Abstract
Cytochrome P450 (CYP) 1A1 and CYP1B1 are inducible by 2,3,7,8-tetrachlorodibenzo-p-dioxin (dioxin) in the human breast cancer cell line, MCF-7. Since CYP1A1 was inducible to a much greater degree than CYP1B1, we hypothesized that there may be differences in coactivator recruitment to the promoter and/or enhancer regions of these genes. Dioxin treatment leads to recruitment of the aryl hydrocarbon receptor to the enhancer regions but not to the proximal promoter regions of both the CYP1A1 and CYP1B1 genes. On the other hand, dioxin treatment facilitated recruitment of RNA polymerase II to the promoters but not the enhancer regions. Dioxin treatment also elicited recruitment of the transcriptional coactivators, steroid receptor coactivator 1 (SRC-1) and steroid receptor coactivator 2 (SRC-2) and p300, which possess intrinsic histone acetyltranferase activities, to both genes, whereas Brahma (BRM)/Switch 2-related gene 1 (BRG-1), a subunit of nucleosomal remodeling factors, was recruited more robustly to CYP1A1 relative to CYP1B1. Small inhibitory RNA-mediated knockdown of p300 and SRC-2 adversely affected dioxin induction of both genes, whereas knockdown of BRM/BRG-1 reduced CYP1A1 induction but had little, if any, effect on CYP1B1 induction. These results suggest that nucleosomal remodeling is less significant for dioxin-mediated induction of CYP1B1 than that of CYP1A1 and may be related to the more modest inducibility of the former. Interestingly, simultaneous knockdown of SRC-2 and BRM/BRG-1 had no greater effect on CYP1A1 induction than knockdown of each coactivator individually, while simultaneous knockdown of p300 and BRM/BRG-1 had a much greater effect than knockdown of each individual gene, suggesting that the recruitment of SRC-2 to CYP1A1 depends upon BRM/BRG-1, while the recruitments of p300 and BRM/BRG-1 are independent of each other. These observations provide novel insights into the functional roles of the endogenous coactivators in dioxin induction of the human CYP1A1 and CYP1B1 genes in their natural chromosomal configurations.
Keywords: CYP1A1, CYP1B1, aryl hydrocarbon receptor, transcription, coactivator
The aryl hydrocarbon receptor (AHR) is a cytosolic, ligand-activated transcription factor that binds a variety of xenobiotics, including polycyclic aromatic hydrocarbons (PAHs) and halogenated aromatic hydrocarbons, such as 2,3,7,8-tetrachlorodibenzo-p-dioxin (dioxin). In the absence of ligand, the AHR is localized to the cytosol, where it is complexed with chaperone proteins including p23, XAP2/ARA9, and Hsp90 (Meyer et al., 1998; Petrulis et al., 2002). Upon binding ligand, the AHR undergoes a conformational change that results in the loss of certain chaperone proteins and exposure of a nuclear localization signal that facilitates translocation to the nucleus. Once in the nucleus, the AHR interacts with the aryl hydrocarbon nuclear translocator to form an activated transcription factor complex, referred to as the aryl hydrocarbon receptor complex (AHRC). The AHRC then binds consensus nucleotide sequences, termed xenobiotic response elements (XREs), in the 5′ untranslated region of responsive genes, thereby modulating the transcription of these genes.
Genes regulated by the AHRC in a dioxin-dependent manner include phase I drug and carcinogen metabolizing enzymes, such as cytochrome P450 (CYP), certain phase II metabolizing enzymes, as well as transforming growth factors (Hankinson, 1995; Rivera et al., 2002; Whitlock et al., 1997). The most well-characterized dioxin-inducible gene is CYP1A1, which is induced in many different organs and tissues. CYP1B1 has been shown to be inducible by dioxin in fibroblast and steroidogenic tissues (Sutter et al., 1994). CYP1A1 and CYP1B1 are the principal CYPs responsible for the metabolic activation of PAHs, which represent important carcinogens in cigarette smoke, smog, and some cooked foods, into their carcinogenic derivatives.
Dioxin is also a potent nongenotoxic carcinogen, acting as a tumor promotor. Modulation of the AHR gene battery is thought to be the primary mechanism by which dioxin exerts its toxic and carcinogenic effects. AHR-null mice are resistant to the toxic and carcinogenic effects of dioxin (Gonzalez, 1990) and are resistant to PAH-induced carcinogenesis at the site of application (Shimizu et al., 2000). The AHR therefore plays a significant role in mediating the toxic and carcinogenic effects of a variety of important environmental carcinogens, most likely via modulation of gene expression.
AHR-mediated transcription of human CYP1A1 and CYP1B1 is mediated by the binding of the AHRC to enhancer regions located ∼1 kb upstream of their transcription start sites, each of which contains a cluster of XREs (Kress et al., 1998; Tsuchiya et al., 2003). In mouse hepatoma cells, the binding of the AHRC to XREs has been shown to stimulate local changes in chromatin structure at the enhancer region of Cyp1a1 and to induce chromatin modification and nucleosomal displacement at the promoter region located just upstream of the transcriptional start site (Okino and Whitlock, 1995). The displacement of the fixed nucleosome exposes a TATA sequence and allows for promoter accessibility by the preinitiation complex and stabilization of RNA polymerase II (pol II). The binding of activators and general transcription factors alone is insufficient to activate most genes (Kim et al., 1994; Meisterernst and Roeder, 1991), and alterations in chromatin structure directed by the action of transcriptional coactivator proteins are also required. Transcriptional regulation by the AHRC is therefore dependent on the coordinated recruitment of coactivator proteins, chromatin remodeling factors, and general transcription factors (Hankinson, 2005).
