Abstract
New HIV therapies are urgently needed to address the growing problem of drug resistance. In this article, we characterize the anti-HIV drug candidate 3-O-(3′,3′-dimethylsuccinyl) betulinic acid (PA-457). We show that PA-457 potently inhibits replication of both WT and drug-resistant HIV-1 isolates and demonstrate that the compound acts by disrupting a late step in Gag processing involving conversion of the capsid precursor (p25) to mature capsid protein (p24). We find that virions from PA-457-treated cultures are noninfectious and exhibit an aberrant particle morphology characterized by a spherical, acentric core and a crescent-shaped, electron-dense shell lying just inside the viral membrane. To identify the determinants of compound activity we selected for PA-457-resistant virus in vitro. Consistent with the effect on Gag processing, we found that mutations conferring resistance to PA-457 map to the p25 to p24 cleavage site. PA-457 represents a unique class of anti-HIV compounds termed maturation inhibitors that exploit a previously unidentified viral target, providing additional opportunities for HIV drug discovery.
The introduction of highly active antiretroviral therapy has led to a significant improvement in the prognosis for HIV-1-infected individuals. However, the emergence of virus isolates resistant to approved drugs can have a significant adverse impact on both treatment options and disease outcome. It is estimated that 40-45% of HIV-infected individuals harbor drug-resistant virus with a rapidly growing subgroup (5-10%) exhibiting resistance to all classes of reverse transcriptase (RT) and protease (PR) inhibitors (1). Treatment issues involving drug resistance are increasingly encountered in newly infected individuals. In a recent study, it was determined that in areas where antiretroviral therapy is widely used >25% of new HIV-1 infections involve viruses resistant to one or more approved drugs (2). Because resistant viruses are slower to respond to therapy, the time to suppression of viral load in patients infected with these isolates is markedly longer than observed for individuals harboring drug-sensitive strains. In addition, the time to virologic failure is significantly shorter among those infected with drug-resistant virus (3). These results highlight the need for new HIV treatment options.
Because of the large number of potential therapeutic targets, HIV assembly and budding have long been a focus of drug development efforts. HIV-1 assembly is driven largely by the Gag precursor protein Pr55Gag. After synthesis, Pr55Gag is transported to the plasma membrane where virus assembly occurs (4, 5). Through a complex combination of Gag-lipid, Gag-Gag, and Gag-RNA interactions, a multimeric budding structure forms at the inner leaflet of the plasma membrane. The budding virus particle is ultimately released from the cell surface in a process that is promoted by an interaction between the late domain in the p6 region of Gag (6, 7) and host proteins, most notably the endosomal sorting factor TSG101 (tumor susceptibility gene 101) (8-12). Concomitant with particle release, the viral PR cleaves Pr55Gag and Pr160GagPol. These processing events generate the mature Gag proteins matrix (MA), capsid (CA), nucleocapsid, and p6, two small Gag spacer peptides (SP1 and SP2), and the mature pol-encoded enzymes PR, RT, and integrase. Gag and GagPol cleavage triggers a structural rearrangement termed maturation, during which the immature particle transitions to a mature virion characterized by an electron-dense, conical core. The efficiencies with which PR cleaves its target sequences vary widely, resulting in a highly ordered Gag and GagPol processing cascade (13-15). The sequential nature of Gag processing can be disrupted by altering the amino acid sequence at cleavage sites within Gag (16-18), and even partial inhibition of Gag processing profoundly impairs virus maturation and infectivity (19). Mutating key residues in the p6 late domain (6-7, 20) or inhibiting the interaction between p6 and TSG101 (9, 11) also delays Gag processing and increases levels of the Gag cleavage intermediates p25 (CA-SP1) and p41 (MA-CA) in virions. It has also been reported that deletions in the dimer initiation site of the viral genomic RNA lead to an accumulation of p25 and a defect in virus maturation (21, 22).
Here, we report results from the characterization of 3-O-(3′,3′-dimethylsuccinyl) betulinic acid (PA-457), a potent HIV drug candidate that acts through a previously unidentified viral target to inhibit virus replication. PA-457 (Fig. 1A) was developed by activity-directed derivitization of betulinic acid, which was originally identified as a weak inhibitor (therapeutic index <5) of HIV-1 replication in a mechanism-blind screening assay (23, 24). We find that PA-457 exhibits a high degree of potency against both prototypic and clinical HIV-1 isolates and, importantly, retains its potent antiviral activity against a panel of viruses resistant to the three classes of approved drugs targeting the viral enzymes RT and PR. Using a series of in vitro experiments, we establish that the compound does not target the activities of the viral RT or PR. Consistent with a previous report, we found that PA-457 blocks HIV-1 replication at a late step in the virus life cycle (25). However, unlike that earlier work, we determined that the compound does not reduce the efficiency of virus particle release; rather, it induces a defect in Gag processing. Specifically, the cleavage of the CA precursor (p25) to mature CA (p24) is disrupted. Finally, by isolating and characterizing a PA-457-resistant isolate we show that the determinants of activity map to the p25 to p24 cleavage site. These observations demonstrate that PA-457 acts through a novel target to inhibit virus replication by disrupting p25 to p24 conversion, resulting in the formation of defective, noninfectious virus particles.
