Abstract
FRAS1 is mutated in some individuals with Fraser syndrome (FS) and the encoded protein is expressed in embryonic epidermal cells, localizing in their basement membrane (BM). Syndactyly and cryptophthalmos in FS are sequelae of skin fragility but the bases for associated kidney malformations are unclear. We demonstrate that Fras1 is expressed in the branching ureteric bud (UB), and that renal agenesis occurs in homozygous Fras1 null mutant blebbed (bl) mice on a C57BL6J background. In vivo, the bl/bl bud fails to invade metanephric mesenchyme which undergoes involution, events replicated in organ culture. The expression of glial cell line-derived neurotrophic factor and growth-differentiation factor 11 was defective in bl/bl renal primordia in vivo, whereas, in culture, the addition of either growth factor restored bud invasion into the mesenchyme. Mutant primordia also showed deficient expression of Hoxd11 and Six2 transcription factors, whereas the activity of bone morphogenetic protein 4, an anti-branching molecule, was upregulated. In wild types, Fras1 was also expressed by nascent nephrons. Foetal glomerular podocytes expressed Fras1 transcripts and Fras1 immunolocalized in a glomerular BM-like pattern. On a mixed background, bl mutants, and also compound mutants for bl and my, another bleb strain, sometimes survive into adulthood. These mice have two kidneys, which contain subsets of glomeruli with perturbed nephrin, podocin, integrin α3 and fibronectin expression. Thus, Fras1 protein coats branching UB epithelia and is strikingly upregulated in the nephron lineage after mesenchymal/epithelial transition. Fras1 deficiency causes defective interactions between the bud and mesenchyme, correlating with disturbed expression of key nephrogenic molecules. Furthermore, Fras1 may also be required for the formation of normal glomeruli.
INTRODUCTION
Fraser syndrome (FS; OMIM #219000) is an autosomal recessive disorder occurring in 11/100 000 stillbirths and 0.4/100 000 livebirths and characterized by cryptophthalmos (eyeball covered by skin), syndactyly (fused digits) and kidney malformations (1,2). Winter (3) suggested that bleb mutant mice are models for human FS, with four lines affected by ocular, digital and renal anomalies. Both blebbed (bl) and myelencephalic blebs (my) homozygous embryos undergo epidermal–dermal dehiscence below the plane of the basement membrane (BM). Incomplete healing of resulting haemorrhagic blisters causes cryptophthalmos and soft tissue syndactyly (4,5). By analogy with bleb mice, human FS surface malformations are considered sequelae of embryonic skin fragility (3,5).
Some FS individuals have mutations in either FRAS1 or FRAS1-related extracellular matrix (ECM) gene 2 (FREM2), and Fras1 and Frem2 are, respectively, implicated in bl and my bleb mice (6–9). Fras1 and Frem2 are transmembrane proteins containing an extracellular repeated chondroitin sulphate proteoglycan domain, and the extracellular, N-terminal domain of Fras1 contains repeated von Willebrand factor type C and cysteine-rich partial furin motifs (5). Fras1 and Frem2 transcripts are expressed in embryonic epidermis, immunolocalizing in sublamina densa of the BM, whereas Frem1, a related protein lacking a transmembrane domain and mutated in head blebs mice, is expressed by adjacent stroma (10,11). Normal mouse skin contains Fras1/Frem1/Frem2 protein complexes, whereas bleb mutants have defective assembly of this ECM complex (4).
The metanephros initiates by interactions between ureteric bud (UB) epithelia and metanephric mesenchyme (MM). MM-generated glial cell line-derived neurotrophic factor (Gdnf) stimulates UB growth via the Ret receptor, and both Gndf and Ret null mutants have renal agenesis (12,13). Growth-differentiation factor 11 (Gdf11) is, like Gdnf, a distant member of the transforming growth factor β (Tgfb) superfamily, and it is expressed in metanephric primordia with its receptor, ActRIIB (14). Gdf11 null embryos have deficient Gdnf expression in MM and renal agenesis (14). Bone morphogenetic protein 4 (Bmp4) expression around the mesonephric duct (MD) prevents ectopic UB formation via pathways phosphorylating Smad1/5/8, and Bmp4 expression activity is minimal where the normal UB emerges (15,16). During UB growth into the MM, the latter’s cells are rescued from apoptosis and become compactly organized around the bud (17). Induced MM changes its pattern of gene expression such that fibronectin is downregulated, whereas Bcl2, an anti-apoptosis molecule, and integrin α8 (Itga8) are upregulated (18–21). Itga8 on MM cell surfaces mediates interaction with the UB and, by an undefined mechanism, maintains MM Gdnf expression (22). MM also expresses transcription factors controlling nephrogenesis, including the homeobox 11 (Hox11) family, paired-box 2 (Pax2), sal-like 1/homologue of Drosophila spalt 1 (Sall1), sine oculis Drosophila homologue (Six) 1 and 2 and Wilms tumour 1 (Wt1) (23–33). Thus, metanephric initiation is mediated by growth factors, ECM molecules and transcription factors.
FS individuals often have absent kidneys (bilateral in 20–40% or unilateral in 20–40%), and 10% also have absence of one or both ureters (1). Postnatally, most FS individuals die in the first year because of kidney failure and/or upper airway atresias, another feature of the syndrome (1). When human FRAS1 and mouse Fras1 mutations were first reported, it was demonstrated that Fras1 transcripts were expressed in mouse MDs and their UB branches, with Fras1 immunolocalized in a BM-like pattern around these epithelia (6,7). It was subsequently found that Fras1 is also expressed as the UB lineage branches into collecting ducts (CDs) (34–36).
Beyond these observations, the bases for renal defects in FS and bl mice were unknown, and there was no information about the expression of nephrogenic genes in bl renal primordia. It was also unclear whether Fras1 was expressed in the nephron lineage. We demonstrate that Fras1 is expressed in UB branches and that renal agenesis occurs with high penetrance in homozygous Fras1 null mutant blebbed (bl) mice maintained in a C57BL6J background. In this context, Fras1 deficiency causes defective interaction between the UB and MM, correlating with disturbed expression of specific nephrogenic molecules. We also show that Fras1 is upregulated as MM differentiates into nephrons, with Fras1 immunolocalizing in nascent glomeruli in a BM-like pattern, and we demonstrate that bl and my mutants maintained in a mixed background sometimes survive into adulthood; these mice have two kidneys, which contain subsets of defective glomeruli. Thus, the data presented in this paper not only explain how Fras1 is needed for the initiation of the metanephric kidney but also suggest that this ECM-related protein has a function at a later stage of development, namely in the formation of glomeruli, filtering units of the kidney.
RESULTS
Fras1 expression
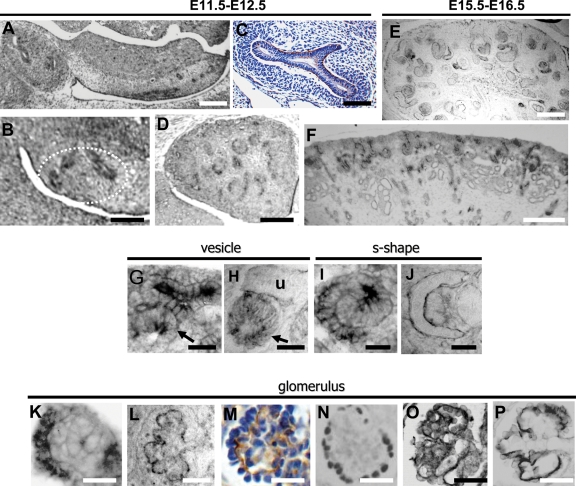
We sought Fras1 expression in wild-type mice by in situ hybridization (ISH) and immunohistochemistry (IHC). At embryonic day (E) 11, Fras1 transcripts were noted in the UB and its first branches (Fig. 1A and B). ISH signals above background levels were not detected in MM (Fig. 1B). Fras1 protein was detected as a fine linear signal on the basal aspect of the UB stalk and its first branches, but the signal did not extend outwards into the MM (Fig. 1C). Views of E12–16 metanephroi (Fig. 1D–F) revealed that Fras1 transcripts and protein were expressed in nephrons and UB derivatives. Fras1 transcripts were not only detected in branching UB tips but also in adjacent vesicles, nephron precursors which have undergone mesenchymal to epithelial transformation (Fig. 1G). Fras1 immunolocalized in a linear pattern around UB branches but in a cytoplasmic pattern in vesicles (Fig. 1H). Fras1 transcripts were expressed in S-shaped bodies (Fig. 1I), where, in the proximal limb, Fras1 was immunodetected in a linear pattern on basal aspects of parietal (Bowman’s capsule) and visceral (podocyte) epithelia in the nascent glomerulus (Fig. 1J). In capillary loop-stage glomeruli, podocytes expressed Fras1 transcripts (Fig. 1K), with protein detected on their basal aspects in a wavy, linear pattern (Fig. 1L and M). This pattern of Fras1 protein contrasts with that of Wt1, located in podocyte nuclei (37), and is similar to IHC patterns of podocin and Itga3 (Fig. 1N–P), both located on basal surfaces of podocytes (38,39).
