Abstract
It has been suggested that autism, like other complex genetic disorders, may benefit from the study of rare or Mendelian variants associated with syndromic or non-syndromic forms of the disease. However, there are few examples in which common variation in genes causing a Mendelian neuropsychiatric disorder has been shown to contribute to disease susceptibility in an allied common condition. Joubert syndrome (JS) is a rare recessively inherited disorder, with mutations reported at several loci including the gene Abelson’s Helper Integration 1 (AHI1). A significant proportion of patients with JS, in some studies up to 40%, have been diagnosed with autism spectrum disorder (ASD) and several linkage studies in ASD have nominally implicated the region on 6q where AHI1 resides. To evaluate AHI1 in ASD, we performed a three-stage analysis of AHI1 as an a priori candidate gene for autism. Re-sequencing was first used to screen AHI1, followed by two subsequent association studies, one limited and one covering the gene more completely, in Autism Genetic Resource Exchange (AGRE) families. In stage 3, we found evidence of an associated haplotype in AHI1 with ASD after correction for multiple comparisons, in a region of the gene that had been previously associated with schizophrenia. These data suggest a role for AHI1 in common disorders affecting human cognition and behavior.
INTRODUCTION
Autism is a neurodevelopmental syndrome characterized by impairments in social behavior, communication, language and the presence of repetitive-restricted behaviors. It is best considered as the most severe form of a spectrum of symptom clusters known as autism spectrum disorders (ASDs) under the clinical diagnostic classification of pervasive developmental disorders (1,2). There is strong evidence of genetic contributions to ASD (3–5), with heritability estimated between 60 and 90% on the basis of twin studies (6). Rates of autism in siblings of those affected are ∼5–10%, which is 20–50 times higher than the rate of autism in the general population (7,8). Prevalence estimates for ASD are one in 166, and autistic disorder, which represents the narrowest diagnostic category, has a prevalence of one to two in 1000 (9). Modeling suggests multiple genes contributing to ASD genetic risk (10,11), which is consistent with recent data from a variety of genetic approaches that demonstrate significant genetic heterogeneity (12–14), similar to that found in many other common diseases.
Studies of rare chromosomal or structural genomic alterations, as well as rare Mendelian causes of more common disorders, ranging from diabetes (15), hyperlipidemia (16) to Alzheimer’s disease (17) and disorders of speech and language (18), have played central roles in understanding disease pathophysiology (19). However, the extent to which common variation in Mendelian disease genes contributes to common diseases in general is not known, and few examples of such contributions have been demonstrated (19). In this regard, it is notable that autism has been described in more than 25 different genetic syndromes including Fragile X syndrome, Rett syndrome, Down syndrome, tuberous sclerosis and Joubert syndrome (JS) (20,21), further supporting the notion that many different etiologies account for ASD. However, common variants in any of the genes causing these syndromic forms of autism have yet to be associated with ASD.
JS is an autosomal recessive disorder characterized by partial or complete agenesis of the cerebellar vermis, and cognitive and behavioral dysfunction. Features of ASD, such as deficits in social behavior, language dysfunction and repetitive behaviors, have been described in up to 40% of JS patients (20–23), and ∼25% of JS patients meet criteria for a DSM-IV diagnosis of strict autistic disorder (20), making it an important syndromic form of the disorder (5,20–23). Recently, mutations were found in the Abelson’s helper integration 1 gene (AHI1), encoding the ‘Jouberin’ protein in a subset of families segregating JS (24,25). Mutations in AHI1 are encountered in 7.3 (26) to 11% (27) of JS patients, predominantly in those with the ‘pure’ form of the disease, that is, in cases with signs and symptoms restricted to the central nervous system (CNS) ± retinal involvement (26). The Jouberin protein (28) domain structure suggests that it functions in signal transduction, perhaps as an adaptor molecule, but little is known about AHI1 and how it might be involved in the pathogenesis of JS.
We reasoned that in addition to the role that JS might have as a rare cause of syndromic autism (20–22,29), the common variation in the AHI1 gene may contribute to ASD risk. The utility of studying genes first identified as causing rare, or syndromic forms of common diseases, such as hyperlipidemia, maternity onset diabetes of the young and type II diabetes, in understanding the genetic basis and pathophysiology of common disorders is becoming increasingly appreciated (16–19). This approach is further supported by recent findings in ASD, in which rare recessive mutations in a gene (CNTNAP2) (30) have been shown to cause a syndromic form of ASD, whereas common variation in the same gene has been shown to contribute to genetic risk for the common, non-syndromic forms of the disorder (31,32).
To investigate the AHI1 locus as a possible contributor to non-syndromic ASD, we performed a three-stage study. To detect any possibility of involvement, we first sequenced the AHI1 gene in 48 independent ASD subjects from sibling pairs ascertained for ASD and having the highest allele sharing for markers in the genomic region around the AHI1 locus. This was done to identify common AHI1 variants in families with autistic probands and identify candidates for single-nucleotide polymorphism (SNP) association studies. Common SNPs were compared with several publicly available control group frequencies to identify whether there were any SNPs with nominal case–control association in this first-stage screening procedure. This was followed by a second stage, where we performed SNP genotyping in an independent sample of 326 ASD parent–child trios from the Autism Genetic Resource Exchange (AGRE) cohort to test the two variants that were nominally associated in stage-1 screening analysis. Stage-2 family-based analyses yielded a significant association of these two SNPs, suggesting that it would be beneficial to more exhaustively study AHI1 with haplotype-tagging SNPs to more completely cover the AHI1 gene. This stage-3 association study covered 99% of the haplotype diversity in AHI1 to more thoroughly assess AHI1 in 337 AGRE trios, 111 of which were independent of those analyzed in stage 2. This family-based analysis identified an association of ASD with an AHI1 haplotype, which was significant after Bonferroni correction for multiple comparisons. This ASD-associated region overlaps with one previously identified as significantly associated with schizophrenia in two independent cohorts.
RESULTS
Sequencing and variant identification
We performed a first-stage evaluation by sequencing all the exons and their flanking regions of the AHI1 gene in 48 unrelated ASD subjects (96 chromosomes) from the subsample of the AGRE cohort ascertained for having the highest estimated marker allele sharing by affected siblings in that region. Thirty-six polymorphisms in both exonic and intronic regions distributed throughout the AHI1 gene were identified and are summarized in Table 1. Eleven changes within coding exons were found, five of which were in non-translated exons. Out of the six remaining exonic variants, three were non-synonymous changes within the conserved WD40 5 repeat and 7 codons downstream of WD40 7 repeat, domains highly conserved in mammals. These polymorphisms were previously reported in parents of JS-related syndrome patients, but did not segregate in their affected offspring (26). An additional maternally inherited insertion–deletion (indel) polymorphism, located 6 bp downstream of exon 20, and predicted to alter loop stability (see Materials and Methods), was also identified in a patient (Supplementary Material, Fig. S1) and was not in HapMap. On the basis of the potential functional consequence of this polymorphism, we screened 189 independent autistic probands and 320 unaffected sibling controls (see Materials and Methods), but did not find a significant difference in its frequency between patients and controls.
Table 1.
