Abstract
The traditional approach to bioinformatics analyses relies on independent task-specific services and applications, using different input and output formats, often idiosyncratic, and frequently not designed to inter-operate. In general, such analyses were performed by experts who manually verified the results obtained at each step in the process. Today, the amount of bioinformatics information continuously being produced means that handling the various applications used to study this information presents a major data management and analysis challenge to researchers. It is now impossible to manually analyse all this information and new approaches are needed that are capable of processing the large-scale heterogeneous data in order to extract the pertinent information. We review the recent use of integrated expert systems aimed at providing more efficient knowledge extraction for bioinformatics research. A general methodology for building knowledge-based expert systems is described, focusing on the unstructured information management architecture, UIMA, which provides facilities for both data and process management. A case study involving a multiple alignment expert system prototype called AlexSys is also presented.
Keywords: expert system, knowledge-based system, data integration, UIMA, AlexSys, multiple sequence alignment
INTRODUCTION
Paradigm shift in bioinformatics
In the last decade, the high-throughput genome sequencing techniques and other large-scale experimental studies, including transcriptomics, proteomics, interactomics or phenotypic analyses, have clearly led to an increased amount of sequence data, but have also resulted in the diversification of molecular biology data, leading to a new biological research paradigm, one that is information-heavy and data-driven. In this context, complex informatics data management and integration systems are now being introduced to collect, store and curate all this heterogeneous information in ways that will allow its efficient retrieval and exploitation. These developments are opening up the possibility of new large-scale studies, aimed at understanding how genetic information is translated to molecular function, networks and pathways, all the way to physiology and even ecological systems.
New challenges
Such system-level studies necessitate a combination of experimental, theoretical and computational approaches and a crucial factor for their success will be the efficient exploitation of the multitude of heterogeneous data resources that include genomic sequences, 3D structures, cellular localisations, phenotype and other types of biologically relevant information. Nevertheless, several limitations of these ‘omics’ data have been highlighted. For example, data emerging from ‘omic’ approaches are noisy (information can be missing due to false negatives, and information can be misleading due to false positives) and it has been proposed that some of these limitations can be overcome by integrating data obtained from two or more distinct approaches [1]. In this context, a major challenge for bioinformaticians in the post-genomic era is clearly the management, validation and analysis of this mass of experimental and predicted data, in order to identify relevant biological patterns and to extract the hidden knowledge [2].
Significant research efforts are now underway to address these problems. One approach has been data warehousing, where all the relevant databases are stored locally in a unified format and mined through a uniform interface. SRS [3] and Entrez [4] are probably the most widely used database query and navigation systems for the life science community. Alternatively, distributed systems implement software to access heterogeneous databases that are dispersed over the internet and provide a query facility to access the data. Examples include IBM's DiscoveryLink [5], BioMOBY [6]. More recently, semantic web based methods have been introduced that are designed to add meaning to the raw data by using formal descriptions of the concepts, terms and relationships encoded within the data. Many of these technologies are reviewed in more detail in ref. [7].
Today's information-rich environment has also led to the development of numerous software tools, designed to analyse and understand the data. The tools can be combined using pipelines, or more recently workflow management systems (WMS), to provide powerful computational platforms for performing in silico experiments [8]. However, the complexity and diversity of the available analysis tools mean that we now require automatic processing by ‘intelligent’ computer systems, capable of automatically selecting the most appropriate tools for a given task. One major insight gained from early work in intelligent problem solving and decision making was the importance of domain-specific knowledge. A doctor, for example, is not only effective at diagnosing illness uniquely because he possesses some general problem-solving skills, but also because he knows a lot about medicine. Similarly, traditional bioinformatics studies were generally performed by experts who had the experience necessary to understand the patterns revealed by the computational analyses and who manually verified the results obtained. Domain scientists used their own expert knowledge to assess the significance of the results, to make reliable conclusions and to make further predictions. Thus, an expert user has expectations and knowledge beyond that applied by the tool, and brings this together with all of the output data to come to an informed conclusion.
KNOWLEDGE-BASED EXPERT SYSTEMS
Human expert knowledge is a combination of a theoretical understanding in a given domain and a collection of heuristic problem-solving rules that experience has shown to be effective. Computer-based expert systems (also known as knowledge-based systems) can be constructed by obtaining this knowledge from a human expert and transforming it into a form that a computer may use to solve similar problems. The ‘expert’ programme does not know what it knows through the raw volume of facts in the computer's memory, but by virtue of a reasoning-like process of applying a set of rules to the knowledge. It chooses among alternatives, not through brute-force calculation, but by using some of the same rules-of-thumb that human experts use.
Thus, an expert system can be described as a computer programme that simulates the judgement and behaviour of experts in a particular field and uses their knowledge to provide problem analysis to users of the software. There are several forms of expert systems that have been classified according to the methodology used [9], including:
rule-based systems use a set of rules to analyse information about a specific class of problems and recommend one or more possible solutions;
case-based reasoning systems adapt solutions that were used to solve previous problems and use them to solve new problems;
neural networks implement software simulations of massively parallel processes involving the processing of elements that are interconnected in a network architecture; and
fuzzy expert systems use the method of fuzzy logic, which deals with uncertainty and is used in areas where the results are not always binary (true or false), but involve grey areas and the term ‘may be’.
Expert systems were first used in the mid-1960s when a few Artificial Intelligence (AI) researchers, who grew tired of searching for the illusive general-purpose reasoning machine, turned their attention toward well-defined problems where human expertise was the cornerstone for solving the problems [10]. But expert systems really took off with the development of the internet in the 1990s, which facilitated access to data and deployment of applications. Today, thousands of systems are in routine use world-wide, particularly in business, industry and government.