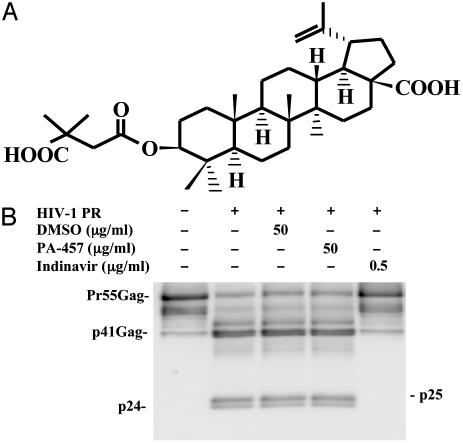
Fig. 1.
(A) The chemical structure of PA-457 (FW 584). (B) In vitro assays show that PA-457 does not affect HIV-1 PR function. After a 30-min incubation, PR-mediated processing of baculovirus-expressed Pr55Gag in the presence of high concentrations of PA-457 (dissolved in DMSO) is identical to that observed with no compound and compound (DMSO only) controls. Contrast these results with the complete block to PR function observed in the presence of the PR inhibitor indinavir at 0.5 μg/ml.
Materials and Methods
Compounds. PA-457 was prepared as described (23). The nucleoside RT inhibitor AZT was purchased from Sigma. The nonnucleoside RT inhibitor nevirapine and the PR inhibitor indinavir were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program. The peptide entry inhibitor, T20, was commercially prepared (New England Peptide, Gardner, MA).
Plasmids and Virus Isolates. The HIV-1 molecular clone pNL4-3 (26) used in this study was obtained from the National Institutes of Health AIDS Research and Reference Reagent Program. The TSG-5′ expression vector pcGNM2/TSG-5′ (11) was a gift from Z. Sun (Stanford University, Stanford, CA). The pNL4-3/CA5 was a gift from H. G. Krausslich (Universitätsklinikum Heidelberg, Heidelberg). All drug-resistant HIV-1 isolates and WT viruses BZ167 (27), 92HT599, US1 (27), and 92US723 were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program. HIV-2ROD and simian immunodeficiency virus Mac251 were provided by A. Langlois (Duke University, Durham, NC).
In Vitro Antiviral Activity Assays. Standard assay formats using either peripheral blood mononuclear cell or MT-2 cell line (28) targets were used to characterize the antiviral activity of PA-457. A multinuclear-activation galactosidase indicator (MAGI) assay (29) was used to determine whether PA-457 targeted an early or late step in viral replication. For detailed procedures, see Supporting Text, which is published as supporting information on the PNAS web site.
RT and PR activity. The effect of PA-457 on RT activity was determined by using the Roche Diagnostics chemiluminescent RT kit (catalogue no. 1828657). To characterize the effect of PA-457 on the activity of the viral PR enzyme, a cell-free fluorometric assay using a synthetic peptide substrate (Molecular Probes, H-2930) and in vitro HIV-1 Gag polyprotein processing experiments (30) were performed. For detailed procedures, see Supporting Text.
Radioimmunoprecipitation Assays, TSG-5′ Incorporation, and Electron Microscopy (EM) Analysis. Transfections, metabolic labeling, and EM analysis for the effect of PA-457 on Gag processing, virus release, TSG-5′ incorporation, and virus morphology were performed as described (11, 31, 32). For detailed procedures, see Supporting Text.
Selection for and Characterization of PA-457-Resistant Virus Isolates. PA-457-resistant isolates were selected by serial passage of WT NL4-3 in the MT-2 cell line in the presence of inhibitory concentrations of the compound. Virus replication during the selection processing was monitored by observing the formation of syncytia. The entire Gag and PR coding regions of the viral genome derived from a PA-457-resistant virus were amplified by using the high-fidelity RT-PCR kit (Pro-STAR Ultra HF, Stratagene) and sequenced. The resistance-conferring mutation was further characterized by introducing the identical change into the parental NL4-3 backbone. This was accomplished by subcloning a 503-bp SpeI-ApaI gag fragment from the PA-457-resistant virus RT-PCR product into WT pNL4-3. For detailed procedures, see Supporting Text.
Results
In Vitro Activity of PA-457 Against WT and Drug-Resistant HIV-1 Isolates. In assays using patient-derived WT virus isolates, PA-457 exhibited a mean IC50 of 10.3 nM (Table 1). The compound retained this activity against virus isolates resistant to the approved RT and PR inhibitors (Table 2). In assays against these viruses, PA-457 exhibited a mean IC50 of 7.8 nM, which is similar to that observed against drug-sensitive HIV-1 strains (Table 2). With an average 50% cytotoxicity value of 25 μM (data not shown), the therapeutic index for PA-457 is >2,500. The compound's antiviral activity was HIV-1 specific. In experiments using the related retroviruses HIV-2ROD and simian immunodeficiency virus MAC251 the IC50 values for PA-457 were >5 μM (data not shown).
