Abstract
Microorganisms maintain the biosphere by catalyzing biogeochemical processes, including biodegradation of organic chemical pollutants. Yet seldom have the responsible agents and their respective genes been identified. Here we used field-based stable isotopic probing (SIP) to discover a group of bacteria responsible for in situ metabolism of an environmental pollutant, naphthalene. We released 13C-labeled naphthalene in a contaminated study site to trace the flow of pollutant carbon into the naturally occurring microbial community. Using GC/MS, molecular biology, and classical microbiological techniques we documented 13CO2 evolution (2.3% of the dose in 8 h), created a library of 16S rRNA gene clones from 13C labeled sediment DNA, identified a taxonomic cluster (92 of 95 clones) from the microbial community involved in metabolism of the added naphthalene, and isolated a previously undescribed bacterium (strain CJ2) from site sediment whose 16S rRNA gene matched that of the dominant member (48%) of the clone library. Strain CJ2 is a β proteobacterium closely related to Polaromonas vacuolata. Moreover, strain CJ2 hosts the sequence of a naphthalene dioxygenase gene, prevalent in site sediment, detected before only in environmental DNA. This investigative strategy may have general application for elucidating the bases of many biogeochemical processes, hence for advancing knowledge and management of ecological and industrial systems that rely on microorganisms.
Identification of the microorganisms and genes responsible for catalyzing biogeochemical reactions in terrestrial and aquatic habitats is one of the major goals of microbial ecology (1, 2). Mechanistic information about microbial processes improves our ability to understand and manage ecological systems (3, 4) Among the many reactions effected by microorganisms is biodegradation of organic chemical pollutants (5-7).
Although a multitude of microorganisms have been isolated from contaminated soils or waters and shown in laboratory assays to have the potential to biodegrade environmental pollutants (8, 9), only rarely [e.g., via field inoculation (10)] have the populations actually responsible for pollutant metabolism in field sites been identified. Many methodological obstacles have traditionally prevented investigators from simultaneously documenting the identity and activity of microorganisms in real-world habitats such as soil. Prominent among these obstacles are: an incomplete understanding of the physical, chemical, and nutritional characteristics of microbial habitats; a large reservoir of dormant but potentially responsive cells in environmental samples; and the related propensity for microbial communities to change both physiologically and in their population structure after removal from their in situ context (5, 6)
Thus, investigations aimed at linking identity with biogeochemical activity of microorganisms in soils and waters have relied on multidisciplinary, often indirect, approaches. If active microorganisms are recognizable microscopically [due to either morphological distinctiveness or phylogenetic probing for their 16S rRNA genes (11, 12)], they can be simultaneously examined for evidence that the gene(s) that catalyze the biogeochemical process of interest are expressed [e.g., mRNA or key enzymes or metabolic intermediates (13, 14)]. Alternatively, some natural systems contain a substrate fortuitously labeled with a unique stable isotopic signature (e.g., methane in anaerobic marine sediments); this has allowed the signature to be directly traced to taxonomically recognizable membrane lipids or other microbial biomarkers in field-derived samples (15, 16).
Another, perhaps compromised, path to simultaneously measure the identity and activity of microorganisms involves the use of model systems (typically samples of the habitat of interest incubated in the laboratory after addition of radioactive or stable isotopically labeled substrates) that allow metabolic activity to be measured and the detection of label incorporated into active cells (17, 18). In the present investigation, we used field-based stable isotopic probing (SIP) (1, 19) to discover a previously undescribed group of bacteria responsible for in situ metabolism of an environmental pollutant, naphthalene. In addition, a representative of the active bacteria was isolated and found to be host to a distinctive biodegradation gene present in the sediment microbial community.
Materials and Methods
Site. This study was conducted near South Glens Falls, NY. The site is a shallow unconfined aquifer (water table at ≈2.5 m depth) near the western edge of the Hudson River. Coal-tar waste, rich in naphthalene and other aromatic hydrocarbons, was deposited 40 years ago in this flat, sandy, forested area where flowing groundwater has created a narrow subsurface plume of coal-tar constituents and has deposited naphthalene in surface sediments in a low-lying seepage area (6, 19-22).
