each immonium ion individually while they frequently occur simultaneously (and likely independently) within the same spectrum. In the data set of doubly-charged spectra, 94% of spectra have two or more immonium ions and 65% have three or more (Supporting Information Table 1). It is likely that the predictive power of the immonium ion peaks can be combined and integrated into database searching algorithms. Possible applications include (1) incorporating immonium ion information as part of database searching based on a probabilistic model, utilizing the immonium ion within the scoring algorithm used to determine the probability of a correct assignment, (2) using the immonium ion information when searching spectra for which search algorithms are not able to make high-confidence assignments, or (3) ranking spectral assignments determined by separate applications to determine the probability of correct assignment.
CONCLUSIONS
We have demonstrated that the presence or absence of immonium ion peaks from phenylalanine, tyrosine, valine, isoleucine, leucine, tryptophan, proline, and histidine are good predictors of peptide composition. This is increasingly important at the current time due to the expanded use of hybrid time-of-flight mass spectrometers. It is likely that if incorporated into search algorithms these findings could improve the state-of-the-art of TOF-based shotgun proteomics.
Supplementary Material
ACKNOWLEDGMENT
This work was partially supported by National Cancer Institute Grant K08 CA097282 to D. B. Martin and Contract N01-HV-28179 from the National Heart, Lung, and Blood Institute. We thank Amelia Peterson for proof-reading this manuscript.
Footnotes
SUPPORTING INFORMATION AVAILABLE
The full data set for doubly- and triply-charged high-confidence peptides (Table 1), fraction of peptides containing each of the studied residues in the yeast database (Table 2), categorization of residue frequency according to normalized immonium ion intensity (Table 3), fragment masses calculated for all combinations of dipeptide ions on the amino terminus of peptides following a–x type and b–y type fragmentation, additional charts indicating masses following the loss of water and ammonia and combinations of water and ammonia (Table 4), fraction of peptides containing each of the studied residues in the yeast database for 100 Da ranges from 800 to 3100 Da (Table 5). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hunt DF, Yates JR, 3rd, Shabanowitz J, Winston S, Hauer CR. Proc. Natl. Acad. Sci. U.S.A. 1986;83:6233–6237. doi: 10.1073/pnas.83.17.6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paizs B, Suhai S. Mass Spectrom. Rev. 2004 doi: 10.1002/mas.20024. [DOI] [PubMed] [Google Scholar]
- 3.Roepstorff P, Fohlman J. Biomed. Mass Spectrom. 1984;11:601. doi: 10.1002/bms.1200111109. [DOI] [PubMed] [Google Scholar]
- 4.Biemann K. Methods Enzymol. 1990;193:886–887. doi: 10.1016/0076-6879(90)93460-3. [DOI] [PubMed] [Google Scholar]
- 5.Martin DB, Eng JK, Nesvizhskii AI, Gemmill A, Aebersold R. Anal. Chem. 2005;77:4870–4882. doi: 10.1021/ac050701k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ambihapathy K, Yalcin T, Leung H, Harrison AG. J. Mass Spectrom. 1997;32:209–215. [Google Scholar]
- 7.Falick AM, Hines WM, Medzihradszky KF, Baldwin MA, Gibson BW. J. Am. Soc. Mass Spectrom. 1993;4:882–893. doi: 10.1016/1044-0305(93)87006-X. [DOI] [PubMed] [Google Scholar]
- 8.Eng J, McCormack A, Yates JR., III J. Am. Soc. Mass Spectrom. 1994:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 9.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 10.Zhang N, Aebersold R, Schwikowski B. Proteomics. 2002;2:1406–1412. doi: 10.1002/1615-9861(200210)2:10<1406::AID-PROT1406>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 11.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 12.Smolka M, Zhou H, Aebersold R. Mol. Cell. Proteomics. 2002;1:19–29. doi: 10.1074/mcp.m100013-mcp200. [DOI] [PubMed] [Google Scholar]
- 13.Yi EC, Lee H, Aebersold R, Goodlett DR. Rapid Commun. Mass Spectrom. 2003;17:2093–2098. doi: 10.1002/rcm.1150. [DOI] [PubMed] [Google Scholar]
- 14.Gygi SP, Han DK, Gingras AC, Sonenberg N, Aebersold R. Electrophoresis. 1999;20:310–319. doi: 10.1002/(SICI)1522-2683(19990201)20:2<310::AID-ELPS310>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 15.Han DK, Eng J, Zhou H, Aebersold R. Nat. Biotechnol. 2001;19:946–951. doi: 10.1038/nbt1001-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kutner MH, Nachtsheim CJ, Neter J, Li W. Applied Linear Statistical Models. 5th ed. McGraw-HillBoston; MA: 2005. [Google Scholar]
- 17.Keller A, Eng J, Zhang N, Li XJ, Aebersold R. Mol. Syst. Biol. 2005;1:0017. doi: 10.1038/msb4100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu C, Tsaprailis G, Breci L, Wysocki VH. Anal. Chem. 2000;72:5804–5813. doi: 10.1021/ac000555c. [DOI] [PubMed] [Google Scholar]
- 19.Kapp EA, Schutz F, Reid GE, Eddes JS, Moritz RL, O'Hair RA, Speed TP, Simpson RJ. Anal. Chem. 2003;75:6251–6264. doi: 10.1021/ac034616t. [DOI] [PubMed] [Google Scholar]
- 20.Yalcin T, Harrison AG. J. Mass Spectrom. 1996;31:1237–1243. doi: 10.1002/(SICI)1096-9888(199611)31:11<1237::AID-JMS416>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 21.Ya Ping., Tu AGH. Rapid Commun. Mass Spectrom. 1998;12:849–851. [Google Scholar]
- 22.Schwartz JC, Senko MW, Syka JE. J. Am. Soc. Mass Spectrom. 2002;13:659–669. doi: 10.1016/S1044-0305(02)00384-7. [DOI] [PubMed] [Google Scholar]
- 23.Wysocki VH, Tsaprailis G, Smith LL, Breci LA. J. Mass Spectrom. 2000;35:1399–1406. doi: 10.1002/1096-9888(200012)35:12<1399::AID-JMS86>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.