Table 1. In vitro activity of PA-457 against clinical HIV-1 isolates.
| Resistance, IC50, nM
|
|||||
|---|---|---|---|---|---|
| Virus | Coreceptor use | PA-457 | AZT | Nev | Ind |
| BZ167 | R5/X4 | 4.5 | 2.2 | 31.1 | 1.2 |
| 92HT599 | X4 | 13.9 | 10.1 | 39.2 | 10.7 |
| US1 | R5 | 6.2 | 0.9 | 22.1 | 1.9 |
| 92US714 | R5 | 11.7 | 1.6 | 7.1 | 20.8 |
| 92US712 | R5 | 20.1 | 10.0 | 65.5 | 14.5 |
| 92US723 | R5/X4 | 5.1 | 1.2 | 26.8 | 3.9 |
| Mean | 10.3 | 4.3 | 40.0 | 8.8 | |
Control compounds included zidovudine (AZT), nevirapine (Nev), and indinavir (Ind). Assays used phytohemagglutinin-stimulated peripheral blood lymphocytes as targets and p24 production on day 8 as an indicator of virus replication.
Table 2. In vitro activity of PA-457 against drug-resistant virus isolates.
| Resistance, IC50, nM
|
||||||
|---|---|---|---|---|---|---|
| Virus | Phenotype | Genotype | PA-457 | AZT | Nev | Ind |
| N119* | NNRTI | Y181C | 5.1 | — | >3,800 (>570×) | — |
| A17* | NNRTI | K103N/Y181C | 5.4 | — | 3,000 (450×) | — |
| RF/41-D2* | PI | V82A | 9.8 | — | — | 28.0 (4×) |
| RF/L/323-9-1* | PI | I84V | 5.8 | — | — | 25.9 (4×) |
| M461/L63P V82T/I84V† | PI | M46I/L63P/V82T/I84V | 12.8 | — | — | 101.9 (12×) |
| 1495-2† | NRTI | K70R/T215Y/F | 2.7 | 29.4 (7×) | — | — |
| G910-11† | NRTI | T215Y/F | 13.3 | 216.0 (50×) | — | — |
Control compounds included zidovudine (AZT), nevirapine (Nev), and indinavir (Ind). Changes in activity from WT for drugs against resistant virus isolates are shown in parentheses.
Assays used MT 2 cell line targets and cell killing as an endpoint for virus replication.
Assays were carried out in a manner identical to that for clinical isolates.
PA-457 Does Not Block Virus Attachment or Entry or Inhibit RT or PR Activity in Vitro. Results from in vitro assays allow us to conclude that PA-457 does not block virus attachment or entry and does not affect the function of the viral RT (data not shown). The lack of effect on RT activity has been reported (24), and results from activity assays using HIV-1 isolates resistant to RT inhibitors support this observation (Table 2).
A series of in vitro assays were carried out to determine the effect of PA-457 on the activity of the viral PR enzyme. In a cell-free fluorometric assay using a synthetic peptide substrate PA-457 had no effect on PR function at concentrations of 50 μg/ml (data not shown). Experiments using a recombinant form of the Gag precursor protein Pr55Gag and assay formats sensitive to small changes in PR activity gave similar results. In one format, partial Gag processing was achieved by limiting PR concentration, allowing slight changes in enzyme activity to be detected by changes in the relative proportions of the intermediate Gag cleavage products. As shown (Fig. 1B), using this approach, after 30 min, Gag processing in samples treated with 50 μg/ml of PA-457 was identical to that observed in untreated controls. The second assay format involved following Pr55Gag processing as a function of time. This approach was particularly sensitive to the effect of test compounds on any single processing step (Fig. 6, which is published as supporting information on the PNAS web site). In this system, PA-457 exhibited no effect on PR activity. In both experiments, results obtained with PA-457 contrasted dramatically with the results seen with the PR inhibitor indinavir, which blocks all stages of Gag processing.
PA-457 Blocks a Late Step in Virus Replication. To characterize the inhibitory activity of PA-457 against early and late replication targets, a multinuclear-activation galactosidase indicator (MAGI) infectivity assay was used (Table 3). In this assay, the targets are HeLa cells stably expressing CD4, CXCR4, and CCR5 and harboring an integrated copy of the β-galactosidase gene under transcriptional control of a truncated HIV-1 LTR. As shown in Table 3, the entry inhibitor T20, the nucleoside RT inhibitor AZT, and the non-nucleoside RT inhibitor nevirapine caused significant reductions in β-galactosidase gene expression in HIV-1-infected MAGI cells, indicating that they disrupt early entry or postentry events. In contrast, the PR inhibitor indinavir targets a late step in viral replication (after Tat expression) and does not inhibit virus infectivity in this system. Similar results were obtained with PA-457 as with indinavir, indicating that PA-457 blocks virus replication at a time point after the completion of viral DNA integration and Tat expression.