Re-sequencing results
| SNP ID/bp positiona | SNP | Location | Exon variants |
|---|---|---|---|
| rs13197301 | A/G | 5′-UTR | |
| rs13197384 | C/A | 5′-UTR | |
| 135860511 | C/T | Exon 1 | |
| rs9402709 | G/T | Intron 3 | |
| rs2757651 | T/C | Intron 6 | |
| 135826225 | G/A | Intron 6 | |
| 135820659 | G/A | Intron 7 | |
| 135818827 | T/C | Intron 8 | |
| rs4896147 | G/A | Intron 8 | |
| 135818503 | T/G | Intron 9 | |
| rs11964449 | A/G | Intron 10 | |
| 135810083 | A/T | Intron 11 | |
| 135805628–5631 | TTTT/- | Intron 12 | |
| rs17053651 | G/A | Intron 12 | |
| rs2757645 | G/A | Intron 12 | |
| rs2614274 | T/C | Intron 13 | |
| 135801436 | A/C | Intron 13 | |
| rs737561 | C/T | Intron 14 | |
| rs2273761 | A/G | Exon 15 | D741D |
| 135792717 | G/A | Exon 17 | R830Wa |
| 135791578 | C/T | Exon 18 | R835R |
| rs13218824 | T/C | Intron 18 | |
| rs2757639 | A/G | Intron 19 | |
| 135774342 | T/C | Exon 20 | Y933Ca |
| 135774153/8-172 | indel | Intron 20 | |
| 135757701 | T/C | Exon 22 | S1005S |
| 135757506 | C/T | Intron 22 | |
| 135681408 | G/A | Exon 25 | S1123Fa |
| rs6914831 | C/T | Intron 25 | |
| 13568282 | G/T | Intron 25 | |
| rs4896141 | C/G | Intron 26 | |
| rs11970282 | T/C | Intron 27 | |
| rs9494209 | C/G | Exon 28 | |
| rs1052502 | C/T | Exon 28 | |
| 135648179–81 | -/CA | Exon 28 | |
| 135647428 | T/A | Exon 28 |
SNP identity and location within the AHI1 gene are listed. The SNP NCBI rs ID is shown in column 1. Where the SNP ID is not reported, position in base pair is listed. Base pair position is according to Ensembl database (http://www.ensembl.org). SNP ID is based on NCBI dbSNP Build 126 database (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=snp). The exon/intron of the AHI1 gene where the SNP is found is reported in column 3. Amino acid changes are indicated where applicable in column 4.
aNon-synonymous variants located in the WD40 or downstream of SH3-conserved domains. See Results section for exact locations in domains.
SNP genotyping and association analysis
In a stage-1 screen to identify SNPs potentially associated with ASD, we analyzed the 13 coding variants that had a minor allele frequency (MAF) > 1% in the autistic sample, and that had been previously genotyped in control populations, such as HapMap. We compared control allele frequencies HapMap (n = 120 chromosomes for CEU sample), Perlegen (n = 48 chromosomes for EUR) and TSC panel (n = 84 chromosomes), with their frequencies in the AGRE probands (Table 2), identifying four SNPs rs4896141, rs11970282, rs9494209 and rs1052502 showing nominal association (P < 0.05, Table 2). Two of these, rs9494209 and rs11970282, were in complete LD with each other (r2 = 1) in the AGRE sample and in HapMap (http://www.hapmap.org), so SNP rs9494209 was chosen to represent these two SNPs in subsequent analyses.
Table 2.
Case–control association and AHI1 SNPs with ASD
| SNP | MAF autism sample | MAF controls | P-value |
|---|---|---|---|
| rs9402709 | G = 0.125 | G = 0.058 (Hmp) | 0.09 |
| rs2757651 | T = 0.05 | T = 0.025 (Hmp) | 0.3 |
| rs11964449 | G = 0.02 | G = 0.008 (Hmp) | 0.6 |
| G = 0.020 (Per) | |||
| rs17053651 | A = 0.02 | A = 0.021 (Per) | 1 |
| rs2757645 | G = 0.09 | G = 0.05 (Hmp) | 0.4 |
| rs737561 | C = 0.11 | C = 0.05 (Hmp) | 0.3 |
| C = 0.06 (TSC) | |||
| rs2273761 | G = 0.02 | G = 0.008 (Hmp) | 0.6 |
| G = 0.020 (Per) | |||
| rs2757639 | A = 0.10 | A = 0.05 (Hmp) | 0.1 |
| A = 0.042 (Per) | |||
| rs6914831 | C = 0.43 | C = 0.405 (Hmp) | 0.5 |
| C = 0.312 (Per) | |||
| rs4896141 | G = 0.11 | G = 0.05 (Hmp) | 0.02* |
| G = 0 (Per) | |||
| rs11970282 | C = 0.13 | C = 0.042 (Hmp) | 0.03* |
| rs9494209 | G = 0.11 | G = 0 (Per) | 0.02* |
| rs1052502 | T = 0.125 | T = 0.05 (Hmp) | 0.009* |
| T = 0 (Per) |
Allele frequencies of each SNP in the AGRE-sequenced sample, compared with HapMap (Hmp), Perlegen (Per) and SNP Consortium Ltd (TSC) control panels. 91.7% AGRE sample is White, 81.8% of this sample was White (not Hispanic or Latino) and the remaining 18.2% was White Hispanic or Latino. The populations used in each panel were the CEU (CEPH Utah residents with ancestry from northern and western Europe in HapMap); the EUR (population of European American descent in Perlegen) and the CSHL (Caucasian individuals in the TSC panel). The number of chromosomes genotyped was 120 for Hmp CEU (except for SNP rs11964449, where 118 chromosomes were typed, and SNP rs6914831 with 116 chromosomes), 48 for EUR in Perlegen and 84 for the TSC-CSHL. All genotypes were in H–W equilibrium. In the cases where more than one panel genotyped the same SNP, the P-values are the result of the combined panels compared with the 96 AGRE chromosome sample.
*Two-tailed P < 0.05 according to Fisher’s exact test.
These three independent SNPs that showed significant differences in frequency between patients and controls in the initial screen, rs1052502, rs9494209 and rs4896141, were then genotyped in 327 independent parent–child trios to permit an independent family-based association analysis to confirm this preliminary, nominal level of association in stage 2. Out of the 327 trios, 326 were successfully genotyped (99.7% successful call rate and 1 Mendelian error). One SNP, rs1052502, was not in Hardy–Weinberg (H–W) equilibrium in the parents (P = 0.0002) and was excluded from further analysis. Association testing, using the single-marker transmission disequilibrium test (TDT) in the WHAP software (33), was significant for SNPs rs9494209 and rs4896141 (Table 3) after Bonferroni correction (p_MAX = 0.02 and p_SUM = 0.01).
Table 3.
Single SNP TDT analysis in stage 2
| SNP ID | Overtransmitted allele | Odds ratio (T:U) | Empirical P-value |
|---|---|---|---|
| rs9494209 | C | 1.8; 95% CIE 1.34–2.26 (54:30) | 0.01 |
| rs4896141 | C | 1.64; 95% CIE 1.22–2.06 (59:36) | 0.02 |
Empirical P-values are obtained by performing 1000 permutations with transmitted versus untransmitted alleles using WHAP. The two individual tagging SNPs are significant at P < 0.05. The global permutation tests yielded significant P-values as well (p_max = 0.019 and p_sum = 0.015).
T:U, transmitted versus untransmitted.