Many coactivator proteins alter the restrictive promoter structure of genes into a more accessible chromatin configuration through the application of their histone acetyltransferase (HAT), or histone methyltransferase, activities that covalently modify the lysine and arginine resides of protruding histone tails, respectively (Bauer et al., 2002). The coactivators generally function in large multisubunit protein complexes that bind responsive genes. The p160 family of coactivators, such as steroid receptor coactivator 1 (SRC-1), steroid receptor coactivator 2 (SRC-2)/nuclear coactivator 2 (NcoA-2), and steroid receptor coactivator 3 (SRC-3)/p300/cAMP response element binding protein-binding protein (CBP)–interacting protein are coactivator proteins that possess HAT activity and have been shown to associate with the mouse AHRC in a ligand-dependent manner, increasing the transcription of an XRE-driven reporter gene (Beischlag et al., 2002). The Brahma (BRM)/Switch 2-related gene 1 (BRG-1) and BRM proteins are the adenosine triphosphatase (ATPase) subunits of the mammalian SWI-fructose non-fermentable ATPase-dependent chromatin remodeling complex and have been identified as ligand-dependent interacting partners and essential components of the AHRC during transcriptional activation of mouse Cyp1a1 (Wang and Hankinson, 2002). Several other coactivator proteins have been identified as interacting proteins of the AHRC (Beischlag et al., 2004; Chen et al., 2006; Hankinson, 2005; Kobayashi et al., 1997; Kumar and Perdew, 1999; Kumar et al., 1999; Rowlands et al., 1996; Swanson and Yang, 1998; Tojo et al., 2002; Wang et al., 2004a; Wei et al., 2004), yet their functions in dioxin-induced transcriptional regulation of CYP1A1 and CYP1B1 have yet to be fully defined.
CYP1A1 and CYP1B1 exhibit differences in tissue expression (Shimada et al., 1996; Uno et al., 2008), and their transcripts are differentially regulated by dioxin in many cell lines. We previously studied the roles of several transcriptional coactivator proteins in the induction of the mouse Cyp1a1 gene in the hepatoma cell line, Hepa-1. Here, we carry out an analysis of the role of some of these same coactivator proteins in the induction of the human isoform, focusing on the role of endogenous coactivator proteins in the transcriptional activation of the CYP1A1 gene in its natural chromosomal setting. Additionally, unlike the Hepa-1 cell line, the human breast cancer cell line, MCF-7, which we have used in these studies, is inducible by dioxin for both CYP1A1 and CYP1B1, which provides an opportunity to compare the roles of the coactivators in the induction of the two genes. This is particularly relevant because CYP1A1 and CYP1B1 are inducible to different degrees in this cell line. Most importantly, we investigate the potential functional interaction of the coactivators in the induction of the two genes, providing new insight into their mechanisms of regulation.
MATERIALS AND METHODS
Cell culture and antibodies.
The MCF-7 human breast carcinoma cells (American Type Culture Collection, Manassas, VA) were grown as a monolayer and maintained in α-minimal essential media containing 10% fetal bovine serum, 1% Fungizone (Amphotercin B), and 1% penicillin-streptomycin (Invitrogen, Carlsbad, CA) at 37°C and 5% CO2. Antibodies used for chromatin immunoprecipitation (ChIP) were as follows: α-AHR (Zhang et al., 1996) for pol II; N-20 for p300; N-15 for BRG-1; H-88 for BRM; C-20 for SRC-1; M341 (all from Santa Cruz Biotechnology, Santa Cruz, CA) for SRC-2 (BD Transduction Laboratories, San Jose, CA).
Reverse transcription and quantitative real-time PCR.
Total RNA was isolated using the RNeasy Mini kit according to the manufacture's protocol (Qiagen, Valencia, CA). Reverse transcription was performed using Superscript III reverse transcriptase according to the manufacture's protocol (Invitrogen). Briefly, 2 μg of total RNA was used in a 20-μl reverse transcriptase reaction under the following program: 25°C, 10 min; 48°C, 30 min; and 95°C, 5 min, using the Icycler Thermal Cycler (Bio-Rad, Hercules, CA). Endogenous CYP1A1, and CYP1B1, mRNAs were quantified by real-time PCR and normalized to that of the 36B4 ribosomal subunit. TaqMan primers/probes for 36B4 were described previously (Hsu et al., 2007). CYP1A1 primers were as follows: forward 5′-CAAGAGGAGCTAGACACAGTGATT-3′ and reverse 5′-AGCCTTTCAAACTTGTGTCTCTTGT-3′. CYP1B1 primer sequences were forward 5′-CACCGTTTTCCGCGAATTC-3′ and reverse 5′-CCTTCTTTTCCGCAGAGAGGAT-3′. The dual-labeled TaqMan probes were 5′-/5Cy5/CATCTGCCCTATATGGAGG/3BHQ_2/-3′ and 5′/HEX/CTGGAAGGTGCAGCGGCGC/3BHQ_1/-3′ for CYP1A1 and CYP1B1, respectively. All primers and probes were designed using Primer Express 3.0 software (Applied Biosystems, Foster City, CA) and synthesized by Integrated DNA Technologies, Inc. (Coralville, IA). A 20× mix of primers (18μM each) and probe (5μM) was diluted to 1× in a 15-μl multiplex reaction with 1× ABI Fast TaqMan reagent and 2 μl of diluted cDNA. TaqMan assays were performed using Applied Biosystems 7500 Fast machine in fast mode. Reactions consisted of an initial 95°C hold for 20 s followed by 40 cycles of the following: 95°C, 3 s, and 60°C, 30 s.
ChIP assay.
Cells were seeded in a 150-mm dish, and the following day (at 85% confluence), they were treated with 100nM dioxin for 60 min, or the indicated time points, at 37°C. DNA-protein complexes were cross-linked by the addition of 1% formaldehyde for 10 min at 37°C. The cells were rinsed twice with ice-cold PBS and collected in 1 ml of ice-cold PBS + 1× protease inhibitor solution (Roche, Palo Alto, CA). Cells were centrifuged at 600 × g for 5.5 min at 4°C in a Beckman tabletop centrifuge. The pellets were then resuspended in 800 μl of lysis buffer #1 (10mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid [HEPES, pH 7.4], 10mM KCl, 0.2mM EDTA, 1mM dithiothreitol [DTT, added just before use], and 1× protease inhibitor solution) and incubated on ice for 15 min. To enrich for nuclear extracts, the lysates were treated with 50 μl of 10% nonidet P40 (NP-40), vortexed for 10 s, and centrifuged at 2000 revolutions per minute (rpm) for 5 min at 4°C. Pellets were then treated with 310 μl of lysis buffer #2 (55.6mM Tris-HCl [pH 8.1], 11.1mM EDTA, 1.11% SDS, and 1× protease inhibitor solution) and incubated on ice for 10 min. Cell lysates were sonicated twice on high power for 8 min, alternating between 30 s on and 30 s off using a Bioruptor cell sonicator (Diagenode, Inc., New York, NY) to shear DNA fragments to sizes between 200 and 900 bp. Cellular debris was removed by centrifugation for 10 min at 13,000 rpm at 4°C. Supernatants were stored at − 80°C overnight.