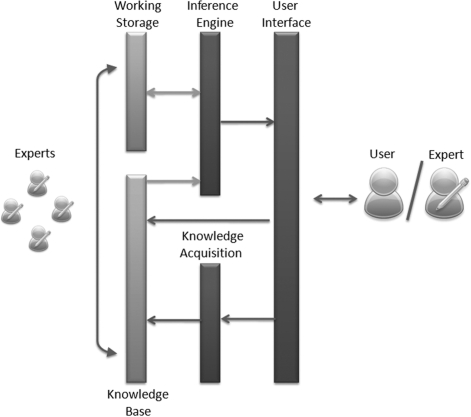
The major components of a typical knowledge-based expert system [11] are shown in Figure 1, and are described below:
The knowledge base contains domain expertise in the form of facts that the expert system will use to make determinations. Dynamic knowledge bases, known as truth maintenance systems, may be used, where missing or incorrect values can be updated as other values are entered;
The working storage is a database containing data specific to a problem being solved;
The inference engine is the code at the core of the system which derives recommendations from the knowledge base and problem-specific data in the working storage;
The knowledge acquisition module is used to update or expand dynamic knowledge bases, in order to include information gained during the expert system experiments; and
The user interface controls the dialog between the user and the system.
Figure 1:
Typical expert system architecture, components and human interface. Working Storage techniques (databases) and Knowledge Bases provide the basis of an expert system. The Inference Engine, which is accessible to the user through a User Interface, obtains facts or rules from the Knowledge Base and problem-specific data from the Working Storage. Each time the system is used, the Knowledge Base may be enriched through Knowledge Acquisition modules that derive new information from the user's experiments.
In this context, we can define an expert system as a framework that manages information dynamically by the integration of dedicated analysis tools. The tools to be used in any particular situation are chosen by special modules that reason about the best algorithms to use according to the information type and features. The reasoning part may be created using current Artificial Intelligence concepts and subsequently incorporated in the expert system which may also include workflows as an elementary module.
The following general points about expert systems and their architecture have been demonstrated [12]:
The sequence of steps used to analyse a particular problem is not explicitly programmed, but is defined dynamically for each new case;
Expert systems allow more than one line of reasoning to be pursued and the results of incomplete (not fully determined) reasoning to be presented; and
Problem solving is accomplished by applying specific knowledge rather than a specific technique. This is a key idea in expert systems technology. Thus, when the expert system does not produce the desired results, the solution is to expand the knowledge base rather than to re-programme the procedures.
As expert system techniques have matured into a standard information technology, the most important recent trend is the increasing integration of this technology with conventional information processing, such as data processing or management information systems. These capabilities reduce the amount of human intervention required during processing of large-scale data, such as genome-scale biological data.
Tools for implementation of expert systems
Many expert systems are built with architectures called expert system shells. The shell is a piece of software which provides a development framework, containing the user interface, a format for declarative knowledge in the knowledge base and an inference engine, e.g. the C Language Integrated Production System (CLIPS) [13] or the Java Expert System Shell (JESS) [14]. The use of a shell can reduce the amount of maintenance required and increase reusability and flexibility of the application. However, the tools are often specialised and may not match the exact requirements of a given problem. An alternative is to build a customised expert system using conventional languages, such as C or specialised languages, such as Prolog (programming in logic). This results in greater portability and performance, but needs increased development and maintenance time. Thus, it has been recommended [15] to use a shell when the system and problem space are small or when building a prototype and to use a programming language when enough is known about the scale and extent of the expert system or when performance becomes a major issue.
Expert systems in bioinformatics
In the biocomputing domain, various expert systems have been built for a number of specific tasks. While an exhaustive list of approaches is not presented here, it is hoped that the reader will gain a sense of the type of work being conducted in this field. For example, artificial neural networks have been used successfully for pattern discovery in many areas, including DNA and protein sequence analysis [16] and microarray data analysis [17]. Fuzzy logic approaches have also been applied to the analysis of gene expression data, e.g. [18]. In recent years, a very active area of research has been the reconstruction of functional networks from various sources of high-throughput experimental data, such as expression data or interaction data, and intelligent systems (fuzzy logic, neural networks, genetic algorithms, etc.) provide useful tools for modelling the network structures, e.g. [19]. These approaches are also finding applications in wider fields, such as drug discovery and design or medical diagnosis [20]. Another important task in bioinformatics is the extraction of knowledge from the biomedical literature and here, a case-based reasoning system has been developed for the classification of biomedical terms [21]. Case-based approaches are also widely used in the medical sciences for diagnosis and treatment planning, reviewed in [22]. Rule-based systems, that generally incorporate an expert knowledge base, have also been widely applied. For example, an inference engine, based on the JESS rule engine, has been built on top of the BioMediator data integration platform, with the aim of elucidating functional annotations for uncharacterised protein sequences [23]. FIGENIX [24] also addresses the problems of automatic structural and functional annotation under the supervision of a rule-based expert system, built using the Prolog language. Another group [25] has applied argumentation theory in the field of protein structure homology modelling, in order to construct arguments for and against the conclusion that the result is a good predictor of protein structure.
Many of these approaches were compared in a study [26] based on four biological data sets, where it was shown that no single method performed consistently well over all the data sets, but that a combination of methods could provide better specificity, sensitivity and accuracy. Unfortunately, there is no standard architecture that is widely used and that would allow exchange of information and code between the many different applications.
A PROPOSED SOLUTION FOR BIOINFORMATICS: UIMA
Because of the heterogeneity of traditional biological data resources, one of the most difficult aspects of building an expert system in this domain is information integration. Biological information is stored in many different formats, often including unstructured data, e.g. the results of certain programmes, database annotations in natural language or the scientific literature. This unstructured information thus represents an important unused source of information and it will be crucial for bioinformatics expert systems to be able to efficiently exploit this resource in the rule-finding and decision-making processes.