Table 3. Results from a multinuclear-activation galactosidase indicator assay indicate that PA-457 blocks late in the virus life cycle.
| Inhibitor | % Decrease (β-galactosidase) |
|---|---|
| DMSO | 0 |
| T20 | 98 |
| AZT | 82 |
| Nevirapine | 85 |
| Indinavir | 10 |
| PA-457 | 12 |
The entry inhibitor T20 and the RT inhibitors AZT and nevirapine block replication before Tat protein expression, resulting in the inhibition of β-galactosidase expression. The PR inhibitor indinavir inhibits virus replication at a step post-Tat expression and has no effect on β-galactosidase expression. PA-457 gives results similar to indinavir, indicating that it blocks virus replication at a point post-Tat expression. All compounds were added at time zero and tested at 5 μg/ml.
PA-457 Blocks a Late Step in Gag Processing: The Conversion of p25 to p24. To define further the target of PA-457 activity, we characterized the production of virus from infected cells treated with the compound. We observed that PA-457 had no effect on virus particle release, as determined by p24 production and Western blot analysis of culture supernatants from treated cultures (data not shown). Quantitative radioimmunoprecipitation analysis indicated that the efficiency of virus release in the presence of PA-457 was 110% (±27.5%) of that observed in the absence of drug. This analysis also indicated that the drug had no effect on gp160 processing (Fig. 2) or Env glycoprotein incorporation into virions (data not shown). However, we did observe abnormal Gag protein processing in virus generated in the presence of compound.
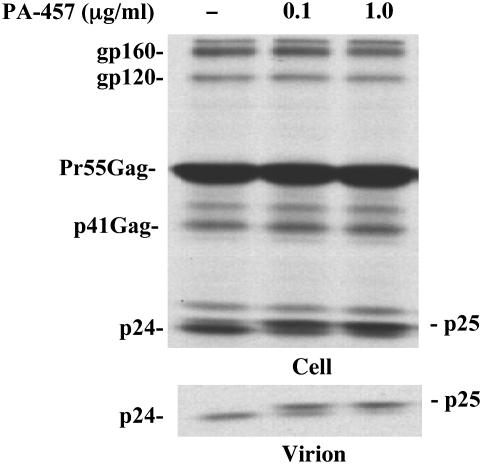
Fig. 2.
Effect of PA-457 on virus particle production and Gag processing. HeLa cells were transfected with pNL4-3 and cultured in the absence or presence of indicated concentrations of PA-457. Two days posttransfection, cells were metabolically labeled for 2 h with 35S-Met/Cys. Cell lysates (Upper) and virus lysates (Lower) were immunoprecipitated with HIV Ig as described (11). The positions of virally encoded proteins gp160, gp120, Pr55Gag, p41Gag, p25, and p24 are indicated. Note the accumulation of p25 in the presence of PA-457.
Radioimmunoprecipitation analyses (Fig. 2) revealed that, in a dose-dependent manner, PA-457 specifically inhibited the conversion of p25 (CA-SP1) to p24 (CA) as increased levels of p25 were detected in both cell and virion lysates (Fig. 2). This defective Gag processing phenotype was also observed by Western blot analysis when NL4-3-infected U87.CD4.CXCR4 cells or phytohemagglutinin-stimulated peripheral blood mononuclear cells were treated with PA-457 (data not shown). We also analyzed the effect of PA-457 by pulse-chase analysis. The results confirmed that the compound does not alter the kinetics of virus particle production but disrupts the conversion of p25 to p24, as elevated levels of p25 were observed in both cell and virion lysates (data not shown). Thus, using several approaches, we demonstrated that PA-457 did not affect the assembly and release of virus particles but rather specifically inhibited the conversion of p25 to p24.
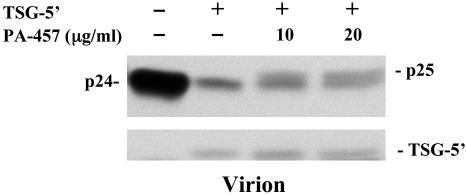
PA-457 Does Not Disrupt the Gag-TSG101 Interaction. As noted in the Introduction, disruption of the interaction between HIV-1 Gag and TSG101 inhibits the conversion of p25 to p24 (6, 7, 9, 11, 20). To test whether PA-457 disrupts p25 processing by preventing the Gag-TSG101 interaction, we measured the incorporation of the N-terminal, Gag-binding domain of TSG101 (TSG-5′) into virions. HeLa cells were cotransfected with pNL4-3 and the TSG-5′ expression vector (11, 32) in the presence and absence of PA-457. The incorporation of TSG-5′ into virions, which requires a specific interaction between the p6 late domain and TSG101 (11), was measured by radioimmunoprecipitation analysis. As reported (11), TSG-5′ disrupts the interaction between Gag and TSG101 resulting in markedly reduced levels of virion-associated Gag. The levels of TSG-5′ incorporation into virions were not affected by PA-457, indicating that the compound does not act by blocking the Gag-TSG101 interaction (Fig. 3).
