Abstract
Osmotic homeostasis in the brain involves movement of water through aquaporin-4 (AQP4) membrane channels. Perivascular astrocyte end-feet contain distinctive orthogonal lattices (square arrays) assembled from 4- to 6-nm intramembrane particles (IMPs) corresponding to individual AQP4 tetramers. Two isoforms of AQP4 result from translation initiation at methionine residues M1 and M23, but no functional differences are known. In this study, Chinese hamster ovary cells were transfected with M1, M23, or M1+M23 isoforms, and AQP4 expression was confirmed by immunoblotting, immunocytochemistry, and immunogold labeling. Square array organization was examined by freeze-fracture electron microscopy. In astrocyte end-feet, >90% of 4- to 6-nm IMPs were found in square arrays, with 65% in arrays of 13-30 IMPs. In cells transfected with M23, 95% of 4- to 6-nm IMPs were in large assemblies (rafts), 85% of which contained >100 IMPs. However, in M1 cells, >95% of 4- to 6-nm IMPs were present as singlets, with <5% in incipient arrays of 2-12 IMPs. In M1+M23 cells, 4- to 6-nm IMPs were in arrays of intermediate sizes, resembling square arrays in astrocytes. Structural cross-bridges of 1 × 2 nm linked >90% of IMPs in M23 arrays (≈1,000 cross-bridges per μm2) but were rarely seen in M1 cells. These studies show that M23 and M1 isoforms have opposing effects on intramembrane organization of AQP4: M23 forms large square arrays with abundant cross-bridges; M1 restricts square array assembly.
Aquaporins are specialized water transport channels in plasma membranes of water-permeable tissues (1). Aquaporins 1 and 4 (AQP1 and AQP4) are most important to fluid movements in mammalian brain. AQP4 exists as two isoforms, differing at their N termini, because of translation initiation at the first methionine (M1, 323 aa) or the second methionine (M23, 301 aa) (2, 3). Both isoforms are present in brain, but M23 is at least 3-fold more abundant (4, 5). Endogenous AQP4 is a tetramer usually containing M1 and M23 subunits. The water permeabilities of M1 and M23 are similar, and functional differences are not known (3, 4).
Fluid movements are precisely orchestrated within the rigid cranium to prevent physical damage from swelling or shrinkage. Interfaces between brain parenchyma and cerebrospinal fluid occur around the ventricles, surrounding blood vessels, and at the brain surface. AQP1 is expressed in rat choroid plexus, the site of cerebrospinal fluid secretion (6), whereas AQP4 is enriched in rat astrocyte end-feet surrounding brain capillaries (7, 8). Astrocyte processes forming the glia limitans at brain surfaces, ependymal cells lining brain ventricles, and Müller cells facing the vitreous body and retinal blood vessels all have abundant AQP4 (9). AQP4 in perivascular membranes of astrocyte end-feet has been implicated in neurological disorders, including acute hyponatremic edema, postischemic injury, and epileptic seizures (10-13).
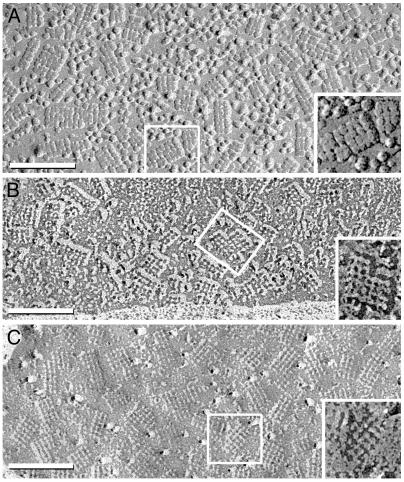
Perivascular membranes of astrocyte end-feet contain numerous strikingly regular arrays of intramembrane particles (IMPs) in freeze-fracture electron micrographs. These IMP arrays have been referred to as square arrays, assemblies, or orthogonally arranged particles (OAPs) (14). In early freeze-fracture images of astrocyte end-feet (15), square arrays were resolved as 6-nm IMP protrusions in P-face images (protoplasmic leaflets) or as smaller pits in E-faces (extraplasmic leaflets). The sizes and shapes of square arrays vary, but the IMPs and pits have uniform 6-nm lattice spacings within arrays (Fig. 1). Square arrays were observed in Chinese hamster ovary (CHO) cells transfected with AQP4 (16, 17) but not in AQP4-null mice (18). Anti-AQP4 directly labeled square arrays in replicas of astrocyte end-feet (19). The atomic structure of the homologous protein AQP1 has a tetramer cross-sectional diameter of 6 nm (20), similar to the 4- to 6-nm IMPs.
Fig. 1.