Overview of Field-Based Stable Isotope Probing and 13CO2 Respiration. Procedures, reported previously (19), involved release of aqueous 13C-naphthalene solutions (≈22 ppm concentration, 1-ml doses at times 0, 24, and 48 h) added to surface sediment, covering the sediment with a small glass chamber, monitoring 13CO2 and 12CO2 evolution in headspace gases by MS, freezing on site (at time = 54 h) the 13C-naphthalene-dosed sediment in dry ice/ethanol, laboratory extraction of nucleic acids from the sediment, separating 12C-DNA from 13C-DNA by CsCl equilibrium centrifugation, and then amplifying (by PCR), cloning, and sequencing 16S rRNA genes from the 13C fraction of DNA.
Field Chambers. The “open-bottom soil cover” treatments were prepared, as described (19), by inserting truncated 250-ml glass canning jars 3-5 cm into the soil, enclosing 28 cm2 of the surface. For the respiration assay, there were four replicate chambers. For DNA extraction, there were three replicates each for treatments dosed with 13C-naphthalene (13C6, 99% purity, Cambridge Isotope Laboratories, Cambridge, MA) and 12C (unlabeled)-naphthalene controls.
Respiration Assay. Headspace sampling (250-μl gas-tight syringe) occurred three times over an 8-h period. All 12 syringes were shuttled to the laboratory, and GC/MS analyses were completed on the sampling day. A Hewlett-Packard 5971A GC/MS equipped with a Hewlett-Packard Pora Plot Q column (25 m × 0.32 mm, 10-μm film thickness and He as the carrier gas) in splitless mode was used to separate gaseous components. The detector was operated at 1.10-5 torr (1 torr = 133 Pa), 70 eV (19). The GC oven program was isothemal 60°C. CO2 eluted at 2.5 min. By using the single ion monitoring mode, the detector was able to simultaneously quantify 12CO2 (m/z 44) and 13CO2 (m/z = 45). Gross 13CO2 release was directly measured. Net 13CO2 evolution produced from the 13C-naphthalene was computed (19) by subtracting background 13CO2 produced from sediment organic matter. The latter was inferred from direct measurement of 12CO2 adjusted according to the known fixed ratio of 12Cto 13C in naturally occurring carbon [1.11% (19, 23)]. Net values for 13CO2 evolved within replicate chambers were pooled; averaged values at different times were compared with Student's t tests.
DNA Extraction and CsCl Fractionation of 13C DNA. Sediment (5 g per chamber) that received substrate was removed by using a sterile spatula, placed on dry ice, transported to the lab on the dry ice, and stored at -80°C (19). DNA extraction was carried out by using the Fast DNASPIN kit with a bead-beating procedure (Qbiogene, Carlsbad, CA).
As a positive control for 13C and 12C DNA, Pseudomonas putida strain G7 was grown in three mineral salts media, one with 100% 13C-glucose, one with 50% 13C-glucose, and the other with nonenriched (“12C”) glucose. DNA was extracted as above. One milliliter of the DNA solution was diluted to 4.5 ml with TE buffer (10 mM Tris/1 mM EDTA, pH 8), and 4.5 g CsCl was added and shaken gently until dissolved. Ethidium bromide (100 μl; 10 mg/ml) was added to each ultracentrifuge tube, which was then sealed. Tubes were centrifuged at 140,000 × g (Vti 81 rotor; 41,900 rpm) for 66 h at 20°C (19). Resultant bands in standards were clearly separated. The 13C band in field treatments could not be seen; instead, we used known standards to guide DNA removal by piercing the centrifuge tubes using standard methods with an 18-g needle (24). In processing DNA from the 13C-naphthalene treatment, we compensated for partial 12C-labeling of DNA (two-thirds of the C atoms) by sampling 2 mm below the 50% of the 13C band. One-half milliliter of CsCl solution containing the DNA was withdrawn. Ethidium bromide was extracted from the DNA by the addition of 10 volumes of TE-saturated 1-butanol and gently mixing. The organic layer was discarded and the extraction repeated five times, then the volume of DNA was brought to 3 ml in TE. DNA precipitation occurred overnight at -20°C by addition of 300 μl of 3 M sodium acetate (pH 4.6) and 2× volume of ethanol. After pelleting at 13,000-15,000 × g for 30 min, the DNA was washed twice with 70% ethanol, centrifuged at the same speed for 10 min, resuspended in 50-100 μl of distilled water, and stored at -20°C.