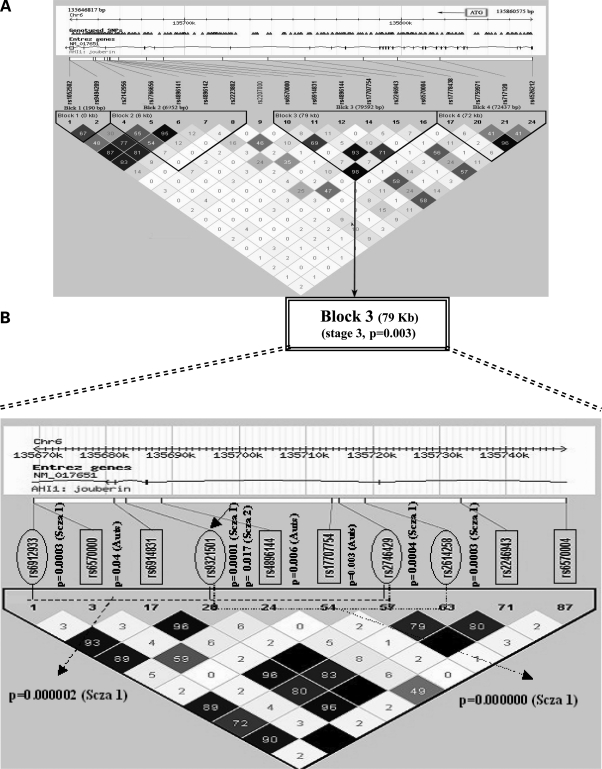
These results suggested that a more thorough investigation of AHI1 was warranted to more clearly define the associated region(s) in AHI1. In this final third stage, 18 SNPs, representing four haplotype blocks defined using Haploview (see Materials and Methods) were analyzed in 337 parent–child trios, 226 of which had been typed for the three SNPs in stage 2, and 111 of which were new. These four blocks capture 99% of the haplotype diversity within the AHI1 gene (Fig. 1A). Block 3, which extends over 79 kb, was significantly associated after a Bonferroni correction (P = 0.003) (Table 4). Remarkably, this block contains two SNPs (rs6914831, rs2246943) that serve as tags in this analysis for five SNPs (rs9321501, rs6912933, rs2614258, rs2746429 and rs6931735) that were previously shown to be associated with schizophrenia (34) (Fig. 1B). For example, SNP rs2246943 typed here is in complete LD (r2 = 1.0; Fig. 1B) with SNP rs9321501, which is at the center of the most significantly associated schizophrenia haplotype identified in two independent samples (1.9 × 10−10, ref 34).
Figure 1.
Fine-scale depiction of the AHI1 ASD-associated haplotype. The genomic region is shown on top, followed by an exon–intron map and NCBI genome release 35 SNP location. Shown below is an LD plot of the region based on r2 (see Materials and Methods). SNPs genotyped in the autism sample in this study are boxed, whereas those genotyped in the published schizophrenia associations (34,45) are circled. The single SNP P-values are shown adjacent to each SNP. Schizophrenia SNPs are marked ‘Scza 1’ or ‘Scza 2’ depending on whether they were typed in the Arab-Israeli (34) or in the Icelandic population (45). Associated SNPs in our study are marked as ‘Auts’. The two schizophrenia haplotypes in the Arab-Israeli study are depicted with discontinuous lines. One can see the ∼30 kb area of overlap of these two haplotypes in the center of this region, which occurs in a region of strong LD within the gene. The region also harbors the two most highly associated SNPs in our current autism study (rs4896144 and rs17707754), which are in the center of this block of low LD.
Table 4.
Haplotype TDT analysis in stage 3
| Haplotype | Frequency | T:U | Block empirical P-value (100K permutations) | Allele empirical P-value (100K permutations) | Odds ratio (95% CIE) |
|---|---|---|---|---|---|
| Block 3; markers: 10, 11, 12, 14, 15, 16 | 0.003 | ||||
| GTCTTT | 0.496 | 175:190 | NS | 0.9 (0.7–1.1) | |
| GCCTAT | 0.343 | 139:169 | NS | 0.8 (0.6–1) | |
| GTCCTT | 0.067 | 58:30 | 0.003 | 1.9 (1.5–2.4) | |
| ACTTAA | 0.056 | 43:25 | 0.02 | 1.7 (1.2–2.2) | |
| GCTTAT | 0.021 | 17:11 | NS | 1.5 (0.8–2.3) | |
| GTCTAT | 0.013 | 7:8 | NS | 0.9 (.01–1.9) |
Allele frequencies, transmitted alleles, odds ratios with 95% CIE and empirical P-values after 100K permutations using WHAP are shown for block 3 in all the trios analyzed in stage 3.
NS, non-significant.
Markers: 10: rs6570000, 11: rs6914831, 12: rs4896144, 14: rs17707754, 15: rs2246943, 16: rs6570004.
The contributions to the association of individual alleles of haplotype block 3 were further assessed in the stage 3 autism sample (337 trios) using the WHAP software (Table 4), revealing that two major alleles of this haplotype, occurring at frequencies of 6.7% and 5.6%, respectively, in ASD individuals, account for a large proportion of the association (empiric P-values of 0.003 and 0.02, respectively, after 100 000 permutations) (Table 4). Both alleles are overtransmitted (Table 4), the more common allele results in an odds ratio of 1.9 [95% confidence interval estimate (CIE) = 1.5–2.4], and the other results in an odds ratio of 1.7 (95% CIE 1.2–2.2). Although the region spanned by this ASD-associated haplotype encompasses the two haplotype blocks formed by three different SNPs associated with schizophrenia in previous studies (34), the same SNPs were not associated with ASD and schizophrenia (Fig. 1B), suggesting that the same region, but not the same allele, contributes to both disorders in the populations studied.
DISCUSSION
Autism, similar to other common diseases, is a complex genetic disorder with contributions from both rare and common genetic variants (13,31,32,35,36). In some genes, for example, neuroligins 3 and 4 (37,38), only rare mutations have been identified. For others, syndromic forms, as well as rare and common variant contributions from the same gene, have been identified, for example, CNTNAP2 (30,39). These published findings emphasize the striking genetic heterogeneity of ASD and support the potential utility of studying relatively rare genetic syndromes to help provide insight into its etiologies (40).
Here, we took a relatively straightforward approach to assess the role of common variants in AHI1, the first gene identified as mutated in JS, within the ASD spectrum. As a significant proportion of JS patients with mutations in AHI1 have autism, this was a biologically focused candidate gene analysis to test the hypothesis that this gene related to a syndromic cause of autism, also harbored variation-mediating common genetic susceptibility to ASD. Our analysis supports the hypothesis that common genetic variants within a gene that frequently results in a rare syndromic form of ASD can contribute to more general ASD susceptibility. These findings emphasize the importance of studying rare forms of neuropsychiatric conditions as a means to inform our understanding of common disease susceptibility and pathophysiology (19).
Here, we took a stepwise approach to our analyses of the gene. First, we re-sequenced it to identify common variants. We did not identify any clear coding mutations. Sequencing a much larger number of cases and controls, which is beyond the scope of the current project, would be more likely to demonstrate a rare variant contributing to ASD, as in Bakkaloglu et al. 2008 (41). However, we were able to identify two common variants that showed association with ASD in a large family-based cohort. Although we chose ethnically matched cohorts for stage 1, we acknowledge that ethnic stratification could be present, a factor eliminated in stage 2 with the family-based approach. We observed a significant association of two SNPs in the AHI1 gene, after correcting for multiple comparisons. In stage 3, we identified and more exhaustively studied all known common haplotypes in AHI1, demonstrating a significant association with ASD in the same circumscribed region that had previously been associated with schizophrenia. Although SNPs within the ASD-associated haplotype are in high LD with some of the schizophrenia-associated SNPs in HapMap, the specific alleles associated with each condition are distinct (Fig. 1).
The AHI1 region in autism and other neuropsychiatric diseases
The chromosome 6q region, where the AHI1 gene resides, is not considered a major ASD linkage region. However, two published studies have suggested an involvement of this locus in ASD (40,42). Phillipe et al. (42) report an excess of identity by descent (IBD) allele sharing on chromosome 6q (MLS = 2.23, IBD = 68.6%) in a small cohort of multiplex families from Sweden, France, Norway, the USA, Italy, Austria and Belgium. McCauley et al. (40) used genome-wide linkage and ordered-subset analysis (43), ranking families according to family-specific LOD scores at peak sites with LOD scores ≥1.5. They used clusters of ADI indices such as (i) to walk unaided; (ii) to sit unaided on flat surface; (iii) age of first single words; (iv) age of first phrase; (v–vi) acquisition of bladder control: daytime, night and (vii) acquisition of bowel control and found support for a suggestive autism locus on chromosome 6q23 in 158 combined Tufts, Vanderbilt and AGRE multiplex families. Large deletions on 6q have also been related to the autism phenotype (44), and a recent study using array comparative genome hybridization reported chromosomal re-arrangements in two autistic individuals from the same family in the same 6q region highlighted by the linkage studies cited earlier (39). However, no association between autism and specific genes in this region has been identified previously.
