Abstract
Various public databases provide Arabidopsis gene information via the internet. It is useful to abstract information obtained from such databases. We have developed the KAGIANA tool, which allows a user to retrieve summary information obtained from selective databases and to access pages for a gene of interest in those databases. The tool is based on Microsoft Excel and provides several macro programs for gene expression analyses. It can assist plant biologists in accessing omics information for plant biology. The KAGIANA tool is freely available at http://pmnedo.kazusa.or.jp/kagiana/.
Keywords: KAnnotation, Arabidopsis, Database, Gene expression, Omics
Since the completion of the genome sequence of the model plant Arabidopsis thaliana (Arabidopsis Genome Initiative 2000), advances in genome and gene expression analysis have resulted in a vast number of data sets generated for Arabidopsis. Data sets of the Arabidopsis genome sequence are available at GenBank (Benson et al. 2008; http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Nucleotide) and DDBJ (Sugawara et al. 2008; http://www.ddbj.nig.ac.jp/). In Arabidopsis, the sequence is separated into five chromosomes, which have 33,282 gene loci according to TAIR8 (Swarbreck et al. 2008; http://www.arabidopsis.org/). Amino acid sequences of proteins encoded by genes have been analyzed for various purposes. On the basis of localization signals included in such sequences, subcellular localization of proteins has been predicted using tools such as WoLF PSORT (Horton et al. 2007; http://wolfpsort.org/) and TargetP (Emanuelsson et al. 2007; http://www.cbs.dtu.dk/services/TargetP/). Domain structures, which show functional properties in proteins, have been predicted on the basis of amino acid sequences using tools such as SCOP (Andreeva et al. 2008; http://scop.berkeley.edu/), and can be found in databases such as InterPro (Mulder et al. 2007; http://www.ebi.ac.uk/interpro/). Analysis of transmembrane domains has been performed using tools such as TMHMM (Moller et al. 2001; http://www.cbs.dtu.dk/services/TMHMM/) and that of hydropathy can be found in databases such as SUBA (Heazlewood et al. 2007; http://www.plantenergy.uwa.edu.au/suba2/). Data sets of gene expression are available from databases such as the Gene Expression Omnibus (GEO) database (Barrett et al. 2006; http://www.ncbi.nlm.nih.gov/geo/). Several databases such as ATTED-II (Obayashi et al. 2007; http://atted.jp/) and Genevestigator (Zimmermann et al. 2004; https//www.genevestigator.ethz.ch/) provide a function to perform gene expression analysis. Using a vast number of gene expression data sets, approaches for detecting co-expressed genes, such as the ARACNE tool (Margolin et al. 2006; http://amdec-bioinfo.cu-genome.org/html/ARACNE.htm), the average clustering coefficient index (Gupta et al. 2006) and the Confeito algorithm (http://pmnedo.kazusa.or.jp/kagiana/coexprocess/) have been developed. On the basis of these analyses, molecular function, subcellular localization and biological processes of genes have finally been consistently assigned to ‘molecular function’, ‘cellular component’ and ‘biological process’, respectively, the three aspects of the Gene Ontology (GO) terminology (Gene Ontology Consortium 2008; http://geneontology.org/).
To obtain genomic and transcriptomic information on genes of interest, a user can visit these databases and access these tools via the internet or download them for personal use. However, to retrieve the information, users generally require knowledge of the omics information published in the databases; for example, how to select an adequate website and how to set an adequate threshold value such as the gene-to-gene correlation coefficient for acquiring data of interest in the website. For biological users, unfamiliar with omics analyses such as genomics and transcriptomics, it is useful to have access to abstracted gene information from such databases and analyses and to use quick links to these databases.
We have developed the KAGIANA (Kazusa Arabidopsis Gene Information And Network Analysis) tool to summarize various Arabidopsis omics analyses from the above-mentioned databases and tools, and to provide links to pages for genes of interest in the databases. The tool is based on Microsoft Excel (version 2003 or higher) and thus requires only enough skill for basic Excel operation. The implementation of this tool is verified using Windows XP or higher for PC, and OS X or higher for Macintosh. The macro programs of the tools are available only for Windows users as of November 2008. Our goal is to assist plant biologists in accessing information from omics analyses so that they can incorporate it into their plant biology research.
