Abstract
Mathematical modelling and computational analysis play an essential role in improving our capability to elucidate the functions and characteristics of complex biological systems such as metabolic, regulatory and cell signalling pathways. The modelling and concomitant simulation render it possible to predict the cellular behaviour of systems under various genetically and/or environmentally perturbed conditions. This motivates systems biologists/bioengineers/bioinformaticians to develop new tools and applications, allowing non-experts to easily conduct such modelling and analysis. However, among a multitude of systems biology tools developed to date, only a handful of projects have adopted a web-based approach to kinetic modelling. In this report, we evaluate the capabilities and characteristics of current web-based tools in systems biology and identify desirable features, limitations and bottlenecks for further improvements in terms of usability and functionality. A short discussion on software architecture issues involved in web-based applications and the approaches taken by existing tools is included for those interested in developing their own simulation applications.
Keywords: systems biology, web-based applications, kinetic modelling, dynamic simulation
INTRODUCTION
The study of metabolic, signalling and regulatory networks is being greatly aided by the development of computational models and the availability of abundant amounts of high-throughput experimental data [1, 2]. One of the key goals of systems biology is to build precise and reliable mathematical models that can be used to predict cellular responses to a range of stimuli, thus allowing biologists to gain a better understanding of how complex biological systems function [3]. Dynamic models, in which the behaviour of a system and its components is studied over time, have been used routinely in a variety of physics and engineering fields as well as in social science areas such as economics and sociology. Applications have been developed for simulating interesting phenomena observed in a wide range of subjects, such as robotics, meteorological phenomena, fluid dynamics, aerospace and vehicular simulations, population dynamics, etc. Several large-scale simulation projects have been undertaken in recent years, including efforts to simulate a mammalian brain [4], the formation and evolution of galaxies [5] and large climate prediction simulations [6]. In addition to temporal considerations, kinetic properties such as reaction rates and mechanisms can also be incorporated into models. Recently, these and several other techniques are being applied to simulating models of various biochemical networks [7]. This approach begins with a hypothetical model describing cellular behaviour and physiology derived from prior biological knowledge and experimental observations. A predictive model is then created, refined and validated in an iterative way until a suitable agreement is reached between the model's predictions and the experimental observations [8]. Indeed, in silico modelling and the associated simulation of biological systems incorporate a synergistic combination of theoretical, computational and experimental efforts. This motivates systems biologists, bioengineers and bioinformaticians to devise suitable tools and environments that facilitate the interactions among researchers with diverse backgrounds; including those who might be unfamiliar with the underlying computational methods or the biological system.
As a result of these efforts, a multitude of kinetic modelling applications have been developed and can be found as listed on the systems biology community (http://sbml.org). Each of these tools has its own capabilities and features, differing in terms of user interface (UI), mathematical framework, model portability and database functionality [9–11]. The modelling process can be composed of mainly three steps: model construction, simulation and systems analysis. Although most kinetic modelling tools focus on one of these steps, it is desirable for non-expert users to get access to the entire model building process in one ‘all-in-one’ package. Based on the location of computational resources, such tools available at this time can be classified into two types: stand-alone and web-based. Stand-alone applications are installed and run locally on users’ computers, whereas web-based ones perform their computations on central servers with the results delivered to the users through web browsers. It should be noted that some advanced web technologies allow portions of the computation to take place on the user's machine, thus reducing the burden on the central server. We still consider these approaches as web-based, since a big part of the system still operates via a web interface. To date, several stand-alone modelling environments such as E-Cell [12], the Systems Biology Workbench [13], COPASI [14], etc. have been developed. Alternatively, a web browser presents the advantage of being readily available to all users without having to install any additional software packages. This makes web-based applications accessible online regardless of the users’ platform, providing advantages over stand-alone applications in terms of accessibility, software updates and computational capabilities. However, web-based applications have the disadvantage of being slower than stand-alone applications due to the shared nature of computational resources and slow response time over the Internet. Perhaps because of these limitations, only a few such applications are now available and thus the area of web-based simulation tools is still relatively sparse compared to other types of simulation tools available. This entails the need for evaluating the capabilities and characteristics of current web-based tools in systems biology and for identifying desirable features and bottlenecks for further improvements.