The following day, samples were thawed on ice for 30 min and centrifuged for 10 min at 4°C to precipitate the SDS. A small aliquot (10 μl) of lysate was removed for input control, the remaining lysates were diluted 1:10 in 1% Triton X-100, 2mM EDTA, 150mM NaCl, 20mM Tris-HCl (pH 8.1), and 1× protease inhibitor solution (Roche). Immunoclearing was achieved by the addition of 40 μl of a 50% slurry of protein-A agarose beads in Tris-EDTA per 2.5 μg of sonicated salmon sperm DNA/bovine serum albumin (TE/SSDNA/BSA) solution (Upstate Biotechnology, Lake Placid, New York) and incubated on a rotator at 4°C for 30 min. Beads were briefly pelleted, and supernatants were placed in a new tube and probed with 2 μg of antibody overnight on a rotator at 4°C. The solutions were then treated with 40 μl of a 50% slurry of protein-A agarose beads in TE/SSDNA/BSA solution and incubated for 1 h at 4°C on a rotator. The beads were pelleted and sequentially washed in buffers A, B, LiCl, and followed by two washes in 10mM Tris-HCl (pH 8.1) and 1mM EDTA (TE). Buffer recipes were as follows—buffer A: 0.1% SDS, 1% Triton X-100, 2mM EDTA, 20mM Tris-HCl (pH 8.1), and 150mM NaCl; buffer B: 0.1% SDS, 1% Triton X-100, 2mM EDTA, 20mM Tris-HCl (pH 8.1), and 500mM NaCl; LiCl buffer: 0.25M LiCl, 1% NP-40, 1% deoxycholate, 1mM EDTA, and 10mM Tris-HCl (pH 8.1). Chromatin complexes were eluted by the addition of 0.5 ml of freshly prepared elution buffer (1% SDS, 0.1M NaHCO3). The cross-linking was reversed by incubating samples at 65°C overnight (18 h) in a Hybaid dry bath (Thermo Fisher Scientific, Inc., Waltham, MA). The solutions were then digested with 20 μg of PCR-grade recombinant proteinase K solution (Roche) for 1 h at 45°C.
DNA was isolated using Qiagen PCR extraction columns and eluted in 50 μl of water. Primers for the CYP1A1 enhancer were 5′-CCGCCACCCTTCGA-3′ and 5′-CAGGCGTTGCGTGAGA-3′. Those for the CYP1A1 promoter were 5′-CGTGGCCACACGTACAA-3′ and 5′-AGCAACTCACCTGAGGTACTG-3′. CYP1B1 enhancer primer sequences were 5′-TGTCAGGTGCCGTGAGAA-3′ and 5′-CGAACTTTATCGGGTTGAA-3′ and CYP1B1 promoter sequences were 5′-GTTTGGCGCTGGGTTAC-3′ and 5′-AGGTCGGAGCTGACTCTCT-3′. Dual-labeled probes for the CYP1A1 enhancer and promoter were 5′-/5HEX/CATGCAGGCTGCCTCTCCTCGC/3BHQ_1/-3′ and 5′-/5TexRd-XN/CAGGGAAGGAGGCGTGGCCA/3IAbRQSp/-3′, respectively, and those for the CYP1B1 enhancer and promoter were 5′-/5Cy5/TTCTCTTAGCTGTCTTGAAAATCCTAT/3BHQ_2/-3′ and 5′-/56-FAM/TCGATGCCCGCAGCGTTGTC/36-TAMSp/-3′, respectively. All primers and probes were designed using Primer Express 3.0 software (Applied Biosystems) and synthesized by Integrated DNA Technologies, Inc. A 20× mix of primers (18μM) and probe (5μM) was diluted to 1× with 1× ABI fast reagent and 1 μl of ChIP DNA. Multiplex conditions were as described earlier.
RNA interference.
To reduce the endogenous expression of the p300 and SRC-2 proteins, MCF-7 cells were infected with a retroviral vector containing a short hairpin small inhibitory RNA (siRNA) duplex targeting either 5′-CCCCUCCUCUUCAGCACCA-3′ of the p300 gene, 5′-AAGAGCAAACUCAUCCGUUC-3′ of the SRC-2 gene, or 5′-UUCUCCGAACGUGUCACG-3′, a scrambled sequence (scx, control). Construction of the retroviral green fluorescent protein puromycin (RVGP)-shP300, -SRC-2, and scrambled control (scx) RNA interference (RNAi) vectors took place as follows: double-strand DNA oligos containing the corresponding gene targeting sequence was inserted into the mU6pro vector (provided by Dr Stephen Smale, University of California Los Angeles), digested with BbsI and EcoRI, and the resulting vectors harboring the DNA oligos were digested with NheI and XbaI. The 0.5-kb DNA fragments produced in these reactions (containing the DNA oligos flanked by a U6 small nuclear RNA promoter at the 5′ ends) were cloned into the RVGP-driven retroviral RNAi vector (also provided by Dr Smale) and digested with NheI. Retrovirus was made by cotransfection of 293T cells with the RNAi vectors and an amphotropic packaging plasmid, pCL-10A1 (provided by Dr Smale), using the Lipofectamine 2000 (Invitrogen). In total, 3 × 105 MCF-7 cells were incubated with 2 ml retrovirus-containing medium under centrifugation at 2500 rpm (rpm or × g? If rpm, should designate centrifuge per rotor) at 32°C for 1.5 h, after which time, 2 ml fresh minimal essential medium (MEM) was added to the cells. Forty-eight hours later, the infected cells were selected in α-MEM complete media plus 3 μg/ml puromycin. An siRNA oligo targeting both the BRG-1 and BRM genes (siBB), as well as the scrambled control oligo (scx), were synthesized by Qiagen as previously described (Wang et al., 2004b). MCF-7 or MCF-7 cells infected with small hairpin RNA were transfected with 100nM siBB using Oligofectamine according to the manufacture's protocol (Invitrogen). Twenty-four hours posttransfection, the cells were treated with 100nM dioxin or dimethyl sulfoxide vehicle for an additional 24 h.