The current findings are notable within the context of the recent association of the AHI1 gene with schizophrenia in an inbred Arab population (34). Moreover, this association has been replicated in a large, case–control study performed in an independent European cohort (45). These association studies were conducted in the context of prior evidence of linkage, which has been described in the 6q AHI1 region for schizophrenia (46–48) and for which fine mapping pointed to 6q23 (49–51). The odds ratios for the risk variants in the European schizophrenia cohort were small, but significant, ranging from 1.15 to 1.29, whereas the odds ratio for the associated haplotypes identified here in this autism sample are larger, ranging from 1.7–1.9 (Table 4).
Remarkably, several of the SNPs that form the haplotype block associated with ASD are in strong LD with those previously associated with schizophrenia (r2 = 0.8–1.0, Fig. 1). For example, one of the tagging SNPs in the associated block reported here (rs6914831) is in strong LD and within 2.0 kb of rs9321501, which was one of those most strongly associated with schizophrenia (34,45). Moreover, this region of overlap within the region of the ASD-associated haplotype contains the most associated single SNPs in ASD, and a number of the most associated SNPs in both studies are within several kilobases of each other. Despite this proximity, the same SNPs and haplotype alleles are not associated with both ASD and schizophrenia (29,49). But, the two associated haplotype blocks identified in the previous schizophrenia study in Arab-Israelis (34) delineate a 30 kb region that is contained within the center of the larger haplotype block that was most significantly associated with ASD here (Block 3, Fig. 1; Table 4). This increases the evidence for a circumscribed region of the gene in both disorders, but not a particular shared allele. That different alleles are associated with each disorder is not surprising and is likely due to the very different ethnic backgrounds in the studies and also suggests that different specific variants within this region of AHI1 contribute to schizophrenia and ASD susceptibility.
Despite the differences between the two studies, which were drawn from distinct populations, and have different ascertainment schemes and diagnoses, the convergence on this locus suggests a possible role for AHI1 in human cognition and behavior that could be relevant to a wide body of psychopathology. This observation is consistent with the notion of significant etiological overlap between neuropsychiatric disorders that were previously considered distinct (5). Overlapping symptom domains have been observed between ASD and schizophrenia (52) and recent studies have even identified mutations that are observed in both autism (53) and childhood onset schizophrenia (54). These data eschew using the categorical clinical diagnosis as a phenotype with which to identify susceptibility genes for neuropsychiatric conditions and support the exploration of endophenotypes related to language, social cognition or brain development that may reflect common susceptibility at a neurobiological level. One can speculate that given their clustering within a small region of the AHI1 gene, these alleles act through a common biologic mechanism. Testing this hypothesis awaits further replication and deep re-sequencing of this candidate gene across allied disorders, an approach that is likely to be productive in elucidating the full extent to which AHI1 contributes to neuropsychiatric susceptibility. In addition, these results support the idea that common variants in genes known to cause rare, Mendelian forms of a neuropsychiatric condition can contribute to common disease susceptibility, as has been observed previously in metabolic disorders (55) and suggested in ASD (41,44).
In this regard, it is interesting to speculate on the manner in which AHI1 might influence ASD susceptibility. The gene is expressed widely, and at high levels in the CNS. Patients with JS have a characteristic hindbrain malformation, involving the cerebellum and brainstem, as well as abnormalities in the cerebral cortex (56). Further study of the anatomical and biological basis of the cognitive and behavioral deficits in JS, including comparison of patients with and without ASD, will likely provide important clues as to the role of specific abnormalities in the CNS and their relationship to autistic behaviors. Parallel assessment of brain structure and function in those carrying AHI1 risk alleles, regardless of their diagnoses, may also aid in defining the manner by which common variants of this gene influence cognition and behavior.
MATERIALS AND METHODS
Subjects
Forty-eight independent autistic individuals from the AGRE sample were selected for sequencing. These individuals were ascertained because they exhibited the highest likelihood of marker allele sharing with their affected siblings in the region harboring the AHI1 gene on the basis of the closest flanking markers D6S1040 (128.93 cM) and D61009 (137.74 cM) used in our previous genome scans (57,58) The individuals selected had the maximum IBD marker allele sharing of both alleles (Genehunter v 2.1).
For association testing at stage 2, we selected 981 individuals representing 327 trios (parents and their ASD-affected proband), which were genotyped for three SNPs at the AHI1 locus. Affected individuals were ascertained for the diagnosis of ASD under the categories of autism, broad spectrum and not quite autism using the Autism Diagnostic Interview—Revised (ADI-R) domain scores (http://www.agre.org) (3,59–61) (Supplementary Material, Table S1). Individuals with non-idiopathic autism, including Fragile X syndrome, chromosomal abnormalities and other medical conditions (http://www.agre.org) were not included in this study.
For stage 3, we genotyped 1011 individuals from 337 AGRE parent–child trios, 111 of which were independent of those analyzed in stage 2 (see Supplementary Material, Table S1 for a list of all the individuals used in all stages). We also screened for a complex indel downstream of exon 20 in 189 autistic probands and 319 unaffected siblings used as controls from the AGRE population (Supplementary Material, Table S2). More than half of these 189 autistic patients (62%) were also genotyped in stage 2.
Both AGRE and UCLA have protocols that are approved by respective Institutional Review Boards.
Sequencing
Sequencing was performed at DOE Joint Genome Institute (JGI) (http://www.jgi.doe.gov/). Briefly, primers were designed to give a maximum product size of 500 bp and a minimum of 40 bp flanking the splice sites using the Exon Locator & Extractor for Resequencing program (EXLR) (http://mutation.swmed.edu/ex-lax/). For a complete list of primers, see Appendix 1. An M13F tag (gttttcccagtcacgacgttgta) and an M13R tag (aggaaacagctatgaccat) were added to forward and reverse primers, respectively. Ten nanograms of DNA from each sample was amplified in a 10 μl PCR reaction using AmpliTaq Gold® (Applied Biosystems) and purified using the PCR product pre-sequencing kit (USB Corporation). Sequencing reactions were performed using the M13 primers along with BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) (http://www.jgi.doe.gov/sequencing/protocols/archive/BigDye3.1auto1.0.doc) and purified with tetra-ethylene glycol before separation on a 3730xl DNA Analyzer (ABI). Base calling, quality assessment and assembly were carried out using the Phred, Phrap, Polyphred, Consed software suite (www.phrap.org). All sequence variants identified were verified by manual inspection of the chromatograms (see Supplementary Material, Table S1 for a list of all the individuals sequenced).
SNP genotyping
SNPs rs1052502, rs9494209 and rs4896141 were genotyped in stage 2 by Illumina, Inc. in a set of 327 independent, parent–child trios from the AGRE sample (http://www.illumina.com). For stage 3, we initially selected 27 tagging SNPs having an MAF = 1% in the CEU population for genotyping using HapMap data (NCBI build 35) and Tagger (two-point and aggressive multimarker tagging options) to cover the entire gene at an r2 = 0.8. Twenty-five of these (93%) were successfully genotyped (call rate range 99.2–100%). For a subsequent statistical analysis, only SNPs with an MAF ≥ 5% in our sample of parents were used. After removing SNPs not in H–W equilibrium in parents (n = 1) and those with MAF < 5% (n = 6), 18 SNPs typed in 337 trios remained (see Supplementary Material, Table S3 for a list of SNPs). These included SNPs rs9494209 and rs4896141 for which we found nominal association in stage 2 and four SNPs (rs2246943, rs4526212, rs6914831 and rs7759971) that tagged distinct regions of this gene that were previously associated with schizophrenia.
