Abstract
The Spx protein of Bacillus subtilis represses activator-stimulated transcription by interacting with the C-terminal domain of RNA polymerase (RNAP) α subunit. Its concentration increases in cells lacking the ATP-dependent protease, ClpXP, resulting in severe effects on growth and developmental processes. Microarray analysis was undertaken to identify genes that are induced or repressed when Spx interacts with RNAP. The induced genes included those encoding products known to function in maintaining thiol homeostasis. Two genes, thioredoxin (trxA) and thioredoxin reductase (trxB), are transcriptionally induced under conditions of thiol-specific oxidative (disulfide) stress by a mechanism involving Spx-RNAP interaction. Disulfide stress also results in an increase in Spx-dependent transcriptional repression. The increase in Spx activity in cells encountering disulfide stress is due in part to a posttranscriptional mechanism of spx control resulting in an increase in Spx concentration. An spx null mutant and a strain bearing an allele of rpoA that prevents Spx-RNAP interaction show hypersensitivity to disulfide stress. From these results, it is proposed that Spx is an activator that mobilizes the operations necessary to reverse the effects of oxidative damage, but it also serves as a negative regulator that causes the postponement of developmental programs and energy-consuming growth-related functions while the cell copes with the period of stress.
Keywords: transcription activation, ClpXP, disulfide stress
Transcription initiation in prokaryotes is controlled at many levels and through a variety of protein-protein and protein-DNA interactions between RNA polymerase (RNAP), promoter DNA, and diverse regulatory factors. The RNAP α subunit, particularly its C-terminal domain (αCTD), is a common target for activator-RNAP interaction that allows RNAP to establish productive contact with promoter DNA (1-4). Recently, the αCTD has been shown to be a target for a form of negative control that is exerted by the Spx protein in the spore-forming bacterium, Bacillus subtilis (5). Spx prevents or disrupts activator-RNAP interaction by binding to αCTD. Spx is highly conserved among a large number of Gram-positive species (5-7) and resembles members of the arsenate reductase (ArsC) (8) family of proteins. In Lactococcus lactis, a gene encoding a homolog of Spx was identified as the site of mutations that conferred heat resistance to recA mutants (6) and that suppressed clpP mutations by causing a general increase in proteolytic activity (7).
The spx (yjbD) gene was identified as the site of mutations that alleviated the defects in developmentally regulated transcription that are conferred by mutations in clpX and clpP in B. subtilis (9). The clpX and clpP genes encode the subunits of the ATP-dependent protease, ClpXP (10-12), which functions in disposal of incomplete translation products that become SsrA-tagged (13, 14), as well as in the control of σ factor RpoS concentration in Escherichia coli (15, 16). A role in the degradation of denatured protein in stressed cells was also proposed for B. subtilis ClpXP (17). The Spx protein accumulated to high concentration in clpX and clpP cells and was subsequently found to be a substrate for ClpXP-catalyzed proteolysis (9, 18).
Certain amino acid substitutions in the part of the rpoA gene encoding the RNAP αCTD resulted in a similar clpX suppressor phenotype as conferred by an spx null mutation (19). It has recently been shown that Spx interacts with αCTD, and the product of the suppressor rpoAcxs (clpX suppressor) allele was unable to bind to Spx (5). This interaction was discovered to be the basis for the negative effect Spx exerted on developmentally regulated transcription. The activity of the response regulatory proteins of the two-component protein family, ComA and ResD, was inhibited when Spx was added to reactions containing RNAP and promoter DNA of ComA- or ResD-controlled genes (5).
These studies provided an understanding of Spx activity but provided very little information about the physiological role of Spx-αCTD interaction. Genomic microarray analysis of cells overproducing Spx, as described in this report, was undertaken to learn the role of Spx in B. subtilis. Genes positively affected by Spx-RNAP interaction were discovered that function in thiol homeostasis, such as thioredoxin (trxA) and thioredoxin reductase (trxB). Evidence is presented that Spx-αCTD interaction functions in both positive and negative control and is associated with cells undergoing disulfide stress.
Materials and Methods
Isolation of the Promoter Regions of trxA and trxB. The promoter region of the trxA gene was amplified by PCR, and the product was inserted into the plasmid pDG793 (20), which yielded plasmid pSN79. The promoter region of trxB gene was amplified by PCR, and the product was inserted into pDG793 to generate pSN78. The DNA sequences of the promoter regions in pSN78 and pSN79 were verified.
Isolation of Total RNA from B. subtilis. Samples (15 ml) of B. subtilis cultures were collected under the indicated conditions, and each was mixed with an equal volume of ice-cold methanol. The culture was centrifuged and the cell pellet was frozen at -80°C. The total RNA was prepared by RNeasy mini kit (Qiagen, Chatsworth, CA).