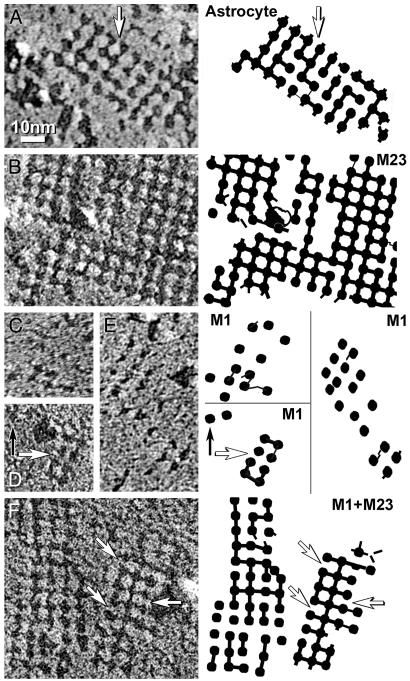
Freeze-fractured astrocyte end-feet membranes in suprachiasmic nucleus from adult rat. (A) The conventional replica has a 2-nm-thick platinum coat and a 1- to 2-nm coat of water vapor (27). Multiple square arrays appear as 6-nm P-face IMPs. (B) In higher-resolution replicas made with ≈1 nm of platinum without detectable water vapor, square arrays consist of 4-nm IMPs linked by 1 × 2 nm cross-bridges. (C) In E-face images, 3- to 4-nm pits are linked by 1 × 2 nm furrows. (Insets) Representative examples are enlarged and printed with black shadows. (Bars = 100 nm.)
Here we report freeze-fracture analyses of AQP4 proteins after stable transfection of CHO cells with M1, M23, or M1+M23 isoforms. M23 homotetramers produce large square lattices with 1 × 2 nm inter-IMP cross-bridges, whereas M1 homotetramers produce dispersed 4- to 6-nm IMPs with few incipient square arrays. When M1 and M23 are coexpressed to form hetero- and homotetramers, square arrays are of intermediate sizes, similar to square arrays in astrocyte end-feet. These studies demonstrate that regulation of the size of AQP4 arrays occurs by interaction of the M1 and M23 isoforms of AQP4 and provide a mechanism for regulating the organization of AQP4 arrays.
Materials and Methods
Plasmid Construction and Cell Lines. AQP4 expression constructs were created by using pCI-neo (Promega) and cDNA for rat AQP4 M1 or M23 (4), pIRESneo3 (Clontech) and cDNA for M23, and pcDNA3.1/Zeo (Invitrogen) and cDNA for M1. CHO-K1 cells (American Type Culture Collection) were cultured in 90% F12-K and 10% fetal bovine serum (Invitrogen). Cells were transfected with AQP4 expression vectors, or empty vector, by using Lipofectamine 2000 (Invitrogen) and the manufacturer's protocol; 10-cm2 dishes were cultured for 2 weeks in 1mg/ml Geneticin (Invitrogen). Cells expressing M1+M23 were obtained from stable M23 cells (pIRESneo3) transfected with pcDNA3.1/Zeo-M1 and cultured for 2 weeks in 1 mg/ml Geneticin and 500 μg/ml Zeocin (Invitrogen). Stable expression of M1 and M23 was confirmed by immunoblotting with anti-AQP4 (Chemicon) at 2 μg/ml.
Antibodies and Immunogold Labels. Affinity-purified antibodies to the C terminus of rat AQP4 (anti-CT) were from Søren Nielsen (University of Aarhus, Aarhus, Denmark) (21) or Chemicon; affinity-purified antibodies raised to the N terminus of rat AQP4 (anti-NT) were reported (4). Goat anti-rabbit IgG antibodies conjugated to 10-nm gold beads were from British Biocell International (Cardiff, U.K.) and Chemicon, and 12-nm gold beads (goat anti-rabbit IgG) were from Jackson ImmunoResearch.
Fluorescence Microscopy and Biochemical Studies. Transfected CHO cells grown on Lab-Tek Permanox chamber slides (Nalge Nunc) were fixed in PBS with 4% formaldehyde, permeabilized in 0.5% Triton X-100, blocked in 1% BSA, and incubated with AQP4 antibody (Chemicon) at 0.5 μg/ml in blocking solution, followed by Alexa-488-conjugated goat anti-rabbit IgG secondary antibody (Molecular Probes) diluted to 2 μg/ml in blocking solution. Confocal microscopy used an UltraView LCI (Perkin-Elmer) on a Zeiss Axiovert 200.