Re-sequencing and digestion of a complex indel polymorphism
We re-sequenced an indel polymorphism identified in two individuals in our first round of rare variant identification in 48 subjects. We screened this indel in 189 independent autistic probands from different families and 319 unaffected siblings by PCR amplification and restriction enzyme digestion. Exon 20 (197 bp) + 366 bp of intronic sequence (181+185 bp on either side of the exon) were amplified from genomic DNA using primers AHI_20F and AHI_20R (see Appendix 1). The 563/568 bp PCR product was digested with HpyCH4V. The digestion pattern for the normal allele, five bands of 22, 49, 89, 147 and 256 bp, changed to four bands of 22, 89, 201 and 256 bp, respectively, in the presence of the complex indel. This banding pattern was resolved in a 4% NuSieve gel by electrophoresis. We then used computational methods to predict possible alternative splicing events [Berkeley Drosophila Genome Project (BDGP) (http://www.fruitfly.org/seq_tools/splice.html) and SpliceView (http://l25.itba.mi.cnr.it/~webgene/wwwspliceview.html)] mediated by this polymorphism. We also checked potential alterations in exonic splicing enhancer (ESE) sequences (http://rulai.cshl.edu/tools/ESE/ESEbkgr.html) and in ACESCAN2 web server (http://genes.mit.edu/acescan2/). Finally, hairpin loop stability changes were predicted using the HairpinFetcher Institute of Genomics & Interative Technology website, http://miracle.igib.res.in/hfinder/.
Statistical methods
Genehunter v 2.1 was used to estimate IBD marker allele sharing in the chromosomal region surrounding the gene AHI1 to flag those individuals most likely to contain AHI1 mutations, in order to select the subjects for AHI1 re-sequencing. In stage 1, we compared the frequencies of the variants identified by re-sequencing to public ‘control’ frequencies available in HapMap (Hmp) (http://www.hapmap.org,), Perlegen (Per) (http://www.perlegen.com) and the SNP Consortium panels (TSC) (http://snp.cshl.org) matching for ethnicity. This allowed us to select for stage 2 those SNPs showing frequency differences in ASD subjects when compared with publicly available controls. To compare frequencies, a two-tailed χ2 test of difference in proportions was conducted. For alleles of low frequency, Fisher’s exact test was conducted. SNPs chosen for stage 2 follow-up had a P-value <0.05. Stage 1 involved a small case–control population that was subject to potential errors induced by population stratification, whereas in stage 2, we used a relatively large family-based design to control for stratification, and the TDT analysis was conducted.
Haploview v 4.ORC2 (62) was used to detect Mendelian errors, assess linkage disequilibrium, test for H–W equilibrium, estimate the frequencies of transmitted and transmitted alleles, form haplotype blocks and conduct the single SNP TDT analyses. Haplotype TDT analyses were conducted using the haplotype-based association analysis package WHAP (33) (http://pngu.mgh.harvard.edu/~purcell/whap/). WHAP allows the estimation of empirical P-values through permutations based on Monte Carlo simulations. Permutations are performed by randomly swapping the within-family components of association (the transmitted versus untransmitted alleles). Significance was assessed by two statistics, p_MAX (maximum) and p_SUM (summed). The p_Max P-value is the number of times that a P-value less than or equal to the maximum P-value for the observed data in the distribution of P-values for all the permutations performed across all the SNPs. p_Sum is similar, but the statistic is the sum of the permutated scores for all the SNPs tested. Both reflect region-wide significance and correct for multiple testing in that region.
ELECTONIC DATABASE INFORMATION
ACESCAN2: http://genes.mit.edu/acescan2/.
Autism Genetic Resource Exchange (AGRE): http://www.agre.org.
Applied Biosystems: http://www.jgi.doe.gov/sequencing/protocols/archive/BigDye3.1auto1.0.doc.
Berkeley Drosophila Genome Project (BDGP): http://www.fruitfly.org/seq_tools/splice.html.
DOE Joing Genome institute (JGI): http://www.jgi.doe.gov/.
ESE finder: http://rulai.cshl.edu/tools/ESE/ESEbkgr.html.
Exon Locator & Extractor for Resequencing program (EXLR): http://mutation.swmed.edu/ex-lax/.
HapMap (Hmp): http://www.hapmap.org.
HairpinFetcher Institute of Genomics & Interative Technology: http://miracle.igib.res.in/hfinder/.
Illumina: http://www.illumina.com.
NCBI dbSNP Build 126 database: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=snp.
Phred, Phrap, Polyphred, Consed software suite: www.phrap.org.
Perlegen (Per): http://www.perlegen.com.
SNP Consortium (TSC): http://snp.cshl.org.
The low-density lipoprotein receptor (LDLR) gene in familial hypercholesterolemia: http://www.ucl.ac.uk/fh/.
WHAP software: http://pngu.mgh.harvard.edu/~purcell/whap/.
Retinal Degeneration Retnet Network Database: http://www.sph.uth.tmc.edu/retnet.
SpliceView: http://l25.itba.mi.cnr.it/~webgene/wwwspliceview.html.
SUPPLEMENTARY MATERIAL
FUNDING
This project was supported by the National Institute of Health research grants R01 (MH64547 to D.H.G. and ACE Genetics Network grant MH081754 to D.H.G.); the National Institute of Mental Health; the National Institute of Child Health and Human Development; the National Institute of Deafness and Other Communication Disorders; the National Institute of Neurological Disorders and Stroke (U54 MH068172 to D.H.G., M. Sigman, PI); Simons Foundation to J.G.G.; Department of Energy Contract, University of California, E.O. Lawrence Berkeley National Laboratory (DE-AC02-05CH11231 to L.A.P.).
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge the resources provided by the Autism Genetic Resource Exchange (AGRE) Consortium given in Appendix 2 (http://www.agre.org) and the participating AGRE families. The AGRE is a program of Cure Autism Now and is supported, in part, by grant MH64547 from the National Institute of Mental Health to D.H.G. (PI). In addition, we would like to thank Drs Jennifer Stone, Maricela Alarcón and Rebecca Mar-Heyming for helpful discussions, as well as Jackie Duvall for software assistance. We also thank the reviewers for their helpful comments.
Conflict of Interest statement. None declared.