Figure 1.
Fras1 expression in wild-type metanephroi. (A), (B), (D), (F), (G), (I) and (K) are of ISH, and (C), (E), (H), (J) and (L–P) are of IHC. Sections in (C) and (M) were counterstained with haematoxylin. (A) Fras1 transcripts (black) in the E11 UB stalk and branches within the metanephros (oval in left of frame). Fras1 was also expressed in the MD and in mesonephric tubules (mesonephros/gonadal ridge is the elongated structure in the centre and right of frame). (B) Higher power of an E11 metanephros: ISH Fras1 signal in UB branches (outlined by white dashes). (C) Fras1 protein was detected as a fine (brown) line on basal surfaces of E11 UB stalk and branches (note the lack of positive signal in cell layers of surrounding MM). (D), (E) and (F) Respective low-power views of Fras1 ISH at E12, IHC at E15 and ISH at E16. Note Fras1 expression (black) in diverse epithelial structures. (G) and (H) High-power views of E16 nephrogenic cortex showing Fras1 transcripts in UB branches and adjacent vesicle [arrowed in (G)], with Fras1 immunolocalized in a linear pattern around the UB branches [u in (H)] and in a cytoplasmic pattern in a nephron vesicle [arrow in (H)]. (I) and (J) High-power views of E16 deeper cortex showing an S-shaped body, with the proximal limb to the left and distal limb to the right. Fras1 transcripts [black in (I)] were detected in both limbs with protein detected in a linear pattern on glomerular epithelia (J): the left-hand crescent is Fras1 on the basal aspect of parietal epithelia, whereas the inner crescent is the basal aspect of immature, columnar podocytes. (K)–(P) Capillary loop-stage glomeruli in E16 metanephros. (K) Fras1 transcripts (black) in glomerular epithelia. (L) and (M) Fras1 immunolocalized [black in (L) and brown in (M)] in a wavy, linear pattern at the basal aspect of podocytes. (N), (O) and (P) Frames, respectively, show ICH for Wt1, podocin and Itga3, with positive signals in black. Note the similarity of the Fras1 ICH pattern to that of Itga3, and to a lesser extent, podocin. Bars in (A), (D), (E) and (F) are 200 µm; bars in (B) and (C) are 100 µm; bars in (G)–(P) are 25 µm.
Renal agenesis in bl mutant embryos in vivo
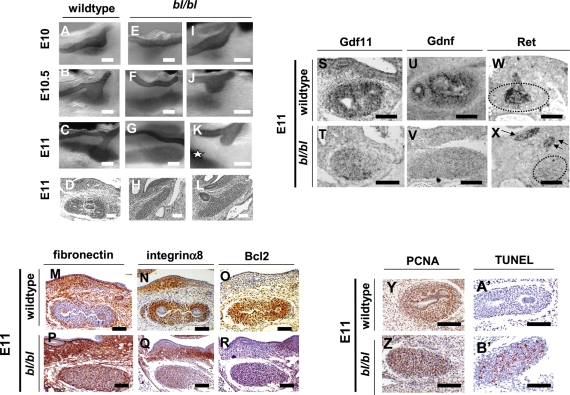
The background of the original bl mutant strain is unknown (40). Using a bl colony maintained in a C56BL6J background for over five generations, we assessed embryos immunostained for Pax2, which is expressed by MDs, UBs and by uninduced and induced MM (25). In all 30 wild-type (+/+) embryos, UB initiation and interaction with MM occurred in E10–11 (Fig. 2A–D). In 23 bl/bl embryos studied in E10–11, UBs usually failed to sprout (Fig. 2E–H) or, if they did, they nearly always failed to enter the MM (Fig. 2I–L). Of 16 E11 bl/bl embryos, seven demonstrated bilateral absent UBs, six had unilateral absent UB and a contralateral UB stump, two had bilateral UB stumps and one had unilateral absent UB and contralateral overtly normal metanephros. Neither physical blebs nor UB ectopia nor duplex UBs were detected, and mesonephroi and adrenal glands were grossly normal (not shown).
Figure 2.
In vivo bl/bl phenotypes, gene expression and cell turnover. (A)–(C), (E)–(G) and (I)–(K) are wholemounts immunostained for Pax2 marking MD, UB and both uninduced and induced MMs in black; (D), (H), (L)–(R), (Y), (Z), (A′) and (B′) were counterstained with haematoxylin. Sections in (M)–(R) and (Y), (Z), (A′) and (B′) underwent IHC (positive brown signal); sections in (S)–(X) underwent ISH (positive black signal). Wild-type E10 (A), E10.5 (B) and E11 [(C) and (D)] renal tracts showing UB initiation from the MD and growth into MM. (E)–(H) bl/bl renal tracts at E10 (E), E10.5 (F) and E11 [(G) and (H)] in which UBs fail to initiate. (I)–(L) bl/bl mutants at E10 (I), E10.5 (J) and E11 [(K) and (L)], where UBs sprout but fail to enter MM [indicated by ‘*’ in (K)]. (M)–(R) Wild-type MMs downregulated fibronectin and upregulated Itga8 and Bcl2. Mutant MMs expressed fibronectin but not significant levels of Itga8 and Bcl2. (S)–(X) Gdnf expression and Gdf11 expression were deficient in bl/bl MMs, whereas mutant MDs and UB stumps expressed Ret [arrows in (X)]. Dotted lines in (W) and (X) outline the MM. (Y) and (Z) IHC for PCNA. Note that this surrogate marker of proliferation was detected in most nuclei of wild-type and mutant MMs (brown nuclei). (A′) and (B′) TUNEL probe. Note numerous apoptotic nuclei (red) in bl/bl MM but not in wild-type MM. Bars in all frames are 100 µm.
Induction markers, expression of Gdnf and Gdf11 and cell turnover in vivo
In wild-type E11 MMs, harmonizing with UB penetration and consistent with MM induction, MM expressed Itga8 and Bcl2 but not fibronectin (Fig. 2M–O). E11 bl/bl mesenchymes, however, expressed fibronectin, whereas Itga8 and Bcl2 signals were barely detectable (Fig. 2P–R). Because Gdnf and Gdf11 null mutants have failed UB progression similar to bl/bl mice, and because Fras1 contains N-terminal chordin-like domains similar to those binding Tgfb family proteins (5), we explored the expression of Gdnf and Gdf11. In E10–11 wild-types, Gdf11 transcripts were detected in MD, UB and MM but signals were barely detectable in bl/bl primordia (Fig. 2S and T; data not shown). E10–11 wild-type MMs expressed Gdnf, whereas bl/bl MMs demonstrated reduction at E10 and profound attenuation at E11, a kidney-specific effect because normal Gdnf expression occurred in gut mesenchyme (Fig. 2U and V; not shown). Ret was expressed in MDs and UBs of wild-type embryos, and bl/bl mutants had similar expression in MDs and in variably present UB stumps (Fig. 1W and X). Most MM nuclei in E11 wild-type and mutant MMs contained proliferating cell nuclear antigen (PCNA) (Fig. 2Y and Z). Although apoptosis was rare in wild-type MMs, up to 20% of nuclei in E11 bl/bl MMs were positive using terminal deoxynucleotidyl transferase-mediated DUTP nick end-labelling (TUNEL) (Fig. 2A′ and B′), and similar proportions of cells expressed activated capsase 3 as assessed by IHC (data not shown).