The KAGIANA tool is downloadable as a ZIP-format file at http://pmnedo.kazusa.or.jp/kagiana/. The KAGIANA tool is formatted as a Microsoft Excel workbook file, composed of five worksheets [one database sheet (‘Data20080524’), two readme sheets (‘ReadMe_1st’ and ‘ReadMe_Tools’) and two retrieval sheets (‘Selected_Link’ and ‘Selected_GO’)] and one macro program (‘Tools’) comprising four analysis tools (‘Confeito’, ‘GX bar chart’, ‘GO pie chart’ and ‘ATTED chart’). In KAGIANA, AGI codes (e.g. At1g01010) are used for the retrieval and performance of the tool.
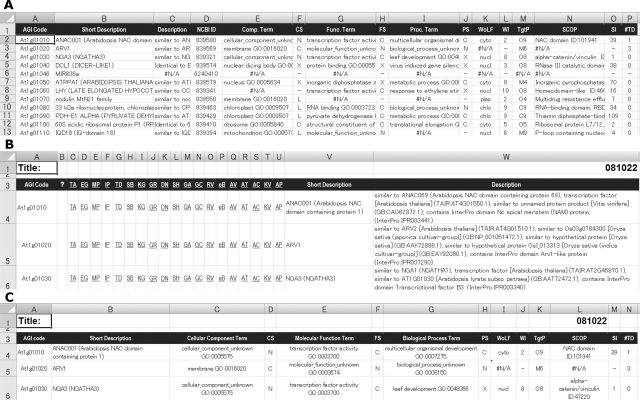
The database sheet is composed of the following information for 33,362 loci (Fig. 1A), which was obtained from the TAIR database. First, the A to D columns represent AGI codes, a short description, description, and identifiers for NCBI, respectively. Secondly, the E to J columns display representative GO terms, which certainly accompany the evidence codes, and their Evidence Code categories, which are abbreviated as ‘X’ (experimental) for EXP, IDA, IPI, IMP, IGI and IEP; ‘S’ (statement) for TAS and IC; ‘C’ (computational) for ISS, ISO, ISA, ISM, IGC and RCA; ‘L’ (electronic) for IEA; and ‘N’ (not available) for NAS and ND, in the three aspects of GO terminology, i.e. cellular component (the E and F columns), molecular function (the G and H columns) and biological process (the I and J columns), respectively. A GO term was selected as the representative term for each aspect for a gene, according to the order of Evidence Code categories, i.e. X, S, C, L and N. The following columns represent information from the analytical tools. The K and L columns represent data from WoLF PSORT, which predicts the subcellular localization of proteins, and the reliability index, whose best score is 14, respectively. The M column represents information from TargetP, which also predicts subcellular localization, and the reliability index, ranging from 0 to 9 at the maximum. The N and O columns represent that from SCOP, which predicts domains of proteins, and the reliability index, which is the negative logarithm of the actual value, respectively. The P column represents TMHMM, which predicts the number of transmembrane domains of proteins.
Fig. 1.
Composition of the KAGIANA worksheets. (A) The database sheet, including summary information of Arabidopsis genes obtained from the selected databases. (B) The sheet for hyperlinks to the selected public databases, as shown in Table 1. (C) The sheet for summary information of the selected omics analyses, i.e. GO terms and results from analyses of WoLF PSORT, TargetP, SCOP and TMHMM.