For immunoprecipitations, CHO cells expressing M23, M1, or M1+M23 were solubilized in solubilization buffer [200 mM NaCl/20 mM Tris·HCl, pH 8/1% Triton X-100/1% sodium deoxycholate/0.1% SDS plus protease inhibitors (Complete mixture tablets, Roche Diagnostics)] and incubated with 3 μg/ml anti-NT for 1 hr on ice. Immune complexes were concentrated by using protein A-Sepharose (Sigma), immunoblotted, and probed with anti-CT. For surface biotinylation assays, cells were rinsed in PBS, incubated in 0.5 mg/ml Sulfo-NHS-LC-Biotin (Pierce) on ice for 30 min, washed with 50 mM Tris·HCl, pH 8.0, for 5 min, rinsed twice with ice-cold PBS, and resuspended in solubilization buffer. After centrifugation for 5 min at 14,000 × g, 1 ml of supernatant was mixed with 200 μl of agaroseimmobilized streptavidin (Pierce) and incubated at 4°C overnight. The beads were washed three times in solubilization buffer and eluted with 50 μl of Laemmli loading buffer (3% SDS/10 mM Tris·HCl, pH 6.8/10% glycerol/0.2% bromophenol blue) for 30 min at 37°C. Biotinylated AQP4 was visualized by immunoblotting 12 μl of eluate and probing with anti-CT (2 μg/ml). Immunoblots were stripped and reprobed with anti-actin (1:10,000, Sigma).
Freeze Fracture and Electron Microscopy. Rat tissues were fixed by transcardiac perfusion with 2% glutaraldehyde in 140 mM Sorensen's phosphate buffer (SPB), pH 7.4, and 150-μm sections were prepared from the suprachiasmatic nucleus by using a refrigerated Lancer 1000 Vibratome. CHO cells for freeze fracture were pelleted and fixed in 2.5% glutaraldehyde. CHO cells for immunolabeling were immersion-fixed in 1% formaldehyde (22) in SPB for 10 min, rinsed, and stored up to 48 hr in SPB. Tissue sections and CHO cells were infiltrated with 30% glycerol, frozen (23), fractured, and replicated in a JEOL RFD-9010C freeze-fracture device with high-resolution shadowing methods revealing details <1 nm in nonaveraged images (24). Samples fractured at -170°C, precoated with 0.8- to 1-nm carbon were shadowed with 1-1.5 nm of platinum at a 60° angle (25). After freeze fracture, samples were bonded in Lexan plastic to gold index grids (22), thawed, photomapped, and cleaned in chromic/sulfuric acid. For freeze-fracture immunolabeling, samples were cleaned with 2.5% SDS before immunogold labeling (22, 26, 27).
A JEOL 2000 EX-II transmission electron microscope was used. Images at ×30,000 to ×150,000 magnification were digitized by ArtixScan 2500 digital scanning (Microtek International, Carson, CA), and processed with photoshop (Adobe Systems, San Jose, CA). Seven sets with 42 replicates of 29 blinded samples were photographed before decoding. Freeze-fracture images were examined from 30 untransfected control CHO cells (3 replicates), 40 vector-transfected cells (4 replicates), 150 M1-transfected cells (10 replicates), 30 M23-transfected cells (3 replicates), and 30 cells transfected with M1 and M23 (3 replicates).
Results
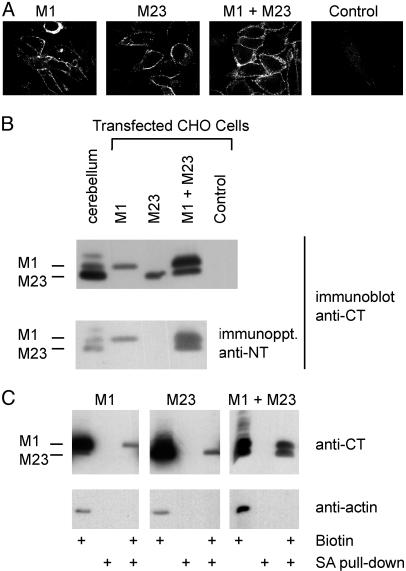
Expression of AQP4 M1 and M23 Isoforms. Stably transfected CHO cells were evaluated by confocal immunofluorescence microscopy (5). More than 95% of cells transfected with M1, M23, or M1+M23 exhibited plasma membrane immunofluorescence. Cells transfected with empty vector exhibited no detectable plasma membrane staining (Fig. 2A). Background staining with anti-NT prevented its use in immunofluorescence. Expression of AQP4 isoforms in transfected cells was compared with endogenous AQP4 expression in rat cerebellum by immunoblotting (Fig. 2B Upper). Anti-CT revealed a 34-kDa band in membranes from M1 cells, a 32-kDa band in M23 cells, and both bands in M1+M23 cells. Immunoprecipitation with anti-NT followed by immunoblotting with anti-CT confirmed that M1 and M23 are associated within AQP4 heterotetramers in M1+M23 cells (Fig. 2B Lower). Surface expression of M1 and M23 was confirmed by surface biotinylation and precipitation with streptavidin (Fig. 2C).
Fig. 2.