Despite the shortage of web-based modelling tools, there is still some diversity in the types of features they provide. Online model repositories, such as BioModels [15], DOQCS (Database of Quantitative Cellular Signalling) [16] and JWS (Java Web Simulator) Online [17] provide users with centralized resources where they can store, annotate and retrieve models. Among these repositories, JWS provides the added functionality of allowing users to simulate the models. Virtual Cell [18] is one of the most well-known kinetic modelling and simulation environments, allowing users to build both compartmental and spatial models of biological systems. As its unique feature, Virtual Cell is capable of describing the complex geometry of a cellular system in two or three dimensions. Another web-based platform is PyBioS [19], supporting the modelling features for simulating cellular processes. WebCell [20] is an integrated online environment for managing qualitative and quantitative information on cellular networks as well as interactively exploring their steady-state and dynamic behaviours. Most recently developed is SYCAMORE [21] that focuses on helping non-expert users to set up biochemical models and run simulations and analyses on them. Instead of implementing all aspects of a modelling package on its own, SYCAMORE integrates several online applications along with its own locally installed tools. An interesting project focusing on the quantitative study of the biology of ageing is the Biology of Ageing e-Science Integration and Simulation System (BASIS) [22]. This system provides web-based services that make it possible for users from different research groups to collaborate and integrate relevant data and hypotheses to the biology of ageing. Another package is the CADLIVE [23, 24] system, where users can convert biochemical network maps into dynamic models and then simulate them online. However, the Java-based graphical user interface (GUI) for constructing large-scale biochemical networks is distributed as a stand-alone application and must be run on the users’ local machine.
In this report, we focus on applications that enable users to build, manage and analyse kinetic models of biological systems through a web browser-based interface. Six representative applications in systems biology are reviewed, viz.: JWS Online, Virtual Cell, PyBioS, WebCell, SYCAMORE and BASIS. We compare the capabilities and features of those web-based applications based on various usability and functionality criteria. Finally, for research groups that are interested in developing their own web-based simulation tools, we include a short discussion on the various issues concerning the system architecture in building such a system. We also discuss some alternative technologies and strategies that can be used to improve the performance of existing tools.
EVALUATING CAPABILITIES AND FEATURES OF WEB-BASED APPLICATIONS
A detailed comparison of the selected web-based applications is presented in the following subsections. Their features and characteristics are compared with respect to usability and functionality.
Usability
Efficiency, user satisfaction and a low learning curve are the primary notions of usability [25]. The usability of an application depends primarily on its UI. A natural and intuitive UI allows non-expert users to easily adapt to the application without having to spend much time consulting user manuals or help files. How much effort users have to make in order to learn a software package and how easily they can build models and get solutions to problems they are interested in solving are other important aspects of the usability of a system. Especially, since users might not have mathematical or computational backgrounds, applications must not be excessively complex. Finally, relevant user guides and documentation must be readily available so that users can easily get the information they need in order to build and simulate their models.
Three major types of interfaces are used for defining, handling and managing models in web-based applications: scripts, wizards/forms and diagrammatic interfaces [26]. Script-based interfaces ask users to define models strictly in text form, allowing them to directly edit the models. Some applications provide wizards or a series of forms and dialogue boxes to define models, guiding users via a series of well-defined and highly structured tasks. Diagrammatic interfaces allow models to be defined by drawing diagrams or networks and subsequently specifying the required parameters through pop-up text entry boxes [9]. Out of all the applications surveyed, Virtual Cell was the only one that combined all three types of interfaces. JWS, PyBioS, SYCAMORE and BASIS use a form-based interface, while WebCell provides both wizard- and script-based interfaces for model definition. Modelling experts may prefer the script-based interface rather than wizards; information, such as kinetic equations, rate expressions and other parameters can be entered within a single text form and easily edited, thus reducing model building efforts. However, this index-free and context-heavy structure raises the probability of typographical errors appearing in the model. Ideally, the combined graphical and text UI of Virtual Cell decreases the occurrence of such problems.
During the model building process, users can either follow predefined rate expressions or define their own rate expressions under Virtual Cell, PyBioS, WebCell and BASIS environments. JWS, however, limits its users by only allowing them to simulate models predefined in the library. Interestingly, SYCAMORE adds a useful feature where it allows users to search the SABIO-RK [27] database for relevant kinetic and reaction data to add to their models instead of having to provide the data themselves.
Visualization of models through a graphical interface renders it possible for users to better understand the overall structure of the model networks along with the relationships among the various components or molecules. This essential feature is basically supported by all of the applications surveyed. However, one desirable feature of this functionality is an interactive graphical interface supporting dynamic visualization using automatic layout algorithms. In this sense, Virtual Cell is the only application to satisfy such functionality, while PyBioS, WebCell, SYCAMORE and BASIS only provide non-interactive displays of model networks. JWS has static and pre-drawn network diagrams in its model library. Uniquely, Virtual Cell and BASIS allow users to perform multi-compartmental graphical visualization by defining different compartments of a given model in their forms-based UI.