Western blot analysis.
MCF-7 cells were lysed in a modified Jun N-terminal kinase buffer (25mM HEPES [pH 7.4], 2.5mM MgCl2, 400mM NaCl, 50mM β-glycophosphate, 10mM p-nitrophenylphosphate, 1mM Na3VO4, 1mM DTT, and 1× complete protease inhibitors [Roche Applied Science, Indianapolis, IN]). The cell lysates (50 μg) were resolved on a 5% (p300) or 7% (SRC-2) SDS-polyacrylamide gel. p300 was detected using a 1:200 dilution of the p300 antibody, and SRC-2 was detected using a 1:250 dilution of the SRC-2 antibody.
Statistical analyses.
Experiments were repeated three or more times, except for those presented in Figures 3 and 6, which were performed twice. The figures depict data from representative experiments. Error bars represent the SDs between the triplicate quantitative real-time (QRT) PCR determinations within the given experiment (Hsu et al., 2007).
FIG. 3.
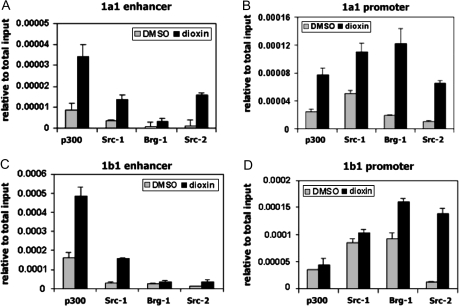
Dioxin-induced coactivator recruitment to CYP1A1and CYP1B1. MCF-7 cells were treated with 100nM dioxin for 60 min and subjected to ChIP assays. Lysates were probed with 2 μg of α-p300, α-BRG-1, α-SRC-1, and α-SRC-2. Reactions were performed using primers for the CYP1A1(A and B) and CYP1B1(C and D) regulatory regions.
FIG. 6.
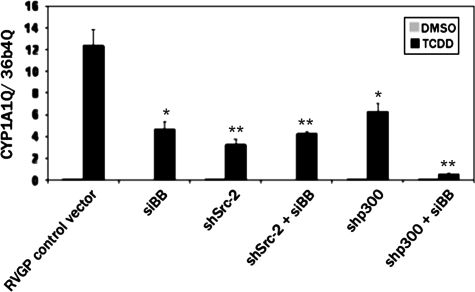
Effect of the loss of multiple coactivators on dioxin-mediated induction of CYP1A1. MCF-7 cells stably expressing shp300, shSRC-2, or the RVGP control vector were treated with 100nM siBB or a scrambled siRNA control (scx) for 24 h and then treated with 100 nM dioxin for an additional 24 h. RNA was isolated and subjected to reverse transcription and real-time PCR. The amount of the CYP1A1mRNA was corrected against the levels of the constitutively expressed ribosomal subunit, 36B4. *p < 0.05; **p < 0.001 relative to the RVGP control.
RESULTS
Differential Induction of CYP1A1 and CYP1B1 by Dioxin
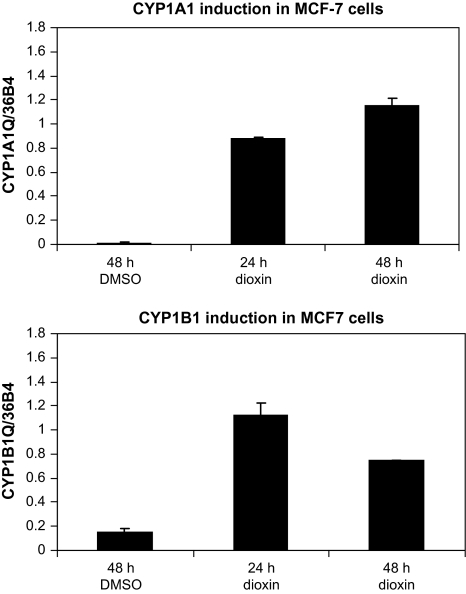
QRTPCR analysis demonstrated that both CYP1A1 and CYP1B1 were induced by 100nM dioxin in the human breast carcinoma cell line, MCF-7. This concentration of dioxin elicited maximal induction of both CYPs (data not shown). A 24-h treatment with dioxin induced CYP1A1 mRNA an average of 370-fold (range: 30- to 1890-fold) in six experiments, whereas CYP1B1 mRNA was considerably less inducible (average of 12-fold, range of 4- to 30-fold), reflecting the significant basal expression of this mRNA in the absence of dioxin. Representative data for the induction of CYP1A1 and CYP1B1 are shown in Figure 1.
FIG. 1.
Dioxin induction of CYP1A1and CYP1B1. MCF-7 cells were treated with 100nM dioxin for 24 or 48 h or dimethyl sulfoxide vehicle for 48 h. Total RNA was isolated, and the relative quantities of CYP1A1and CYP1B1mRNAs were normalized to that of the constitutively expressed 36b4 ribosomal subunit by QRTPCR.