PCR Cloning, Restriction Digestion and Sequencing. PCR amplification of 16S rRNA genes in the 13C-DNA fraction used universal eubacterial primers (27f and 1492r) by methods described previously (25). The product was ligated into the vector pCR2.1 (TA cloning, Invitrogen) following the manufacturer's recommended protocol. After transformation of plasmids into host cells and blue/white screening, colonies with inserts were verified by PCR with vector-specific primers (5′-GTAACGGCCGCCAGTGTGCT and 5′-CAGTGTGATGGATATCTGCA) that flank the cloning region. The amplicons were digested with HaeIII and HhaI. Restriction fragment length polymorphism (RFLP) patterns were analyzed on 3% MetaPhore agarose (BioWhittaker, Molecular Applications, Rockland, ME) gels with a 100-bp ladder (Promega) as marker. Clones containing unique RFLP patterns were selected for sequencing, grown overnight in 5 ml of LB with appropriate antibiotics (Kanomycin, Sigma), pelleted, and plasmids were purified (QiaPrep spin miniprep kit, Qiagen, Santa Clarita, CA). PCR primers (M13 forward, M13 reverse, and 16S rDNA universal) were used by the Cornell DNA sequencing facility (Ithaca, NY). Raw sequence data from both strands were assembled into full length sequences by using the program seqman II (DNASTAR, Madison, WI). After assembly, the consensus sequence was verified manually by referring to the corresponding ABI chromatograms of the sequencing reactions. The computational tools of the Ribosomal Database II project (http://rdp.cme.msu.edu/html) were used to check chimeras and to calculate the similarity values for individual rDNA sequences by using the SEQUENCE_MATCH program. A BLAST search (http://ncbi.nlm.nih.gov/BLAST) was also used to identify the additional related sequences. The closest relatives identified from both searches were included in dendrograms. To evaluate dendrogrm topology, sequence alignment and phylogenies were done with clustal w and phylip [dnaml (maximum likelihood) and dnadist (Kimura model); http://evolution.gs.washington.edu/phylip.html] software packages after deleting regions of sequence ambiguity from the analyses.
Population Profiling in DNA Extracts by Terminal (T)-RFLP Analysis of 16S rDNA Genes. For T-RFLP analysis, standard procedures (26) were followed. PCR primer 27F-FAM (5′ end-labeled with phosphoramidite fluorochrome 5-carboxy-fluorescein) was used instead of the above forward primer of 16S rDNA. Fluorescently labeled PCR products were digested for 6 h with HhaI and electrophoretically separated in an Applied Biosystems 3730 DNA analyzer (Applied Biosystems), and the data were analyzed by using genemapper software (Applied Biosystems).
Isolation of Naphthalene-Degrading Bacterium. To isolate the bacterium responsible for naphthalene degradation, previously described methods (25, 27) were used. One gram of the coal tar-contaminated sediment was transferred to tubes containing PBS. Serial dilutions were prepared, and 0.1-ml aliquots were spread onto mineral salts base (MSB) (28) agar plates, incubated in the presence of naphthalene vapors at 10°C. The colonies were repeatedly transferred on agar media to isolate the microorganisms able to grow in the presence, but not absence, of the naphthalene vapor.
Naphthalene Biodegradation Assays. Metabolism of naphthalene by strain CJ2 (cells grown on MSB-pyruvate medium and washed) was confirmed after extraction by GC/MS measurements of aqueous naphthalene added to MSB media (25). An uninoculated control accounted for abiotic losses, which were negligible. In the slurried sediment assay, mixtures of groundwater (30 ml) mixed with 10 g (wet weight) of seep sediment were prepared with and without inoculation of 107 cells per ml strain CJ2 (three replicates per treatment) and incubated at 10°C. Periodically, the slurries were sampled (1 ml) with a syringe. After mixing with 1 ml of methanol, the samples were analyzed by GC/MS.
Analysis of Napthalene Dioxygenase Genes. As described (20), diluted nucleic acids from sediment or strain CJ2 (also water as a negative control) were used as templates. PCR amplification of a 482-bp fragment of nahA was achieved with the following PCR primers: Ac114F 5′-CTGGC(T/A)(T/A)TT(T/C)CTCAC(T/C)CAT-3′ and Ac596R 5′-C(G/A)GGTG(C/T)CTTCCAGTTG-3′ (20). PCR amplification was carried out in 50-μl reaction volumes, using previously described (20, 25) thermal cycler and reagents with primer concentration of 1 μM and a cycling regime of 94°C for 5 min (1 cycle); 94°C for 1 min, 56°C for 45 sec, and 72°C for 45 sec (35 cycles); 72°C for 10 min (1 cycle). The amplicons were cloned, sequenced, and analyzed as described above for 16S rRNA genes.