APPENDIX 1
COMPLETE LIST OF PRIMERS FOR AHI1
| AHI1_1F | CTGGAAGAAGGCGGTGG |
| AHI1_1R | CGGTCACTACGCTATCAGCC |
| AHI1_2F | CGCTTCTTTCACTGGTCTCC |
| AHI1_2R | TAAACGCAGCACAACCTTTG |
| AHI1_3F | TTTCCCTTGATTTTAGAGCCC |
| AHI1_3R | TTGTCCATCTGAGTCCCAAAC |
| AHI1_4F | AAAGGTCAGAGGGATACAGGTG |
| AHI1_4R | TGTTATTGACCATGACATAGTGTTAC |
| AHI1_5F | CAAAATGAATCCAAAGTGTTAATCC |
| AHI1_5R | GAGCAAGAAAACCCAACTGC |
| AHI1_6_1F | GAAAATAGCATTTGAGATTTGAGC |
| AHI1_6_1R | CTTCTCTTCCCTCATTTGCC |
| AHI1_6_2F | GGTGCCCCAGTTGACTACAC |
| AHI1_6_2R | GGGTAGGGACAAGGATCTGG |
| AHI1_7F | ACCACCTATGATACCTATTTGACAC |
| AHI1_7R | TCAGAGAAGACTCAAGTGATACAAAC |
| AHI1_8F | ATGGTTTGCTGTTGTCTGGC |
| AHI1_8R | AGATGTTCTCCATTTCTTCTACAAC |
| AHI1_9F | TCCCAGCTTTATATGGCAAAG |
| AHI1_9R | TCACTTGATTCCACAGCATTG |
| AHI1_10F | TCAGAAAATGGACCCTCCC |
| AHI1_10R | GAATGAGCATAACCTGAGCTTG |
| AHI1_11F | GCCATTTGGGCTACCTTTTG |
| AHI1_11R | TTTTCCTAATACTAAAAGCCACATTTC |
| AHI1_12F | ACAGGACTGTAGTTTTAAGCAGC |
| AHI1_12R | TGCTTATATACACATGCTAGGCAC |
| AHI1_13F | TGTGGCAGTGATGGCTTTAG |
| AHI1_13R | TCCCCATGAGATTTATTCATCC |
| AHI1_14F | TGTGCTGCAAATGTCTTTGG |
| AHI1_14R | TTATGACAGTCCCTTCTTGGG |
| AHI1_15F | TGGTTTACCATTGGTCGTTG |
| AHI1_15R | GACCTGCAAAATAAGCATAAACC |
| AHI1_16F | ACCTTTTAATAGCCCTTCAATTC |
| AHI1_16R | ATCAGTCAGCCATCAGGAGG |
| AHI1_17F | GACGTTACTCAGAATCCTTTTGC |
| AHI1_17R | ATGTTCCTTGGTGAGTGTGG |
| AHI1_18F | CTGGGCTCTTGGGACTAGC |
| AHI1_18R | AAACATAATTGAAAACCCAAAATC |
| AHI1_19F | AGAAGGAGGAGGGTCAGTGG |
| AHI1_19R | TGATTCCAACATAAGGGCAC |
| AHI1_20F | TGCTTCCTTATGATTTTATAGCATTG |
| AHI1_20R | CCTAAATTCAGGTTGTCTGTTGC |
| AHI1_21F | AATGGATAAAGAAACTTCCAAATG |
| AHI1_21R | TGAGCTCCAATAATGAAAGTACAC |
| AHI1_22F | TGTACATCATTACCACTCCTTTTG |
| AHI1_22R | AACTTACTTTTGAAATAACCTTGGTG |
| AHI1_23F | CATCTTAAAAGGATCTTGCCTTC |
| AHI1_23R | TGCAGGGATATAACTTTTGGC |
| AHI1_24F | CCTCATCCCATTAGCAAACC |
| AHI1_24R | CAGCTTCTAATGTACTAGTGCCTCC |
| AHI1_25F | TTGCAAATTGCCGTAACAAG |
| AHI1_25R | TGGCTATAACGTTTGCCTTATG |
| AHI1_26F | GACAATATATGTTCAGGACAGGC |
| AHI1_26R | AAAATCGTAAACAAGCTAAACTCC |
| AHI1_27F | TCATATCCTCACATTTATCCCC |
| AHI1_27R | CCGGTTACAGATCCCAAATG |
| AHI1_28_1F | GAGGCAAACTAGAAGATAGGAAATTAG |
| AHI1_28_1R | GCACCGTGCCCAGTCAG |
| AHI1_28_2F | TGTGAATAAAAGGTGTTTGCG |
| AHI1_28_2R | GAACATTGTTGCCAGTCCAG |
| AHI1_28_3F | ATGGCGCATTCAAAACAGTC |
| AHI1_28_3R | CAAAATTTACACCTCAATGCCC |
| AHI1_28_4F | GCTGATAAAATATTTAACCCCAAG |
| AHI1_28_4R | TAGCAAATGCTTAGCTCCCC |
| AHI1_25ALTF | AGGGCAGTCATGTTCTTTGG |
| AHI1_25ALTR | CACTGGCATACCCAAATCCT |
APPENDIX 2
AGRE CONSORTIUM
Daniel H. Geschwind, University of California at Los Angeles, Los Angeles; Maja Bucan, University of Pennsylvania, Philadelphia; W. Ted Brown, New York State Institute for Basic Research in Developmental Disabilities, Long Island; Joseph D. Buxbaum, Mt Sinai School of Medicine, New York; Rita M. Cantor, University of California, Los Angeles; John N. Constantino, Washington University School of Medicine, St Louis; T. Conrad Gilliam, Columbia Genome Center, New York; Clara Lajonchere, Cure Autism Now, Los Angeles; David H. Ledbetter, Emory University, Atlanta; Christa Lese-Martin, Emory University, Atlanta; Janet Miller, Cure Austism Now, Los Angeles; Stanley F. Nelson, University of California at Los Angeles School of Medicine, Los Angeles; Gerard D. Schellenberg, University of Washington and Veterans Affairs Medical Center, Seattle; Carole Samango-Sprouse, Children’s National Medical Center, Baltimore; Sarah J. Spence, University of California, Los Angeles; Rudolph E. Tanzi, Massachusetts General Hospital, Boston.
REFERENCES
- 1.WHO. The international conference for the tenth revision of the International Classification of Diseases. Strengthening of Epidemiological and Statistical Services Unit. World Health Organization, Geneva. World Health Stat. Q. 1990;43:204–245. [PubMed] [Google Scholar]
- 2.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 4th edn. Washington, DC: American Psychiatric Association; 1994. [Google Scholar]
- 3.Rutter M. Genetic studies of autism: from the 1970s into the millennium. J. Abnorm. Child Psychol. 2000;28:3–14. doi: 10.1023/a:1005113900068. [DOI] [PubMed] [Google Scholar]
- 4.Folstein S.E., Rosen-Sheidley B. Genetics of autism: complex aetiology for a heterogeneous disorder. Nat. Rev. Genet. 2001;2:943–955. doi: 10.1038/35103559. [DOI] [PubMed] [Google Scholar]
- 5.Abrahams B.S., Geschwind D.H. Advances in autism genetics: on the threshold of a new neurobiology. Nat. Rev. Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailey A., Le Couteur A., Gottesman I., Bolton P., Simonoff E., Yuzda E., Rutter M. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- 7.Ritvo E.R., Jorde L.B., Mason-Brothers A., Freeman B.J., Pingree C., Jones M.B., McMahon W.M., Petersen P.B., Jenson W.R., Mo A. The UCLA–University of Utah epidemiologic survey of autism: recurrence risk estimates and genetic counseling. Am. J. Psychiatry. 1989;146:1032–1036. doi: 10.1176/ajp.146.8.1032. [DOI] [PubMed] [Google Scholar]
- 8.Bailey A., Phillips W., Rutter M. Autism: towards an integration of clinical, genetic, neuropsychological, and neurobiological perspectives. J. Child Psychol. Psychiatry. 1996;37:89–126. doi: 10.1111/j.1469-7610.1996.tb01381.x. [DOI] [PubMed] [Google Scholar]
- 9.Fombonne E. Epidemiology of autistic disorder and other pervasive developmental disorders. J. Clin. Psychiatry. 2005;66(Suppl. 10):3–8. [PubMed] [Google Scholar]
- 10.Pickles A., Bolton P., Macdonald H., Bailey A., Le Couteur A., Sim C.H., Rutter M. Latent-class analysis of recurrence risks for complex phenotypes with selection and measurement error: a twin and family history study of autism. Am. J. Hum. Genet. 1995;57:717–726. [PMC free article] [PubMed] [Google Scholar]
- 11.Risch N., Spiker D., Lotspeich L., Nouri N., Hinds D., Hallmayer J., Kalaydjieva L., McCague P., Dimiceli S., Pitts T., et al. A genomic screen of autism: evidence for a multilocus etiology. Am. J. Hum. Genet. 1999;65:493–507. doi: 10.1086/302497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schellenberg G.D., Dawson G., Sung Y.J., Estes A., Munson J., Rosenthal E., Rothstein J., Flodman P., Smith M., Coon H., et al. Evidence for multiple loci from a genome scan of autism kindreds. Mol. Psychiatry. 2006;11:1049–1060. doi: 10.1038/sj.mp.4001874. 979. August 1, Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 13.Sebat J., Lakshmi B., Malhotra D., Troge J., Lese-Martin C., Walsh T., Yamrom B., Yoon S., Krasnitz A., Kendall J., et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szatmari P., Paterson A.D., Zwaigenbaum L., Roberts W., Brian J., Liu X.Q., Vincent J.B., Skaug J.L., Thompson A.P., Senman L., et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat. Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCarthy M.I. Progress in defining the molecular basis of type 2 diabetes mellitus through susceptibility-gene identification. Hum. Mol. Genet. 2004;13(Spec no. 1):R33–R41. doi: 10.1093/hmg/ddh057. [DOI] [PubMed] [Google Scholar]