Microarray Hybridization Analysis. Strain culturing and RNA sample collection were performed in triplicate. RNA from the triplicate set of samples was purified and used to generate dye-labeled cDNA probes for microarray hybridization (20). The microarrays were prepared as described (21) and contained PCR fragments made by using oligonucleotide primers specifying 4,100 genes of the B. subtilis genome. Dye-labeled cDNA probes made from RNA of samples collected at each time point from isopropyl β-d-thiogalactoside (IPTG)-treated and untreated cultures were hybridized simultaneously with the microarray slides. Fluorescent intensities were measured, and ratios of expression +IPTG/-IPTG were calculated. Table 1, which is published as supporting information on the PNAS web site, lists the genes that show >3-fold induction or repression by SpxDD production after 30 min of IPTG treatment in WT but not in rpoAcxs-1 cells. Fold induction and fold repression compared with RNA levels in cells not treated with IPTG are indicated.
Primer Extension Analysis. The amounts of trxA, trxB, srfA, and yocJ transcripts were quantified by primer extension analysis. Ten micrograms (for trxA, srfA, and yocJ) or 20 μg (for trxB) of the total RNA from each strain was hybridized to each corresponding primer at 55°C for 20 min after incubation at 94°C for 2 min in the AMV reverse transcriptase buffer (Promega). Then 1 mM (final concentration) dNTP and AMV reverse transcriptase (Promega) was added to each reaction. After 30-min incubation at 42°C, synthesized cDNA was precipitated with ethanol and loaded onto a 6% or 8% polyacrylamide gel containing 8 M Urea. The dideoxynucleotide sequence ladder was obtained by using the Thermo Sequenase Cycle Sequencing Kit (United States Biochemical) with pSN78 (trxA), pSN79 (trxB), pMMN89 [srfA (22)], and a PCR product specifying yocJ as templates. The yocJ primer extension product served as a control to test the quality of the RNA purified from diamide-treated cells.
Site-Directed Mutagenesis. Plasmid pMMN464, a derivative of pPS34 (23) that harbors 9.3 kbp of chromosomal DNA from yjbG to oppB, was digested with EcoRV, thus releasing a 3.9-kbp DNA fragment. This was digested with EcoRI to release a 1.4-kbp EcoRV-EcoR1 DNA fragment that contains the intact spx gene. This fragment was inserted into pUC19 to create pSN49. The spxDD mutant gene was generated by PCR-based site-directed mutagenesis (24). The first round PCR was performed by using JH642 chromosomal DNA as the template. The second round PCR product was then inserted into plasmid pSN49, thus generating pSN53. The nucleotide sequence of spxDD region in pSN53 was confirmed.
Overexpression of spx in B. subtilis. DNA containing the WT spx coding sequence and that of its spxDD derivative were amplified by PCR by using JH642 chromosomal DNA and pSN53 as templates, respectively. The PCR products were inserted into pDR111[Phyperspank(spank-hy) fusion vector (21)], thus generating pMMN521(WT spx) and pSN56(spxDD). The DNA sequences of spx gene in pMMN521 and pSN56 were verified.
Overproduction and Purification of Proteins. The coding sequence of the mutant spxDD allele was amplified by PCR, and the product was inserted into pPROEX (Life Science Technologies, Grand Island, NY/GIBCO). The nucleotide sequence of the spxDD coding sequence in the resulting plasmid, pSN60, was verified. SpxDD, WT Spx, ClpX, and ClpP proteins were purified as described (18).
Western Blot Analysis. The total protein from cells of B. subtilis cultures grown in TSS minimal medium (25) was prepared by passage through a French press. Protein (30 μg) from each sample was applied to a 15% SDS-polyacrylamide gel. Western blot analysis by using anti-Spx antiserum was performed as described (9).
In Vitro ClpXP-Catalyzed Proteolysis Reaction. In vitro degradation reaction was performed under conditions as described (5).
Assay of β-Galactosidase Activity. β-Galactosidase activity was determined as described (26) and is presented as Miller units (27).