The principal challenge with unstructured information is that it needs to be analysed in order to identify, locate and relate the entities and relationships of interest, i.e. to discover the vital knowledge contained therein. The bridge from the unstructured world to the structured is enabled by the software agents that perform this analysis. These can scan a text document, for example, and pull out chemical names and their interactions, or identify events, locations, products, problems, methods, etc. A number of systems have been developed to address this problem. The best known in the public domain are OpenNLP [27], GATE [28] and UIMA [29]. OpenNLP (Open Natural Language Processing) is an umbrella structure for open source projects for the treatment of natural language, and also defines a set of Java interfaces and implements some basic infrastructure for NLP components. GATE (General Architecture for Text Engineering) provides a comprehensive infrastructure dedicated to language processing software development. Although OpenNLP and GATE both contain many powerful and robust algorithms for the processing of natural language texts, UIMA (Unstructured Information Management Architecture) [30] is more general and allows the analysis of many different types of information, including both structured and unstructured data. UIMA also supplies an execution environment in which developers can integrate their implementations of components in order to build and run complex applications.
UIMA thus presents a number of advantages for the development of expert systems in bioinformatics. First, UIMA is a lightweight Java platform and is very easy to deploy on any machine architecture. It nonetheless provides powerful capabilities for distributed computing through services. It is a scalable and extensible software architecture for the development of unstructured information management applications from combinations of multimodal semantic analysis and search components, including rule-based and statistical machine learning algorithms. Second, it is open source and is actively supported by a wide community of developers and users. UIMA was originally developed by IBM but has since been accepted by the Apache Incubator Project Management Committee [31]. Moreover, UIMA is not restricted to text analysis but also provides unique opportunities for the integration of any kind of data, such as 3D structural data, microscopy photographs, transcription profiles, etc. Associated with knowledge and rule discovery tools, UIMA can thus be used for rule definition as well as for data-driven decision making by the expert system.
Overview of UIMA
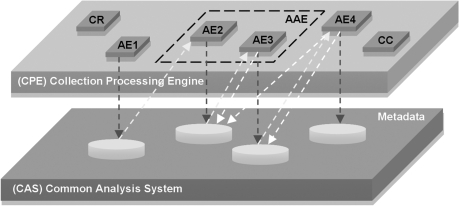
UIMA is an architecture in which primitive processing units called Analysis Engines (AEs) are combined to analyse data containing structured or unstructured information (Figure 2). The AE's core is called an Annotator and contains the actual analysis software. The AEs can then be organised using Flow Controllers (FCs) inside more complex structures called Aggregate Analysis Engines (AAEs). AAEs are used to group AEs that perform similar tasks, or that are elementary parts of a specific workflow. The AEs share data via a common representation system called the Common Analysis System (CAS), which is the primary data structure used to represent and share analysis results. Within the CAS, an object is specified by a Type, which is roughly equivalent to a class in an object-oriented programming language, or a table in a database. A collection of related Types then forms a Type System (TS). A complete UIM application can be built by combining AAEs in a Collection Processing Engine (CPE), consisting of:
A Collection Reader (CR), which allows the CPE to treat all the data in a given collection;
A corpus: AEs and AAEs, possibly managed by FCs; and
A CAS Consumer (CC), which takes the results from the CAS and stores them in an exploitable format (XML file, database, knowledge base …).
Figure 2:
UIMA components and organisation. A typical UIMA based system contains a CR which reads the input data and stores them into a temporary memory known as the CAS. The CAS is accessible by all AEs, which are ‘agent’ like modules that perform a specific task. A CC is a special AE that retrieves the results from the CAS at the end of the processing and exports them. AEs that perform similar tasks, or that are elementary parts of a workflow can be collected into an AAE.
UIMA thus provides all the functionality required to build a knowledge-based expert system, as defined above. The first layer of a UIMA application (Figure 2), represents the Working Storage and contains the system's memory and the structures used to store the metadata associated with each AE. The second layer, or application layer, consists of all elementary and AAEs and acts as the Inference Engine. Each module in the application layer can then read, write or update the metadata in the CAS at each step of the data analysis. The metadata are not frozen and are always accessible during the analysis process. Depending on the accumulated knowledge in the metadata, decisions can be made to execute the most pertinent AE. This data-driven decision process is the essential distinction between an Expert System and a workflow. In an expert system, the analysis pipeline is not pre-defined by the developer or user, but depends on the data and the previous ‘experience’ gained by the system. As a consequence, the more annotations and information that are generated, the better will be the final analysis. The third and last layer (not shown in Figure 2) corresponds to the User Interface between the final user and the system. In the case of UIMA applications, this can be built using either the Eclipse Interactive Development Environment or a Graphical User Interface, for example in Java or the Eclipse Rich Client Platform.
UIMA is now exploited by numerous applications in many diverse fields, including several systems related to the biological domain. We can cite, for example, the BioNLP [32] UIMA Component Repository which provides UIMA wrappers for novel and well-known 3rd-party NLP tools used in biomedical text processing, such as tokenizers, parsers, named entity taggers, and tools for evaluation.
A CASE STUDY: THE ALEXSYS MULTIPLE ALIGNMENT EXPERT SYSTEM
Based on expert knowledge gained over many years
Multiple sequence alignment (MSA) is a domain of research that has been widely studied for over 20 years. The first formal algorithm for MSA [33] was computationally expensive and therefore, most programmes in use today employ some kind of heuristic approach, such as the progressive alignment procedure [34], which exploits the fact that homologous sequences are evolutionarily related. This method involves three main steps: (i) pairwise sequence alignment and distance matrix calculation, (ii) guide tree construction and (iii) multiple alignment following the branching order in the guide tree. A number of different alignment programmes based on this method exist, using either a global alignment method to construct an alignment of the complete sequences (e.g. ClustalW/X [35]), or a local algorithm to align only the most conserved segments of the sequences (e.g. Pima [36]).