Results
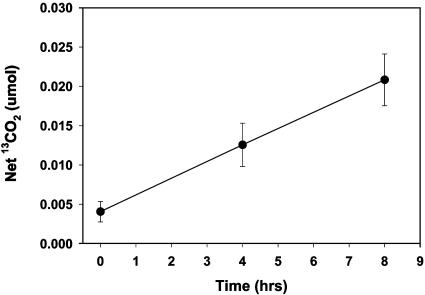
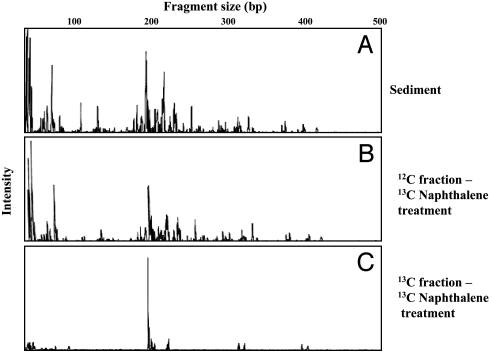
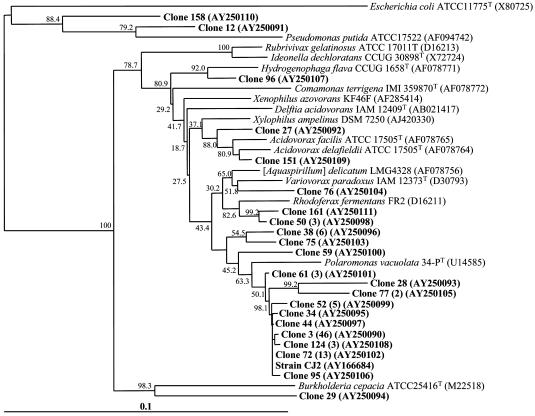
Previous reports have demonstrated that microorganisms native to a coal tar waste-contaminated field study site have been engaged in naphthalene biodegradation (e.g., refs. 6, 20, and 21). To discover the identity of the active populations, we implemented field respiration and SIP procedures (19), which released 13C-labeled naphthalene where flowing groundwater has deposited naphthalene in organic matter-rich surface sediment (22). The small volumes of naphthalene-dosed sediment (≈50 ml) were covered by open-bottom glass chambers that allowed headspace gases produced by the native microbial community to be periodically gathered and analyzed for evolution of 13CO2 above background produced from sediment organic matter (19). In an initial 8-hr experiment examining in situ respiration, net 13CO2 rose steadily, amounting to net mineralization of 2.3% of the added naphthalene (Fig. 1). We then revisited the site, adding 13C-naphthalene to sediment in three doses over a 54-hr period. Parallel control treatments added 12C (unlabeled) naphthalene to adjacent sediments. The chambers were removed, and DNA was extracted for CsCl density gradient ultracentrifugation to separate 12C- from 13C-DNA. As found in previous studies (19), when PCR primers designed to amplify the 16S rRNA gene were applied to the location of the CsCl gradient that corresponded with a 13C-DNA standard, a robust amplicon was obtained from the 13C-naphthalene-treated sediment but not from the 12C-naphthlene-treated sediment (Fig. 6, which is published as supporting information on the PNAS web site). The strongly amplified signal from 13C-labeled 16S rRNA genes provides a means to identify microbial populations that metabolized the 13C-naphthalene. Confirmation that the 13C-DNA fraction that was amplified by PCR was not simply a relic from the 12C-band seemed prudent; therefore, we also conducted 16S rDNA-based T-RFLP analysis to allow comparison of population profiles between bulk soil DNA and both of the CsCl-separated (12C- and 13C-) DNA fractions from the soil that had been dosed with 13C naphthalene. The T-RFLP population fingerprints (Fig. 2) showed that the 13C-DNA fraction was readily distinguishable from and far simpler than (Fig. 2C) the other two population profiles, which matched one another as expected (Fig. 2 A and B). After examining enzyme restriction digests of 95 clones derived from the 13C-labeled DNA (Fig. 2C) and determining 22 full 16S rRNA gene sequences, we prepared a phylogenetic tree to examine the relationships between the cloned sequences and those in GenBank from related bacteria (Fig. 3). The vast majority of the clones (92 of 95) clustered in an unusual group associated with Acidovorax, Variovorax, Rhodoferax, and Polaromonas, members of the β proteobacteria. Three outlying sequences were split between other Gram-negative species (Pseudomonas and Burkolderia). Interestingly, there was surprisingly little variation among the vast majority of cloned 16S rDNA sequences: 84 of the 92 fell into the same tight clade whose closest cultured relatives were Polaromonas vacuolata and dichloroethene-degrading bacterium JS666 ≈93% identity (29, 30). Forty-six clones from the 13C-DNA were identical (clone 3 in Fig. 3).