Results
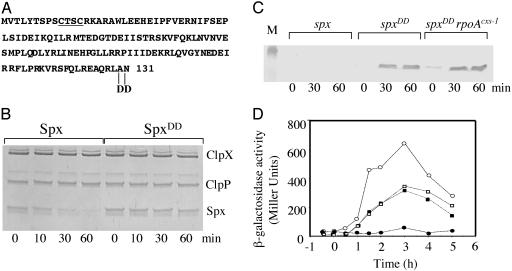
Amino Acid Substitutions at the Extreme C Terminus of Spx Reduce Sensitivity to ClpXP-Catalyzed Proteolysis. The Spx protein of B. subtilis is a substrate for the ATP-dependent protease ClpXP, which maintains Spx concentration at low levels under normal growth conditions (9, 18). It was determined that Spx could repress transcription by binding to the αCTD of RNAP, thereby preventing the α subunit from interacting with certain transcriptional regulators (5). However, the repressor activity of Spx in vivo had been observed only in the highly pleiotropic clpX or clpP mutant cells in which Spx concentration is high. To begin to understand the conditions under which Spx protein functions in clpX+ cells, a microarray analysis was undertaken to identify the genes that are induced or repressed when Spx protein establishes contact with RNAP. To overexpress spx in a WT background, thus avoiding the pleiotropy of a clpX mutant, it was necessary to generate a mutant form of Spx that was resistant to ClpXP-catalyzed proteolysis. Some evidence was obtained that the amino acid sequence at the extreme C terminus of Spx was necessary for Spx to serve as a ClpXP substrate (5, 18). This sequence bears some resemblance to that of the C-terminal end of the SsrA-encoded peptide tag, which is the substrate sequence for ClpXP in both B. subtilis and E. coli (13, 14). A mutant allele of spx in which the terminal Ala and Asn codons were replaced by two Asp codons (spxDD) was constructed (Fig. 1A). Such a mutation in ssrA prevents the proteolysis of SsrA peptide-tagged polypeptides by ClpXP (28). An N-terminal His-6-tag spxDD product, which was expressed in and purified from E. coli, was shown to be resistant to ClpXP-catalyzed proteolysis in vitro (Fig. 1B). The WT and mutant alleles of spx were fused to the IPTG-inducible promoter Pspank-hy and introduced into B. subtilis cells, where, on IPTG induction, the SpxDD protein accumulated, whereas little WT Spx protein was detected (Fig. 1C). Thus, it was observed that the spxDD allele encoded a stable product.
Fig. 1.
Protease-resistant SpxDD and its effect on srf-lacZ expression. (A) Amino acid sequence of Spx showing the location of the 2-Asp (DD) substitution at the C terminus. The CXXC motif is underlined. (B) Coomasssie-stained SDS-polyacrylamide gel of ClpXP proteolysis reactions containing either WT Spx protein or mutant SpxDD. Time of incubation is shown at the bottom. (C) Western blot analysis of whole-cell extracts obtained from cultures of the following strains: ORB4271 (amyE::pMMN521 [Pspank-hy-spx]), lanes marked spx; ORB4342 (amyE::pSN56 [Pspank-hy-spxDD]), lanes marked spxDD; ORB4343 (amyE::pSN56 rpoAcxs-1), lanes marked spxDD rpoAcxs-1. Marker (M) band is lysozyme (20.8 kDa). (D) Expression of transcriptional srf-lacZ over time in ORB4349 (srfA-lacZ [pMMN84] amyE::pSN56, circle) and ORB4353 (srfA-lacZ [pMMN84] amyE::pSN56, rpoAcxs-1, square) grown in DSM. The cultures were split at OD 600 = 0.5 at time 0 and grown in the absence (open symbols) or presence (filled symbols) of 1 mM IPTG. The x axis represents the time (h) of incubation after the addition of IPTG.
SpxDD Retains the Spx Repressor Activity That Requires the WT Allele of rpoA. The Pspank-hy-spxDD construct was introduced into two strains, one WT and one rpoAcxs-1, each containing a srf-lacZ transcriptional fusion. The product of rpoAcxs-1 bears a Tyr to Cys substitution in the αCTD that renders it unable to interact with Spx (5, 19). Addition of IPTG to the Pspank-hy-spxDD srf-lacZ cells resulted in a severe repression of srf-lacZ expression late in the growth curve, when srf transcription is normally induced. Induction of spxDD in rpoAcxs-1 srf-lacZ cells, however, had no effect on srf transcription (Fig. 1D). IPTG treatment resulted in accumulation of SpxDD in the rpoAcxs-1 cells (Fig. 1C), showing that the rpoA mutation did not affect the production of SpxDD. These data showed that the stable SpxDD mutant retains its repressor activity, which requires productive interaction with αCTD. The IPTG-inducible spxDD expression construct provided the means to conduct a genome-wide examination of the effect the Spx-RNAP complex has on gene expression.
Microarray Analysis Identified Genes Induced by Spx-RNAP Interaction That Function in Disulfide Stress Response. To better understand the role of Spx-RNAP interaction in B. subtilis, we sought to identify the genes whose expression was affected by SpxDD production, but not in a rpoAcxs-1 background. Cultures of strains bearing the Pspank-hy-spxDD construct and containing either the cxs-1 or WT alleles of rpoA were grown in minimal TSS medium. Samples for dye-labeled cDNA synthesis were collected before IPTG treatment, as well as 30 and 60 min after IPTG addition. Table 1 lists 106 genes that showed 3-fold greater (Induced) and 176 genes showing 3-fold or greater reduced (Repressed) expression 30 min after IPTG induction of spxDD in rpoA+ cells but not in rpoAcxs-1 cells. All of the genes listed showed induction or repression, as defined above, in all three samples of the triplicate set and were ordered according to the extent of induction or repression that was measured.