Explanted embryonic renal tracts
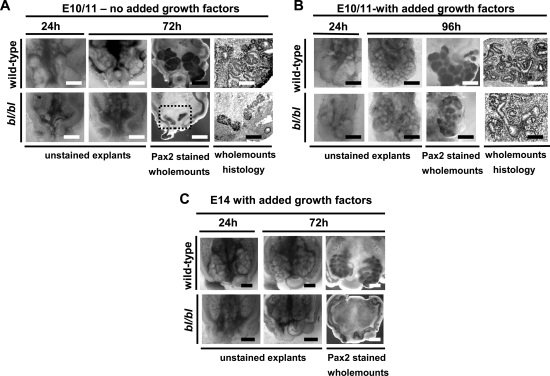
Overviews of explanted nephrogenic fields (paired MDs and MMs) are shown in Figures 3 and 4, and gene expression in Figure 4. In E10–11 wild-type explants (n = 5) cultured for 48 h, UBs invariably grew into MMs (n = 10), which became induced as assessed by the compaction of MM cells around the incoming bud and the facts that MM expressed Bcl2 and Itga8 but little fibronectin (Fig. 4A–E). In contrast, all time-matched bl/bl explants (n = 5) recapitulated their in vivo fates; although a UB sometimes grew towards its MM, it never entered it, and MMs (n = 10) contained unorganized cells expressing fibronectin but not Bcl2 or Itga8 (Fig. 4F–J). Using these outcomes, there were significant differences between wild-types and mutants (Fisher’s exact test, two-tail P = 0.008 for explants and P = 0.00001 for metanephroi). When E10–11 wild-types (two explants/four metanephroi) and time-matched mutants (three explants/six metanephroi) were cultured to 72 h, all wild-type metanephroi formed organs with branched UBs and nascent nephrons, whereas all mutants showed progressive involution (Fig. 3A), thus showing that mutant MM differentiation was seriously disrupted, rather than being delayed. When wild-type E10–11 explants were cultured with recombinant human (h) GDNF (100 ng/ml) for 48 h, two UBs often arose from the MD (not shown), with similar results in mutants, indicating that their MDs, which express Ret, respond to this growth factor (Fig. 4K and L) even in the absence of functional Fras1. hGDNF-treated E10–11 mutant renal tract explants (n = 4) contained MM cells condensed around the incoming UB in all eight metanephroi and immunoprobing for Itga8, Bcl2 and fibronectin suggested that MMs had initiated a normal programme of gene expression (Fig. 4M–O). In bl/bl explants (n = 4) cultured for 48 h with hGDF11 (50 ng/ml), a single-UB grew from each MD into each of the MMs, forming eight discrete metanephroi with condensed MM expressing Bcl2 and Itga8 but not fibronectin (Fig. 4P–T). For treatment with either factor, there was a significantly improved outcome versus untreated mutants (P = 0.03 for explants and P = 0.0001 for metanephroi). When E10–12 mutant explants (n = 4) were cultured for extended periods (72–96 h) with both growth factors, all eight metanephroi underwent development beyond the stage of UB in-growth, with histology showing some early nephron formation. However, mutant metanephroi appeared smaller versus time-matched wild-types (n = 4 explants/8 metanephroi) treated similarly (Fig. 3B). When we explanted three older (E13–14) bl/bl explants, exogenous growth factors failed to rescue kidneys (Fig. 3C), a predictable result given that MMs would have already involuted in vivo (6).
Figure 3.
bl/bl renal tract explants. (A) Representative examples of E10/11 renal tracts cultured without added hGDNF or hGDF11 (E10/11—no added growth factors). Each explant contained paired MDs and adjacent MMs. In the wild type, note progressive growth of two metanephroi containing Pax2-expressing UB branches and immature nephrons. Contrast this with the failure to initiate discrete metanephroi in the bl/bl mutant explant from the same litter. In the Pax2-immunostained bl/bl wholemount, although the MDs and disorganized MMs are detected, no UBs are visible after 72 h culture. The area in the box in the Pax2-stained bl/bl wholemount picture is depicted, in a histology section, in the adjacent frame. (B) Representative examples of E10/11 renal tracts cultured with both growth factors (E10/11—with added growth factors; see main text for details). Metanephroi formed in both wild-type and mutant explants, and both contained UB branches and nephron vesicles; however, the mutant kidneys were smaller. (C) When older (i.e. E13–14) explants were explanted in the presence of both growth factors (E14 with added growth factors), mutant metanephroi could not be rescued. As depicted in the Pax2-stained wholemount frame, although the two MDs were present, no UBs or MMs were detected after 72 h culture. In (A), bars in wholemount frames are 100 µm and bars in histology frames are 25 µm; in (B), bars in wholemount frames are 200 µm and bars in histology frames are 25 µm; in (C), all bars are 200 µm.
Figure 4.
Gene expression in renal tract explants. This panel depicts E10 explants after 48 h serum-free culture. All were wholemount-immunostained with antibody to Pax2 which labels MDs, UBs and both uninduced and induced MMs. (A), (F), (K) and (P) are wholemounts with Pax2 signal appearing black. (B)–(E), (G)–(J), (L)–(O), (Q)–(T) frames are histology sections of whole mounts; in (B), (G), (L) and (Q), the Pax2 signal is black, whereas the signal appears brown in remaining frames. (A)–(E) Wild types show UB penetration into condensing, induced MM expressing Itga8/red and Bcl2/blue, but lacking MM fibronectin/red (note that surrounding non-renal mesoderm does express fibronectin). (F)–(J) bl/bl explants show failure of both UB growth and MM induction with minimal expression of MM Itga8 (red) and Bcl2 (blue), but prominent fibronectin (red). Addition of hGDNF [(K)–(O)] or hGDF11 [(P)–(T)] to bl/bl explants allowed the formation of discrete metanephroi with penetrated UBs and aggregated and which expressed Itga8 (red) and Bcl2 (blue) but little fibronectin (red). In double-immunostained sections, nuclear signals for Pax2 are brown, and these signals are detected in both induced and uninduced MMs, and also in MDs and UBs. In (A), (F), (K) and (L), bars are 200 µm; in all other frames, bars are 75 µm.
Expression of transcription factors and Bmp4 signalling in vivo
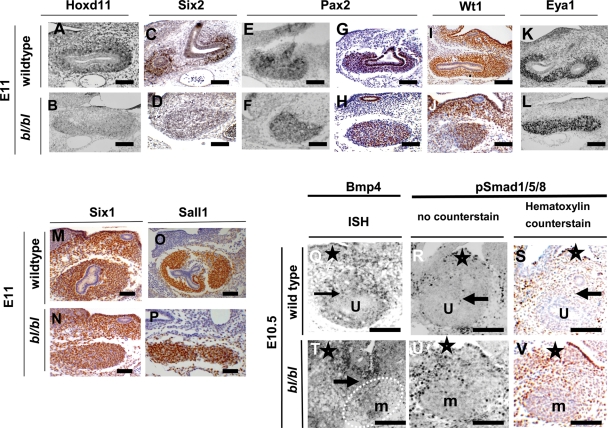
We sought the expression of diverse other genes implicated in metaneprhic induction, as outlined in the Introduction. E10–11 bl/bl MMs showed defective expression of Hoxd11 and Six2 (Fig. 5A–D), whereas the expression of Pax2, Wt1, Eya1, Six1 and Sall1 was normal (Fig. 5E–P). In E10 wild-types, we detected Bmp4 transcripts and weak immunohistochemical signals for phospho-Smad1/5/8 in sparse cells around UB stalks (Fig. 5Q–S). In contrast, in bl/bl embryos, several layers of cells between the MD and MM expressed Bmp4 accompanied by prominent phospho-Smad1/5/8 (Fig. 5T–V).