Dioxin-Induced Cofactor Recruitment to CYP1A1 and CYP1B1
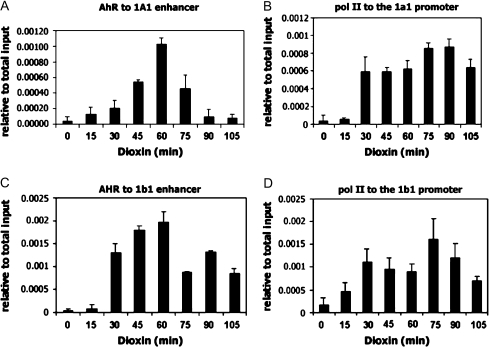
The ChIP assay was employed to characterize dioxin-induced coactivator recruitment over the CYP1A1 and CYP1B1 enhancers (located approximately 1 kb upstream of their transcriptional start sites) and proximal promoter regions (located just upstream of their transcriptional start sites). Dioxin induced a rapid association of AHR to the enhancer regions of both genes. Binding of the AHR to the enhancer regions reached maximal levels at approximately 60 min and then declined (Figs. 2A and 2C). Unlike some other published reports which describe a cycling of the AHR on and off the CYP1A1 gene with other AHR ligands (Hestermann and Brown, 2003), we observed no cycling in MCF-7 cells after extended periods of dioxin treatment (data not shown). We did not detect AHR binding to the proximal promoter regions of either gene, clearly demonstrating specificity of binding of AHR to the enhancer regions of these genes. Furthermore, we demonstrate dioxin-dependent recruitment of RNA pol II to the promoter regions of both genes (Figs. 2B and 2D) but were not able to detect pol II at the enhancer regions of either gene (data not shown).
FIG. 2.
Dioxin-induced transcription factor recruitment to CYP1A1and CYP1B1. MCF-7 cells were treated with 100nM dioxin for the indicated time points and subjected to ChIP analysis using α-AHR or α-pol II antibodies. Dioxin-dependent association of AHR and pol II to the regulatory regions of CYP1A1(A and B) and CYP1B1(C and D) were quantitated by real-time PCR.
We found dioxin to stimulate the recruitment of several coactivators to CYP1A1 and CYP1B1. Our ChIP assays demonstrate a dioxin-dependent recruitment of members of the p160 family of coactivator proteins (SRC-1 and SRC-2), the p300 coactivator protein, as well as BRG-1, an ATPase-containing subunit of the SWI-SNF complex, to the CYP1A1 and CYP1B1 genes (Figs. 3A–D). In contrast to the situation with AHR and pol II, dioxin induced recruitment of the coactivators to both the enhancers and proximal promoters of each gene. This may be explained by the fact that AHR and pol II contact DNA directly and are therefore efficiently cross-linked at their sites of DNA binding, whereas coactivators associate with chromatin principally via protein-protein interactions. Under our cross-linking conditions, the coactivators were probably efficiently cross-linked, either directly or indirectly, to other proteins that bind directly to DNA at the enhancers and proximal promoters of the CYP1A1 and CYP1B1 genes. Of interest, dioxin induced a considerably greater degree of association of BRG-1 over the CYP1A1 gene than over the CYP1B1 gene, whereas the effects of dioxin on binding of the other coactivators were not so different for the two genes. This observation indicates that BRG-1 exhibits a more marked differential recruitment to the CYP1A1 gene compared with the CYP1B1 gene after dioxin treatment.
Knockdown of Individual Coactivators
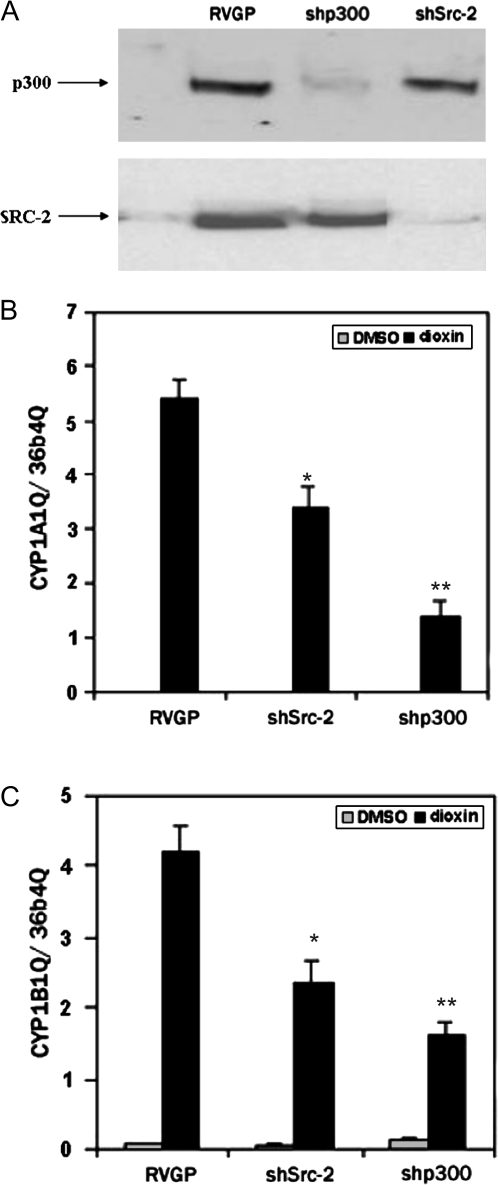
RNAi was used to identify coactivator proteins that are essential for CYP induction elicited by dioxin. MCF-7 cells were infected with retroviral vectors expressing short hairpin siRNAs designed against the SRC-2 and p300 coactivator proteins, and cell lines stably expressing the vectors were isolated. Western blot analysis confirmed the specificity and effectiveness of the shRNA vectors. Cells infected with shp300 showed marked reduction in the p300 protein while cells infected with shSRC-2 show marked reduction in levels of the SRC-2 protein (Fig. 4A). We found that the loss of either p300 or SRC-2 adversely affected induction of both CYP1A1 (Fig. 4B) and CYP1B1 (Fig. 4C).
FIG. 4.
p300 and SRC-2 are required for maximal induction of CYP1A1and CYP1B1by dioxin. MCF-7 cells were infected with a retrovirus containing a short hairpin siRNA directed against p300 or SRC-2 or with the parental virus, RVGP (negative control). Whole-cell extracts from MCF-7 cells were probed with α-p300 or α-SRC-2 antibodies (A). To quantify the expression of CYP1A1and CYP1B1, stable knockdown cells were treated with 100nM dioxin for 24 h. Total RNA was isolated, reverse transcribed, and subjected to real-time PCR (B). The mRNA levels of both genes were normalized to that of the constitutively expressed ribosomal subunit, 36b4. Student t-test was performed to evaluate statistical significance from the RVGP control. *p < 0.05; **p < 0.001.