Consistency of display is another important aspect of usability, which ensures uninterrupted operation of a web-based application. For example, the result display may be confusing to users if results of different analyses are shown in pop-up windows. Another important aspect of consistency is the internal page navigation through which users can browse and find the resources or services they are looking for. PyBioS, WebCell, SYCAMORE and BASIS provide an adequate internal navigation schema, which allows users to easily understand the application structure.
The final issue of importance from a usability standpoint is the availability of tutorials and user manuals. All of the applications surveyed provide at least basic tutorials or help pages that explain the purpose and functionality of the tools. However, the information provided by JWS, PyBioS and BASIS was quite sparse without including any screenshot of the systems in use. On the other hand, Virtual Cell, WebCell and SYCAMORE provide case studies and tutorials with screenshots showing the users how to build and simulate the models in a step-by-step format. Table 1 highlights several key usability features of the applications.
Table 1:
Usability features of web-based applications
| Web-based applications | JWS Onlinea | Virtual Cell | PyBios | WebCell | SYCAMORE | BASIS | |
|---|---|---|---|---|---|---|---|
| User interface for model building | Wizard/ form-based | – | ✓ | ✓ | ✓ | ✓ | ✓ |
| Script-based | – | ✓ | ✓ | ||||
| Diagrammatic | – | ✓ | |||||
| Kinetic rate expression for model building | Predefined | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| User-defined | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| Multi-compartmental graphical representation | ✓ | ✓ | |||||
| Parameters and initial conditions changeable | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Internal page navigation | ✓ | ✓ | ✓ | ✓ | |||
| Simulation results | Graph | Graph/Text | Graph | Graph/Text | Graph/Text | Graph/Text | |
| User guides and documentation | Text | Text/ Screenshots | Text | Text/ Screenshots | Text/ Screenshots | Text | |
| URL (version) | http://jjj.biochem.sun.ac.za | http://www.vcell.org (v4.3.1) | http://pybios.molgen.mpg.de (v1.0) | http://webcell.org (v2.0) | http://sycamore.eml.org | http://www.basis.ncl.ac.uk |
aJWS Online provides a library of predefined models and as such does not allow users to build their own models.
Functionality
Functionality is a multi-faceted measure that defines the features and capabilities of modelling applications. This includes support for different types of analysis, such as steady-state, sensitivity and pathway analyses of reaction networks as well as time-course analysis of kinetic models. It also considers supporting features that applications should provide, e.g. model validation, and different types of mathematical frameworks and solvers. External features such as links to public databases and data/model exchange formats are also included as major attributes in the functionality provided by web-based applications.
There are several functions that need to be performed as part of modelling a dynamic system and its subsequent analysis. One of the first steps in model building is to validate the model by checking the kinetic parameters for completeness and correctness. WebCell and SYCAMORE provide support for model validation. Since experimental data are often lacking, some parameter values may be missing from a model. Virtual Cell and SYCAMORE can use parameter estimation [28] methods to get estimated values for the missing parameters. In SYCAMORE, these are obtained through an interface to the qPIPSA [29] software which relates protein structure information to kinetic parameters.
After the model has been built, one area of interest is to explore the systems responses to changes in parameters. Parameter scanning is one method that determines the effects of changing the values of a single parameter on the levels of different variables, such as metabolic fluxes or metabolite concentrations. Virtual Cell and PyBioS are able to assess the robustness and performance of the system through parameter scanning. Sensitivity analysis is a similar technique that identifies which parameters a given metabolic or cellular phenomenon is most sensitive to. SYCAMORE supports sensitivity analysis and displays a colour-coded table, where most sensitive parameters are highlighted. JWS, PybioS and WebCell provide metabolic control analysis (MCA), in which the sensitivity of network components such as fluxes or concentrations in response to perturbations in enzyme activity or other processes is analysed by computing control, elasticity and response coefficients [30]. The structural and functional properties of biological reaction networks can be investigated by carrying out topological pathway analysis, e.g. identifying independent pathways, futile cycles and elementary modes [31]. WebCell provides explicit functionality for this analysis.
The mathematical framework of an application determines the structure of the kinetic formulation for a given network model. Models can either be deterministic, where there is no randomness in the system and the same inputs will always lead to the same output; or stochastic where randomness is incorporated into the model. All of the applications except BASIS simulate biological systems deterministically. Thus, models are described in the form of ordinary differential equations (ODE), which are then solved to explore time-variant dynamic behaviour of the cellular system. In addition to ODE, WebCell is able to handle differential algebraic equations (DAE) in which constraints, such as mass conservation constraints are placed in addition to ODE. Since Virtual Cell deals with time- and space-variant properties of system components, its models are expressed using both ODE and partial differential equations (PDE) to represent systems with different spatial components. In order to solve the system of equations for a specific model, Virtual Cell and WebCell provide various solvers for stiff and non-stiff systems, based on both variable and fixed step sizes. These include Fast Newton, Modified Newton, Runge–Kutta, Euler–Newton, LSODI, etc. SYCAMORE uses the COPASI software solvers to simulate its models. Unlike all the other applications, BASIS supports discrete stochastic simulation based on Gillespie's direct method [32]. Stochastic simulations may be more realistic in terms of biological accuracy, but the popular algorithm for the stochastic simulation has some limitations and in general, there are many more analytical methods available for deterministic simulation that are not available for stochastic models [10].