Expression of M1 and M23 isoforms of AQP4 in stably transfected CHO cells. (A) Confocal AQP4 immunofluorescence of cells transfected with M1, M23, M1+M23, or control (empty vector) incubated with anti-CT (reacts with M1 and M23). (×100.) (B Upper) Anti-CT immunoblot of membrane proteins prepared from rat cerebellum or transfected CHO cells (10 μg of protein per lane). (Lower) Solubilized cell lysate immunoprecipitated with anti-NT (specific for N terminus of M1) analyzed by anti-CT immunoblot. (C) Biochemical demonstration of M1 and M23 at the cell surface. Cells were treated with a membrane-impermeant biotinylating agent, precipitated by streptavidin (SA), and analyzed by anti-CT or anti-actin immunoblot (see Materials and Methods).
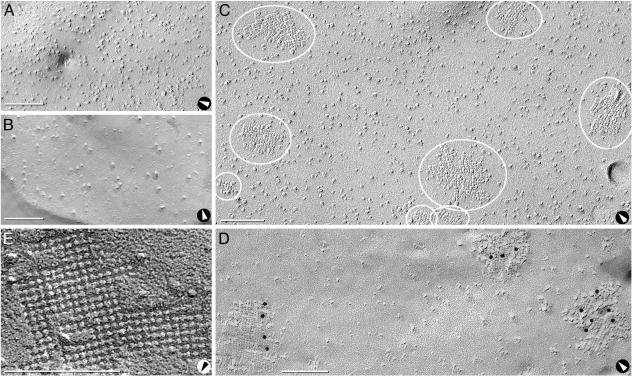
Freeze-Fracture Studies. Freeze-fracture analysis of untransfected control CHO cells revealed 7- to 9-nm IMPs at 2,000 per μm2 in P-faces (Fig. 3A) and 7- to 9-nm pits at >500 per μm2 in E-faces (Fig. 3B). Neither square arrays nor 4- to 6-nm IMPs or pits were observed.
Fig. 3.
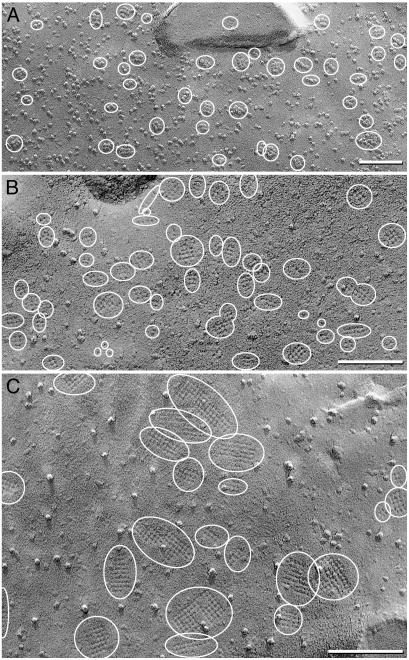
Freeze-fractured control CHO cells and cells expressing M23. (A and B) P-face (A) and E-face (B) images of cells transfected with empty vector reveal P-face IMPs and E-face pits of 7- to 9-nm diameter but no square arrays. No 4- to 6-nm-diameter pits were detected. E-faces have fewer IMPs than P-faces (A). (C) P-face image of M23 cell reveals three square arrays (small ovals) and five large rafts, all with 6-nm spacing (large ovals). (D) Anti-CT immunogold labeling of rafts in P-face of M23 cell. (E) High magnification of E-face image of raft in M23 cell. D was from formaldehyde-fixed cells (24), but the other images were from glutaraldehyde-fixed cells. Arrows denote direction of shadowing. (Bars = 100 nm.)
Freeze-fracture images of CHO cells stably transfected with M23 had large, raft-like square arrays with uniform lattice spacings of 6 nm. At 3-5 rafts per μm2, these arrays varied in size but generally contained >100 P-face IMPs (Fig. 3 C and D) or E-face pits (Fig. 3E). Many of these rafts appeared to be formed from side-to-side association of smaller square arrays. AQP4 protein was confirmed in M23 rafts by immunogold labeling with anti-CT multiple immunogold beads (Fig. 3D). The largest rafts were labeled by up to 20 immunogold beads (not shown). Immunogold beads were rarely observed over other areas of plasma membrane, suggesting that M23 polypeptides exist predominantly within square arrays.
Freeze-fracture analysis of M1 cells revealed 4- to 6-nm P-face IMPs or E-face pits dispersed primarily as singlets (see below) and were increased by 550 per μm2. In a third of M1 cells, up to 5% of IMPs or pits were found in incipient square arrays consisting of 2-12 IMPs or pits (Fig. 4). When present, square arrays in M1 cells were much smaller than rafts in M23 cells or square arrays in astrocyte end-feet. Immunogold labeling of P-faces in M1 was above background but could not be uniquely associated with any IMP or IMP cluster (not shown).