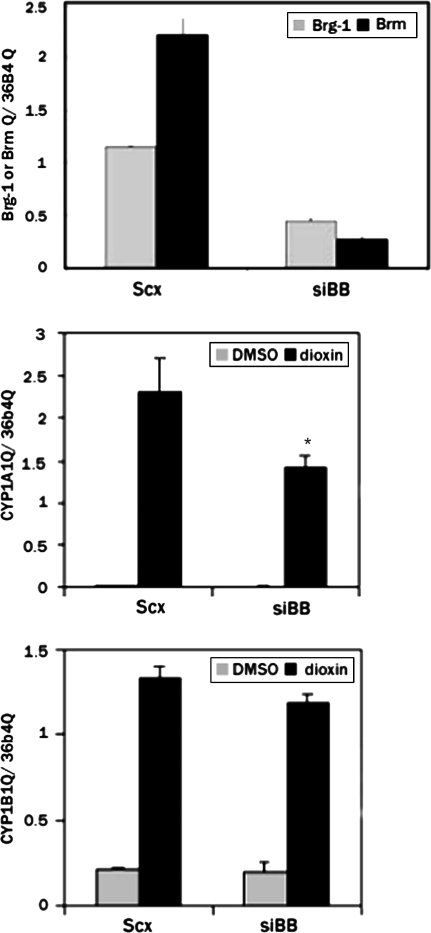
An siRNA oligonucleotide directed against both the BRG-1 and BRM genes (siBB) was transfected into MCF-7 cells or MCF-7 cells stably expressing shp300 or shSRC-2. QRTPCR revealed a 61% reduction in the BRG-1 mRNA and an 88% reduction in BRM mRNA (Fig. 5A). We previously demonstrated that this oligonucleotide elicits equivalent dimunitions in the amounts of the BRG-1 and BRM proteins in human cells (Wang et al., 2004a). Here, we observed that the loss of BRG-1/BRM has an adverse effect on CYP1A1 induction after dioxin treatment, while displaying very little effect on CYP1B1 induction (Fig. 5B).
FIG. 5.
Gene-specific requirement for BRG-1/BRM. MCF-7 cells were treated with 100nM siBB for 24 h followed by a 24-h treatment with 100nM dioxin. RNA was isolated and subjected to reverse transcription and real-time PCR. The relative amount of the BRG-1 and BRM mRNAs were corrected against the levels of the constitutively expressed ribosomal subunit, 36B4 (A). CYP1A1(B) and CYP1B1(C) mRNAs were quantitated in the transfected cells. *p < 0.05 relative to the scrambled siRNA (scx) control.
Effect of Combined Coactivator Knockdown on Dioxin Induction of CYP1A1 and CYP1B1
When siBB was introduced into cells that were stably knocked down for endogenous p300, an additive or even synergistic adverse effect on CYP1A1 dioxin induction was observed (Fig. 6). In contrast, when SRC-2 and BRM/BRG-1 were depleted together, the effect on induction of CYP1A1 was no greater than when each of SRC-2 and BRM/BRG-1 were depleted on their own.
DISCUSSION
Transcriptional activation in mammalian cells is a highly organized event that involves interaction between transcription factors, coactivators, corepressors, and chromatin. In vitro, general transcription factors such as the TATA-binding protein and the CBP along with the core RNA polymerase II complex are able to activate transcription of “naked” DNA containing a TATA sequence (Li et al., 1994). However, in response to stimulators that induce gene-specific activation, additional coactivator proteins are needed for active gene transcription, particularly on a chromatin template. The activation of xenobiotic metabolizing genes by the AHRC is hindered by the presence of compacted chromatin in the form of nucleosomes over regulatory regions of target DNA. In order to overcome this challenge, the AHRC recruits coactivator proteins, which aid in the remodeling or covalent modification of chromatin.
Utilization of the MCF-7 cell line provided us with the opportunity to compare the role of transcriptional coactivators in the dioxin induction of CYP1A1 and CYP1B1, which are both induced in this cell line, but to considerably different degrees. Furthermore, utilization of this cell line allowed us to extend our studies to the human isoforms. We found that in response to dioxin, AHR and pol II are specifically recruited to the enhancer and promoter regions, respectively, of these genes. Recruitment of AHR to the promoters and pol II to the enhancers of CYP1A1 and CYP1B1 was only observed at background levels (data not shown). We also made this observation in our studies on the mouse Cyp1a1 gene in Hepa-1 cells (Wang et al., 2004b) and is in contrast to one report which demonstrated binding of the AHR to the promoter region of CYP1A1 after dioxin treatment of MCF-7 cells (Matthews et al., 2005). We believe that this difference may be related to variations in experimental details in the ChIP assay, such as cross-linking efficiency, sonication efficiency, or other factors.
We previously demonstrated that SRC-1, SRC-2, p300, and BRG-1 are recruited in a dioxin-dependent fashion to the mouse Cyp1a1 enhancer, but not detectably to its promoter, in the hepatoma cell line, Hepa-1. We now show that these same recruitments also occur on the CYP1A1 gene in human cells. This result is consistent with a previous report demonstrating that the first three of the above coactivators are recruited to the CYP1A1 gene in MCF-7 cells, although no distinction was made between binding at the enhancer region or the proximal promoter in these previous studies (Matthews et al., 2005). We now also demonstrate that the same coactivators are recruited to the human CYP1B1 gene in MCF-7 cells but that dioxin has a markedly lesser effect on BRG-1 association with the CYP1B1 gene relative to the CYP1A1 gene. By using RNAi, we demonstrate that SRC-2, p300, and BRG-1 are all required for maximal induction of human CYP1A1, consistent with our previous observations for the mouse Cyp1a1 (and also in the case of BRG-1, for human CYP1A1) using different experimental approaches (Beischlag et al., 2002; Wang and Hankinson, 2002; Wang et al., 2004a). We also show here that SRC-2 and p300 are required for maximal induction of CYP1B1 in the human MCF-7 cell line. Of interest, however, we found that although BRG-1 is required for maximal induction of CYP1A1, it is apparently not required for CYP1B1 induction. BRG-1 is a component of the SWI-SNF complex, which functions to displace or remove nucleosomes in chromatin, and we speculate that the differential requirement for BRG-1 may be a consequence of different nucleosomal configurations over the CYP1A1 and CYP1B1 genes in MCF-7 cells and, furthermore, may be related to their different degrees of induction by dioxin.