Another important facility that web-based applications should provide is links to public databases. Researchers from different fields can look up relevant data on various compounds, enzymes, proteins and even import entire biological models if they are available publicly. Currently, Virtual Cell provides connectivity to the KEGG and other databases while SYCAMORE allows users to search the BRENDA [33] and SABIO-RK [27] databases for enzymatic kinetic data. Finally, cross-compatibility is a very important issue for web-based applications in terms of exchanging data and models among different applications. The systems biology markup language (SBML) is one widely accepted format for representing models of biochemical reaction networks [34]. All of the applications surveyed can support SBML import and/or export, while other formats such as MATLAB and CellML are also exchangeable in some applications. Table 2 summarizes the key functional features of the web-based applications.
Table 2:
Functionalities provided by different web applications
| Web-based applications | JWS online | Virtual cell | PyBios | WebCell | SYCAMORE | BASIS |
|---|---|---|---|---|---|---|
| Network visualization | Static | Dynamic/Interactive | Dynamic | Dynamic | Dynamic | Dynamic |
| Reaction network validation | ✓ | ✓ | ||||
| Moiety conservation analysis | ✓ | ✓ | ||||
| Time-course analysis | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Metabolic control analysis (MCA) | ✓ | ✓ | ✓ | |||
| Pathway analysis | ✓ | |||||
| Parameter scanning | ✓ | ✓ | ||||
| Parameter estimation | ✓ | ✓ | ||||
| Mathematic framework | ODE | ODE/PDE | ODE | ODE/DAE | ODE | SDE |
| Link to public databases | KEGG/Swissprot | BRENDA/SABIO-RK | ||||
| Data exchange formats | ||||||
| Import | SBML | SBML | SBML | SBML | SBML | |
| Export | SBML | SBML/CellML/MATLAB | SBML | SBML/MATLAB | SBML |
SDE, stochastic differential equations.
DISCUSSION
System architecture
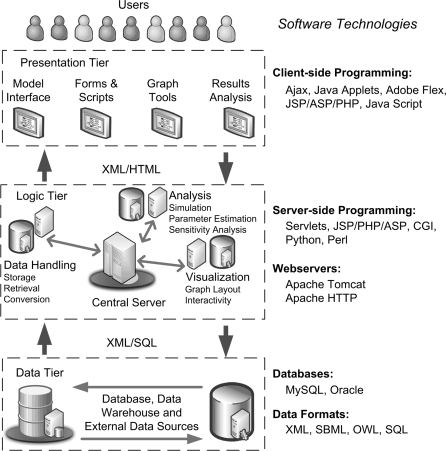
So far, we have compared web-based applications from the users’ point of view on the basis of usability and functionality. For developers, critical issues include what combination of software and hardware is considered, how the various system components are organized and how the features are implemented internally. Traditionally, web-based applications have adopted a three-tier architecture where three independent layers or tiers, i.e. presentation, logic and data tiers, are configured. Figure 1 depicts the structural overview of three-tier architecture, along with examples of available programming technologies to use in the different tiers. The UI and any component of how the user interacts with the application are handled by the presentation tier, while the data tier manages the internal and/or external storage of application-related data and provides access to it. The logic tier acts as a connector between the other two layers, handling their communication and performing any logical processing/analysis of data using various computational resources. The three-tier structure is dependent on connectivity existing between all tiers to exchange models, information and analysis results. All of the web-based simulation tools follow this three-tier architecture.
Figure 1:
Three-tier architecture used in most web applications. Several software technologies can be used to implement the configured layers including data, logic and presentation.