The ‘Selected_Link’ sheet provides hyperlinks to 19 selected public databases for information retrieval of genes of interest and their Short Description and Description (Fig. 1B). These hyperlinks lead a user to the pages for individual genes in the individual databases by the following steps: (i) input AGI code(s) in the A column from the A4 cell to the lower cells (e.g. input ‘At1g01010’ in the A4 cell and ‘At1g01020’ in the A5 cell); (ii) select the range of the B4 to the W4 cells; and (iii) double-click the right lower corner (a black square) to copy the equations in the fourth row into the lower rows in the same columns (e.g. copy the B4-W4 into the B5-W5). Then, a user can access a database of interest from among the C to U columns (e.g. click the T4 cell for access to the page for the query gene in the KaPPA-View tool). The tool provides access to the databases shown in Table 1. The way to use this sheet is also described in the ‘ReadMe_1st’ sheet.
Table 1.
Abstract of databases linkable from the KAGIANA tool
In the ‘Selected_GO’ sheet, a user can retrieve information on genes of interest from various omics analyses (Fig. 1C), i.e. the three GO term aspects, WoLF PSORT, TargetP, SCOP and TMHMM as mentioned above. Steps for retrieval are similar to those in the ‘Selected_Link’ sheet. The terms in the third row are the same as those in the database sheet mentioned above, and the ‘ReadMe_1st’ sheet has the explanation for such retrieval. By selecting the ‘Selected_Link’ and the ‘Selected_GO’ sheets, a user can manage to operate them simultaneously, e.g. when inputting AGI codes.
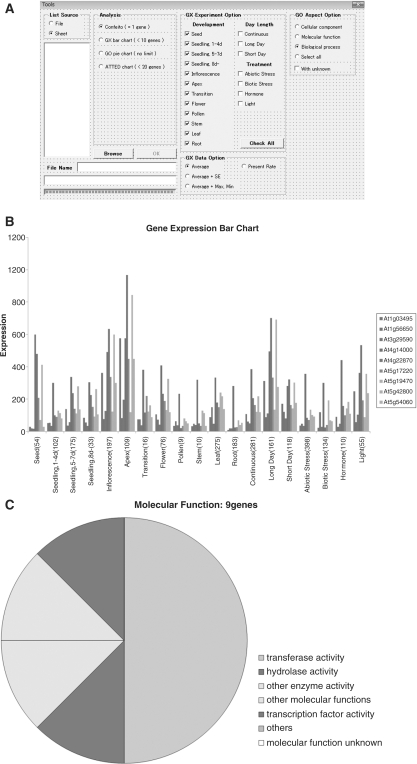
KAGIANA provides ‘Tools’ macro programs including the four analyses (Fig. 2A), i.e. including ‘Confeito’, ‘GX bar chart’, ‘GO pie chart’ and ‘ATTED chart’. The ‘Confeito’ tool allows a user to extract co-expressed genes using the Confeito algorithm on the basis of a co-expression network approach (http://pmnedo.kazusa.or.jp/kagiana/coexprocess/). The way to use the tools is described in the ‘ReadMe_Tools’ sheet. The ‘GX bar chart’ tool allows a user to depict bar charts of gene expression profiles for multiple genes of interest (Fig. 2B). Bar charts are depicted using 1,245 DNA microarray data from the AtGenExpress project, which are available at http://www.weigelworld.org/resources/microarray/AtGenExpress/. The ‘GO pie chart’ tool allows a user to depict a pie chart of the distribution of GO-SLIM terms associated with multiple genes of interest (Fig. 2C). GO-SLIM terms are available at the TAIR database. This tool counts all multiple GO-SLIM terms assigned to a gene. For this version of KAGIANA, such terms were obtained at May 2008. The ‘ATTED chart’ tool helps users download the charts of AtGenExpress gene expression profiles for individual genes from the ATTED database onto a worksheet in KAGIANA per gene.
Fig. 2.
Composition of the KAGIANA tools. (A) The window of the ‘Tools’ macro program, including analyses of ‘Confeito’, ‘GX bar chart’, ‘GO pie chart’ and ‘ATTED chart’. (B) The result from the ‘GX bar chart’ analysis. (C) The result from the ‘GO pie chart’ analysis, showing the aspect of molecular function.