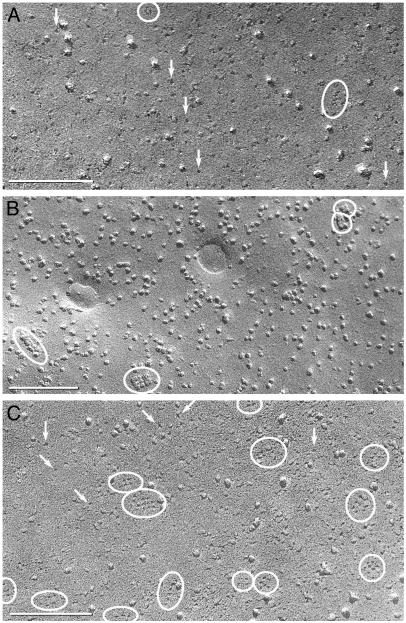
Fig. 4.
Freeze-fracture images of cells expressing M1. (A and B) E-face (A) and P-face (B) images reveal abundant 4- to 6-nm pits or IMPs and occasional incipient square arrays with ≤12 pits or IMPs in square lattices with 6-nm spacing (ovals). (C) E-face image of an occasional cell with a larger number of incipient arrays (ovals) and singlet 4- to 6-nm pits (arrows). (Bars = 100 nm.)
Four populations of M1+M23 cells were observed. Only ≈10% of cells had no detectable square arrays (not shown), whereas ≈10% of cells contained abundant 4- to 6-nm IMPs and rudimentary square arrays composed of 2-8 IMPs or pits (Fig. 5A). Most cells coexpressing M1 and M23 contained 20-50 square arrays per μm2 (Fig. 5B), with 4-24 IMPs or pits per array. Approximately 10% of cells had large rafts consisting of dozens of IMPs or pits (Fig. 5C).
Fig. 5.
Freeze-fracture images of cells expressing M1 and M23. (A and B) P-face (A) and E-face (B) images reveal multiple small square arrays of up to 20 IMPs or pits in 6-nm spacing (ovals). (C) E-face of occasional cell with larger square arrays of up to 100 pits (ovals). (Bars = 100 nm.)
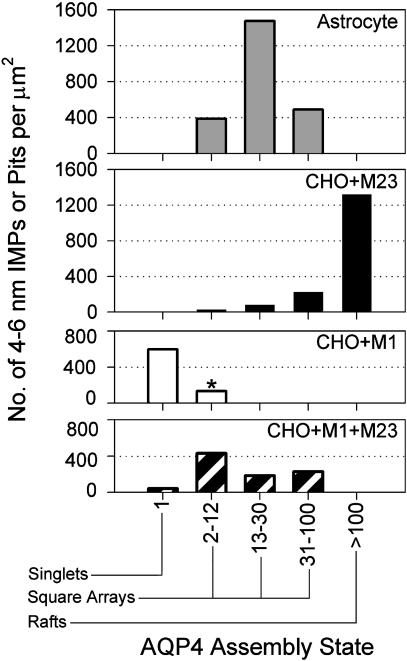
Morphometric Analyses of Square Arrays. Differences in square array formation were quantified from high-magnification freeze-fracture images. The average assembly state of 4- to 6-nm IMPs or pits was calculated by dividing total number of 4- to 6-nm IMPs or pits by the sum of the number of assemblies (singlets + square arrays + large rafts). For technical reasons, individual 4- to 6-nm IMPs or pits were difficult to count (28). The number of IMPs or pits in different assembly states was determined with bins corresponding to the following assembly states (Fig. 6): singlets (isolated IMP or pit); incipient arrays (2-12 IMPs or pits, M1 only); square arrays (2-12, 13-30, or 31-100 IMPs or pits); and rafts (>100 IMPs or pits). In astrocytes, the average array contained 17 IMPs or pits (ranging up to 53). Arrays in M23 cells were larger, containing an average of 78 IMPs or pits (up to 450), with 85% of 4- to 6-nm IMPs or pits in large rafts, 10% in square arrays and small rafts, and <5% as singlets. In contrast, 4- to 6-nm IMPs or pits in M1 cells were predominantly dispersed as singlets, with the few incipient arrays containing an average of 2 IMPs or pits (up to 12). M1+M23 cells contained assemblies in a variety of states corresponding to incipient arrays, square arrays, and rafts. M1+M23 assemblies contained an average of 6 IMPs or pits (up to 72), intermediate between the values for M1 cells and M23 cells. Large areas (>10 μm2) were examined, but evidence did not suggest that membrane domains had different distributions within a cell. The images are representative from >1,000 micrographs (all samples).
Fig. 6.