All web-based applications are hosted on a central server, with software handling requests from users and serving content back to them. Popular open source web servers such as the Apache (JWS, Basis) and Tomcat (WebCell, SYCAMORE) servers can be used to host applications. Based on user requests, scripts are executed on the web servers to generate HTML pages for the user's web browser. A wide variety of server/client-side languages and tools are available for web publishing and service, and often applications employ a combination of them. WebCell generates its web pages by combining JSP scripts with Jython and Java Servlets. JSP pages are also used by SYCAMORE. JWS provides Java Applets to run their simulations on the client's machines. PyBioS uses the Python language and is implemented on the Zope web application server environment. Virtual Cell uses a Java Enterprise solution for its architecture with separate dedicated messaging, remote method invocation, simulation and database API servers. BASIS follows a different approach compared to the other applications and is based on a service-oriented architecture (SOA). Various web services are written in Python or Java with information between machines being passed by sending XML data over the simple object access protocol (SOAP). The final aspect of system architecture, storage and management of model data, is handled by database management software, which is accompanied by a system to communicate with the applications. Popular open source database management systems such as MySQL and PostgreSQL are used by some of the applications surveyed, whereas Virtual Cell uses the popular commercial database software Oracle.
Dedicated hardware should be provided since simulating cellular models is a computationally expensive process, compared with other data processing [35]. Hybrid (both grid and cluster) structure of distributed computing ensures WebCell and BASIS a reliable and faster operation on the server side. Web servers also have to provide data storage facilities; both for internal data and also for any data generated by users. User generated data can be classified as being either private or public. One unique feature of BASIS is that any model declared as ‘public’ can be stored along with the simulation and analysis results. Therefore, any user can access not only that model but also its results.
Pros and cons of web-based approaches
Web-based applications have several advantages over stand-alone tools. They operate through a web browser and are, therefore, easily accessible on different platforms. Since they are accessed through a browser, the user does not have to install a copy of the software themselves and can access their data on different machines. There is also no need to install any subsequent upgrades or bug fixes that may become available, though users must realize that some software upgrades may make their previous simulations incompatible with future runs. As an attractive feature, web-based applications can allow researchers from different locations to work together towards a common goal of developing models using better computational resources than would normally be available to independent researchers. As a result, web-based applications usually have a larger model repository developed through this collaborative approach, providing more processor-intensive functionality at better speeds through more powerful and robust servers than ordinary desktop computers.
However, web-based applications do suffer from a significant disadvantage in terms of speed of response. Since they have to usually deal with several simultaneous requests and transfer large amounts of data between machines in server and client sides, response time is significantly slower than stand-alone tools. One possible solution to tackle this problem is to employ Ajax (Asynchronous JavaScript and XML), which is a combination of several existing web technologies [36]. Ajax uses requests that ask for small amounts of data from the server asynchronously and the HTML of the web page is then updated to reflect any changes instead of reloading the whole page [37]. In this manner, Ajax applications can provide a much richer interface along with an uninterrupted user experience. It should be noted that Ajax cannot improve the speed of simulations; this can only be achieved through more optimized algorithms or hardware improvements.
To date, no commercial web-based application is available. Despite their high cost and restrictive license and installation issues, commercial simulation tools do provide some advantages over freely available software. These advantages mainly come from advanced features and better usability, but the major aspect lies in dedicated user support via thorough testing and upgrades that at the same time guarantee backwards compatibility with previous versions of the software. As mentioned above, versioning support is a very important feature to ensure that previous simulations remain compatible with future experiments. Maintaining data privacy is another issue since storing data in a public repository may not be feasible in some research projects. Thus, much remains to be done to render web-based application commercially popular in both industry and academia.
Future outlook for web-based modelling tools
Although web-based applications can provide many capabilities for analysing biological systems that are available in stand-alone tools, there are still several features that could be implemented to better meet the requirements of researchers. In order to further enhance the usability, application programming interfaces (APIs) can be created that allow users to develop their own modules which can be added to the application for extended functionality. In addition to existing analysis techniques in web-based applications, desirable functional features include bistability analysis [38], simulation of hybrid systems that contain continuous as well as discrete events [39], multi-scale modelling [40] and flux analysis [41], which are currently not available in most web-based applications. It would be of great help for researchers to facilitate modelling and analysis of their systems under investigation if those features are supported in the future.
From the view point of system architecture, following a SOA similar to the one used in BASIS will be very beneficial. In SOA, the functionality of the application is divided into several small services which can be combined and reused, thereby rendering such services interoperable in the system [42]. Another issue pertaining to interoperability is how kinetic models can be effectively combined with high-throughput experimental data within the context of systems biology [43]. For example, kinetic modelling can be combined with metabolome data that is one of the front-end types of data describing phenotypic cellular behaviour [44]. However, models and data are represented in different formats, which need to be integrated under a common framework. This can be achieved by using semantic web technology that allows for effective exchange of knowledge through explicit description of structured data [45]. In this framework, the metabolome data and constructed kinetic models are standardized into XML-based data formats. Subsequently, their relationship can be clearly described by developing resource description framework (RDF) and using ontology web language (OWL) from a semantic perspective [46]. Thereby, such a semantic association of XML-based knowledge renders both metabolomic data and kinetic models interoperable. One final consideration for further improvement is to provide more links to public databases, thus making the modelling process easier and allowing users to combine information from different resources.