Detailed steps for using these tools are described in the ‘ReadMe_Tools’ sheet in the KAGIANA workbook. Briefly, the steps are (i) click ‘Tools’ in the menu bar; (ii) select ‘Macro’ and click ‘Macros’; (iii) select ‘Tools’ in the macro box and click ‘Execute’ (open the ‘Tools’ window); (iv) select a tool in the ‘Analysis’ frame in the window; (v) input AGI codes into different lines in the textbox left of the frame; (vi) select the option frame when selecting ‘GX bar chart’ and ‘GO pie chart’ tools; and then (vii) click the ‘OK’ button if the character color on the button is black (otherwise, there is insufficient information for retrieval).
Funding
The New Energy and Industrial Technology Development (NEDO) program, part of the ‘Development of Fundamental Technologies for Controlling the Material Production Process of Plants’ project.
Glossary
Abbreviations:
- AGI
Arabidopsis Genome Initiative
- GO
Gene Ontology
- NCBI
The National Center for Biotechnology Information
- TAIR
The Arabidopsis Information Resource.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, et al. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch-Hoffmann M, Yalovsky S, et al. Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science. 2008;320:938–941. doi: 10.1126/science.1157956. [DOI] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Willhite SE, Ledoux P, Rudnev D, Evangelista C, et al. NCBI GEO: mining tens of millions of expression profiles—database and tools update. Nucleic Acids Res. 2006;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund AC, Sjölund E, Östlund G, Sonnhammer EL. InParanoid 6: eukaryotic ortholog clusters with inparalogs. Nucleic Acids Res. 2008;36:D263–D266. doi: 10.1093/nar/gkm1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S, Williams SR, Vermaas EH, Storck T, Moon K, McCollum C, et al. In vitro cloning of complex mixtures of DNA on microbeads: physical separation of differentially expressed cDNAs. Proc. Natl. Acad. Sci. USA. 2000;18:630–634. doi: 10.1073/pnas.97.4.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigon DJ, James N, Okyere J, Higgins J, Jotham J, May S. NASCArrays: a repository for microarray data generated by NASC’s transcriptomics service. Nucleic Acids Res. 2004;32:D575–D577. doi: 10.1093/nar/gkh133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium. The Gene Ontology project in 2008. Nucleic Acids Res. 2008;36:D440–D444. doi: 10.1093/nar/gkm883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Maranas CD, Albert R. Elucidation of directionality for co-expressed genes: predicting intra-operon termination sites. Bioinformatics. 2006;22:209–214. doi: 10.1093/bioinformatics/bti780. [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH. SUBA: the Arabidopsis Subcellular Database. Nucleic Acids Res. 2007;35:D213–D218. doi: 10.1093/nar/gkl863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Jaiswal P, Hebbard C, Avraham S, Buckler ES, Casstevens T, et al. Gramene: a growing plant comparative genomics resource. Nucleic Acids Res. 2008;36:D947–D953. doi: 10.1093/nar/gkm968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Dalla-Favera R, et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7:S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller S, Croning MDR, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, et al. New developments in the InterPro database. Nucleic Acids Res. 2007;35:D224–D228. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Kinoshita K, Nakai K, Shibaoka M, Hayashi S, Saeki M, et al. ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 2007;35:D863–D869. doi: 10.1093/nar/gkl783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai N, Shibata D. KaPPA-View for integrating quantitative transcriptomic and metabolomic data on plant metabolic pathway maps. J. Pestic. Sci. 2006;31:293–295. [Google Scholar]
- Sugawara H, Ogasawara O, Okubo K, Gojobori T, Tateno Y. DDBJ with new system and face. Nucleic Acids Res. 2008;36:D22–D24. doi: 10.1093/nar/gkm889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An ‘electronic fluorescent pictograph’ browser for exploring and analyzing large-scale biological data sets. PLoS ONE. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Foerster H, Tissier CP, Mueller L, Paley S, Karp PD, et al. MetaCyc and AraCyc. Metabolic pathway databases for plant research. Plant Physiol. 2005;138:27–37. doi: 10.1104/pp.105.060376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]