Fig. 3.
PA-457 does not inhibit the Gag-TSG101 interaction. HeLa cells were transfected with pNL4-3 alone or cotransfected with a 1:1 DNA ratio of pNL4-3 and the TSG-5′ expression vector, pcGNM2/TSG-5′, which expresses an hemagglutinin (HA)-tagged N-terminal TSG101 fragment (11, 32). Cells, either not treated or treated with the indicated concentration of PA-457, were metabolically labeled overnight with 35S-Met/Cys. Virus lysates were immunoprecipitated with HIV Ig (Upper). Virus lysates were also immunoprecipitated with anti-HA antiserum (11) (Lower). The positions of virally encoded proteins p25, and p24 and TSG-5′ are indicated.
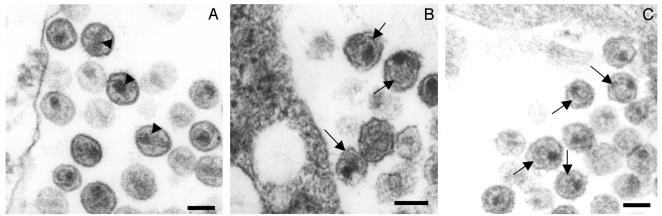
PA-457 Blocks Normal Virion Maturation. To determine whether the PA-457-induced defect in p25 to p24 processing affected virus morphology we performed EM analysis. In the absence of PA-457, cells transfected with pNL4-3 produced virus particles with the classical mature morphology characterized by the presence of condensed, conical cores (Fig. 4A). In contrast, virus from cells treated with PA-457 lacked conical cores. Instead, these virions displayed spherical, acentric cores and were further distinguished from the untreated particles by the presence of an additional electron-dense layer inside the viral membrane (Fig. 4B). This morphology, particularly the additional electron-dense layer, is also found in particles produced by the pNL4-3/CA-5 mutant, which contains substitutions that block the processing of CA-SP1 to CA (17) (Fig. 4C). Taken together, these results indicate that PA-457 blocks proper virion maturation.
Fig. 4.
Thin-section EM analysis of virions produced from PA-457-treated cells. HeLa cells were transfected with pNL4-3 (A and B) or pNL4-3/CA-5 (C) and were not treated (A and C) or treated (B) with PA-457. Two days posttransfection, cells were fixed and analyzed by thin-section EM. Arrowheads in A indicate mature, conical cores; arrows in B and C indicate the crescent-shaped, electron-dense layer inside the viral membrane that results from inhibition of p25 processing. (Bar: ≈100 nm.)
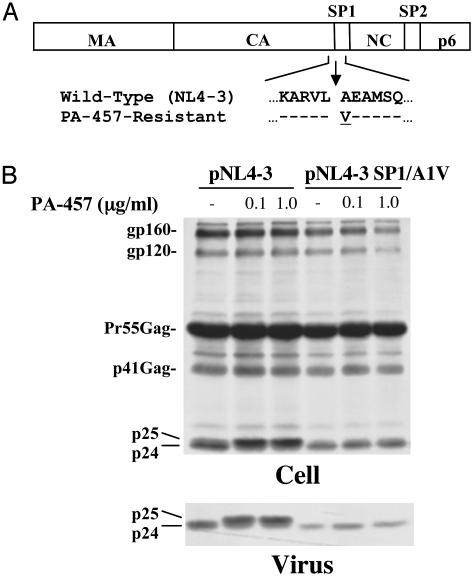
Selection for and Characterization of PA-457-Resistant Virus Isolates. To elucidate in more detail the mechanism of action of PA-457, resistant isolates were selected by serial passage of WT NL4-3 in the presence of inhibitory concentrations of compound. After ≈8 weeks in culture, a virus resistant to concentrations of >1 μg/ml PA-457 was isolated. To identify mutations responsible for PA-457 resistance the Gag and PR coding regions from the resistant virus were sequenced. We identified a single mutation encoding an amino acid change (A to V) at the N terminus of SP1 (Fig. 5A). To determine whether the A-to-V mutation could confer PA-457 resistance, we introduced this substitution into the NL4-3 backbone to generate the NL4-3 SP1/A1V mutant. We then compared the effect of PA-457 treatment on WT vs. SP1/A1V Gag processing. As shown earlier (Fig. 2), PA-457 disrupted the processing of WT p25 to p24, leading to an accumulation of p25 in both cell and virion fractions (Fig. 5B). In striking contrast, PA-457 had no effect on the processing of the mutant Gag, as no p25 was detected even at a concentration of 1 μg/ml. In addition, and consistent with the biochemical results (Fig. 5B), we observed no effect of PA-457 on the morphology of SP1/A1V mutant virions (data not shown). These results indicate that the A-to-V substitution at the p25 cleavage site confers PA-457 resistance.
Fig. 5.