CONCLUSIONS
The continuing combination of high-throughout experimental data with better modelling tools will lead to an improved understanding of biological functions and predicting the effects internal or external perturbations may have on a system. Web-based tools are an attractive solution to modelling biological systems, especially to those new to the area of systems biology. In this report, we reviewed six applications that allow users to build and analyse computational models. Although all of the tools surveyed provide similar and basic functionality, there are several differences among them that may appeal to different types of users. For beginners, SYCAMORE may be the best tool to use: it focuses on helping users to set up their models by providing guidance through tutorials and links to external databases. More experienced users looking for advanced analysis features may turn into Virtual Cell, PybioS and WebCell, while users interested in building models containing additional spatial components may favour Virtual Cell. JWS provides an ever-growing repository of kinetic models that users can run simulations on quite easily. Finally, BASIS provides unique functionality for stochastic simulations.
In conclusion, web-based applications are still not as widely available for systems biology research as compared to other areas, but they are gradually becoming more sophisticated. The main advantages of web-based applications are easy accessibility on various platforms, potential for collaborative research and larger libraries or repositories of biological models. However, increasing the response speed between the web server and clients, building richer UIs and improving interoperability remain main challenges. Thus, web-based approaches may not be the most suitable and effective at this moment and considered as alternative solution for systems biological research and pedagogy. Nevertheless, we believe that the increased use of emerging technologies like Ajax, SOA and semantic web along with enhanced functionality, such as multi-stage modelling and links with public databases will make web-based applications more appealing to researchers for their respective research purposes.
Key Points.
Mathematical and computational simulation tools have found increasing use in the study of metabolic, signalling and regulatory networks.
More recently, web-based versions of these simulation tools have been developed, providing several advantages over stand-alone tools in terms of platform independence, accessibility, software updates and computational capabilities.
Six representative simulation tools were surveyed and compared from both the users’ and developers’ points of view on the basis of their usability, functionality and system architecture.
Web-based simulation tools suffer from a few disadvantages compared to stand-alone tools in terms of speed and UIs. These problems can be solved by using emerging technologies, such as Ajax, distributed computing and SOA.
The integration of various heterogeneous experimental data into mathematical models is also an ongoing topic of research. The use of XML-based standards through semantic web technology is one possible solution towards making the processes of model building, storage, simulation and exchange interoperable.
FUNDING
Academic Research Fund (R-279-000-258-112) from the National University of Singapore.
Biographies
Dong-Yup Lee is an assistant professor of Chemical and Biomolecular Engineering at the National University of Singapore and leads the Bioinformatics group at the Bioprocessing Technology Institute, A*STAR. His research interests include Systems Biology/Biotechnology/Bioinformatics, Synthetic and Engineering Biology and Drug and Disease Modelling.
Rajib Saha is currently a PhD student in Chemical and Biomolecular Engineering at the National University of Singapore. His research interests include Systems Biology and Microbial Fuel Cells.
Faraaz Noor Khan Yusufi is a Research Officer at the Bioprocessing Technology Institute. His research interests include omics data analysis and biological network analysis.
Wonjun Park is a Research Officer at the Bioprocessing Technology Institute.
Iftekhar A. Karimi is full professor of Chemical and Biomolecular Engineering at the National University of Singapore. His research interests include Systems Biology and Supply Chain Optimization.