The genes of the srf operon were repressed (10.4- to 20.9-fold repression) by SpxDD-RNAP interaction, as were a number of genes that are associated with normal growth and metabolism. These included genes whose products function in purine, pyrimidine, and amino acid biosynthesis.
Genes That Function in Maintenance of Thiol Homeostasis Are Induced by Spx-RNAP Interaction. Among the genes that were induced (Table 1), several encode products that are associated with thiol-redox homeostasis, such as trxA (14.8-fold induction), trxB (9.3-fold), tpx (probable thiol peroxidase, 4.2-fold), msrA (peptide methionine sulfoxide reductase, 4.0-fold), as well as genes ycgT (4.2-fold), ydbP (3.1-fold), and ytpP (3.2-fold), which encode thioredoxin-like proteins. All were induced in spxDD rpoA+ cells but showed no or poor induction in rpoAcxs-1 cells. Recently, a genomic expression analysis was carried out to determine which genes become active when disulfide stress is induced by treatment with the thiol-specific oxidant, diamide (29). We compared the genomic expression profile of spxDD expressing cells with that reported for diamide-treated cells and found a close correlation, 47 of the 106 genes listed (Table 1), including those mentioned above, being induced under both conditions. Of those repressed, 81 of the 176 were also repressed in diamide-treated cells.
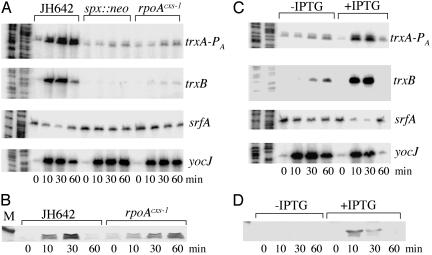
Transcriptional Induction of trxA and trxB in Diamide-Treated Cells Depends on Spx and the WT Allele of rpoA. The microarray data suggested that Spx caused the induction of trxA and -B transcription under disulfide stress. To further examine the control of trxA and -B transcription and the possible involvement of Spx, primer extension analyses were carried out to determine the level of trxA and -B transcripts after treatment of WT, spx mutant, and rpoAcxs-1 cells with diamide (Fig. 2).
Fig. 2.
Effects of diamide, spx, and rpoAcxs-1 on the levels of trxA, trxB, srf, and yocJ transcripts. (A) Primer extension analysis of trxA, trxB, srf, and yocJ transcripts in RNA from cultures grown in TSS and treated with 1 mM diamide after midlogarithm phase. RNA was purified from strains JH642 (WT), ORB3834 (spx::neo), and ORB3621 (rpoAcxs-1). Time after diamide addition in minutes is shown at the bottom. The left lanes show gel profile of A and T specific sequencing reactions. (B) Western analysis, by using anti-Spx antiserum, of Spx protein concentration in cells of strains JH642 (WT) and ORB3621 (rpoAcxs-1) treated with diamide as described in A. Marker (M) band is lysozyme (20.8 kDa). (C) Primer extension analysis of trxA, trxB, srf, and yocJ transcripts in RNA from cultures of strain ORB4533 (spx::neo amyE::pMMN521 [Pspank-hy-spx]) grown in TSS with and without 0.1 mM IPTG and treated with 1 mM diamide at midlogarithm phase. Time after diamide treatment in minutes is indicated at the bottom. Left lanes show gel profiles of A and T specific sequencing reactions. (D) Western blot analysis of Spx protein from cultures of strain ORB4533 grown in TSS with or without IPTG and treated with diamide. Time after diamide treatment in minutes is indicated at bottom.
Transcription of the trxA gene initiates from two promoters, PA (initiating 24 bp from start codon), which is likely a σA-used promoter, and PB (243 bp upstream from start codon), transcription from which requires the σB form of RNAP (30). Fig. 2 A shows that transcription from trxA PA is induced by diamide, but significantly less transcript is detected in the spx null mutant. Lower transcript levels are also observed in rpoAcxs-1 cells, suggesting that induction of trxA transcription under disulfide stress requires interaction between Spx and RNAP αCTD. Utilization of the upstream PB promoter was not affected by diamide treatment (data not shown).
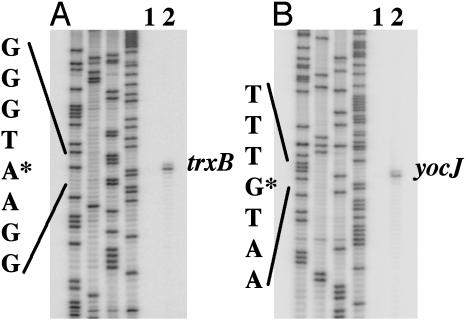
Spx had a similar effect on trxB RNA levels. Primer extension (Fig. 3A) detected a single transcriptional start site for trxB that is located 78 bp upstream of the start codon. There is a sequence upstream (TAGGAT) that bears some resemblance to a -10 region that is recognized by the σA form of RNAP, but there is no -35 sequence similarity. The diamide-dependent induction of trxB transcription observed in WT cells was eliminated by the spx null mutation and the rpoAcxs-1 allele (Fig. 2 A).