Figure 5.
Expression of transcription factors and Bmp4. (A)–(P) are sections from E11 animals when, in wild types, the UB has undergone one round of branching within the MM. (Q)–(V) are E10.5 embryos, where the wild-type UB had just penetrated the MM. Positive expression signals are black in (A), (B), (E), (F), (K), (L), (Q), (R), (T) and (U); in other frames, positive IHC signals are brown. (C), (D), (G)–(J), (M)–(P), (S) and (V) were counterstained with haematoxylin. (A) and (B) ISH for Hoxd11 and (C) and (D) IHC for Six2 showing downregulated expression in bl/bl MMs. (E)–(P) Several other genes showed similar intensity of signal in wild-type and mutant MMs: Pax2 ISH [(E) and (F)]; Pax2 IHC [(G) and (H)]; Wt1 IHC [(I) and (J)]; Eya1 ISH [(K) and (L)]; Six1 IHC [(M) and (N)]; Sall1 IHC [(O) and (P)]. (Q)–(V) Bmp4 ISH depicted in (Q) and (T), whereas other frames are IHC for phospho-Smad1/5/8. Note that (S) and (V), respectively, are frames (R) and (U) counterstained with haematoxylin. In wild-types, Bmp4 transcripts were detected around UB stalks, with weak signal for phospho-Smad1/5/8 in this location. In bl/bl littermates, Bmp4-expressing cells were detected between the MM and unbranched MD, with several cell layers expressing phospho-Smad1/5/8 in this location. Stars have been placed next to MDs, and arrows indicate normal UB stalk in wild types and an abnormal UB stump in the bl/bl embryo; in the wild-type frames, ‘u’ indicates the UB within the MM and, in the bl/bl fames, ‘m’ indicates the un-penetrated MM. Bars in all frames are 100 µm.
bl and my mutants in a mixed background
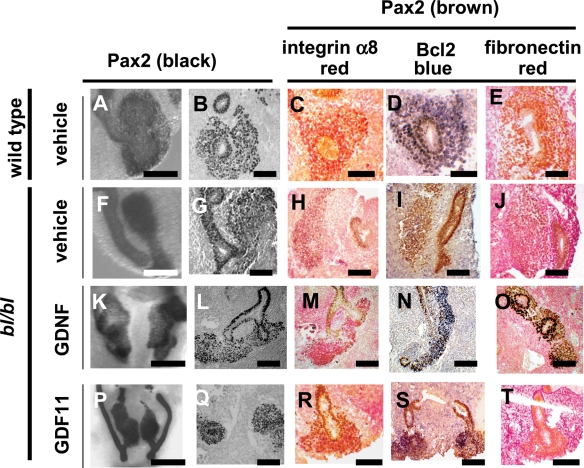
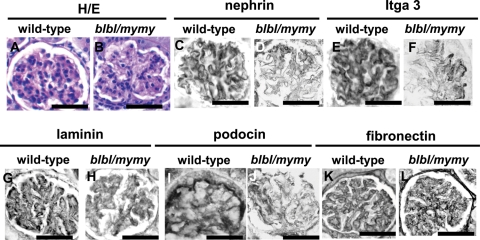
When bl and my alleles were maintained in a mixed background, by interbreeding the original bl colony with the my allele which was in an NMRI background, some mice homozygous for bl or my or my/bl (e.g. 45% bl/bl and 72% my/my mice) survived to adulthood (8,40). We previously reported that the kidneys of these mutants, at 6–9 months of age, contained sporadic cysts usually arising from CDs (8). Having now found that Fras1 was expressed by foetal podocytes, mutant kidneys were re-examined with a focus on glomeruli. Subsets of glomeruli (∼10–20%/section) in kidneys of three of four compound homozygous bl/my mutants were abnormal, whereas glomeruli in wild-type or heterozygous bl and/or my mice (n = 7) were normal (Fig. 6). Segments of bl/my glomeruli lacked nuclei (Fig. 6A and B) and the normal patterns of nephrin (Fig. 6C and D), Itga3 (Fig. 6E and F), (pan)laminin (Fig. 6G and H) and podocin (Fig. 6I and J) were lost. Furthermore, the delicate looped pattern of fibronectin was replaced by a mesangial signal (Fig. 6K and L). In this collection (8), the two bl/bl mutants which were wild type in the my locus also had subsets of glomeruli with abnormal patterns of Itga3, (pan)lamin and fibronectin (Supplementary Material, Fig. S1). Finally, similar lesions were noted in the kidneys of an extraordinary bl/bl adult survivor in the C56BL6J colony (not shown); it had syndactyly and cryptophthalmos and was sacrificed at 4 months of age owing to failure to thrive. Notably, all mutants in the mixed background had two kidneys.
Figure 6.
Histology from wild-type and double-mutant bl/my mice in a mixed background. (A), (C), (E), (G), (I) and (K) are wild-type kidneys, whereas (B), (D), (F), (H) and (J) are double-mutants. Sections in (A) and (B) were counterstained with haematoxylin and eosin. Other sections were not counterstained and positive IHC signals are black. (A) and (B) Note the segmental, relatively acellular areas in the mutant. (C)–(H) Note the loss of the normal patterns of nephrin [(C) and (D)], Itga3 [(E) and (F)], (pan)laminin [(G) and (H)] and podocin [(I) and (J)] in bl/my mutant glomeruli. (K) and (L) Note the delicate loop-like pattern of fibronectin in the wild-type glomerulus and the loss of this pattern and accentuation in the mesangial core in the mutant. Bars in all frames are 100 µm.
DISCUSSION
Fras1 is expressed by both the UB and the nephron lineages in the mammalian kidney
Our study supports reports that state Fras1 is expressed in the UB lineage (6,7,34–36). At E11, Fras1 IHC signal closely surrounded the UB stalk and its branches and did not extend outwards into the several MM cell layers. Fras1 does, however, become prominently expressed in the nephron lineage after the MM/epithelial transformation. Fras1 was prominently expressed in vesicles, which are epithelial spheroids arising from MM aggregates. We interrogated the GenitoUrinary Development Molecular Anatomy Project (http://www.gudmap.org/) and noted that Fras1 had been detected in certain expression microarray experiments using microdissected MM from E11–12 metanephroi. In one set, however, Ret was also expressed, perhaps most consistent with UB contamination. The ISH and IHC techniques we used would be unable to detect very low levels of Fras1 transcripts and protein, and thus it remains possible that Fras1 expression initiates in the MM prior to epithelial transformation. Fras1 protein was detected in a cytoplasmic pattern in vesicles, whereas it was immunolocalized on the basal aspects of epithelia in S-shaped bodies and foetal glomeruli. In the UB and its branches, Fras1 protein was also detected as a line on cells’ basal aspect. Hence, by analogy with foetal skin (4,11), Fras1 most likely exists as a BM-associated protein in both UB/CD and nephron epithelia. Fras1 complexes with Frem2 in embryonic skin (4), and Frem2 is also expressed by UB branches, vesicles, S-shaped bodies and nascent glomeruli (our unpublished data and 8,36,40).
Lack of Fras1 causes renal agenesis in the C57BL6J background, correlating with defective expression of key nephrogenic molecules
The bl strain contains a 7313CC→AA Fras1 mutation, resulting in a premature stop codon and rendering this a null mutation; indeed, similar phenotypes occur in mice with genetically engineered null Fras1 mutation (6,7). We found that Fras1 deficiency is associated with failed UB growth between E10–11 in vivo and, consistent with failed UB penetration, bl/bl MMs remained uninduced and were apoptotic. Indeed, apoptotic involution is the default pathway of uninduced MM (17). The observations explain the facts that kidneys and ureters are often undetectable in late gestation Fras1 null mutant mice and in FS individuals (1,6,7). The lack of UB growth in bl/bl embryos in vivo was associated with Gdnf deficiency. Our experiments show that Ret remains expressed in Fras1 null mutant MDs, and that these structures can be stimulated to produce UBs by hGDNF, even in the absence of Fras1 protein which normally coats the MD and UB (6). In future, a more prolonged rescue might be obtained in vivo by upregulating Gdnf/Ret signalling in Fras1 mutant mice by introducing a heterozygous Sprouty1 mutation, as used to rescue Itga8 mutants (22).