Histogram of IMP or pit assembly state in membranes of astrocyte end-feet and CHO cells expressing M23, M1, or M1+M23. Morphometric analyses of P-face and E-face images compiled the number of singlet 4- to 6-nm IMPs or pits, incipient arrays with 2-12 IMPs or pits (*, M1 only), square arrays with 2-12, 13-30, and 31-100 IMPs or pits, and rafts with >100 IMPs or pits. The survey compiled 4- to 6-nm IMPs or pits from astrocytes (1,832 IMPs, 0.52 μm2), M23 cells (1,552 IMPs, 0.94 μm2), M1 cells (252 IMPs, 0.46 μm2), and M1+M23 cells (1,319 IMPs, 1.49 μm2).
Cross-Bridges and Furrows. Freeze-fracture analyses of astrocyte end-feet revealed 1 × 2 nm cross-bridges connecting IMPs in P-face images or furrows between pits in E-face images (Fig. 1 B and C). Because pits and furrows are less vulnerable to plastic deformation, E-face images of astrocytes and transfected cells were studied (Fig. 7). Technical limitations precluded firm conclusions, but some differences were observed. Astrocyte end-feet contained furrows between ≈60% of E-face pits (Fig. 7A). In contrast, furrows connected >90% of E-face pits (>500 per μm2) in M23 rafts (Fig. 7B). M1 cells contained very few incipient arrays and their IMPs/pits were rarely linked by cross-bridges or furrows (<20 μm2) (Fig. 7 C-E). Most IMPs or pits in M1+M23 cells were in square arrays, and ≈50% were linked by furrows (Fig. 7F).
Fig. 7.
High-magnification E-face images and companion diagrams illustrating 2-nm furrows linking adjacent pits. (A) Astrocyte square arrays with missing furrows (arrow). (B) M23 raft has furrows connecting >90% of pits. (C-E) Incipient arrays in M1 cells have fewer furrows and irregular lattice spacings, ranging from 5 nm to 11 nm. (F) Square array in M1+M23 cell has furrows linking ≈50% of pits. All shadows are from right to left. Companion diagrams denote positions of pits and furrows. Arrows indicate missing furrows. B-E are shadowed at similar local declination angles and at ≈45° azimuth to the orthogonal lattices; each was selected for identical platinum granularity and image resolution. The effects of shadow angle and azimuth on ability to resolve cross-bridges have been described (23). (≈×450,000.)
Discussion
Astrocyte membrane end-feet have long been known to contain strikingly regular square arrays when examined by freeze fracture (14, 15). The arrays were reported to disassemble under various metabolic and osmotic conditions (14, 29, 30) and may vary with tissue preservation conditions and even between adjacent astrocytes (24, 31). Because mechanisms regulating AQP4 array assembly are unknown, we explored the possibility that AQP4 isoforms M1 and M23 may be determinants.
Freeze fracture revealed major differences between CHO cells stably transfected with M1 and M23 isoforms of AQP4. M23 cells contained large rafts, many with 10 times more 4- to 6-nm IMPs or pits as square arrays in normal astrocyte end-feet (Figs. 1 and 3). The small number of large rafts suggests that M23 self-assembles into a highly stable configuration. In contrast, cells expressing M1 contained dispersed 4- to 6-nm IMPs or pits and a few incipient arrays in rudimentary square lattices (Fig. 4). Although additional molecules may participate in AQP4 array assembly, the remarkable differences in assembly appear to be inherent properties of M23 and M1 polypeptides, because similar morphologies were seen after both formaldehyde and glutaraldehyde fixation (Fig. 3 D vs. C). On the basis of lack of 4- to 6-nm IMPs or pits in untransfected CHO cells (Fig. 3 A and B), their appearance in arrays with 6-nm lattice spacing in M23 rafts (Fig. 3 C-E), and presence as dispersed singlets in M1 cells (Fig. 4), we conclude that the 4- to 6-nm IMPs and pits represent individual AQP4 tetramers. This conclusion is supported by atomic structures of homologous proteins AQP1 (20, 32, 33) and GlpF (34), with tetramer cross-sectional diameters of 6 nm. The slightly smaller diameter of the replicated IMPs and pits may reflect the diameter of each tetramer within the lipid bilayer.
Our study indicates that M1 tetramers directly interfere with assembly of M23 tetramers into large arrays. Cross-bridges or furrows linked nearly all 4- to 6-nm IMPs or pits in M23 cells, but were rare in M1 cells (Fig. 7), suggesting that M1 and M23 differ at the site of square array assembly. In contrast, cells cotransfected with M1 and M23 had square arrays with a range of sizes, often resembling arrays in astrocyte end-feet (Figs. 1 and 5). Individual AQP4 tetramers in rat brain (4) and in CHO cells transfected with M1 and M23 polypeptides were shown by immunoprecipitation to contain both M1 and M23 subunits. Nevertheless, stoichiometry in brain and skeletal muscle showed that M23 polypeptides outnumber M1 polypeptides by at least 3:1 (4, 5). M23 and M1 polypeptides are identical except the 22-residue peptide at the N terminus of M1. It is unknown how this additional segment interferes with square array assembly, because the atomic structures of homologous proteins did not resolve N and C termini (20, 32-34).