Interestingly, the combined inhibition of SRC-2 and BRM/BRG-1 did not diminish induction of CYP1A1 to any greater degree than inhibition of SRC-2 or BRM/BRG-1 alone. These two types of coactivators act via histone acetylation and noncovalent histone displacement/translation, respectively, and therefore have very different mechanisms of action. A possible explanation for the nonadditive effects of their depletion on CYP1A1 induction is that the recruitment of each type of activator is mutually inclusive. It should also be noted that the SRC-1 and SRC-3 coactivators are closely related in structure and function to SRC-2, and it is possible that SRC-1 and/or SRC-3 may act redundantly with SRC-2 during induction of CYP1A1 in MCF-7 cells. If this is the case, the deleterious effect of diminishing SRC-2 on CYP1A1 induction may be due principally to the resultant reduction in BRM/BRG-1 recruitment to the gene. In contrast, inhibiting the expression of the p300 and BRM/BRG-1 together reduced CYP1A1 induction much more than inhibiting each alone, nearly eliminating CYP1A1 induction altogether. These last results suggest that recruitment of p300 and BRM/BRG-1 occur independently of each other.
Our observations, focusing on the roles of the endogenous coactivator proteins in regulation of the human CYP1A1 and CYP1B1 genes in their natural chromosomal settings, therefore provide new insights into the functional roles of these coactivators in the induction by dioxin of two key enzymes involved in chemical carcinogenesis.
FUNDING
National Institutes of Health (R01CA28868 [to O.H.]; an underrepresented minority supplement to R01CA28868 [to R.T.T.]); and a University of California Toxic Substances Research and Teaching Program fellowship (to R.T.T.).
References
- Bauer UM, Daujat S, Nielsen SJ, Nightingale K, Kouzarides T. Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep. 2002;3:39–44. doi: 10.1093/embo-reports/kvf013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beischlag TV, Taylor RT, Rose DW, Yoon D, Chen Y, Lee WH, Rosenfeld MG, Hankinson O. Recruitment of thyroid hormone receptor/retinoblastoma-interacting protein 230 by the aryl hydrocarbon receptor nuclear translocator is required for the transcriptional response to both dioxin and hypoxia. J. Biol. Chem. 2004;279:54620–54628. doi: 10.1074/jbc.M410456200. [DOI] [PubMed] [Google Scholar]
- Beischlag TV, Wang S, Rose DW, Torchia J, Reisz-Porszasz S, Muhammad K, Nelson WE, Probst MR, Rosenfeld MG, Hankinson O. Recruitment of the NCoA/SRC-1/p160 family of transcriptional coactivators by the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator complex. Mol. Cell. Biol. 2002;22:4319–4333. doi: 10.1128/MCB.22.12.4319-4333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Beischlag TV, Kim JH, Perdew GH, Stallcup MR. Role of GAC63 in transcriptional activation mediated by the aryl hydrocarbon receptor. J. Biol. Chem. 2006;281:12242–12247. doi: 10.1074/jbc.M512537200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez FJ. Molecular genetics of the P-450 superfamily. Pharmacol. Ther. 1990;45:1–38. doi: 10.1016/0163-7258(90)90006-n. [DOI] [PubMed] [Google Scholar]
- Hankinson O. The aryl hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- Hankinson O. Role of coactivators in transcriptional activation by the aryl hydrocarbon receptor. Arch. Biochem. Biophys. 2005;433:379–386. doi: 10.1016/j.abb.2004.09.031. [DOI] [PubMed] [Google Scholar]
- Hestermann EV, Brown M. Agonist and chemopreventative ligands induce differential transcriptional cofactor recruitment by aryl hydrocarbon receptor. Mol. Cell. Biol. 2003;23:7920–7925. doi: 10.1128/MCB.23.21.7920-7925.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu EL, Yoon D, Choi HH, Wang F, Taylor RT, Chen N, Zhang R, Hankinson O. A proposed mechanism for the protective effect of dioxin against breast cancer. Toxicol. Sci. 2007;98:436–444. doi: 10.1093/toxsci/kfm125. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77(4):599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Numayama-Tsuruta K, Sogawa K, Fujii-Kuriyama Y. CBP/p300 functions as a possible transcriptional coactivator of Ah receptor nuclear translocator (Arnt) J. Biochem. 1997;122:703–710. doi: 10.1093/oxfordjournals.jbchem.a021812. [DOI] [PubMed] [Google Scholar]
- Kress S, Reichert J, Schwarz M. Functional analysis of the human cytochrome P4501A1 (CYP1A1) gene enhancer. Eur. J. Biochem. 1998;258(2):803–812. doi: 10.1046/j.1432-1327.1998.2580803.x. [DOI] [PubMed] [Google Scholar]
- Kumar MB, Perdew GH. Nuclear receptor coactivator SRC-1 interacts with the Q-rich subdomain of the AhR and modulates its transactivation potential. Gene Expr. 1999;8:273–286. [PMC free article] [PubMed] [Google Scholar]
- Kumar MB, Tarpey RW, Perdew GH. Differential recruitment of coactivator RIP140 by Ah and estrogen receptors. Absence of a role for LXXLL motifs. J. Biol. Chem. 1999;274:22155–22164. doi: 10.1074/jbc.274.32.22155. [DOI] [PubMed] [Google Scholar]
- Li Y, Flanagan PM, Tschochner H, Kornberg RD. RNA polymerase II initiation factor interactions and transcription start site selection. Science. 1994;263(5148):805–807. doi: 10.1126/science.8303296. [DOI] [PubMed] [Google Scholar]