Gdf11 ablation causes metanephric Gdnf deficiency in vivo (14) and we found that Gdf11 expression was downregulated from E10 in bl/bl renal tracts; furthermore, the addition of hGDF11 in organ culture stimulated UB growth in mutants. One can therefore postulate that Gdf11 deficiency is upstream to the Gdnf deficit in bl/bl renal primordia (Fig. 7). It is, however, unknown why a lack of Fras1 is associated with Gdf11 deficiency. Perhaps an Fras1/Frem1/Frem2 complex (4) has ‘outside-in’ signalling functions controlling the expression of Gdf11 and/or Gdnf; indeed, Frem1 has an integrin-binding motif (41). We noted that, in vivo, bl/bl MMs also had deficient expression of transcription factors Hoxd11 and Six2, both regulators of metanephrogenesis (31,33). At least in non-metanephric contexts, Gdf11 upregulates Hoxd11 (42). In addition, a Hox11–Eya1–Pax2 complex activates Six2 and Gdnf in MM (32), Six2 itself can upregulate Gdnf (30) and Hox11 upregulates Itga8 in MM (28). Bmp4 expression and signalling, as assessed by phospho-Smad1/5/8 expression, was upregulated between MDs and MMs. Given that Bmp4 inhibits UB outgrowth (15,16), this provides another reason, as Gdnf deficiency, for failed UB initiation in Fras1 mutants; upregulated Bmp4 signalling is also consistent with the fact that we never observed duplex UB outgrowths. The data are also consistent with the hypothesis that chordin-like domains in Fras1 bind to Bmp4 and downregulate its activity (5). Indeed, the overexpression of Fras1 chordin-like cysteine-rich repeats downregulate phospho-Smad1 in NIH3T3 cells, without changing total Smad1 (40). Gdf11 downregulates Smad1 phosphorylation when expressed in Xenopus ectoderm (43), so Gdf11 deficiency in bl/bl embryos provides another possible explanation for phospho-Smad1 upregulation.
Figure 7.
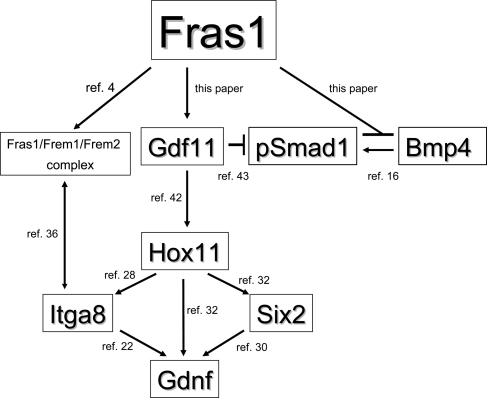
Fras1 protein may enhance metanephric initiation via several pathways. (A) Putative network of metanephric effector molecules which may be ultimately regulated by Fras1, an ECM-related protein which coats the surface of the UB. Beyond Fras1 itself, the expression of the molecules are altered in bl/bl renal primordia, as demonstrated in the current paper, and several of them have additionally been shown to regulate other molecules depicted in the scheme. See the main text for details and references cited for this figure.
Hence, one can construct a putative network of molecules directly or indirectly controlled by Fras1 coating the UB (Fig. 7). In future, a comprehensive picture of gene expression of bl primordia could be sought using microarrays, as performed with Wnt4 mutant metanephroi (44). Furthermore, investigating the effects of added growth factors, such as Gdnf and Gdf11, on isolated Fras1 mutant MDs stripped of their surrounding mesenchyme (45) may in future add extra mechanistic insights beyond those obtain from the renal tract cultures, as described in this study.
The possible roles of Fras1 in renal epithelia beyond induction of the metanephros
The widespread expression patterns of Fras1 lead to the hypothesis that epithelial-derived Fras1 plays roles in the biology of not just the UB but also CDs and glomeruli. Interestingly, poorly differentiated, hypoplastic and/or cystic kidneys have been reported in non-agenesis FS renal tracts (1) and we reported that, in Fras1/Frem2 mutants in a mixed background, kidneys sometimes formed and they contain cysts, some reacting with markers of CDs (8). We can now extend these observations to show that these kidneys contain subsets of glomeruli which have segmental lesions with the diminished expression of nephrin, podocin and Itga3, podocyte proteins implicated in glomerulogenesis (38,46,47) and increased mesangial fibronectin. The data are compatible with the theory that Fras1 and Frem2 are essential for glomerulogenesis; alternatively, they may be required to maintain normally formed glomeruli. Support for the former contention comes from a study (48) in which increased mesangial matrix in adult my mice was preceded by increased glycosaminoglycans in developing glomeruli. In our current study, however, the coincidence of cystic and glomerular lesions means that we cannot draw final conclusions concerning roles of Fras1 in CDs or glomeruli. In future, targeted CD (49) or podocyte (50) ablation of Fras1 in a uniform background will be used to understand the genesis of CD cysts and glomerular lesions.
Human genetic diseases which cause both renal agenesis and glomerular defects
The facts that Fras1 is a BM-related protein coating the surface of the UB and is also expressed nascent podocytes are highly reminiscent of the expression patterns of anosmin-1, a BM-related protein which is mutated in X-linked Kallmann syndrome (51). Affected individuals with Kallmann syndrome can have unilateral or bilateral renal agenesis, and in the former case, progressive renal failure accompanied by proteinuria can occur in adulthood (52). More rarely, affected individuals can have cystic kidney malformations (53), consistent with defective epithelial morphogenesis. Reports of adults with FS are very rare, although we note that one individual who survived 96 years has been described (54). She was found to have a solitary kidney and moderate renal failure with both proteinuria and haematuria, the latter finding perhaps consistent with a glomerular lesion beyond that simply associated with hyperfiltration nephropathy as might occur in a solitary kidney with ‘overworked’ glomeruli (55). Having said this, the FS mice with glomerular lesions described in this paper had two kidneys.
We consider that FS and X-linked Kallmann syndrome should be regarded as founding members of a group of human diseases in which the mutated gene codes for a BM-associated protein which plays critical roles both at the start and also towards the end of kidney development, by virtue of the encoded proteins having a widespread distribution within the kidney. In Table 1 we compare features of FS and X-linked Kallmann syndrome, and we add a third disorder to the group, namely the recently described syndrome of haematuria and kidney cortical and medullary cysts caused by the mutation of COL4A1, the gene which codes for the α1 chain of collagen IV located in both glomerular and kidney tubular BMs (56,57). Finally, we suggest that variants of the FS genes such as FRAS1 and FREM2 should be considered as candidates in humans with congenital glomerular defects which cannot be explained by mutations of known podocyte genes such as those encoding nephrin and podocin proteins (47).
Table 1.
Inherited diseases caused by mutations of genes coding for proteins localized both in glomeruli and in tubule BMs
| Disease | Mutation (inheritance) | Spectrum of kidney disease | Extra-renal disease |
|---|---|---|---|
| FS |
FRAS1 FREM2 (autosomal recessive) |
Uni- or bilateral kidney agenesis, cystic dysplastic kidneys and (in rare adult survivors) proteinuria and haematuria | Cryptophthalmos, cutaneous syndactyly, ambiguous genitalia, larynx atresia and anal atresia/stenosis |
| X-linked Kallmann syndrome | KAL-1 (X-linked recessive) | Uni- or bilateral kidney agenesis, cystic dysplastic kidneys and proteinuria in adults with solitary functioning kidneys | Anosmia and hypogonadotrophic hypogonadism |
| HANAC (hereditary angiopathy, nephropathy, aneurysms and cramps) syndrome | COL4A1 (autosomal dominant) | Haematuria and renal cortical and medullary cysts | Myopathies with cramps and intracranial aneurysms |
Conclusions
Fras1 protein coats branching UB epithelia and is also strikingly upregulated in the nephron lineage after mesenchymal/epithelial transition. Fras1 deficiency causes defective interactions between the bud and mesenchyme, correlating with disturbed expression of key nephrogenic molecules. Furthermore, Fras1 not only initiates kidney development but may also be required for the formation of normal glomeruli.