- 16.Naukkarinen J., Ehnholm C., Peltonen L. Genetics of familial combined hyperlipidemia. Curr. Opin. Lipidol. 2006;17:285–290. doi: 10.1097/01.mol.0000226121.27931.3f. [DOI] [PubMed] [Google Scholar]
- 17.Tanzi R.E., Bertram L. 20 years of the Alzheimer’s disease amyloid hypothesis: a genetic perspective. Cell. 2005;120:545–555. doi: 10.1016/j.cell.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 18.Fisher S.E., Lai C.S., Monaco A.P. Deciphering the genetic basis of speech and language disorders. Annu. Rev. Neurosci. 2003;26:57–80. doi: 10.1146/annurev.neuro.26.041002.131144. [DOI] [PubMed] [Google Scholar]
- 19.Peltonen L., Perola M., Naukkarinen J., Palotie A. Lessons from studying monogenic disease for common disease. Hum. Mol. Genet. 2006;15(Spec no. 1):R67–R74. doi: 10.1093/hmg/ddl060. [DOI] [PubMed] [Google Scholar]
- 20.Ozonoff S., Williams B.J., Gale S., Miller J.N. Autism and autistic behavior in Joubert syndrome. J. Child Neurol. 1999;14:636–641. doi: 10.1177/088307389901401003. [DOI] [PubMed] [Google Scholar]
- 21.Artigas-Pallares J., Gabau-Vila E., Guitart-Feliubadalo M. Syndromic autism. II. Genetic syndromes associated with autism. Rev. Neurol. 2005;40(Suppl. 1):S151–S162. [PubMed] [Google Scholar]
- 22.Holroyd S., Reiss A.L., Bryan R.N. Autistic features in Joubert syndrome: a genetic disorder with agenesis of the cerebellar vermis. Biol. Psychiatry. 1991;29:287–294. doi: 10.1016/0006-3223(91)91291-x. [DOI] [PubMed] [Google Scholar]
- 23.Kumandas S., Akcakus M., Coskun A., Gumus H. Joubert syndrome: review and report of seven new cases. Eur. J. Neurol. 2004;11:505–510. doi: 10.1111/j.1468-1331.2004.00819.x. [DOI] [PubMed] [Google Scholar]
- 24.Dixon-Salazar T., Silhavy J.L., Marsh S.E., Louie C.M., Scott L.C., Gururaj A., Al-Gazali L., Al-Tawari A.A., Kayserili H., Sztriha L., et al. Mutations in the AHI1 gene, encoding jouberin, cause Joubert syndrome with cortical polymicrogyria. Am. J. Hum. Genet. 2004;75:979–987. doi: 10.1086/425985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferland R.J., Eyaid W., Collura R.V., Tully L.D., Hill R.S., Al-Nouri D., Al-Rumayyan A., Topcu M., Gascon G., Bodell A., et al. Abnormal cerebellar development and axonal decussation due to mutations in AHI1 in Joubert syndrome. Nat. Genet. 2004;36:1008–1013. doi: 10.1038/ng1419. [DOI] [PubMed] [Google Scholar]
- 26.Valente E.M., Brancati F., Silhavy J.L., Castori M., Marsh S.E., Barrano G., Bertini E., Boltshauser E., Zaki M.S., Abdel-Aleem A., et al. AHI1 gene mutations cause specific forms of Joubert syndrome-related disorders. Ann. Neurol. 2006;59:527–534. doi: 10.1002/ana.20749. [DOI] [PubMed] [Google Scholar]
- 27.Parisi M.A., Doherty D., Eckert M.L., Shaw D.W., Ozyurek H., Aysun S., Giray O., Al Swaid A., Al Shahwan S., Dohayan N., et al. AHI1 mutations cause both retinal dystrophy and renal cystic disease in Joubert syndrome. J. Med. Genet. 2006;43:334–339. doi: 10.1136/jmg.2005.036608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang X., Hanna Z., Kaouass M., Girard L., Jolicoeur P. Ahi-1, a novel gene encoding a modular protein with WD40-repeat and SH3 domains, is targeted by the Ahi-1 and Mis-2 provirus integrations. J. Virol. 2002;76:9046–9059. doi: 10.1128/JVI.76.18.9046-9059.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Braddock B.A., Farmer J.E., Deidrick K.M., Iverson J.M., Maria B.L. Oromotor and communication findings in Joubert syndrome: further evidence of multisystem apraxia. J. Child Neurol. 2006;21:160–163. doi: 10.1177/08830738060210020501. [DOI] [PubMed] [Google Scholar]
- 30.Strauss K.A., Puffenberger E.G., Huentelman M.J., Gottlieb S., Dobrin S.E., Parod J.M., Stephan D.A., Morton D.H. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N. Engl. J. Med. 2006;354:1370–1377. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- 31.Alarcon M., Abrahams B.S., Stone J.L., Duvall J.A., Perederiy J.V., Bomar J.M., Sebat J., Wigler M., Martin C.L., Ledbetter D.H., et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am. J. Hum. Genet. 2008;82:150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arking D.E., Cutler D.J., Brune C.W., Teslovich T.M., West K., Ikeda M., Rea A., Guy M., Lin S., Cook E.H., et al. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am. J. Hum. Genet. 2008;82:160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purcell S., Daly M.J., Sham P.C. WHAP: haplotype-based association analysis. Bioinformatics. 2007;23:255–256. doi: 10.1093/bioinformatics/btl580. [DOI] [PubMed] [Google Scholar]
- 34.Amann-Zalcenstein D., Avidan N., Kanyas K., Ebstein R.P., Kohn Y., Hamdan A., Ben-Asher E., Karni O., Mujaheed M., Segman R.H., et al. AHI1, a pivotal neurodevelopmental gene, and C6orf217 are associated with susceptibility to schizophrenia. Eur. J. Hum. Genet. 2006;14:1111–1119. doi: 10.1038/sj.ejhg.5201675. [DOI] [PubMed] [Google Scholar]
- 35.Comoletti D., De Jaco A., Jennings L.L., Flynn R.E., Gaietta G., Tsigelny I., Ellisman M.H., Taylor P. The Arg451Cys-neuroligin-3 mutation associated with autism reveals a defect in protein processing. J. Neurosci. 2004;24:4889–4893. doi: 10.1523/JNEUROSCI.0468-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ylisaukko-oja T., Rehnstrom K., Auranen M., Vanhala R., Alen R., Kempas E., Ellonen P., Turunen J.A., Makkonen I., Riikonen R., et al. Analysis of four neuroligin genes as candidates for autism. Eur. J. Hum. Genet. 2005;13:1285–1292. doi: 10.1038/sj.ejhg.5201474. [DOI] [PubMed] [Google Scholar]
- 37.Jamain S., Quach H., Betancur C., Rastam M., Colineaux C., Gillberg I.C., Soderstrom H., Giros B., Leboyer M., Gillberg C., et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat. Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laumonnier F., Bonnet-Brilhault F., Gomot M., Blanc R., David A., Moizard M.P., Raynaud M., Ronce N., Lemonnier E., Calvas P., et al. X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am. J. Hum. Genet. 2004;74:552–557. doi: 10.1086/382137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koochek M., Harvard C., Hildebrand M.J., Van Allen M., Wingert H., Mickelson E., Holden J.J., Rajcan-Separovic E., Lewis M.E. 15q duplication associated with autism in a multiplex family with a familial cryptic translocation t(14;15)(q11.2;q13.3) detected using array-CGH. Clin. Genet. 2006;69:124–134. doi: 10.1111/j.1399-0004.2005.00560.x. [DOI] [PubMed] [Google Scholar]