Fig. 3.
Localization of the transcriptional start sites of trxB and yocJ. Cells of JH642 were grown in TSS medium to midlogarithm, and the cultures were split. Diamide was added to one culture. Cells were harvested after 30 min, and RNA was purified for primer extension reaction. Primer extension products by using the primer specifying the trxB transcript and the yocJ transcript, respectively, are shown. Lane 1, -diamide; lane 2, +diamide. Four lanes on the left in A and B are the gel profiles of sequencing reactions A, G, C, and T.
Several genes, such as yocJ, that are induced by diamide treatment do not require Spx for transcription (Fig. 2 A and C). The yocJ gene encodes a phage-like product and resides in the SKIN element of the chromosome (31). Primer extension analysis identified a transcription start site 37 bp upstream of the start codon (Fig. 3B). The yocJ primer extension product served as a control of RNA quality.
As previously reported, the Spx protein exhibits repressor activity as an “anti-α” factor by interfering with RNAP-activator interaction (5). It does this by binding to the RNAP αCTD, but not to regulatory DNA. One operon that is negatively controlled by Spx is srf, which requires the activator ComA, in its phosphorylated form, for transcription initiation. By primer extension analysis, the level of srf transcript was observed to decrease after diamide treatment in WT cells but was unchanged in cells bearing either the spx null mutation or the rpoAcxs-1 mutation (Fig. 2 A). These results indicate that, as in the case of Spx-dependent transcriptional activation, Spx-dependent repression, by interaction of Spx with RNAP αCTD, is also enhanced under disulfide stress.
Both induction and repression were reduced after 60 min (Fig. 2 A and C), which would suggest that a process of recovery from the stressed state is taking place. This could be the result of consumption of diamide and/or repair of disulfide damage, either of which brought about by the transient Spx-dependent increase in thioredoxin/thioredoxin reductase activity.
Disulfide Stress Results in an Increase in Spx Concentration. Previous studies had shown that the yjbC-spx operon transcript concentration increased after diamide addition (29). The accelerated transcription of spx and consequent increase in spx product concentration could account for the induction of trxA and -B transcription. As shown in Fig. 2B, Spx concentration increases after diamide treatment, which is in keeping with this premise. Likewise, Spx levels also increase in the rpoAcxs-1 mutant treated with diamide. To further examine the role of spx transcriptional control in diamide-dependent induction, the WT spx gene was placed under the control of an IPTG-inducible promoter and the construct was introduced into the chromosome of B. subtilis cells bearing the spx::neo mutation (Fig. 2C). Cultures were grown in the presence or absence of IPTG, and diamide was added at midlogarithm phase. At 10, 30, and 60 min, RNA samples were collected for primer extension analysis. Only the IPTG-treated cells exposed to diamide showed the increase in trxA and -B transcript levels, indicating that spx in trans complemented the spx null mutation. Repression of srf transcription was also observed in diamide-treated cells bearing the IPTG-inducible spx construct (Fig. 2C). An increase in Spx protein concentration was observed in diamide-treated cells bearing the IPTG-inducible spx construct (Fig. 2D). Therefore, it is not likely that increased spx transcription alone explains the diamide-dependent induction of the spx-controlled trxA and -B genes, and there might be a mechanism to increase Spx concentration and/or activity in response to disulfide stress (see Discussion). As observed in the case of Spx-dependent transcriptional activation and repression, there is a reduction of Spx concentration 60 min after diamide addition (Fig. 2 B and D), again an indication of an adaptation or recovery from the stressed state. Examination of the Spx level in rpoAcxs-1 cells shows high levels of protein after 60 min (Fig. 2B), suggesting further that the cell's recovery from the disulfide stressed state might depend on the Spx-RNAP interaction.
Failure of Spx to Interact with RNAP Confers Sensitivity to Diamide. The requirement of Spx and the WT allele of rpoA for induction of trxA and -B under disulfide stress suggested that Spx and its contact with RNAP are important for conferring resistance to oxidative stress. Cells of the WT strain JH642 and the two mutant derivatives, one bearing spx::neo and the other rpoAcxs-1, were streaked onto a disco sporulation medium agar plate containing 50 μM diamide (Fig. 4). The WT parent, but not the mutants, grew on the plate containing diamide.
Fig. 4.
Sensitivity of spx and rpoAcxs-1 cells to diamide. Cells of strains JH642 (WT), ORB3834 (spx::neo), and ORB3621(rpoAcxs-1) were streaked onto DSM plates with (top) or without (bottom) 50 μM diamide.