Fig. 1.
Net respiration of 13CO2 beneath glass chambers enclosing 28 cm2 of sediment in the contaminated field site that was dosed with aqueous 13C-naphthalene. Data reflect averages of four replicate chambers. Background 13CO2 respired was inferred from evolved 13CO2 (19). Error bars show standard deviations. Values at successive sampling times were significantly different (Student's t test, P < 0.008).
Fig. 2.
Microbial community composition reflected by T-RFLP profiles of 16S rRNA genes digested with HhaI, in bulk coal tar-contaminated sediment (A) and CsCl gradient-ultracentrifuged preparations from the sediment 12C-DNA (B) and 13C-DNA (C) fractions.
Fig. 3.
Phylogenetic analysis of 95 cloned bacterial 16S rRNA genes from the sediment-derived 13C-DNA fraction and isolated strain CJ2. Sequences found in this study (bold) are contrasted with those from reference strains (italic). The clones were screened by their enzyme restriction patterns, and 22 full 16S rRNA genes representing these patterns were fully sequenced. Numbers in parentheses indicate frequencies of clones exhibiting the same restriction pattern. Sequence alignment and phylogenic relationships were completed with clustal w and phylip software packages (Kimura model; Department of Genomics, University of Washington). Values at each node are the percentage of 1,000 boot-strap trees. (Bar = 0.1 changes per nucleotide position.) T, type strain.
In an attempt to grow and isolate the host of the 16S rRNA gene sequence that we found was dominant in the clone library, we plated dilutions of seep samples on naphthalene-based mineral salts (MSBN) media and incubated them at the in situ groundwater temperature of 10°C. Large (≈4-mm diameter) mucoid colonies appeared after 2-4 weeks. When restreaked and purified, this culture (strain CJ2) produced large colonies in the presence of naphthalene and not in its absence. Strain CJ2 was unable to grow on MSBN media at temperatures above 20°C because at such temperatures, the vapor pressure of naphthalene was toxic (Table 1, which is published as supporting information in the PNAS web site); thus, the 10°C incubation was crucial for isolation of strain CJ2. Remarkably, the 16S rRNA gene sequence of strain CJ2 fell within the tight taxonomic cluster dominated by clone 3 found in the SIP 13C-DNA library (Fig. 3) of microorganisms involved in naphthalene metabolism in situ. Furthermore, the T-RFLP pattern for strain CJ2's 16S rRNA gene revealed a single major fragment (193 bp) that matched the major fragment from the 13C-DNA fraction shown in Fig. 2.
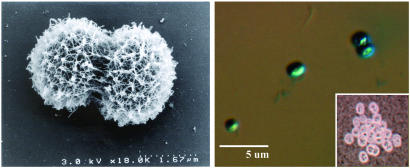
Naphthalene utilization by strain CJ2 was confirmed in mineral salts media analyzed after 10 days by GC/MS. Furthermore, assays that used GC/MS to monitor biodegradation of the naphthalene component of the contaminated sediment in laboratory-incubated sediment slurries (10°C) showed that inoculation of strain CJ2 accelerated naphthalene loss (Fig. 7, which is published as supporting information on the PNAS web site). Thus, strain CJ2 has the potential to be metabolically active in a laboratory approximation of the field habitat. Strain CJ2 is a Gram-negative cocoid bacterium (Fig. 4) with several phenotypic and genotypic characteristics [including absence of flagella, extracellular polysaccharide production, inability to grow on rich media, and intracellular accumulation of polyhydroxyalkanoic acids, polyphosphates, and glycogen (27)] unlike those of taxonomically related bacteria listed in Fig. 3.