Today, MSA methods are evolving in response to the challenges posed by the new large-scale applications [37], and numerous different alignment algorithms have been developed. A comparison of many of these methods based on a widely used alignment benchmark data set, BAliBASE [38], highlighted the fact that no single algorithm was capable of constructing high-quality alignments for all test cases and led to the introduction of new alignment approaches that combined both global and local information in a single alignment programme (e.g. DbClustal [39], TCoffee [40], MAFFT [41] and Muscle [42]), resulting in more reliable alignments for a wide range of alignment problems. Different approaches have also been developed that exploit other information to improve sequence alignments (e.g. 2D/3D structure in 3DCoffee [43] and PRALINE [44] or known domain organisation in Refiner [45]). Yet another approach is the cooperative use of a number of different algorithms, e.g. PipeAlign [46], a multiple alignment processing pipeline, ranging from homology database searches to the construction of a high-quality multiple alignment of complete sequences.
AlexSys prototype: objectives
With the emergence of the new systems biology approaches, comparative sequence studies and evolutionary inferences are playing an essential role in the analysis and comprehension of complex biological networks. In particular, multiple sequence comparisons or alignments now provide the basis for most of the computational methods used in genomic analyses or proteomics projects, from gene identification and annotation to studies of promoters, interactomes, transcriptomes and proteomes. Although much progress has been achieved, none of the MSA programmes available today are capable of aligning difficult, divergent sets of sequences with consistently high quality. In this context, we have exploited the data integration and reasoning capabilities of UIMA, in order to develop a prototype of an ALignment EXpert SYStem, called AlexSys, whose goal is the evaluation and optimisation of all the steps involved in the construction and analysis of a multiple alignment.
Design and implementation
An essential step in the design of the AlexSys prototype was the definition of the TSs, which represent the prototype's ‘memory’ (the UIMA CAS) and correspond to the first framework layer shown in Figure 2. The prototype currently uses five TS: Sequence, Matrix, Tree, Parameter and Alignment. An appropriate specification of the TS is an essential first step, since the expert system is data driven, and thus more enriched features will lead to a more accurate analysis of the problems and eventually, to a more appropriate decision-making process.
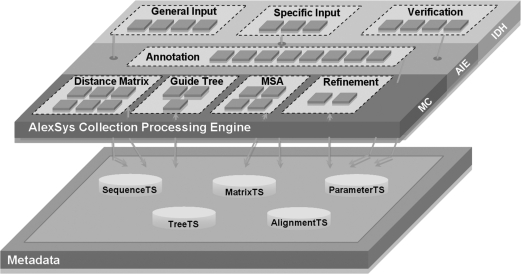
With these TS in hand, we can start to build the second application layer containing the AEs, grouped together into AAEs. The prototype is designed to perform three main tasks: input data handling, annotation and information extraction and MSA construction, shown in Figure 3 and described in more detail below.
Figure 3:
The modular structure of the AlexSys prototype. Dotted rectangles designate AAE. The AlexSys Collection Processing Engine is divided into three interconnected parts; the IDH (Input Data Handling) part consists of three AAEs which treat the input data: General Input contains AEs to read sequences, matrices, trees and alignment files; Specific Input contains AEs to handle parameters, algorithms and eventually predefined scenarios; Verification represents a single AAE containing AEs for checking errors, sequence number and type, and user-defined choices. The AIE (Annotation and Information Extraction) is a set of AEs that treat the data and update the CAS by writing new metadata or updating existing ones, including percent identity, composition, hydrophobicity, transmembrane predictions, secondary structure predictions, GO terms and PFAM domain predictions. The AIE will be extended in the future with additional modules. The MC (MSA Construction) is the most complex part and contains several algorithms grouped into: Distance Matrix calculation, Guide Tree generation, MSA and a set of Refinement algorithms that iteratively improve the accuracy of alignment.
Input data handling
A number of AAE were defined depending on the functionality of the primitive AEs: a General Input AAE includes four AEs dedicated to reading the data in XML format (sequences, matrices, trees or initial alignments); a Specific Input AAE includes three AEs dedicated to parameter, algorithm and scenario specification; a Verification AAE includes four AEs dedicated to error detection, sequence verification and validation of user options.
Annotation and information extraction
This task contains a single AAE, which is used to annotate the input data. The prototype system takes into account information related to biological sequence features, such as sequence number, length and percent identity, residue composition, secondary structures, functional domains and Gene Ontology (GO) terms. In the future, this information will be exploited for the creation of association rules and decision trees that will allow us to optimise the MSA construction, by selecting the most appropriate algorithms and parameters depending on the set of sequences to be aligned.
MSA construction
Here the multiple alignment is constructed using several possible algorithms depending on the information generated previously. The approach used here is based on the progressive schema and involves four main steps:
distance matrix calculation using one of K-tuple or dynamic programming from the ClustalW programme, BLAST 2 Sequences [47], MCWPA (Moving Contracting Window Algorithm) [48], Fast Fourier Transform from the MAFFT programme or a discrete wavelet transform [49];
guide tree construction using either BioNJ [50], Clearcut [51] or FastME [52];
multiple alignment, using either ClustalW, MAFFT [41], Muscle [42] or ProbCons [53] and validation with the NorMD [54] objective function; and
MSA refinement with an iterative optimisation of the Weighted Sum of Pairs alignment score as used in the MAFFT programme. The optimisation is done using an approximate group-to-group alignment algorithm and the tree-dependent restricted partitioning technique [55]. This procedure is referred to as FFT-NS-i.
One of our major goals in developing the AlexSys prototype was the investigation of how these different algorithms might be combined in order to improve the final multiple alignment accuracy.