MATERIALS AND METHODS
Animals
Experiments were conducted with the UK Home Office permission. bl mice, originally obtained from MRC Mammalian Genetics Unit (Harwell), were studied in a C57BL/6 background apart from bl/my compound mutants which were from a local colony, as described (8). Genotypes were determined by PCR (details available on request). Heterozygous parents were timed-mated and embryos collected in PBS and fixed in 4% paraformaldehyde (PFA) for histology, wholemount IHC or ISH. Dehydrated embryos were embedded in paraffin or stored at −20°C in methanol for wholemount immunostaining. Embryos used to derive tissues for in vitro culture were collected into ice-cold L15 medium (Invitrogen).
Immunostaining, ISH and apoptosis detection
Primary antibodies used were rabbit anti-Bcl2 (Calbiochem), rabbit anti-cleaved caspase (Cell Signalling), rabbit anti-fibronectin (DAKO), rabbit anti-Fras1 (7), rabbit anti-Itga3 (Pharmingen), rabbit anti-Itga8 (a gift from U. Muller, Scripps Research Institute, La Jolla, CA, USA), mouse anti-pan laminin (Chemicon), rabbit anti-nephrin (a gift from H. Holthoffer, University of Helsinki, Finland), rabbit anti-Pax2 (Zymed), mouse anti-podocin (Santa Cruz), mouse anti-Sall1 (R&D Systems), mouse anti-PCNA (BD Pharmingen), rabbit anti-Six1 (Cemines), rabbit anti-Six2 (Affinity Bioscience), rabbit anti-pSMAD1/5/8 (Cell Signalling), rabbit anti-Wt1 (Santa Cruz). All antibodies required HIR with citric acid buffer, or with Retriewagen A or B (Pharmingen). Secondary antibodies were HRP- or AP-conjugated and signals generated with DAB chromogen (DAKO), or for alkaline phosphatase with Fast-Red (DAKO) or with BCIP/NBT (Roche). Wholemount immunostaining was performed as described (58). Specimens were rehydrated into PBS and endogenous peroxidase inactivated with 3% hydrogen peroxide. Specimens were incubated with primary antibody and then washed and incubated with HRP-conjugated secondary antibody. Colourimetric reaction was with DAB (DAKO). ISH was performed on paraffin sections as described (59). Briefly, sections were digested with proteinase K (10 µg/ml), hybridized overnight at 68°C in a humidified chamber with probes labelled with digoxigenin and detected with sheep anti-DIG antibody conjugated to AP. Colour signals were generated with BCIP/NBT. Probes used were Bmp4 (a gift from B. Hogan, Duke University Medical Center, USA), Eya1 (a gift from Richard Maas, Brigham and Women’s Hospital and Harvard Medical School, USA), Gdf11 (a gift from S.J. Lee, Johns Hopkins University School of Medicine, Baltimore, USA), Gdnf (a gift from A.L. Zimmer, University of Bonn), Hoxd11 (IMAGE clone; RZPD Germany), Pax2 (a gift from G. Dressler, Howard Hughes Medical Institute, University of Michigan) and Ret (a gift from F. Costantini, Columbia University, USA). Apoptotic cells were detected by TUNEL using sections from paraffin-embedded tissues: we used the Promega Dead-End colourimetric kit with proteinase K digestion for 15 min at room temperature and colour development with AEC.
Explant culture
Nephrogenic fields (i.e. paired mesonephroi, MDs and adjacent MMs) were dissected in ice-cold PBS, and heads used for genotyping. Tissues were placed on cellulose filters and fed with DMEM/F12 (Invitrogen) supplemented with 5% ITS (Sigma) and antifungal and antibiotic mixture (Sigma). They were cultured at 37°C in a 5% CO2/air atmosphere for up to 96 h, and the medium was changed every day. In some experiments, hGDNF (Promega) or mature form of recombinant hGDF11 (R&D Systems) were added at respective concentrations of 100 and 50 ng/ml, similar to that used previously for GDNF (12) or suggested by pilot experiments for GDF11. Explants were photographed through a dissecting microscope and fixed with 4% PFA for wholemount Pax2 immunostaining. Explants were embedded in paraffin and sections were immunostained (using a different colour signal) for either fibronectin, Bcl2 or Itga8.
Numbers of embryos studied
For in vivo morphological analyses between E10 and E11, 30 wild types (+/+) and 23 bl/bl mutants were studied. Of the latter, 16 were at E11. For each ICH/ISH/TUNEL analysis, between two and five wild-type and bl/bl mutant embryos were examined at each stage, allowing the reporting of reproducible data. In a typical explant experiment, all renal tracts from one embryonic litter from bl/+ parents would be explanted and then grown with or without exogenous hGNDF and/or hGDF11. Such litters usually contained 6–10 embryos. Thus, after genotyping, one would find around two +/+ or bl/bl explants in each experiment, and outcomes of all these two genotypes were used to generate data sets. Explant outcomes were compared using Fisher’s exact test. For these comparisons, we grouped E10 and E11 explants together because they behaved uniformly in terms of whether the MMs regressed or not, depending on the absence or presence of exogenous growth factors.
SUPPLEMENTARY MATERIAL
FUNDING
The work was supported by the Wellcome Trust (grants 073624 and 085077), Kidney Research UK (grant RP35/2008), Kids Kidney Research and the Great Ormond Street Hospital Special Trustees. Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust and Kidney Research UK.
Supplementary Material
ACKNOWLEDGEMENTS
For reagents, we thank F. Costantini, G. Dressler, B. Hogan, H. Holthofer, S. Lee, R. Maas, A. McMahon, U. Muller and L. Zimmer.
Conflict of Interest statement. None declared.