Virus selected for resistance to PA-457 contains a single amino acid change in the CA/SP1 cleavage site. (A) Single amino acid mutation was identified in the resistant virus that was not present in the virus passaged in the absence of PA-457, an alanine to valine substitution at the N terminus of SP1 (SP1/A1V). Arrows indicate the site of CA-SP1 cleavage by HIV-1 PR. (B) Effect of PA-457 on Gag processing of the PA-457-resistant mutant SP1/A1V. HeLa cells were transfected with pNL4-3 or the pNL4-3 SP1/A1V mutant and cultured in the absence or the presence of the indicated concentrations of PA-457. Two days posttransfection, cells were metabolically labeled for 2 h with 35S-Met/Cys. Cell lysates (Upper) and virus lysates (Lower) were immunoprecipitated with HIV Ig. The positions of virally encoded proteins gp160, gp120, Pr55Gag, p41Gag, p25, and p24 are indicated.
Discussion
PA-457 was shown to be a potent in vitro inhibitor of HIV-1 replication. In assays using patient-derived WT virus isolates, PA-457 exhibited a mean IC50 of 10.3 nM. This value compared well with the approved drugs AZT and indinavir (4.3 and 8.8 nM, respectively) and was significantly lower than the mean IC50 observed for the non-nucleoside RT inhibitor nevirapine (40.0 nM). With an average 50% cytotoxicity value of 25 μM, the therapeutic index for PA-457 is >2,500. Importantly, PA-457 exhibited a similar level of activity against a panel of virus isolates resistant to the three classes of drugs targeting the viral RT and PR enzymes. In these experiments, PA-457 retained its low nanomolar activity, whereas the approved drugs exhibited losses in activity ranging up to >500-fold against the resistant isolates. Interestingly, the compound's antiviral activity was HIV-1-specific. PA-457 was inactive against the related retroviruses HIV-2 and simian immunodeficiency virus in cell-based replication assays.
The results of this study allow us to conclude that PA-457 acts through a previously unidentified target to block virus replication. We, and others, have demonstrated that the compound does not effect virus attachment or entry and does not disrupt activity of the viral RT or PR enzymes (25). Kanamoto et al. (25) reported that PA-457 inhibited virus release from infected cells, a result we were unable to reproduce in this study. In that report the authors based their conclusion on EM results from PA-457-treated infected cells, an approach that does not allow quantitative assessment of virus release. In contrast, using a variety of assays, including quantitative radioimmunoprecipitation analysis, we demonstrate that PA-457 does not affect virus particle production, but rather inhibits p25 processing and virion maturation.
We showed that virus generated in the presence of PA-457 exhibited a defect late in the Gag processing cascade. Specifically, the conversion of the CA precursor (p25) to mature CA protein (p24) was blocked. As mentioned in the Introduction, correct processing of Pr55Gag is critical to the formation of mature infectious viral particles. The p25 CA processing intermediate consists of the CA protein linked to a 14-aa SP1 that separates CA from nucleocapsid. It has been demonstrated that mutations that block this processing step result in the production of noninfectious viral particles with aberrant cores (16-18). An intriguing observation involved the specificity of the effect. The block to Gag processing was not global but rather appeared to be limited to the p25 to p24 cleavage event. Thin-section EM indicated that the defect in the conversion of p25 to p24 induced by PA-457 led to the formation of virions that displayed aberrant particle morphology. This abnormal core morphology contrasts with the cone-shaped CA structure normally associated with mature infectious HIV-1 particles.
In an effort to identify the molecular determinants of PA-457 activity we selected for and genotyped PA-457-resistant virus. In these experiments we identified a single mutation encoding an amino acid change (A to V) at the N terminus of SP1 (Fig. 5A). The change at this position (flanking the CA/SP1 cleavage site) is consistent with the observation that PA-457 disrupts p25 processing. Although the mechanism of PA-457 resistance remains to be determined, we propose two possibilities. If the N terminus of SP1 serves as a target for PA-457 the A-to-V change could disrupt the ability of the compound to interact with this region of Gag thereby reducing activity. Alternatively, if PA-457 activity involves interactions with a higher-order Gag structure, the A-to-V substitution could alter that structure in a way that affects PA-457 binding and activity. As we gain insight into the molecular target for this compound the mechanism of resistance should become clear. Interestingly, the A-to-V mutation has been reported to reduce HIV-1 replicative fitness compared with WT (33). Significantly, no mutations in the PR coding region were found.