Prototype optimisation
Enhancement of the ClustalW algorithm
The AlexSys architecture is designed to facilitate the evaluation and optimisation of the different steps involved in the construction of a MSA. To demonstrate this, we describe here an in-depth study of the ClustalW programme (version 1.83), which is based on the traditional progressive alignment approach and is one of the most popular MSA programmes in use today. For each of the main steps in the ClustalW method, different algorithms have been developed by various authors with the goal of improving the final alignment accuracy. Many of these algorithms have been incorporated in AlexSys, allowing us to evaluate and optimise their efficiency and accuracy. For example, replacing the dynamic programming algorithm used in the distance matrix calculation with different approaches, such as a modified version of the Moving Contracting Window Pattern Algorithm (MCWPA), resulted in a drastic decrease in the running time of ClustalW (Table 1). Although some alignment accuracy was lost in the process, this could be subsequently corrected using a refinement step, described subsequently. We also tested different approaches for the construction of the guide tree, although the effect of changing the guide tree proved to be less crucial for the resulting MSA, in agreement with previous studies [56].
Table 1:
ClustalW alignment optimisation using UIMA
| MSA programme | Reference 1 Equidistant sequences |
Reference 2 Family with orphans | Reference 3 Divergent subfamilies | Reference 4 Large extensions | Reference 5 Large insertions | Total running time (min) | |
|---|---|---|---|---|---|---|---|
| V1: <20% | V2: 20–40% | ||||||
| ClustalW version 1.83 | 0.46 | 0.85 | 0.86 | 0.62 | 0.75 | 0.61 | 98.6 |
| Optimised ClustalW | 0.50 | 0.87 | 0.86 | 0.65 | 0.77 | 0.63 | 40.4 |
Mean SP scores for each reference set in the BAliBASE multiple alignment benchmark
The final enhancement to the ClustalW algorithm involved the addition of a post-processing refinement step. Many approaches have been developed to improve an MSA, generally based on an optimisation of the Sum of Pairs objective function or its variants. Here, we incorporated the Weighted Sum of Pairs score as used in the MAFFT programme, but other approaches could be considered, such as a Remove First iterative optimisation [57] or the Refiner or RASCAL [58] methods.
The alignment accuracy obtained with each modification was measured using the BAliBASE benchmark database. Table 1 shows the scores obtained by ClustalW version 1.83, compared to the highest scoring, optimised version (distance matrix calculation with MCWPA algorithm, guide tree calculation with BioNJ algorithm, multiple alignment based on the guide tree using ClustalW and MSA refinement using the Weighted Sum of Pairs objective function). The modifications made to the original ClustalW algorithm allowed us to improve alignment accuracy in all reference sets while significantly reducing the computation time required.
Incorporating other algorithms
Numerous studies have been performed to compare the accuracy of diverse MSA programmes, e.g. [59]. These studies, generally based on benchmark data sets, provide scores that are averaged over a large number of alignments, and no one method has been shown to be more accurate in all cases. Although some algorithms are highly efficient for specific purposes, none of the programmes existing today can accurately model the reality of complex biological data. Protein sequences should not be considered as simple strings or characters, rather they reflect a physical object with a 3D structure and molecular and cellular functions that depend on their interactions with other components. The keywords then are optimisation and the intelligent choice of the most appropriate algorithm for the set of sequences to be aligned. With this in mind, we have incorporated a number of other algorithms in the AlexSys prototype, namely Muscle (version 3.51), MAFFT (version 6.24, fast option FFT-NS-2) and ProbCons (version 1.1). These well-known and well-studied programmes provide alternative approaches that address either the scalability or the accuracy of multiple alignments of large, complex sequences.
Currently, the programme used to align a given set of sequences must be selected manually. However, it is important to stress that the objective of the AlexSys expert system is to identify the characteristic features of a set of sequences that determine whether a given alignment method will succeed or fail. Therefore, when we perform a multiple alignment using AlexSys, a history of the analysis is stored in the CAS, including specific sequence features and the best-scoring algorithms involved in the alignment construction.
The results shown in Table 2 confirm the knowledge we have from previous studies and in particular, it is clear that ProbCons generally performs better than the other methods. Nevertheless, none of the programmes tested always provided the most accurate alignment and in certain cases, other alignment approaches should be considered. For example, MAFFT obtained the highest scores for 44.7% of the alignments in ref. [1], as illustrated by the example shown in Figure 4.
Table 2:
Multiple alignment programme scores for the alignment reference sets in Balibase 3.0
| MSA programme | Reference 1 Equidistant sequences |
Reference 2 Family with orphans | Reference 3 Divergent sub families | Reference 4 Large extensions | Reference 5 Large insertions | Reference 7 Trans-membrane proteins | Reference 9 Linear motifs | |
|---|---|---|---|---|---|---|---|---|
| V1: <20% | V2: 20–40% | |||||||
| ProbCons | 0.65 | 0.93 | 0.90 | 0.79 | 0.86 | 0.88 | 0.81 | 0.68 |
| 36.8% | 68.2% | 34.1% | 43.3% | 63.4% | 62.5% | 50.0% | 50.7% | |
| MAFFT | 0.45 | 0.88 | 0.88 | 0.74 | 0.83 | 0.79 | 0.81 | 0.66 |
| 44.7% | 20.4% | 51.2% | 46.7% | 41.5% | 31.2% | 37.5% | 22.4% | |
| Muscle | 0.56 | 0.90 | 0.88 | 0.76 | 0.84 | 0.83 | 0.79 | 0.66 |
| 13.2% | 4.5% | 9.8% | 10.0% | 9.8% | 6.2% | 12.5% | 4.5% | |
| ClustalW | 0.46 | 0.85 | 0.86 | 0.62 | 0.75 | 0.61 | 0.69 | 0.63 |
| 5.3% | 6.8% | 4.9% | 0.0% | 4.9% | 0.0% | 0.0% | 22.4% | |
For each programme, the mean SP scores are shown, as well as the number of times the programme achieved the best alignment score, expressed as a percentage of the total number of reference alignments. The programme with the highest percentage of best scores is highlighted in bold. References 6 and 8 were excluded, since the tests contained non-linear sequences that cannot be aligned by most multiple alignment methods.