Discussion
Although highly conserved among Gram-positive species, Spx bears little resemblance to the many well characterized prokaryotic transcription factors, in terms of both its primary structure and its mode of action. A survey of the entire genome was necessary to learn which genes are controlled by Spx to understand its function. By genomic microarray hybridization, genes either negatively or positively affected by Spx-RNAP interaction were identified. As expected, genes of the ComPA regulon [such as the srf operon, rapF, and pel (32)] were repressed in cells overproducing Spx and having the WT allele of rpoA. Notably, genes whose products function in primary metabolism were also repressed, including both pur and pyr genes required for purine and pyrmidine biosynthesis, amino acid biosynthesis genes, and vitamin biosynthesis. It is not clear how Spx-RNAP interaction negatively affects the levels of these transcripts, but down-regulation of genes required for biosynthetic functions could partly explain the poor growth phenotype in minimal medium of clpX and clpP mutants (8) in which Spx is present in high concentrations.
Of particular significance in the work presented herein was the identification of the genes that were induced by Spx-RNAP interaction. Among the most highly induced were the trxA and -B genes, as well as other genes with functions related to thiol homeostasis. This result suggested a role for Spx in the cell's response to disulfide stress. Indeed, a test for resistance to the thiol-specific oxidant, diamide, showed that spx and rpoAcxs-1 mutants were hypersensitive to diamide. A previously conducted microarray study of genes induced and repressed by diamide treatment illustrated how disulfide stress causes genome-wide changes in transcription patterns (29). A comparison of these data with the microarray data herein shows that a significant number of genes, both induced and repressed by diamide treatment, are controlled by Spx.
Positive and negative mechanisms of Spx-dependent transcriptional control involve previously identified Spx-αCTD interactions. Spx binds to the RNAP α subunit, but not to RpoA mutant proteins bearing amino acid substitutions in the helix 1 region of αCTD, which has been implicated in positive transcriptional control (33, 34) and interaction with UP elements in promoter DNA (35-38). Although a mechanism of negative control can be easily imagined, Spx positive control is difficult to envision because there is no evidence of DNA-binding activity, which would seem necessary to direct RNAP-promoter interaction at a specific gene. Transcription activation of trxA and -B could be an indirect consequence of Spx-negative transcriptional control, which might be acting on a gene encoding an unstable repressor of trxA and -B. Alternatively, Spx could directly participate in positive control as a mediator that establishes contact between αCTD and a DNA-bound activator. Diamide could also cause a conformational change in Spx that uncovers DNA-binding activity. Spx contains a putative CXXC redox active Cys motif (Fig. 1 A) that is highly conserved and might be the site where the effects of oxidative stress can modulate Spx activity and/or stability. A similar motif exists in the C-terminal end of the transcriptional regulator PerR, which controls transcription initiation in response to peroxide stress (39). In this case, the CXXC motif is thought to participate in coordinating a metal ion, which is released on oxidation of a Cys residue. Previous work provided some evidence that Spx is a metal-binding protein, but it is not known whether a metal ion is involved in Spx activity (40).
Disulfide stress causes an increase in Spx intracellular concentration (Fig. 2), which is accompanied by a consequent elevation of trxA and -B transcript levels, along with repression of srf. The diamide-induced increase in Spx concentration cannot be attributed solely to control exerted at the spx gene promoters but is likely also due to posttranscriptional forms of control, perhaps increased Spx stability. One explanation for the increase in Spx concentration is a reduction in the activity of ClpXP. ClpX contains a zinc-finger domain at its N terminus that is required for activity (41). This domain contains two Zn-coordinating CXXC motifs, and oxidation of one or more of the Cys residues by diamide could inactivate the protease, resulting in enhanced Spx stability. Oxidation of Cys residues has been shown to affect the activity of the Zn-fingered chaperonin Hsp33 by causing the release of bound Zn ion (in a manner similar to that of PerR) (42). Alternatively, ClpXP protease could be titrated by denatured or deformed protein aggregates that accumulate during diamide treatment and thus diverted from its role in maintaining low Spx concentration.
Recovery from the stressed state, characterized by a decrease in trxA and -B transcription and srf transcriptional repression, takes place 60 min after onset of disulfide stress. Likewise, a concomitant decrease in Spx concentration is observed but not in cells bearing the rpoAcxs-1 mutation, suggesting that recovery depends on Spx-RNAP interaction. This supports a model that the Spx-dependent induction of a disulfide stress response is also necessary for the restoration of the prestressed state in which Spx activity and/or concentration is reduced to that observed under normal growth conditions. This could be brought about by the action of thioredoxin (43), which in elevated concentrations, restores the reduced state of its target proteins that might also include Spx and/or ClpX.