- 40.McCauley J.L., Li C., Jiang L., Olson L.M., Crockett G., Gainer K., Folstein S.E., Haines J.L., Sutcliffe J.S. Genome-wide and ordered-subset linkage analyses provide support for autism loci on 17q and 19p with evidence of phenotypic and interlocus genetic correlates. BMC Med. Genet. 2005;6:1. doi: 10.1186/1471-2350-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bakkaloglu B., O’Roak B.J., Louvi A., Gupta A.R., Abelson J.F., Morgan T.M., Chawarska K., Klin A., Ercan-Sencicek A.G., Stillman A.A., et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am. J. Hum. Genet. 2008;82:165–173. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Philippe A., Martinez M., Guilloud-Bataille M., Gillberg C., Rastam M., Sponheim E., Coleman M., Zappella M., Aschauer H., Van Maldergem L., et al. Genome-wide scan for autism susceptibility genes. Paris Autism Research International Sibpair Study. Hum. Mol. Genet. 1999;8:805–812. doi: 10.1093/hmg/8.5.805. [DOI] [PubMed] [Google Scholar]
- 43.Hauser E.R., Watanabe R.M., Duren W.L., Bass M.P., Langefeld C.D., Boehnke M. Ordered subset analysis in genetic linkage mapping of complex traits. Genet. Epidemiol. 2004;27:53–63. doi: 10.1002/gepi.20000. [DOI] [PubMed] [Google Scholar]
- 44.Sukumar S., Wang S., Hoang K., Vanchiere C.M., England K., Fick R., Pagon B., Reddy K.S. Subtle overlapping deletions in the terminal region of chromosome 6q24.2–q26: three cases studied using FISH. Am. J. Med. Genet. 1999;87:17–22. [PubMed] [Google Scholar]
- 45.Ingason A., Sigmundsson T., Steinberg S., Sigurdsson E., Haraldsson M., Magnusdottir B.B., Frigge M.L., Kong A., Gulcher J., Thorsteinsdottir U., et al. Support for involvement of the AHI1 locus in schizophrenia. Eur. J. Hum. Genet. 2007;15:988–991. doi: 10.1038/sj.ejhg.5201848. [DOI] [PubMed] [Google Scholar]
- 46.Martinez M., Goldin L.R., Cao Q., Zhang J., Sanders A.R., Nancarrow D.J., Taylor J.M., Levinson D.F., Kirby A., Crowe R.R., et al. Follow-up study on a susceptibility locus for schizophrenia on chromosome 6q. Am. J. Med. Genet. 1999;88:337–343. [PubMed] [Google Scholar]
- 47.Levinson D.F., Holmans P., Straub R.E., Owen M.J., Wildenauer D.B., Gejman P.V., Pulver A.E., Laurent C., Kendler K.S., Walsh D., et al. Multicenter linkage study of schizophrenia candidate regions on chromosomes 5q, 6q, 10p, and 13q: schizophrenia linkage collaborative group III. Am. J. Hum. Genet. 2000;67:652–663. doi: 10.1086/303041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bailer U., Leisch F., Meszaros K., Lenzinger E., Willinger U., Strobl R., Heiden A., Gebhardt C., Doge E., Fuchs K., et al. Genome scan for susceptibility loci for schizophrenia and bipolar disorder. Biol. Psychiatry. 2002;52:40–52. doi: 10.1016/s0006-3223(02)01320-3. [DOI] [PubMed] [Google Scholar]
- 49.Lerer B., Segman R.H., Hamdan A., Kanyas K., Karni O., Kohn Y., Korner M., Lanktree M., Kaadan M., Turetsky N., et al. Genome scan of Arab Israeli families maps a schizophrenia susceptibility gene to chromosome 6q23 and supports a locus at chromosome 10q24. Mol. Psychiatry. 2003;8:488–498. doi: 10.1038/sj.mp.4001322. [DOI] [PubMed] [Google Scholar]
- 50.Duan J., Martinez M., Sanders A.R., Hou C., Saitou N., Kitano T., Mowry B.J., Crowe R.R., Silverman J.M., Levinson D.F., et al. Polymorphisms in the trace amine receptor 4 (TRAR4) gene on chromosome 6q23.2 are associated with susceptibility to schizophrenia. Am. J. Hum. Genet. 2004;75:624–638. doi: 10.1086/424887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Levi A., Kohn Y., Kanyas K., Amann D., Pae C.U., Hamdan A., Segman R.H., Avidan N., Karni O., Korner M., et al. Fine mapping of a schizophrenia susceptibility locus at chromosome 6q23: increased evidence for linkage and reduced linkage interval. Eur. J. Hum. Genet. 2005;13:763–771. doi: 10.1038/sj.ejhg.5201406. [DOI] [PubMed] [Google Scholar]
- 52.Sporn A.L., Addington A.M., Gogtay N., Ordonez A.E., Gornick M., Clasen L., Greenstein D., Tossell J.W., Gochman P., Lenane M., et al. Pervasive developmental disorder and childhood-onset schizophrenia: comorbid disorder or a phenotypic variant of a very early onset illness? Biol. Psychiatry. 2004;55:989–994. doi: 10.1016/j.biopsych.2004.01.019. [DOI] [PubMed] [Google Scholar]
- 53.Weiss L.A., Shen Y., Korn J.M., Arking D.E., Miller D.T., Fossdal R., Saemundsen E., Stefansson H., Ferreira M.A., Green T., et al. Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 54.Walsh T., McClellan J.M., McCarthy S.E., Addington A.M., Pierce S.B., Cooper G.M., Nord A.S., Kusenda M., Malhotra D., Bhandari A., et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 55.Orth M., Schapira A.H. Mitochondria and degenerative disorders. Am. J. Med. Genet. 2001;106:27–36. doi: 10.1002/ajmg.1425. [DOI] [PubMed] [Google Scholar]
- 56.Parisi M.A., Doherty D., Chance P.F., Glass I.A. Joubert syndrome (and related disorders) (OMIM 213300) Eur. J. Hum. Genet. 2007;15:511–521. doi: 10.1038/sj.ejhg.5201648. [DOI] [PubMed] [Google Scholar]
- 57.Badner J.A., Gershon E.S. Regional meta-analysis of published data supports linkage of autism with markers on chromosome 7. Mol. Psychiatry. 2002;7:56–66. doi: 10.1038/sj.mp.4000922. [DOI] [PubMed] [Google Scholar]
- 58.Yonan A.L., Alarcon M., Cheng R., Magnusson P.K., Spence S.J., Palmer A.A., Grunn A., Juo S.H., Terwilliger J.D., Liu J., et al. A genomewide screen of 345 families for autism-susceptibility loci. Am. J. Hum. Genet. 2003;73:886–897. doi: 10.1086/378778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Le Couteur A., Bailey A., Goode S., Pickles A., Robertson S., Gottesman I., Rutter M. A broader phenotype of autism: the clinical spectrum in twins. J. Child Psychol. Psychiatry. 1996;37:785–801. doi: 10.1111/j.1469-7610.1996.tb01475.x. [DOI] [PubMed] [Google Scholar]
- 60.Lord C., Pickles A., McLennan J., Rutter M., Bregman J., Folstein S., Fombonne E., Leboyer M., Minshew N. Diagnosing autism: analyses of data from the Autism Diagnostic Interview. J. Autism Dev. Disord. 1997;27:501–517. doi: 10.1023/a:1025873925661. [DOI] [PubMed] [Google Scholar]
- 61.Geschwind D.H., Sowinski J., Lord C., Iversen P., Shestack J., Jones P., Ducat L., Spence S.J. The autism genetic resource exchange: a resource for the study of autism and related neuropsychiatric conditions. Am. J. Hum. Genet. 2001;69:463–466. doi: 10.1086/321292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.