The relevance of AQP4 square array assembly to the physiology and pathophysiology of brain is unknown. Our studies suggest that M1 and M23 may restrict AQP4 square array assembly to a preferred size, but it is not clear why this is important. AQP4 has a highly polarized distribution in brain. It is greatly enriched in the perivascular membrane of astroglial cells and at the glia limitans at brain surfaces (7, 8) and in retina (9). AQP4 may release water from brain metabolism, and may restore osmotic equilibrium during potassium siphoning after high neuronal activity (11, 12). Recent studies indicate that AQP4 is tethered at perivascular membranes by association with dystrophin-associated proteins through an interaction with α-syntrophin (5, 12, 13, 35). A single linking protein could tether a square array containing many AQP4 proteins to the cytoskeleton, whereas dispersed AQP4 tetramers might each require a tether. Dissociation of square arrays may occur during cerebral ischemia or other neural stress (14, 36). If dissociation of square arrays needs to occur rapidly, organization of AQP4 into large rafts may interfere with disassembly. Balancing the opposing actions of M1 and M23 isoforms of AQP4 in astrocyte end-feet may provide the optimal size of arrays for efficient tethering at the perivascular membrane without impairing the ability to rapidly disassemble.
Acknowledgments
We thank Arvid Maunsbach and Kaoru Mitsuoka for critical evaluations of the manuscript and Joerg Kistler for insightful comments. This work was supported by National Institutes of Health Grants NS-38121, NS-44010, and NS-44395 (to J.E.R.) and HL-33991, HL-48268, and EY-11239 (to P.A.) and by the Human Frontier Science Program (to P.A.).
Abbreviations: AQP1, aquaporin-1; AQP4, aquaporin-4; CHO, Chinese hamster ovary; IMP, intramembrane particle; P-face, protoplasmic leaflet; E-face, extraplasmic leaflet.
References
- 1.Agre, P., King, L. S., Yasui, M., Guggino, W. B., Ottersen, O. P., Fujiyoshi, Y., Engel, A. & Nielsen, S. (2002) J. Physiol. 542, 3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu, M., Lee, M. D., Smith, B. L., Jung, J. S., Agre, P., Verdijk, M. A., Merkx, G., Rijss, J. P. & Deen, P. M. (1996) Proc. Natl. Acad. Sci. USA 93, 10908-10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jung, J. S., Bhat, R. V., Preston, G. M., Guggino, W. B., Baraban, J. M. & Agre, P. (1994) Proc. Natl. Acad. Sci. USA 91, 13052-13056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neely, J. D., Christensen, B. M., Nielsen, S. & Agre, P. (1999) Biochemistry 38, 11156-11163. [DOI] [PubMed] [Google Scholar]
- 5.Neely, J. D., Amiry-Moghaddam, M., Ottersen, O. P., Froehner, S. C., Agre, P. & Adams, M. E. (2001) Proc. Natl. Acad. Sci. USA 98, 14108-14113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nielsen, S., Smith, B. L., Christensen, E. I. & Agre, P. (1993) Proc. Natl. Acad. Sci. USA 90, 7275-7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nielsen, S., Nagelhus, E. A., Amiry-Moghaddam, M., Bourque, C., Agre, P. & Ottersen, O. P. (1997) J. Neurosci. 17, 171-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frigeri, A., Gropper, M. A., Umenishi, F., Kawashima, M., Brown, D. & Verkman, A. S. (1995) J. Cell Sci. 108, 2993-3002. [DOI] [PubMed] [Google Scholar]
- 9.Nagelhus, E. A., Veruki, M. L., Torp, R., Haug, F. M., Laake, J. H., Nielsen, S., Agre, P. & Ottersen, O. P. (1998) J. Neurosci. 18, 2506-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manley, G. T., Fujimura, M., Ma, T., Noshita, N., Filiz, F., Bollen, A. W., Chan, P. & Verkman, A. S. (2000) Nat. Med. 6, 159-163. [DOI] [PubMed] [Google Scholar]