REFERENCES
- 1.van Haelst M.M., Scambler P.J., Hennekam R.C. Fraser Syndrome Collaboration Group. Fraser syndrome: a clinical study of 59 cases and evaluation of diagnostic criteria. Am. J. Med. Genet. A. 2007;143A:3194–3203. doi: 10.1002/ajmg.a.31951. [DOI] [PubMed] [Google Scholar]
- 2.Martínez-Frías M.L., Bermejo Sánchez E., Félix V., Calvo Celada R., Ayala Garcés A., Hernández Ramón F. Fraser syndrome: frequency in our environment and clinical–epidemiological aspects of a consecutive series of cases. An. Esp. Pediatr. 1998;48:634–638. [PubMed] [Google Scholar]
- 3.Winter R.M. Fraser syndrome and mouse ‘bleb’ mutants. Clin. Genet. 1990;37:494–495. doi: 10.1111/j.1399-0004.1990.tb03539.x. [DOI] [PubMed] [Google Scholar]
- 4.Kiyozumi D., Sugimoto N., Sekiguchi K. Breakdown of the reciprocal stabilization of QBRICK/Frem1, Fras1 and Frem2 at the basement membrane provokes Fraser syndrome-like defects. Proc. Natl Acad. Sci. USA. 2006;103:11981–11986. doi: 10.1073/pnas.0601011103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smyth I., Scambler P. The genetics of Fraser syndrome and the blebs mouse mutants. Hum. Mol. Genet. 2005;14:R264–R274. doi: 10.1093/hmg/ddi262. [DOI] [PubMed] [Google Scholar]
- 6.McGregor L., Makela V., Darling S.M., Vrontou S., Chalepakis G., Roberts C., Smart N., Rutland P., Prescott N., Hopkins J., et al. Fraser syndrome and mouse blebbed phenotype caused by mutations in FRAS1/Fras1 encoding a putative extracellular matrix protein. Nat. Genet. 2003;34:203–208. doi: 10.1038/ng1142. [DOI] [PubMed] [Google Scholar]
- 7.Vrontou S., Petrou P., Meyer B.I., Galanopoulos V.K., Imai K., Yanagi M., Chowdhury K., Scambler P.J., Chalepakis G. Fras1 deficiency results in cryptophthalmos, renal agenesis and blebbed phenotype in mice. Nat. Genet. 2003;34:209–214. doi: 10.1038/ng1168. [DOI] [PubMed] [Google Scholar]
- 8.Jadeja S., Smyth I., Pitera J.E., Taylor M.S., van Haelst M., Bentley E., McGregor L., Hopkins J., Chalepakis G., Philip N., et al. Identification of a new gene mutated in Fraser syndrome and mouse myelencephalic blebs. Nat. Genet. 2005;37:520–525. doi: 10.1038/ng1549. [DOI] [PubMed] [Google Scholar]
- 9.Timmer J.R., Mak T.W., Manova K., Anderson K.V., Niswander L. Tissue morphogenesis and vascular stability require the Frem2 protein, product of the mouse myelencephalic blebs gene. Proc. Natl Acad. Sci. USA. 2005;102:11746–11750. doi: 10.1073/pnas.0505404102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smyth I., Du X., Taylor M.S., Justice M.J., Beutler B., Jackson I.J. The extracellular matrix gene Frem1 is essential for the normal adhesion of the embryonic epidermis. Proc. Natl Acad. Sci. USA. 2004;101:13560–13565. doi: 10.1073/pnas.0402760101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalezios Y., Papasozomenos B., Petrou P., Chalepakis G. Ultrastructural localization of Fras1 in the sublamina densa of embryonic epithelial basement membranes. Arch. Dermatol. Res. 2007;299:337–343. doi: 10.1007/s00403-007-0763-8. [DOI] [PubMed] [Google Scholar]
- 12.Towers P.R., Woolf A.S., Hardman P. Glial cell line-derived neurotrophic factor stimulates ureteric bud outgrowth and enhances survival of ureteric bud cells in vitro. Exp. Nephrol. 1998;6:337–351. doi: 10.1159/000020541. [DOI] [PubMed] [Google Scholar]
- 13.Costantini F., Shakya R. GDNF/Ret signaling and the development of the kidney. Bioessays. 2006;28:117–127. doi: 10.1002/bies.20357. [DOI] [PubMed] [Google Scholar]
- 14.Esquela A.F., Lee S.J. Regulation of metanephric kidney development by growth/differentiation factor 11. Dev. Biol. 2003;257:356–370. doi: 10.1016/s0012-1606(03)00100-3. [DOI] [PubMed] [Google Scholar]
- 15.Miyazaki Y., Oshima K., Fogo A., Hogan B.L., Ichikawa I. Bone morphogenetic protein 4 regulates the budding site and elongation of the mouse ureter. J. Clin. Invest. 2000;105:863–873. doi: 10.1172/JCI8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michos O., Gonçalves A., Lopez-Rios J., Tiecke E., Naillat F., Beier K., Galli A., Vainio S., Zeller R. Reduction of BMP4 activity by gremlin 1 enables ureteric bud outgrowth and GDNF/WNT11 feedback signalling during kidney branching morphogenesis. Development. 2007;134:2397–2405. doi: 10.1242/dev.02861. [DOI] [PubMed] [Google Scholar]
- 17.Koseki C., Herzlinger D., al-Awqati Q. Apoptosis in metanephric development. J. Cell Biol. 1992;119:1327–1333. doi: 10.1083/jcb.119.5.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ekblom P. Formation of basement membrane in the embryonic kidney: an immunohistological studies. J. Cell Sci. 1981;91:1–10. doi: 10.1083/jcb.91.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stuart R.O., Bush K.T., Nigam S.K. Changes in global gene expression patterns during development and maturation of the rat kidney. Proc. Natl Acad. Sci. USA. 2001;98:5649–5654. doi: 10.1073/pnas.091110798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winyard P.J., Risdon R.A., Sams V.R., Dressler G.R., Woolf A.S. The PAX2 transcription factor is expressed in cystic and hyperproliferative dysplastic epithelia in human kidney malformations. J. Clin. Invest. 1996;98:451–459. doi: 10.1172/JCI118811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muller U., Wang D., Denda S., Meneses J.J., Pederson R.A., Reichardt L.F. Integrin α8β1 is critically important for epithelial-mesenchymal interactions during kidney morphogenesis. Cell. 1997;88:603–613. doi: 10.1016/s0092-8674(00)81903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linton J.M., Martin G.R., Reichardt L.F. The ECM protein nephronectin promotes kidney development via integrin α8β1-mediated stimulation of Gdnf expression. Development. 2007;134:2501–2509. doi: 10.1242/dev.005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donovan M.J., Natoli T.A., Sainio K., Amstutz A., Jaenisch R., Sariola H., Kreidberg J.A. Initial differentiation of the metanephric mesenchyme is independent of WT1 and the ureteric bud. Dev. Genet. 1999;24:252–262. doi: 10.1002/(SICI)1520-6408(1999)24:3/4<252::AID-DVG8>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 24.Xu P.X., Adams J., Peters H., Brown M.C., Heaney S., Maas R. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat. Genet. 1999;23:113–117. doi: 10.1038/12722. [DOI] [PubMed] [Google Scholar]
- 25.Brophy P.D., Ostrom L., Lang K.M., Dressler G.R. Regulation of ureteric bud outgrowth by Pax2-dependent activation of the glial derived neurotrophic factor gene. Development. 2001;128:4747–4756. doi: 10.1242/dev.128.23.4747. [DOI] [PubMed] [Google Scholar]
- 26.Nishinakamura R., Matsumato Y., Nakao K., Nakamura K., Sato A., Copeland N.G., Gilbert D.J., Jenkins N.A., Scully S., Lacey D.L., et al. Murine homolog of SALL1 is essential for ureteric bud invasion in kidney development. Development. 2001;128:3105–3115. doi: 10.1242/dev.128.16.3105. [DOI] [PubMed] [Google Scholar]