Several models could explain the mechanism by which PA-457 blocks p25 processing. (i) The compound could alter the enzymatic activity of PR such that it inefficiently recognizes the CA/SP1 cleavage site. Because cleavage at this site is not inhibited in our in vitro assays, the effect would have to be specific for PR in the context of an assembled virion. (ii) PA-457 could affect RNA encapsidation, dimerization, or dimer maturation (34, 35) such that p25 processing is inhibited. According to this model, the compound would induce an effect analogous to that imposed by deletions in the RNA dimer initiation site, which have been reported to inhibit p25 processing (21, 22). (iii) PA-457 could bind directly to Pr55Gag or Gag processing intermediates to disrupt CA/SP1 cleavage. Such an interaction could result in changes to the global conformation of Gag such that the ability of PR to cleave at the CA/SP1 processing site is inhibited. Alternatively, the compound could bind directly to the CA/SP1 cleavage site and disrupt p25 processing by blocking PR access to this site. Because PA-457 does not appear to block p25 processing in vitro, the effect would have to be specific for Gag in the context of a higher-order structure. The specificity of the compound for the p25 processing event, coupled with the location of the mutation that confers PA-457 resistance, provide support for this last model.
Unlike a number of other novel HIV-1 inhibitors, including a recently described CA-binding compound (36), PA-457 exhibits many of the properties considered necessary for a drug-development candidate. An in vitro IC50 value similar to that of approved drugs coupled with activity against drug-resistant virus isolates satisfies the first set of HIV therapeutic development criteria. Additional in vitro studies have established that PA-457 is not rapidly metabolized in the presence of human liver microsomes and does not significantly inhibit the activity of cytochrome P450 liver isoforms (unpublished data). Specifically, the IC50 value for the inhibition of CYP3A4 catalytic activity was >120 μM (data not shown). These results suggest that the compound should exhibit a suitable in vivo half-life when administered to humans and a reduced likelihood for the types of drug-drug interaction problems often observed in multidrug HIV therapeutic regimens. Importantly, experimental formulations have been identified that result in high levels of oral bioavailability. In rodent models >50% oral bioavailability has been achieved with plasma concentrations in excess of 25 μM after a single 25 mg/kg oral dose (unpublished work). Overall, these results support further development of this drug candidate.
In summary, our results have established that PA-457 inhibits HIV-1 replication by a unique mechanism of action and represents a member of an emerging class of compounds that block virus maturation. Importantly, in characterizing the mechanism of action of this compound, we have identified a highly conserved target for HIV-1 therapeutic development. This observation is particularly significant as the percentage of HIV-infected individuals harboring drug-resistant strains grows and the need for new drugs active against these virus isolates increases. The preclinical profile of PA-457 places it on the short list of therapeutic development candidates that closely fit the needs of today's treatment-experienced HIV-1-infected patient population. Results of ongoing and future studies with PA-457 and similar compounds will determine the therapeutic potential for this class of inhibitors that target Gag CA-SP1 processing and block HIV-1 maturation.
Supplementary Material
Acknowledgments
We acknowledge K. H. Lee at the University of North Carolina for his contribution to the original discovery of PA-457 and for providing material used in this work. We thank Z. Sun for plasmid pcGNM2/TSG-5′, H. Krausslich for plasmid pNL4-3/CA-5, and K. Strebel, A. Ono, D. Demirov, and M. Shehu-Xhilaga for critical review of the manuscript. This study was funded in part by National Institutes of Health Grant R43 AI51047 (to G.P.A.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PA-457, 3-O-(3′,3′-dimethylsuccinyl) betulinic acid; TSG101, tumor susceptibility gene 101; MA, matrix; CA, capsid; SP, spacer peptide; PR, protease; RT, reverse transcriptase; EM, electron microscopy.
References
- 1.LaBonte, J., Lebbos, J. & Kirkpatrick, P. (2003) Nat. Rev. Drug Discov. 2, 345-346. [DOI] [PubMed] [Google Scholar]
- 2.Grant, R. M., Hecht, F. M., Warmerdam, M., Liu, L., Liegler, T., Petropoulos, C. J., Hellmann, N. S., Chesney, M., Busch, M. P., Kahn, J. O., et al. (2002) J. Am. Med. Assoc. 288, 181-188. [DOI] [PubMed] [Google Scholar]
- 3.Little, S. J., Holte, S., Routy, J. P., Daar, E. S., Markowitz, M., Collier, A. C., Koup, R. A., Mellors, J. W., Connick, E., Conway, B., et al. (2002) N. Engl. J. Med. 347, 385-394. [DOI] [PubMed] [Google Scholar]
- 4.Swanstrom, R. & Wills, J. W. (1997) in Retroviruses, eds. Weiss, R., Teich, N., Varmus, H. & Coffin, J. M. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 263-334.