Fig. 4.
Photomicrographs of strain CJ2. Note coccoid shape of two dividing cells, rough surface texture from extracellular polysaccharide, absence of flagella, and size (1-2 μm) visible in scanning electron microscopy image (Left). Alcian-blue staining at low pH reveals acidic nature of extracellular polysaccharide (Right). (Inset) India ink staining of capsular material.
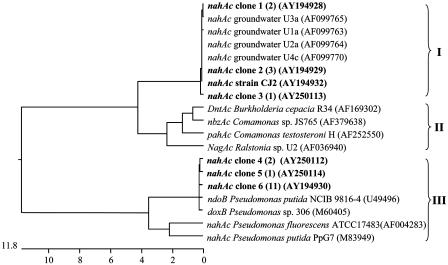
To further explore the environmental significance of strain CJ2, we focused on a gene involved in naphthalene metabolism. We used PCR primers specific for the terminal iron-sulfur component of naphthalene dioxygenase [nahAc (20)] to amplify, clone, and sequence a 482-bp fragment of nahAc. The dendrogram portraying the relationship of strain CJ2's nahAc to related sequences (Fig. 5) reveals that strain CJ2 is host of a distinctive sequence (clade I in Fig. 5). This clade, with ≈7% amino acid dissimilarity with clade II, was previously discovered among dioxygenase genes expressed in situ as mRNA sequences in groundwaters from the study site (20). Until now, there has been no known host for this nahAc allele. Comparison between nahAc sequences of strain CJ2 and the closest previously cultured host, Comamonas testosteroni H (31), showed an identity of 92.8%. To verify that the nahAc-like amplicon from stain CJ2 was present in the study site, we extracted DNA from strain CJ2's sediment of origin and used the nahAc-specific primers to amplify, clone, and sequence related genes in the microbial community. Six of the 20 sediment-derived nahAc clones that we obtained fell within clade I, matching the sequence of strain CJ2 (Fig. 5).
Fig. 5.
Cluster analysis examining the percent dissimilarity of 446-bp naphthalene dioxygenase (nahAc)-like sequences from cultured microorganisms and amplicons from site sediment. Shown are partial nahAc sequences for eight reference strains, new isolate CJ2 (bold), 20 clones from site sediment-derived DNA extracts (bold), and four GenBank sequences previously cloned from well-water mRNA taken from this study site. Dendrograms were constructed with GenBank DNA sequences by using the clustal procedure in dnastar with the default parameters. The horizontal scale denotes the percentage of divergence of the sequences. In parentheses, numbers indicate frequencies of clones found representing the same sequence; GenBank accession nos. are also given. Roman numerals indicate three clades mentioned in the text; note that the sequence labeled “strain CJ2” is the only one in Clade I whose host bacterium has been cultured.
Discussion
This investigation relied on the SIP technique to trace labeled C atoms from an added microbial substrate (naphthalene) into the DNA of naturally occurring populations involved in substrate metabolism. If a high-substrate concentration had been used, SIP may have revealed opportunistic microorganisms other than those active under ambient site conditions. Because the mass of 13C-naphthalene added (≈60 μg/5 g sediment) roughly matched ambient concentrations (22), we feel that potential concentration-based artifacts were avoided. Data interpretation presumed that the 16S rRNA gene sequences found in 13C-labeled DNA were indicative of microbial populations directly responsible for naphthalene biodegradation. But potential indirect incorporation of the 13C (e.g., via excreted metabolites or other C transfers from biodegrading populations to adjacent community members) must be considered. In this implementation of SIP, indirect C incorporation was unlikely because the incubation period in the field was brief (≈2 days) and because our isolated bacterium, host of the predominant SIP-based 16S RNA gene sequence, possessed genotypic and phenotypic traits consistent with its role as an active member of the site's microbial community. Thus, this investigation allows us to link strain CJ2 to in situ metabolism of an environmental pollutant. The presence of the distinctive nahAc allele in both strain CJ2 and its sediment of origin strongly suggests that the enzyme encoded by this gene catalyzes naphthalene biodegradation in situ. Elimination of metabolism by creating a nahAc-deletion mutant of strain CJ2 will solidify this latter conjecture.