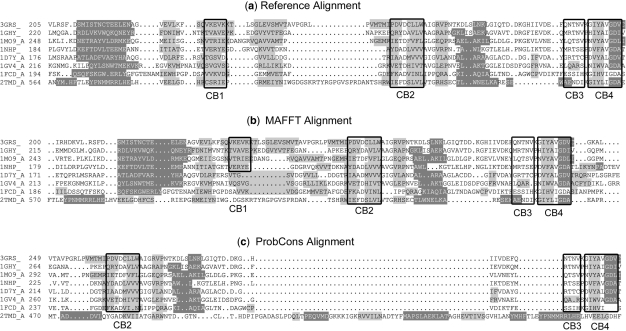
Figure 4:
(A) Part of BAliBASE reference alignment BB11034, aligned by (B) MAFFT and (C) ProbCons. Secondary structure elements are shown in dark grey (helix) or light grey (β-strand). Boxed regions indicate the alignment core blocks. ProbCons totally misaligned the last sequence (2tmd_A) and the core blocks for this sequence are outside the region shown.
In this alignment, consisting of sequences sharing <20% identity, ProbCons has misaligned the 2tmd_A protein, a trimethylamine dehydrogenase, introducing a large gap in the alignment. In contrast, MAFFT has successfully aligned at least some of the core blocks, e.g. CB2-CB4 in Figure 4, although there are still some errors, for example CB1. In the future, the information gained from these in depth studies of the different alignment approaches will be incorporated in the AlexSys knowledge base and will be used to automatically select the most suitable method for a given alignment problem. Where necessary, additional information such as 2D/3D structure or functional sites, will also be used to try to improve such difficult alignments.
Perspectives
The next stage in the development of AlexSys will be the creation of an accurate model of the strengths and weaknesses of the different alignment algorithms, depending on the characteristics of the sequences to be aligned. The generation of a posteriori information represents only a preliminary stage in the development of a complete expert system. Using this ‘use your enemy's hand to catch a snake’ technique, we can identify the most pertinent alignment algorithms based on benchmark tests. The resulting alignment model will play a crucial role in the creation of association rules that will drive the final expert process. The rules will be generated using knowledge extraction techniques, such as those available in the DB2 warehouse (new version of IBM Intelligent Miner). The final goal is to provide a priori knowledge that will guide the alignment procedure automatically, selecting one or more appropriate workflows and adjusting parameters, according to the input data. Thus, the development of the expert system can be considered to mimic the behaviour of a human expert. In the same way that human memory is enriched by events that occur during a lifetime, experiments conducted by the expert system contribute to its working storage and will hopefully lead to objective and intelligent decision making based on the encoded knowledge.
Another important aspect of a fully functional expert system, that has not been addressed here, is the development of a Graphical User Interface, allowing non-expert users to easily interact with the system, to input their data and to extract the pertinent information for subsequent studies.
The use of UIMA, in combination with dependency models and standard ontologies, will facilitate the enrichment of the expert system with more diverse components, covering aspects of genomic and protein data mining, validation and integration of structural/functional data, as well as diverse algorithms ensuring the construction, the refinement, the analysis and the efficient exploitation of MSAs.
CONCLUSION
With the rapid accumulation of genomic data, knowledge-based expert systems will be indispensable for the new integrative systems biology. Development of such expert systems is clearly a problem-oriented domain and requires in-depth knowledge of the domain. In the future, integration of qualitative, quantitative and scientific methods with powerful decision-making capabilities will improve the applicability of expert systems. As a result of this study, we have identified a number of requirements for a general expert system for systems biology:
easy integration of heterogeneous, distributed data: structured, semi-structured and unstructured;
easy integration of different analysis modules and reuse of existing modules;
complex workflow capabilities;
support for decision rules and automatic reasoning; and
facilities for implementation in a distributed grid computing environment.
At its prototype's stage, AlexSys represents a proof-of-concept test case for the suitability of UIMA for building expert systems. Using basic information, we have shown that it should be possible to improve alignment quality by combining different algorithms ‘intelligently’. The highly modular architecture of the AlexSys prototype allows us to intervene at any stage without altering the whole architecture. Using UIMA, it is relatively easy to develop additional analysis modules that can be plugged into the system, providing an ideal facility for system extension and evolution. In its final version, the system will include an intelligent miner, constituting a data-driven inference engine that will automatically define an appropriate workflow for a given multiple alignment problem without the need for benchmark testing.
Key Points.
With the rapid accumulation of genomic data, knowledge-based expert systems will be indispensable for the new integrative systems biology.
Expert system development is a problem-oriented domain and requires integration of in-depth knowledge and large data sets, as well as powerful analysis modules and decision-making capabilities.
UIMA provides sophisticated, unstructured and structured data management capabilities and allows data-driven, rule-based information analysis and knowledge extraction.
The AlexSys prototype represents a proof-of-concept test case for the suitability of UIMA for building expert systems.
FUNDING
Institut National de la Santé et de la Recherche Médicale; the Centre National de la Recherche Scientifique; the Université Louis Pasteur de Strasbourg.
Acknowledgements
We would like to thank Luc Moulinier, Odile Lecompte and Raymond Ripp for valuable discussions and Jean-Claude Thierry and Dino Moras for their continued support.
Biographies
M.R. Aniba is a PhD student at the Université Louis Pasteur and the Institut de Génétique et de Biologie Moléculaire et Cellulaire (CNRS/Inserm/ULP), France.
S. Siguenza is a research engineer at the Institut de Génétique et de Biologie Moléculaire et Cellulaire (CNRS/Inserm/ULP), France.