Several transcriptional regulatory proteins have been identified that function in the cell's response to oxidative stress, and some of these have been characterized in exquisite detail. OxyR (44), SoxRS (45, 46), OhrR (47), CatR (48), and PerR (39) all have redox-sensitive activity and regulate genes whose specific purpose is to counter the effects of oxidative attack. Other regulators exert more global effects on gene expression, such as the RNAP σ factors σS of E. coli and Salmonella (49) and σB of Bacillus (50), and become active in response to the consequences of oxidative stress. Spx represents a previously undescribed and highly conserved transcriptional regulator that exerts global control when cells encounter a thiol-specific oxidant. Under conditions causing disulfide stress, both negative and positive global control of gene expression is exerted by Spx through its interaction with RNAP.
Supplementary Material
Acknowledgments
We are grateful to Megan Rokop for suggesting the construction of the spxDD mutant for use in overproducing Spx. We also thank David Rudner for providing plasmid pDR111 and Michiko M. Nakano for plasmid construction, helpful advice, and critical reading of the manuscript. This research is supported by National Institutes of Health Grants GM45898 (to P.Z.) and GM50895 (to A.D.G.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RNAP, RNA polymerase; αCTD, C-terminal domain of RNAP α subunit; IPTG, isopropyl β-d-thiogalactoside.
References
- 1.Busby, S. & Ebright, R. H. (1999) J. Mol. Biol. 293, 199-213. [DOI] [PubMed] [Google Scholar]
- 2.Igarashi, K., Hanamura, A., Makino, K., Aiba, H., Aiba, H., Mizuno, T., Nakata, A. & Ishihama, A. (1991) Proc. Natl. Acad. Sci. USA 88, 8958-8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Igarashi, K. & Ishihama, A. (1991) Cell 65, 1015-1022. [DOI] [PubMed] [Google Scholar]
- 4.Mencia, M., Monsalve, M., Rojo, F. & Salas, M. (1998) J. Mol. Biol. 275, 177-185. [DOI] [PubMed] [Google Scholar]
- 5.Nakano, S., Nakano, M. M., Zhang, Y., Leelakriangsak, M. & Zuber, P. (2003) Proc. Natl. Acad. Sci. USA 100, 4233-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duwat, P., Ehrlich, S. D. & Gruss, A. (1999) Mol. Microbiol. 31, 845-858. [DOI] [PubMed] [Google Scholar]
- 7.Frees, D., Varmanen, P. & Ingmer, H. (2001) Mol. Microbiol. 41, 93-103. [DOI] [PubMed] [Google Scholar]
- 8.Martin, P., DeMel, S., Shi, J., Gladysheva, T., Gatti, D. L., Rosen, B. P. & Edwards, B. F. (2001) Structure (London) 9, 1071-1081. [DOI] [PubMed] [Google Scholar]
- 9.Nakano, M. M., Hajarizadeh, F., Zhu, Y. & Zuber, P. (2001) Mol. Microbiol. 42, 383-394. [DOI] [PubMed] [Google Scholar]
- 10.Gottesman, S., Clark, W. P., de Crecy-Lagard, V. & Maurizi, M. R. (1993) J. Biol. Chem. 268, 22618-22626. [PubMed] [Google Scholar]
- 11.Wawrzynow, A., Wojtkowiak, D., Marszalek, J., Banecki, B., Jonsen, M., Graves, B., Georgopoulos, C. & Zylicz, M. (1995) EMBO J. 14, 1867-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wojtkowiak, D., Georgopoulos, C. & Zylicz, M. (1993) J. Biol. Chem. 268, 22609-22617. [PubMed] [Google Scholar]
- 13.Gottesman, S., Roche, E., Zhou, Y. & Sauer, R. T. (1998) Genes Dev. 12, 1338-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiegert, T. & Schumann, W. (2001) J. Bacteriol. 183, 3885-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Becker, G., Klauck, E. & Hengge-Aronis, R. (1999) Proc. Natl. Acad. Sci. USA 96, 6439-6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou, Y. & Gottesman, S. (1998) J. Bacteriol. 180, 1154-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kruger, E., Witt, E., Ohlmeier, S., Hanschke, R. & Hecker, M. (2000) J. Bacteriol. 182, 3259-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakano, S., Zheng, G., Nakano, M. M. & Zuber, P. (2002) J. Bacteriol. 184, 3664-3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakano, M. M., Zhu, Y., Liu, J., Reyes, D. Y., Yoshikawa, H. & Zuber, P. (2000) Mol. Microbiol. 37, 869-884. [DOI] [PubMed] [Google Scholar]
- 20.Guerout-Fleury, A. M., Frandsen, N. & Stragier, P. (1996) Gene 180, 57-61. [DOI] [PubMed] [Google Scholar]