References
- Kitano H. Computational systems biology. Nature. 2002;420:206–10. doi: 10.1038/nature01254. [DOI] [PubMed] [Google Scholar]
- Di Ventura B, Lemerle C, Michalodimitrakis K, et al. From in vivo to in silico biology and back. Nature. 2006;443:527–533. doi: 10.1038/nature05127. [DOI] [PubMed] [Google Scholar]
- van Riel NAW; Dynamic modelling and analysis of biochemical networks: mechanism-based models and model-based experiments. Brief Bioinform. 2006;7(4):364–74. doi: 10.1093/bib/bbl040. [DOI] [PubMed] [Google Scholar]
- Markram H. The Blue Brain Project. Nat Rev Neurosci. 2006;7:153–60. doi: 10.1038/nrn1848. [DOI] [PubMed] [Google Scholar]
- Springel V, White SDM, Jenkins A, et al. Simulations of the formation, evolution and clustering of galaxies and quasars. Nature. 2005;435:629–36. doi: 10.1038/nature03597. [DOI] [PubMed] [Google Scholar]
- Stainforth DA, Aina T, Christensen C, et al. Uncertainty in predictions of the climate response to rising levels of greenhouse gases. Nature. 2005;433:403–6. doi: 10.1038/nature03301. [DOI] [PubMed] [Google Scholar]
- Gilbert D, Fuβ H, Gu X, et al. Computational methodologies for modelling, analysis and simulation of signalling networks. Brief Bioinform. 2006;7(4):339–53. doi: 10.1093/bib/bbl043. [DOI] [PubMed] [Google Scholar]
- Lee SY, Lee D-Y, Kim TY. Systems biotechnology for strain improvement. Trends Biotechnol. 2005;23:349–58. doi: 10.1016/j.tibtech.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Hucka M, Finney A, Bornstein BJ, et al. Evolving a lingua franca and associated software infrastructure for computational systems biology: the systems biology markup language (SBML) project. Syst Biol. 2004;1:41–53. doi: 10.1049/sb:20045008. [DOI] [PubMed] [Google Scholar]
- Alves R, Antunes F, Salvador A. Tools for kinetic modelling of biochemical networks. Nat Biotechnol. 2006;24:667–72. doi: 10.1038/nbt0606-667. [DOI] [PubMed] [Google Scholar]
- Klipp E, Liebermeister W, Helbig A, et al. Systems biology standards-the community speaks. Nat Biotechnol. 2007;25:390–1. doi: 10.1038/nbt0407-390. [DOI] [PubMed] [Google Scholar]
- Tomita M, Hashimoto K, Takahashi K, et al. E-Cell: software environment for computational cell biology. Bioinformatics. 1999;15(1):72–84. doi: 10.1093/bioinformatics/15.1.72. [DOI] [PubMed] [Google Scholar]
- Sauro HM, Hucka M, Finney A, et al. Next generation simulation tools: the systems biology workbench and BioSPICE integration. OMICS. 2003;7(4):355–72. doi: 10.1089/153623103322637670. [DOI] [PubMed] [Google Scholar]
- Hoops S, Sahle S, Gauges R, et al. COPASI - a COmplex PAthway SImulator. Bioinformatics. 2006;22(24):3067–74. doi: 10.1093/bioinformatics/btl485. [DOI] [PubMed] [Google Scholar]
- Le Novère N, Bornstein B, Broicher A, et al. BioModels database: a free, centralized database of curated, published, quantitative kinetic models of biochemical and cellular systems. Nucleic Acids Res. 2006;34:D689–91. doi: 10.1093/nar/gkj092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumaran S, Hariharaputran S, Mishra J, et al. The database of quantitative cellular signalling: management and analysis of chemical kinetic models of signalling networks. Bioinformatics. 2003;19(3):408–15. doi: 10.1093/bioinformatics/btf860. [DOI] [PubMed] [Google Scholar]
- Olivier BG, Snoep JL. Web-based kinetic modelling using JWS online. Bioinformatics. 2004;20(13):2143–4. doi: 10.1093/bioinformatics/bth200. [DOI] [PubMed] [Google Scholar]
- Loew LM, Schaff JC. The virtual cell: a software environment for computational cell biology. Trends Biotechnol. 2001;19:401–6. doi: 10.1016/S0167-7799(01)01740-1. [DOI] [PubMed] [Google Scholar]
- Klipp E, Herwig R, Kowald A, et al. Systems biology in practice. Weinheim: Wiley-VCH; 2005. pp. 426–8. [Google Scholar]
- Lee D-Y, Yun C, Cho A, et al. WebCell: a web-based environment for kinetic modeling and dynamic simulation of cellular networks. Bioinformatics. 2006;22(9):1150–1. doi: 10.1093/bioinformatics/btl091. [DOI] [PubMed] [Google Scholar]
- Weidemann A, Richter S, Stein M, et al. SYCAMORE - a systems biology computational analysis and modelling research environment. Bioinformatics. 2008;24(12):1463–4. doi: 10.1093/bioinformatics/btn207. [DOI] [PubMed] [Google Scholar]
- Kirkwood TB, Boys RJ, Gillespie CS, et al. Towards an e-biology of ageing: integrating theory and data. Nat Rev Mol Cell Biol. 2003;4:243–9. doi: 10.1038/nrm1051. [DOI] [PubMed] [Google Scholar]