A. Friedrich is a post-doctoral fellow at the Institut de Génétique et de Biologie Moléculaire et Cellulaire (CNRS/Inserm/ULP), France.
F. Plewniak is a senior research engineer at the Institut de Génétique et de Biologie Moléculaire et Cellulaire (CNRS/Inserm/ULP), France.
O. Poch is a senior scientist at the Institut de Génétique et de Biologie Moléculaire et Cellulaire (CNRS/Inserm/ULP), France.
A. Marchler-Bauer, is a staff scientist at the National Center for Biotechnology Information NLM/NIH.
J.D. Thompson is a staff scientist at the Institut de Génétique et de Biologie Moléculaire et Cellulaire (CNRS/Inserm/ULP), France.
References
- Ge H, Walhout AJ, Vidal M. Integrating ‘omic’ information: a bridge between genomics and systems biology. Trends Genet. 2003;19:551–60. doi: 10.1016/j.tig.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Roos DS. Computational biology. Bioinformatics—trying to swim in a sea of data. Science. 2001;291:1260–1. doi: 10.1126/science.291.5507.1260. [DOI] [PubMed] [Google Scholar]
- Etzold T, Ulyanov A, Argos P. SRS: information retrieval system for molecular biology data banks. Meth Enzymol. 1996;266:114–28. doi: 10.1016/s0076-6879(96)66010-8. [DOI] [PubMed] [Google Scholar]
- Schuler GD, Epstein JA, Ohkawa H, Kans JA. Entrez: molecular biology database and retrieval system. Methods Enzymol. 1996;266:141–62. doi: 10.1016/s0076-6879(96)66012-1. [DOI] [PubMed] [Google Scholar]
- Hass L, Schwartz P, Kodali P, et al. DiscoveryLink: a system for integrating life sciences data. IBM Syst J. 2001;40:489–511. [Google Scholar]
- Wilkinson MD, Links M. BioMOBY: an open source biological web services proposal. Brief Bioinform. 2002;3:331–41. doi: 10.1093/bib/3.4.331. [DOI] [PubMed] [Google Scholar]
- Romano P. Automation of in-silico data analysis processes through workflow management systems. Brief Bioinform. 2008;9:57–68. doi: 10.1093/bib/bbm056. [DOI] [PubMed] [Google Scholar]
- Halling-Brown M, Shepherd AJ. Constructing computational pipelines. Methods Mol Biol. 2008;453:451–70. doi: 10.1007/978-1-60327-429-6_24. [DOI] [PubMed] [Google Scholar]
- Liao SH. Expert system methodologies and applications—a decade review from 1995 to 2004. Expert Syst Appl. 2005;28:93–103. [Google Scholar]
- Durkin J. Expert systems: a view of the field. IEEE Expert. 1996;11:56–63. [Google Scholar]
- Dhaliwal JS, Benbasat I. The use and effects of knowledge-based system explanations: theoretical foundations and a framework for empirical evaluation. Inf Sys Res. 1996;7:342–62. [Google Scholar]
- Giarratano JC, Riley G. Expert systems, principles and programming. course technology. 2005 [Google Scholar]
- C Language Integrated Production System (CLIPS). http://clipsrules.sourceforge.net/ (9 May 2008, date last accessed)
- Java Expert System Shell (JESS) http://herzberg.ca.sandia.gov/jess/ (6 August 2008, date last accessed)
- Garrett R. In: Proceedings of IEEE/ACM International Conference on Developing and Managing Expert System Programs, 30th September – 2nd October, 1991. Out of the lab into the field: system design of large expert systems. pp. 273–8. [Google Scholar]
- Baldi P, Brunak S. Bioinformatics: A Machine Learning Approach. Cambridge, MA: MIT Press; 2002. pp. 91–142. [Google Scholar]
- Linder R, Richards T, Wagner M. Microarray data classified by artificial neural networks. Methods Mol Biol. 2007;382:345–72. doi: 10.1007/978-1-59745-304-2_22. [DOI] [PubMed] [Google Scholar]
- Woolf PJ, Wang Y. A fuzzy logic approach to analyzing gene expression data. Physiol Genomics. 2000;3:9–15. doi: 10.1152/physiolgenomics.2000.3.1.9. [DOI] [PubMed] [Google Scholar]
- Bosl WJ. Systems biology by the rules: hybrid intelligent systems for pathway modeling and discovery. BMC Syst Biol. 2007;1:13. doi: 10.1186/1752-0509-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapetanovic IM, Rosenfeld S, Izmirlian G. Overview of commonly used bioinformatics methods and their applications. Ann NY Acad Sci. 2004;1020:10–21. doi: 10.1196/annals.1310.003. [DOI] [PubMed] [Google Scholar]
- Spasic I, Ananiadou S, Tsujii J. MaSTerClass: a case-based reasoning system for the classification of biomedical terms. Bioinformatics. 2005;21:2748–58. doi: 10.1093/bioinformatics/bti338. [DOI] [PubMed] [Google Scholar]
- Bichindaritz I, Marling C. Case-based reasoning in the health sciences: what's next? Artif Intell Med. 2006;36:127–35. doi: 10.1016/j.artmed.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Cadag E, Louie B, Myler PJ, et al. Biomediator data integration and inference for functional annotation of anonymous sequences. Pac Symp Biocomput. 2007;12:343–54. [PubMed] [Google Scholar]
- Gouret P, Vitiello V, Balandraud N, et al. FIGENIX: intelligent automation of genomic annotation: expertise integration in a new software platform. BMC Bioinformatics. 2005;6:198. doi: 10.1186/1471-2105-6-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferys BR, Kelley LA, Sergot MJ, et al. Capturing expert knowledge with argumentation: a case study in bioinformatics. Bioinformatics. 2006;22:924–33. doi: 10.1093/bioinformatics/btl018. [DOI] [PubMed] [Google Scholar]
- Tan AC, Gilbert D. An empirical comparison of supervised machine learning techniques in bioinformatics. In: Proceedings of the First Asia-Pacific bioinformatics conference on Bioinformatics. 2003;19:219–22. [Google Scholar]
- Open Natural Language Processing (OpenNLP). http://opennlp.sourceforge.net/ (31 August 2008, date last accessed)