- 11.Amiry-Moghaddam, M., Williamson, A., Palomba, M., Eid, T., de Lanerolle, N. C., Nagelhus, E. A., Adams, M. E., Froehner, S. C., Agre, P. & Ottersen, O. P. (2003) Proc. Natl. Acad. Sci. USA 100, 13615-13620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amiry-Moghaddam, M., Otsuka, T., Hurn, P. D., Traystman, R. J., Haug, F.-M., Froehner, S. C., Adams, M. E., Neely, J. D., Agre, P., Ottersen, O. P. & Bhardwaj, A. (2003) Proc. Natl. Acad. Sci. USA 100, 2106-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vajda, Z., Pedersen, M., Fuchtbauer, E. M., Wertz, K., Stodkilde-Jorgensen, H., Sulyok, E., Doczi, T., Neely, J. D., Agre, P., Frokiaer, J. & Nielsen, S. (2002) Proc. Natl. Acad. Sci. USA 99, 13131-13136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolburg, H. (1995) J. Hirnforsch. 36, 239-258. [PubMed] [Google Scholar]
- 15.Landis, D. M. & Reese, T. S. (1974) J. Cell Biol. 60, 316-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Hoek, A. N., Yang, B., Kirmiz, S. & Brown, D. (1998) J. Membr. Biol. 165, 243-254. [DOI] [PubMed] [Google Scholar]
- 17.Yang, B., Brown, D. & Verkman, A. S. (1996) J. Biol. Chem. 271, 4577-4580. [PubMed] [Google Scholar]
- 18.Verbavatz, J. M., Ma, T., Gobin, R. & Verkman, A. S. (1997) J. Cell Sci. 110, 2855-2860. [DOI] [PubMed] [Google Scholar]
- 19.Rash, J. E., Yasumura, T., Hudson, C. S., Agre, P. & Nielsen, S. (1998) Proc. Natl. Acad. Sci. USA 95, 11981-11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sui, H., Han, B. G., Lee, J. K., Walian, P. & Jap, B. K. (2001) Nature 414, 872-878. [DOI] [PubMed] [Google Scholar]
- 21.Terris, J., Ecelbarger, C. A., Marples, D., Knepper, M. A. & Nielsen, S. (1995) Am. J. Physiol. 269, F775-F785. [DOI] [PubMed] [Google Scholar]
- 22.Rash, J. E. & Yasumura, T. (1999) Cell Tissue Res. 296, 307-321. [DOI] [PubMed] [Google Scholar]
- 23.Phillips, T. E. & Boyne, A. F. (1984) J. Electron Microsc. Tech. 1, 9-29. [Google Scholar]
- 24.Rash, J. E., Duffy, H. S., Dudek, F. E., Bilhartz, B. L., Whalen, L. R. & Yasumura, T. (1997) J. Comp Neurol. 388, 265-292. [DOI] [PubMed] [Google Scholar]
- 25.Rash, J. E. & Yasumura, T. (1992) Microsc. Res. Tech. 20, 187-204. [DOI] [PubMed] [Google Scholar]
- 26.Fujimoto, K. (1995) J. Cell Sci. 108, 3443-3449. [DOI] [PubMed] [Google Scholar]
- 27.Fujimoto, K. (1997) Histochem. Cell Biol. 107, 87-96. [DOI] [PubMed] [Google Scholar]
- 28.Rash, J. E. & Giddings, F. D. (1989) J. Electron Microsc. Tech. 13, 204-215. [DOI] [PubMed] [Google Scholar]
- 29.Landis, D. M. & Reese, T. S. (1981) J. Cell Biol. 88, 660-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neuhaus, J., Schmid, E. M. & Wolburg, H. (1990) Neurosci. Lett. 109, 163-168. [DOI] [PubMed] [Google Scholar]
- 31.Landis, D. M. & Reese, T. S. (1981) J. Exp. Biol. 95, 35-48. [DOI] [PubMed] [Google Scholar]
- 32.de Groot, B. L., Engel, A. & Grubmuller, H. (2001) FEBS Lett. 504, 206-211. [DOI] [PubMed] [Google Scholar]
- 33.Murata, K., Mitsuoka, K., Hirai, T., Walz, T., Agre, P., Heymann, J. B., Engel, A. & Fujiyoshi, Y. (2000) Nature 407, 599-605. [DOI] [PubMed] [Google Scholar]
- 34.Fu, D., Libson, A., Miercke, L. J., Weitzman, C., Nollert, P., Krucinski, J. & Stroud, R. M. (2000) Science 290, 481-486. [DOI] [PubMed] [Google Scholar]
- 35.Nico, B., Frigeri, A., Nicchia, G. P., Corsi, P., Ribatti, D., Quondamatteo, F., Herken, R., Girolamo, F., Marzullo, A., Svelto, M., et al. (2003) Glia 42, 235-251. [DOI] [PubMed] [Google Scholar]
- 36.Cuevas, P., Gutierrez Diaz, J. A., Dujovny, M., Diaz, F. G. & Ausman, J. I. (1985) Anat. Embryol. (Berlin) 172, 171-175. [DOI] [PubMed] [Google Scholar]