Although aerobic biodegradation of naphthalene has long been described (32), the majority of cultured hosts that have contributed to understanding both the biochemistry and genetics (33) of naphthalene biodegradation fall into a readily cultivatable broad class of Pseudomonas-like Gram-negative bacteria (34). It is significant that the bacterium that we found to be active in situ and to contain the distinctive sediment-borne naphthalene dioxygenase gene is not related to Pseudomonas. Rather, strain CJ2 is phylogenetically remote and features several unusual traits that argue for its novelty and, perhaps, explain why this strain has not been isolated before. Strain CJ2 joins the growing minority of cultured nonpsuedomonads (e.g., ref. 35) able to biodegrade naphthalene.
An emergent theme in current microbial ecology is the repartee and feedback that develop between culture- and non-culture-based inquiries (36). This dialogue has routinely proven to be heuristic, leading to the discovery of both new taxonomic diversity in the microbial world (37) and clues that allow initially mysterious hosts of traits observable from real-world habitats (e.g., 16S rRNA gene sequences or physiological activity) to be pursued and eventually brought into cultivation (38, 39), thus enabling further physiological, genetic, genomic, and ecological inquiries (3, 40). An eventual goal would be to understand habitat biogeochemistry sufficiently well to be able to quantitatively attribute changes in habitat resources to individual members of microbial communities.
Supplementary Material
Acknowledgments
Constructive criticism from two anonymous reviewers was helpful. We thank W. C. Ghiorse for preparing micrographs. This research was supported by National Science Foundation Microbial Observatories Grant no. MCB-0084175 with cooperation from Niagara Mohawk Power Corporation and the New York State Department of Environmental Conservation.
Abbreviations: SIP, stable isotopic probing; RFLP, restriction fragment length polymorphism; T-RFLP, terminal RFLP; MSB, mineral salts base.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AY166684, AY250090-AY250111 (16S rDNA), AY194928-AY194932, and AY250112-AY250114 (nahAc)].
References
- 1.Radajewski, S., Ineson, R., Parekh, N. R. & Murrell, J. C. (2000) Nature 403, 646-649. [DOI] [PubMed] [Google Scholar]
- 2.Beja, O., Aravind, L., Koonin, E. V., Suzuki, M. T., Hadd, A., Nguyen, L. P., Jovanovich, S. B., Gates, C., Feldman, R. A. & DeLong, E. F. (2000) Science 289, 1902-1906. [DOI] [PubMed] [Google Scholar]
- 3.Newman, D. K. & Banfield, J. F. (2002) Science 296, 1071-1077. [DOI] [PubMed] [Google Scholar]
- 4.Jetten, M. S. M, Strous, M., van de Pas-Schoonen, K. T., Schalk, J., van Dongen, J. G. J. M., Van de Graaf, A. A., Logemaann, S., Muyzer, G., van Loosedrecht, M. C. M. & Kuenen, J. G. (1998) FEMS Microbiol. Rev. 22, 421-437. [DOI] [PubMed] [Google Scholar]
- 5.Madsen, E. L. (1998) Environ. Sci. Technol. 32, 429-439. [Google Scholar]
- 6.Madsen, E. L., Sinclair, J. L. & Ghiorse, W. C. (1991) Science 252, 830-833. [DOI] [PubMed] [Google Scholar]
- 7.National Research Council (2000) Natural Attenuation for Groundwater Remediation (Natl. Acad. Press, Washington, DC).
- 8.Alexander, M. (1994) Biodegradation and Bioremediation (Academic, San Diego), 2nd Ed.
- 9.Wackett, L. P. & Hershberger, D. C. (2001) Biocatalysis and Biodegradation (Am. Soc. Microbiol., Washington, DC).