- Kurata H, Matoba N, Shimizu N. CADLIVE for constructing a large-scale biochemical network based on a simulation-directed notation and its application to yeast cell cycle. Nucleic Acids Res. 2003;31(14):4071–84. doi: 10.1093/nar/gkg461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata H, Masaki K, Sumida Y, et al. CADLIVE dynamic simulator: direct link of biochemical networks to dynamic models. Genome Res. 2005;15:590–600. doi: 10.1101/gr.3463705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettinen A, Aho T, Smolander O-P, et al. Simulation tools for biochemical networks: evaluation of performance and usability. Bioinformatics. 2005;21:357–63. doi: 10.1093/bioinformatics/bti018. [DOI] [PubMed] [Google Scholar]
- Vass MT, Shaffer CA, Ramakrishnan N, et al. The JigCell model builder: a spreadsheet interface for creating biochemical reaction network models. IEEE/ACM Trans. Comput Biol Bioinform. 2006;3:155–64. doi: 10.1109/TCBB.2006.27. [DOI] [PubMed] [Google Scholar]
- Wittig U, Golebiewski M, Kania R, et al. In: Proceedings of the 3rd International workshop on Data Integration in the Life Sciences (DILS’06),Lecture Notes in Bioinformatics. Vol. 4075. Springer, Hinxton, UK: 2006. SABIO-RK: integration and curation of reaction kinetics data; pp. 94–103. [Google Scholar]
- Moles CG, Mendes P, Banga JR. Parameter estimation in biochemical pathways: a comparison of global optimization methods. Genome Res. 2003;13:2467–74. doi: 10.1101/gr.1262503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabdoulline RR, Stein M, Wade RC. qPIPSA: Relating enzymatic kinetic parameters and interaction fields. BMC Bioinformatics. 2007;8:373. doi: 10.1186/1471-2105-8-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacser H, Burns J. The control of flux. Symp Soc Exp Biol. 1973;27:65–104. [PubMed] [Google Scholar]
- Papin JA, Stelling J, Price ND, et al. Comparison of network-based pathway analysis methods. Trends Biotechnol. 2004;22:400–5. doi: 10.1016/j.tibtech.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Gillespie DT. Exact stochastic simulation of coupled chemical reactions. J Phys Chem. 1977;81:2340–61. [Google Scholar]
- Schomburg I, Chang A, Schomburg D. BRENDA, enzyme data and metabolic information. Nucleic Acids Res. 2002;30:47–9. doi: 10.1093/nar/30.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hucka M, Finney A, Sauro HM, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–31. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- Shah AA, Barthel D, Lukasiak P, et al. Web and grid technologies in bioinformatics, computational and systems biology: a review. Curr Bioinformatics. 2008;3:10–31. [Google Scholar]
- Garrett JJ. [(22 April 2008, date last accessed)];Ajax: a new approach to web applications. Adaptive Path. http://www.adaptivepath.com/publications/essays/archives/000385.php.
- Bruno EJ. Ajax: Asynchronous JavaScript and XML, creating dynamic web pages. [(22 April 2008, date last accessed)]; Dr. Dobb's Journal. http://www.ddj.com/web-development/184406430.
- Craciun G, Tang Y, Feinberg M. Understanding bistability in complex enzyme-driven reaction networks. Proc Natl Acad Sci USA. 2006;103:8697–702. doi: 10.1073/pnas.0602767103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald R, Maus C, Rolfs A, et al. Discrete event modelling and simulation in systems biology. J Simulation. 2007;1:81–96. [Google Scholar]
- Hetherington J, Bogle IDL, Saffrey P, et al. Addressing the challenges of multiscale model management in systems biology. Comput Chem Eng. 2007;31:962–79. [Google Scholar]
- Lee D-Y, Yun H, Park S, et al. MetaFluxNet: the management of metabolic reaction information and quantitative metabolic flux analysis. Bioinformatics. 2003;19:2144–6. doi: 10.1093/bioinformatics/btg271. [DOI] [PubMed] [Google Scholar]
- Erl T. Service-oriented architecture: a field guide to integrating XML and web services. New York: Prentice Hall; 2004. [Google Scholar]
- Arita M, Robert M, Tomita M. All systems go: launching cell simulation fueled by integrated experimental biology data. Curr Opin Biotechnol. 2005;16:344–9. doi: 10.1016/j.copbio.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Wu L, van Winden WA, van Gulik WM, et al. Application of metabolome data in functional genomics: a conceptual strategy. Metab Eng. 2005;7:302–10. doi: 10.1016/j.ymben.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Neumann EK, Miller E, Wilbanks J. What the semantic web could do for the life sciences. Drug Discov Today Bio Silico. 2004;2:228–36. [Google Scholar]
- Wang X, Gorlitsky R, Almeida JS. From XML to RDF: how semantic web technologies will change the design of ‘omic’ standards. Nat Biotechnol. 2005;9:1099–103. doi: 10.1038/nbt1139. [DOI] [PubMed] [Google Scholar]