- 27.Wellik D.M., Hawkes P.J., Capecchi M.R. Hox11 paralogous genes are essential for metanephric kidney induction. Genes Dev. 2002;16:1423–1432. doi: 10.1101/gad.993302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valerius M.T., Patterson L.T., Feng Y., Potter S.S. Hoxa11 is upstream of integrin α8 expression in the developing kidney. Proc. Natl Acad. Sci. USA. 2002;99:8090–8095. doi: 10.1073/pnas.122229199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu P.X., Zheng W., Huang L., Maire P., Laclef C., Silvius D. Six1 is required for the early organogenesis of mammalian kidney. Development. 2003;130:3085–3095. doi: 10.1242/dev.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brodbeck S., Besenbeck B., Englert C. The transcription factor Six2 activates expression of the Gdnf gene as well as its own promoter. Mech. Dev. 2004;121:1211–1122. doi: 10.1016/j.mod.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 31.Self M., Lagutin O.V., Bowling B., Hendrix J., Cai Y., Dressler G.R., Oliver G. Six2 is required for suppression of nephrogenesis and progenitor renewal in the developing kidney. EMBO J. 2006;25:5214–5128. doi: 10.1038/sj.emboj.7601381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gong K.Q., Yallowitz A.R., Sun H., Dressler G.R., Wellik D.M. A Hox–Eya–Pax complex regulates early kidney developmental gene expression. Mol. Cell. Biol. 2007;27:7661–7668. doi: 10.1128/MCB.00465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mugford J.W., Sipilä P., Kobayashi A., Behringer R.R., McMahon A.P. Hoxd11 specifies a program of metanephric kidney development within the intermediate mesoderm of the mouse embryo. Dev. Biol. 2008;319:396–405. doi: 10.1016/j.ydbio.2008.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiotaki R., Petrou P., Giakoumaki E., Pavlakis E., Sitaru C., Chalepakis G. Spatiotemporal distribution of Fras1/Frem proteins during mouse embryonic development. Gene Expr. Patterns. 2007;7:381–388. doi: 10.1016/j.modgep.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 35.Petrou P., Chiotaki R., Dalezios Y., Chalepakis G. Overlapping and divergent localization of Frem1 and Fras1 and its functional implications during mouse embryonic development. Exp. Cell Res. 2007;313:910–920. doi: 10.1016/j.yexcr.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 36.Kiyozumi D., Sugimoto N., Nakano I., Sekiguchi K. Frem3, a member of the 12 CSPG repeats-containing extracellular matrix protein family, is a basement membrane protein with tissue distribution patterns distinct from those of Fras1, Frem2, and QBRICK/Frem1. Matrix Biol. 2007;26:456–462. doi: 10.1016/j.matbio.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 37.Patek C.E., Fleming S., Miles C.G., Bellamy C.O., Ladomery M., Spraggon L., Mullins J., Hastie N.D., Hooper M.L. Murine Denys–Drash syndrome: evidence of podocyte de-differentiation and systemic mediation of glomerulosclerosis. Hum. Mol. Genet. 2003;12:2379–2394. doi: 10.1093/hmg/ddg240. [DOI] [PubMed] [Google Scholar]
- 38.Roselli S., Gribouval O., Boute N., Sich M., Benessy F., Attie T., Gubler M.C., Antignac C. Podocin localizes in the kidney to the slit diaphragm area. Am. J. Pathol. 2002;160:131–139. doi: 10.1016/S0002-9440(10)64357-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen C.A., Hwang J.C., Guh J.Y., Chang J.M., Lai Y.H., Chen H.C. Reduced podocyte expression of α3β1 integrins and podocyte depletion in patients with primary focal segmental glomerulosclerosis and chronic PAN-treated rats. J. Lab. Clin. Med. 2006;147:74–82. doi: 10.1016/j.lab.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 40.Jadeja S. Fraser Syndrome and Mouse blebbed Mutants. UK: University of London; 2006. pp. 1–289. Ph.D. Thesis. [Google Scholar]
- 41.Kiyozumi D., Osada A., Sugimoto N., Weber C.N., Ono Y., Imai T., Okada A., Sekiguchi K. Identification of a novel cell-adhesive protein spatiotemporally expressed in the basement membrane of mouse developing hair follicle. Exp. Cell Res. 2005;306:9–23. doi: 10.1016/j.yexcr.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 42.Gamer L.W., Cox K.A., Small C., Rosen V. Gdf11 is a negative regulator of chondrogenesis and myogenesis in the developing chick limb. Dev. Biol. 2001;229:407–420. doi: 10.1006/dbio.2000.9981. [DOI] [PubMed] [Google Scholar]
- 43.Oh S.P., Yeo C.Y., Lee Y., Schrewe H., Whitman M., Li E. Activin type IIA and IIB receptors mediate Gdf11 signaling in axial vertebral patterning. Genes Dev. 2002;16:2749–2754. doi: 10.1101/gad.1021802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valerius M.T., McMahon A.P. Transcriptional profiling of Wnt4 mutant mouse kidneys identifies genes expressed during nephron formation. Gene Expr. Patterns. 2008;8:297–306. doi: 10.1016/j.gep.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maeshima A., Sakurai H., Choi Y., Kitamura S., Vaughn D.A., Tee J.B., Nigam S.K. Glial cell-derived neurotrophic factor independent ureteric bud outgrowth from the Wolffian duct. J. Am. Soc. Nephrol. 2007;18:3147–3155. doi: 10.1681/ASN.2007060642. [DOI] [PubMed] [Google Scholar]
- 46.Kreidberg J.A., Donovan M.J., Goldstein S.L., Rennke H., Shepherd K., Jones R.C., Jaenisch R. α3β1 integrin has a crucial role in kidney and lung organogenesis. Development. 1996;122:3537–3547. doi: 10.1242/dev.122.11.3537. [DOI] [PubMed] [Google Scholar]
- 47.Tryggvason K., Patrakka J., Wartiovaara J. Hereditary proteinuria syndromes and mechanisms of proteinuria. N. Engl. J. Med. 2006;354:1387–1401. doi: 10.1056/NEJMra052131. [DOI] [PubMed] [Google Scholar]
- 48.Center E.M., Emery K.E. Acidic glycosaminoglycans and laminin-1 in renal corpuscles of mutant blebs (my) and control mice. Histol. Histopathol. 1997;12:901–917. [PubMed] [Google Scholar]
- 49.Shao X., Somlo S., Igarashi P. Epithelial-specific Cre/lox recombination in the developing kidney and genitourinary tract. J. Am. Soc. Nephrol. 2002;13:1837–1846. doi: 10.1097/01.asn.0000016444.90348.50. [DOI] [PubMed] [Google Scholar]
- 50.Juhila J., Roozendaal R., Lassila M., Verbeek S.J., Holthofer H. Podocyte cell-specific expression of doxycycline inducible Cre recombinase in mice. J. Am. Soc. Nephrol. 2006;17:648–654. doi: 10.1681/ASN.2005050547. [DOI] [PubMed] [Google Scholar]
- 51.Hardelin J.P., Julliard A.K., Moniot B., Soussi-Yanicostas N., Verney C., Schwanzel-Fukuda M., Ayer-Le Lievre C., Petit C. Anosmin-1 is a regionally restricted component of basement membranes and interstitial matrices during organogenesis: implications for the developmental anomalies of X chromosome-linked Kallmann syndrome. Dev. Dyn. 1999;215:26–44. doi: 10.1002/(SICI)1097-0177(199905)215:1<26::AID-DVDY4>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 52.Duke V., Quinton R., Gordon I., Bouloux P.M., Woolf A.S. Proteinuria, hypertension and chronic renal failure in X-linked Kallmann’s syndrome, a defined genetic cause of solitary functioning kidney. Nephrol. Dial. Transplant. 1998;13:1998–2003. doi: 10.1093/ndt/13.8.1998. [DOI] [PubMed] [Google Scholar]
- 53.Deeb A., Robertson A., MacColl G., Bouloux P.M., Gibson M., Winyard P.J., Woolf A.S., Moghal N.E., Cheetham T.D. Multicystic dysplastic kidney and Kallmann’s syndrome: a new association? Nephrol. Dial. Transplant. 2001;16:1170–1175. doi: 10.1093/ndt/16.6.1170. [DOI] [PubMed] [Google Scholar]
- 54.Impallomeni M., Subramanian D., Mahmood N., Joseph I. Fraser syndrome in a 96-year-old female. Age Ageing. 2006;35:642–643. doi: 10.1093/ageing/afl109. [DOI] [PubMed] [Google Scholar]
- 55.Argueso L.R., Ritchey M.L., Boyle E.T., Jr, Milliner D.S., Bergstralh E.J., Kramer S.A. Prognosis of patients with unilateral renal agenesis. Pediatr. Nephrol. 1992;6:412–416. doi: 10.1007/BF00873996. [DOI] [PubMed] [Google Scholar]
- 56.Miner J.H. Developmental biology of glomerular basement membrane components. Curr. Opin. Nephrol. Hypertens. 1998;7:13–19. doi: 10.1097/00041552-199801000-00003. [DOI] [PubMed] [Google Scholar]
- 57.Plaisier E., Gribouval O., Alamowitch S., Mougenot B., Prost C., Verpont M.C., Marro B., Desmettre T., Cohen S.Y., Roullet E., et al. COL4A1 mutations and hereditary angiopathy, nephropathy, aneurysms, and muscle cramps. N. Engl. J. Med. 2007;357:2687–2695. doi: 10.1056/NEJMoa071906. [DOI] [PubMed] [Google Scholar]
- 58.Pitera J.E., Smith V.V., Thorogood P., Milla P.J. Coordinated expression of 3′ hox genes during murine embryonal gut development: an enteric Hox code. Gastroenterology. 1999;117:1339–1351. doi: 10.1016/s0016-5085(99)70284-2. [DOI] [PubMed] [Google Scholar]
- 59.Pitera J.E., Milla P.J., Scambler P., Adjaye J. Cloning of HOXD1 from unfertilised human oocytes and expression analyses during murine oogenesis and embryogenesis. Mech. Dev. 2001;109:377–381. doi: 10.1016/s0925-4773(01)00530-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.