- 10.Ellis, D. E., Lutz, E. J., Odom, J. M., Buchanan, R. J., Jr., Bartlett, C. L., Lee, M. D., Harkness, M. R. & DeWeerd, K. A. (2002) Environ. Sci. Technol. 34, 2254-2260. [Google Scholar]
- 11.Capone, D. A., Zehr, J. P., Paerl, H. W., Bergman, B. & Carpenter, E. J. (1997) Science 276, 1221-1229. [Google Scholar]
- 12.Pernthaler, A., Pernthaler, J. & Amann, R. (2002) Appl. Environ. Microbiol. 68, 3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paerl, H. W., Priscu, J. C. & Brawner, D. L. (1989) Appl. Environ. Microbiol. 55, 2965-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, F., González, J., M., Dustman, W. A., Moran, M. A. & Hodson, R. E. (1997) Appl. Environ. Microbiol. 63, 4907-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinrichs, K. U., Hayes, J. M., Sylva, S. P., Brewer, P. G. & DeLong, E. F. (1999) Nature 398, 802-805. [DOI] [PubMed] [Google Scholar]
- 16.Orphan, V. J., House, C. H., Hinrichs, K. U., Mckeegan, K. D. & DeLong, E. F. (2001) Science 293, 484-487. [DOI] [PubMed] [Google Scholar]
- 17.Ward, B. B. (1984) Limnol. Oceanogr. 29, 402-410. [Google Scholar]
- 18.Ouverney, C. C. & Fuhrman, J. A. (1999) Appl. Environ. Microbiol. 65, 1746-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Padmanabhan, P., Padmanabhan, S., DeRito, C., Gray, A., Gannon, D., Snape, J. R., Tsai, C. S., Park, W., Jeon, C. & Madsen, E. L. (2003) Appl. Environ. Microbiol. 69, 1614-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson, M. S., Bakermans, C. & Madsen, E. L. (1999) Appl. Environ. Microbiol. 65, 80-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bakermans, C., Hohnstock-Ashe, A. M. Padmanabhan, S., Padmanabhan, P. & Madsen, E. L. (2002) Microb. Ecol. 44, 107-117. [DOI] [PubMed] [Google Scholar]
- 22.Madsen, E. L., Mann, C. L. & Bilotta, S. (1996) Environ. Toxicol. Chem. 15, 1876-1882. [Google Scholar]
- 23.Hoefs, J. (1987) Stable Isotope Geochemistry (Springer, New York), 3rd Ed.
- 24.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 25.Bakermans, C. & Madsen, E. L. (2002) Microb. Ecol. 44, 95-106. [DOI] [PubMed] [Google Scholar]
- 26.Liu, W.-T., Marsh, T. L., Cheng, H. & Forney, L. J. (1997) Appl. Environ. Microbiol. 63, 4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeon, C. O. Ghiorse, W. C. & Madsen, E. L. (2003) Intl. J. Syst. Evol. Microbiol., in press. [DOI] [PubMed]
- 28.Stanier, R. Y., Palleroni, N. J. & Douderoff, M. T. (1966) J. Gen. Microbiol. 43, 159-271. [DOI] [PubMed] [Google Scholar]
- 29.Irgens, R. L., Gosink, J. J. & Staley, J. T. (1996) Intl. J. Syst. Bacteriol. 46, 822-826. [DOI] [PubMed] [Google Scholar]
- 30.Coleman, N. V., Mattes, T. E., Gossett, J. M. & Spain J. C. (2002) Appl. Environ. Microbiol. 68, 2726-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moser, R. & Stahl, U. (2001) Appl. Microbiol. Biotechnol. 55, 609-618. [DOI] [PubMed] [Google Scholar]
- 32.Davies, J. I. & Evans, W. C. (1964) Biochem. J. 91, 251-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gibson, D. T. & Parales, R. E. (2000) Curr. Opin. Biotechnol. 11, 236-243. [DOI] [PubMed] [Google Scholar]
- 34.Williams, P. A. & Sayers, J. R. (1994) Biodegradation 5, 195-217. [DOI] [PubMed] [Google Scholar]
- 35.Larkin, M. J., Allen, C. C., Kulakov, L. A. & Lipscomb, D. A. (1999) J. Bacteriol. 181, 6200-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karner, M. B., DeLong, E. F. & Karl, D. M. (2001) Nature 409, 507-510. [DOI] [PubMed] [Google Scholar]
- 37.Pace, N. R. (1997) Science 276, 734-740. [DOI] [PubMed] [Google Scholar]
- 38.Rappe, M. S., Connon, S. A., Vergin, K. L. & Giovannoni, S. J. (2002) Nature 418, 630-633. [DOI] [PubMed] [Google Scholar]
- 39.Maymo-Gatell, X., Chien, Y. T., Gossett, J. M. & Zinder, S. H. (1997) Science 276, 1568-1571. [DOI] [PubMed] [Google Scholar]
- 40.DeLong, E. F. (2002) Curr. Opin. Microbiol. 5, 520-524. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.