- 5.Freed, E. O. (1998) Virology 251, 1-15. [DOI] [PubMed] [Google Scholar]
- 6.Gottlinger, H. G., Dorfman, T., Sodroski, J. G. & Haseltine, W. A. (1991) Proc. Natl. Acad. Sci. USA 88, 3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang, M., Orenstein, J. M., Martin, M. A. & Freed, E. O. (1995) J. Virol. 69, 6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.VerPlank, L., Bouamr, F., LaGrassa, T. J., Agresta, B., Kikonyogo, A., Leis, J. & Carter, C. A. (2001) Proc. Natl. Acad. Sci. USA 98, 7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garrus, J. E., von Schwedler, U. K., Pornillos, O. W., Morham, S. G., Zavitz, K. H., Wang, H. E., Wettstein, D. A., Stray, K. M., Cote, M., Rich, R. L., et al. (2001) Cell 107, 55-65. [DOI] [PubMed] [Google Scholar]
- 10.Martin-Serrano, J., Zang, T. & Bieniasz, P. D. (2001) Nat. Med. 7, 1313-1319. [DOI] [PubMed] [Google Scholar]
- 11.Demirov, D. G., Ono, A., Orenstein, J. M. & Freed, E. O. (2002) Proc. Natl. Acad. Sci. USA 99, 955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freed, E. O. (2002) J. Virol. 76, 4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krausslich, H. G., Schneider, H., Zybarth, G., Carter, C. A. & Wimmer, E. (1988) J. Virol. 62, 4393-4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mervis, R. J., Ahmad, N., Lillehoj, E. P., Raum, M. G., Salazar, F. H., Chan, H. W. & Venkatesan, S. (1988) J. Virol. 62, 3993-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erickson-Viitanen, S., Manfredi, J., Viitanen, P., Tribe, D. E., Tritch, R., Hutchison, C. A., Loeb, D. D. & Swanstrom, R. (1989) AIDS Res. Hum. Retroviruses 5, 577-591. [DOI] [PubMed] [Google Scholar]
- 16.Krausslich, H. G., Facke, M., Heuser, A. M., Konvalinka, J. & Zentgraf, H. (1995) J. Virol. 69, 3407-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiegers, K., Rutter, G., Kottler, H., Tessmer, U., Hohenberg, H. & Krausslich H. G. (1998) J. Virol. 72, 2846-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Accola, M. A., Hoglund, S. & Gottlinger, H. G. (1998) J. Virol. 72, 2072-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaplan, A. H., Zack, J. A., Knigge, M., Paul, D. A., Kempf, D. J., Norbeck, D. W. & Swanstrom, R. (1993) J. Virol. 67, 4050-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Demirov, D. G., Orenstein, J. M. & Freed, E. O. (2002) J. Virol. 76, 105-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang, C., Rong, L., Laughrea, M., Kleiman, L. & Wainberg, M. A. (1998) J. Virol. 72, 6629-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang, C., Rong, L., Cherry, E., Kleiman, L., Laughrea, M. & Wainberg, M. A. (1999) J. Virol. 73, 6147-6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujioka, T., Kashiwada, Y., Kilkuskie, R. E., Cosentino, L. M., Ballas, L. M., Jiang, J. B., Janzen, W. P., Chen, I. S. & Lee, K. H. (1994) J. Nat. Prod. 57, 243-247. [DOI] [PubMed] [Google Scholar]
- 24.Kashiwada, Y., Hashimoto, F., Cosentino, L. M., Chen, C. H., Garrett, P. E. & Lee, K. H. (1996) J. Med. Chem. 39, 1016-1017. [DOI] [PubMed] [Google Scholar]
- 25.Kanamoto, T., Kashiwada, Y., Kanbara, K., Gotoh, K., Yoshimori, M., Goto, T., Sano, K. & Nakashima, H. (2001) Antimicrob. Agents. Chemother. 45, 1225-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adachi, H., Gendelman, H. E., Koenig, S., Folks, T., Willey, R., Rabson, A. & Martin, M. A. (1986) J. Virol. 59, 284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michael, N. L., Herman, S. A., Kwok, S., Dreyer, K., Wang, J., Christopherson, C., Spadoro, J. P., Young, K. K., Polonis, V., McCutchan, F. E., et al. (1999) J. Clin. Microbiol. 37, 2557-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roehm, N. W., Rodgers, G. H., Hatfield, S. M. & Glasebrook, A. L. (1991) J. Immunol. Methods 142, 257-265. [DOI] [PubMed] [Google Scholar]
- 29.Kimpton, J. & Emerman, M. (1992) J. Virol. 66, 2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morikawa, Y., Shibuya, M., Goto, T. & Sano, K. (2000) Virology 272, 366-374. [DOI] [PubMed] [Google Scholar]
- 31.Freed, E. O., Orenstein, J. M., Buckler-White, A. J. & Martin, M. A. (1994) J. Virol. 68, 5311-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun, Z., Pan, J., Hope, W. X., Cohen, S. N. & Balk, S. P. (1999) Cancer 86, 689-696. [DOI] [PubMed] [Google Scholar]
- 33.Liang, C., Hu, J., Russell, R. S., Roldan, A., Kleiman, L. & Wainberg, M. A. (2002) J. Virol. 76, 11729-11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu, W. & Rein, A. (1993) J. Virol. 67, 5443-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu, W., Gorelick, R. J. & Rein, A. (1994) J. Virol. 68, 5013-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang, C., Loeliger, E., Kinde, I., Kyere, S., Mayo, K., Barklis, E., Sun, Y., Huang, M. & Summers, M. F. (2003) J. Mol. Biol. 327, 1013-1020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.