- Cunningham H, Maynard D, Bontcheva K, et al. In: Proceedings of the 40th Anniversary Meeting of the Association for Computational Linguistics (ACL’02) Philadelphia: 2002. GATE: a framework and graphical development environment for robust NLP tools and applications. [Google Scholar]
- Ferrucci D, Lally A. UIMA: an architectural approach to unstructured information processing in the corporate research environment. Nat Lang Eng. 2004;10:327–48. [Google Scholar]
- Unstructured Information Management Architecture (UIMA). http://uima.framework.sourceforge.net. (15 November 2006, date last accessed)
- Apache Incubator Project Management Committee. http://incubator.apache.org/uima (12 August 2008, date last accessed)
- Baumgartner WA Jr, Cohen KB, Hunter L. An open-source framework for large-scale, flexible evaluation of biomedical text mining systems. J Biomed Discov Collab. 2008;3:1. doi: 10.1186/1747-5333-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankoff D. Minimal mutation trees of sequences. SIAM J Appl Math. 1975;28:35–42. [Google Scholar]
- Feng DF, Doolittle RF. Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J Mol Evol. 1987;25:351–60. doi: 10.1007/BF02603120. [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, et al. Multiple sequence alignment with the clustal series of programs. Nucleic Acids Res. 2003;31:3497–500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RF, Smith TF. Pattern-induced multi-sequence alignment (PIMA) algorithm employing secondary structure-dependent gap penalties for use in comparative protein modelling. Protein Eng. 1992;5:35–41. doi: 10.1093/protein/5.1.35. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Poch O. Multiple sequence alignment as a workbench for molecular systems biology. Curr Bioinformatics. 2006;1:95–104. [Google Scholar]
- Thompson JD, Koehl P, Ripp R, et al. BAliBASE 3.0: latest developments of the multiple sequence alignment benchmark. Proteins. 2005;61:127–36. doi: 10.1002/prot.20527. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Plewniak F, Thierry JC, et al. Rapid and reliable global multiple alignments of protein sequences detected by database searches. Nucleic Acids Res. 2000;28:2919–26. doi: 10.1093/nar/28.15.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–17. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, et al. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan O, Suhre K, Abergel C, et al. 3DCoffee: combining protein sequences and structures within multiple sequence alignments. J Mol Biol. 2004;340:385–95. doi: 10.1016/j.jmb.2004.04.058. [DOI] [PubMed] [Google Scholar]
- Simossis VA, Heringa J. PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res. 2005;33:W289–94. doi: 10.1093/nar/gki390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S, Lanczycki CJ, Panchenko AR, et al. Refining multiple sequence alignments with conserved core regions. Nucleic Acids Res. 2006;34:2598–606. doi: 10.1093/nar/gkl274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plewniak F, Bianchetti L, Brelivet Y, et al. PipeAlign: a new toolkit for protein family analysis. Nucleic Acids Res. 2003;31:3829–32. doi: 10.1093/nar/gkg518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–50. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- Yang QX, Yuan SS, Zhao L, et al. Faster algorithm of string comparison. Pattern Anal Applic. 2003;6:122–33. [Google Scholar]
- Wen ZN, Wang KL, Li ML, et al. Analyzing functional similarity of protein sequences with discrete wavelet transform. Comput Biol Chem. 2005;29:220–8. doi: 10.1016/j.compbiolchem.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Gascuel O. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol. 1997;14:685–95. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]
- Sheneman L, Evans J, Foster JA. Clearcut: a fast implementation of relaxed neighbor joining. Bioinformatics. 2006;22:2823–4. doi: 10.1093/bioinformatics/btl478. [DOI] [PubMed] [Google Scholar]
- Desper R, Gascuel O. Fast and accurate phylogeny reconstruction algorithms based on the minimum-evolution principle. J Comp Biol. 2002;9:687–705. doi: 10.1089/106652702761034136. [DOI] [PubMed] [Google Scholar]
- Do CB, Mahabhashyam MS, Brudno M, et al. ProbCons: probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330–40. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Plewniak F, Ripp R, et al. Towards a reliable objective function for multiple sequence alignments. J Mol Biol. 2001;314:937–51. doi: 10.1006/jmbi.2001.5187. [DOI] [PubMed] [Google Scholar]
- Hirosawa M, Totoki Y, Hoshida M, Ishikawa M. Comprehensive study on iterative algorithms of multiple sequence alignment. Comput Appl Biosci. 1995;11:13–8. doi: 10.1093/bioinformatics/11.1.13. [DOI] [PubMed] [Google Scholar]
- Nelesen S, Liu K, Zhao D, et al. The effect of the guide tree on multiple sequence alignments and subsequent phylogenetic analyses. Pac Symp Biocomput. 2008;13:25–36. doi: 10.1142/9789812776136_0004. [DOI] [PubMed] [Google Scholar]
- Wallace IM, O'Sullivan O, Higgins DG. Evaluation of iterative alignment algorithms for multiple alignment. Bioinformatics. 2005;21:1408–14. doi: 10.1093/bioinformatics/bti159. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Thierry JC, Poch O. RASCAL: rapid scanning and correction of multiple sequence alignments. Bioinformatics. 2003;19:1155–61. doi: 10.1093/bioinformatics/btg133. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Plewniak F, Poch O. A comprehensive comparison of multiple sequence alignment programs. Nucleic Acids Res. 1999;27:2682–90. doi: 10.1093/nar/27.13.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]