- 21.Britton, R. A., Eichenberger, P., Gonzalez-Pastor, J. E., Fawcett, P., Monson, R., Losick, R. & Grossman, A. D. (2002) J. Bacteriol. 184, 4881-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakano, M. M., Xia, L. & Zuber, P. (1991) J. Bacteriol. 173, 5487-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuno, K. & Sonenshein, A. L. (1999) J. Bacteriol. 181, 3392-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogura, M., Liu, L., LaCelle, M., Nakano, M. M. & Zuber, P. (1999) Mol. Microbiol. 32, 799-812. [DOI] [PubMed] [Google Scholar]
- 25.Fouet, A. & Sonenshein, A. L. (1990) J. Bacteriol. 172, 835-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakano, M. M., Marahiel, M. A. & Zuber, P. (1988) J. Bacteriol. 170, 5662-5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, J. H. (1972) Experiments in Molecular Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 28.Flynn, J. M., Levchenko, I., Seidel, M., Wickner, S. H., Sauer, R. T. & Baker, T. A. (2001) Proc. Natl. Acad. Sci. USA 98, 10584-10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leichert, L. I., Scharf, C. & Hecker, M. (2003) J. Bacteriol. 185, 1967-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scharf, C., Riethdorf, S., Ernst, H., Engelmann, S., Volker, U. & Hecker, M. (1998) J. Bacteriol. 180, 1869-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stragier, P., Kunkel, B., Kroos, L. & Losick, R. (1989) Science. 243, 507-512. [DOI] [PubMed] [Google Scholar]
- 32.Ogura, M., Yamaguchi, H., Yoshida, K., Fujita, Y. & Tanaka, T. (2001) Nucleic Acids Res. 29, 3804-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murakami, K., Fujita, N. & Ishihama, A. (1996) EMBO J. 15, 4358-4367. [PMC free article] [PubMed] [Google Scholar]
- 34.Tang, H., Severinov, K., Goldfarb, A., Fenyo, D., Chait, B. & Ebright, R. H. (1994) Genes Dev. 8, 3058-3067. [DOI] [PubMed] [Google Scholar]
- 35.Aiyar, S. E., Gourse, R. L. & Ross, W. (1998) Proc. Natl. Acad. Sci. USA 95, 14652-14657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Estrem, S. T., Ross, W., Gaal, T., Chen, Z. W., Niu, W., Ebright, R. H. & Gourse, R. L. (1999) Genes Dev. 13, 2134-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaal, T., Ross, W., Blatter, E. E., Tang, H., Jia, X., Krishnan, V. V., Assa-Munt, N., Ebright, R. H. & Gourse, R. L. (1996) Genes Dev. 10, 16-26. [DOI] [PubMed] [Google Scholar]
- 38.Ross, W., Gosink, K. K., Salomon, J., Igarashi, K., Zou, C., Ishihama, A., Severinov, K. & Gourse, R. L. (1993) Science 262, 1407-1413. [DOI] [PubMed] [Google Scholar]
- 39.Herbig, A. F. & Helmann, J. D. (2001) Mol. Microbiol. 41, 849-859. [DOI] [PubMed] [Google Scholar]
- 40.Nakano, M. M., Nakano, S. & Zuber, P. (2002) Mol. Microbiol. 44, 1341-1349. [DOI] [PubMed] [Google Scholar]
- 41.Banecki, B., Wawrzynow, A., Puzewicz, J., Georgopoulos, C. & Zylicz, M. (2001) J. Biol. Chem. 276, 18843-18848. [DOI] [PubMed] [Google Scholar]
- 42.Jakob, U., Eser, M. & Bardwell, J. C. (2000) J. Biol. Chem. 275, 38302-38310. [DOI] [PubMed] [Google Scholar]
- 43.Carmel-Harel, O. & Storz, G. (2000) Annu. Rev. Microbiol. 54, 439-461. [DOI] [PubMed] [Google Scholar]
- 44.Storz, G., Tartaglia, L. A. & Ames, B. N. (1990) Science 248, 189-194. [DOI] [PubMed] [Google Scholar]
- 45.Wu, J. & Weiss, B. (1991) J. Bacteriol. 173, 2864-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu, J. & Weiss, B. (1992) J. Bacteriol. 174, 3915-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fuangthong, M., Atichartpongkul, S., Mongkolsuk, S. & Helmann, J. D. (2001) J. Bacteriol. 183, 4134-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hahn, J. S., Oh, S. Y., Chater, K. F., Cho, Y. H. & Roe, J. H. (2000) J. Biol. Chem. 275, 38254-38260. [DOI] [PubMed] [Google Scholar]
- 49.Fang, F. C., Libby, S. J., Buchmeier, N. A., Loewen, P. C., Switala, J., Harwood, J. & Guiney, D. G. (1992) Proc. Natl. Acad. Sci. USA 89, 11978-11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Helmann, J. D., Wu, M. F., Gaballa, A., Kobel, P. A., Morshedi, M. M., Fawcett, P. & Paddon, C. (2003) J. Bacteriol. 185, 243-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.