- Matthews J, Wihlen B, Thomsen J, Gustafsson JA. Aryl hydrocarbon receptor-mediated transcription: Ligand-dependent recruitment of estrogen receptor alpha to 2,3,7,8-tetrachlorodibenzo-p-dioxin-responsive promoters. Mol. Cell. Biol. 2005;25(13):5317–5328. doi: 10.1128/MCB.25.13.5317-5328.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisterernst M, Roeder RG. Family of proteins that interact with TFIID and regulate promoter activity. Cell. 1991;67(3):557–567. doi: 10.1016/0092-8674(91)90530-c. [DOI] [PubMed] [Google Scholar]
- Meyer BK, Pray-Grant MG, Vanden Heuvel JP, Perdew GH. Hepatitis B virus X-associated protein 2 is a subunit of the unliganded aryl hydrocarbon receptor core complex and exhibits transcriptional enhancer activity. Mol. Cell. Biol. 1998;18(2):978–988. doi: 10.1128/mcb.18.2.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okino ST, Whitlock JP., Jr Dioxin induces localized, graded changes in chromatin structure: Implications for Cyp1A1 gene transcription. Mol. Cell. Biol. 1995;15(7):3714–3721. doi: 10.1128/mcb.15.7.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrulis JR, Kusnadi A, Ramadoss P, Hollingshead B, Perdew GH. The hsp 90 co-chaperone XAP2 alters importin {beta} recognition of the Ah receptor's bipartite nuclear localization signal and represses transcriptional activity. J. Biol. Chem. 2002;278:2677–2685. doi: 10.1074/jbc.M209331200. [DOI] [PubMed] [Google Scholar]
- Rivera SP, Saarikoski ST, Hankinson O. Identification of a novel dioxin-inducible cytochrome P450. Mol. Pharmacol. 2002;61(2):255–259. doi: 10.1124/mol.61.2.255. [DOI] [PubMed] [Google Scholar]
- Rowlands JC, McEwan IJ, Gustafsson JA. Trans-activation by the human aryl hydrocarbon receptor and aryl hydrocarbon receptor nuclear translocator proteins: Direct interactions with basal transcription factors. Mol. Pharmacol. 1996;50:538–548. [PubMed] [Google Scholar]
- Shimada T, Hayes CL, Yamazaki H, Amin S, Hecht SS, Guengerich FP, Sutter TR. Activation of chemically diverse procarcinogens by human cytochrome P-450 1B1. Cancer Res. 1996;56(13):2979–2984. [PubMed] [Google Scholar]
- Shimizu Y, Nakatsuru Y, Ichinose M, Takahashi Y, Kume H, Mimura J, Fujii-Kuriyama Y, Ishikawa T. Benzo[a]pyrene carcinogenicity is lost in mice lacking the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. U.S.A. 2000;97(2):779–782. doi: 10.1073/pnas.97.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter TR, Tang YM, Hayes CL, Wo YY, Jabs EW, Li X, Yin H, Cody CW, Greenlee WF. Complete cDNA sequence of a human dioxin-inducible mRNA identifies a new gene subfamily of cytochrome P450 that maps to chromosome 2. J. Biol. Chem. 1994;269:13092–13099. [PubMed] [Google Scholar]
- Swanson HI, Yang JH. The aryl hydrocarbon receptor interacts with transcription factor IIB. Mol. Pharmacol. 1998;54:671–677. [PubMed] [Google Scholar]
- Tojo M, Matsuzaki K, Minami T, Honda Y, Yasuda H, Chiba T, Saya H, Fujii-Kuriyama Y, Nakao M. The aryl hydrocarbon receptor nuclear transporter is modulated by the SUMO-1 conjugation system. J. Biol. Chem. 2002;277:46576–46585. doi: 10.1074/jbc.M205987200. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Nakajima M, Yokoi T. Critical enhancer region to which AhR/ARNT and Sp1 bind in the human CYP1B1 gene. J. Biochem. 2003;133:583–592. doi: 10.1093/jb/mvg075. [DOI] [PubMed] [Google Scholar]
- Uno S, Dragin N, Miller ML, Dalton TP, Gonzalez FJ, Nebert DW. Basal and inducible CYP1 mRNA quantitation and protein localization throughout the mouse gastrointestinal tract. Free Radic. Biol. Med. 2008;44:570–583. doi: 10.1016/j.freeradbiomed.2007.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Zhang R, Beischlag TV, Muchardt C, Yaniv M, Hankinson O. Roles of Brahma and Brahma/SWI2-related gene 1 in hypoxic induction of the erythropoietin gene. J. Biol. Chem. 2004a;279:46733–46741. doi: 10.1074/jbc.M409002200. [DOI] [PubMed] [Google Scholar]
- Wang S, Ge K, Roeder RG, Hankinson O. Role of mediator in transcriptional activation by the aryl hydrocarbon receptor. J. Biol. Chem. 2004b;279:13593–13600. doi: 10.1074/jbc.M312274200. [DOI] [PubMed] [Google Scholar]
- Wang S, Hankinson O. Functional involvement of the Brahma/SWI2-related gene 1 protein in cytochrome P4501A1 transcription mediated by the aryl hydrocarbon receptor complex. J. Biol. Chem. 2002;277:11821–11827. doi: 10.1074/jbc.M110122200. [DOI] [PubMed] [Google Scholar]
- Wei YD, Tepperman K, Huang MY, Sartor MA, Puga A. Chromium inhibits transcription from polycyclic aromatic hydrocarbon-inducible promoters by blocking the release of histone deacetylase and preventing the binding of p300 to chromatin. J. Biol. Chem. 2004;279:4110–4119. doi: 10.1074/jbc.M310800200. [DOI] [PubMed] [Google Scholar]
- Whitlock JP, Jr, Chichester CH, Bedgood RM, Okino ST, Ko HP, Ma Q, Dong L, Li H, Clarke-Katzenberg R. Induction of drug-metabolizing enzymes by dioxin. Drug Metab. Rev. 1997;29(4):1107–1127. doi: 10.3109/03602539709002245. [DOI] [PubMed] [Google Scholar]
- Zhang J, Watson AJ, Probst MR, Minehart E, Hankinson O. Basis for the loss of aryl hydrocarbon receptor gene expression in clones of a mouse hepatoma cell line. Mol. Pharmacol. 1996;50(6):1454–1462. [PubMed] [Google Scholar]