Abstract
Metabolomics is an ‘omics’ approach that aims to analyze all metabolites in a biological sample comprehensively. The detailed metabolite profiling of thousands of plant samples has great potential for directly elucidating plant metabolic processes. However, both a comprehensive analysis and a high throughput are difficult to achieve at the same time due to the wide diversity of metabolites in plants. Here, we have established a novel and practical metabolomics methodology for quantifying hundreds of targeted metabolites in a high-throughput manner. Multiple reaction monitoring (MRM) using tandem quadrupole mass spectrometry (TQMS), which monitors both the specific precursor ions and product ions of each metabolite, is a standard technique in targeted metabolomics, as it enables high sensitivity, reproducibility and a broad dynamic range. In this study, we optimized the MRM conditions for specific compounds by performing automated flow injection analyses with TQMS. Based on a total of 61,920 spectra for 860 authentic compounds, the MRM conditions of 497 compounds were successfully optimized. These were applied to high-throughput automated analysis of biological samples using TQMS coupled with ultra performance liquid chromatography (UPLC). By this analysis, approximately 100 metabolites were quantified in each of 14 plant accessions from Brassicaceae, Gramineae and Fabaceae. A hierarchical cluster analysis based on the metabolite accumulation patterns clearly showed differences among the plant families, and family-specific metabolites could be predicted using a batch-learning self-organizing map analysis. Thus, the automated widely targeted metabolomics approach established here should pave the way for large-scale metabolite profiling and comparative metabolomics.
Keywords: Automated, Comparative metabolomics, Liquid chromatography–mass spectrometry, Metabolite accumulation pattern, Multiple reaction monitoring, Targeted metabolomics
Introduction
Metabolomics is a novel experimental methodology categorized as an ‘omics’ approach along with genomics, transcriptomics and proteomics (Fiehn et al. 2001, Fiehn 2002, Nicholson et al. 2002, Sumner et al. 2003, Saito et al. 2008). Metabolomics is often used in combination with the other omics approaches for deeper understanding of biological processes, especially metabolism. The number of metabolites in the plant kingdom is considered to be far greater than that in the animal kingdom, and is estimated to exceed 200,000 (Fiehn et al. 2001). This is due to the great diversity of metabolic pathways that each plant species has evolved to survive under varying environmental conditions.
Metabolomics deals with diverse metabolites that differ greatly in their physical and chemical properties. The other omics approaches are used to analyze molecules with similar chemical properties (namely DNA, RNA and proteins) using standardized analytical platforms (e.g. the DNA sequencer in genomics). In contrast, metabolomics requires multiple instruments based on different analytical principles. In the past decade, chromatography-coupled mass spectrometry (MS)-based metabolomics and nuclear magnetic resonance (NMR)-based metabolomics have been developed in order to establish non-targeted analysis procedures (Werner et al. 2008a, Werner et al. 2008b). The ‘ultimate’ metabolomics, i.e. quantification of all metabolites in a biological sample, can only be achieved through the integration of data obtained using these various techniques. Non-targeted metabolomics enables us to identify the broad metabolite profiles of samples and to find novel metabolites that can be used as biomarkers (Glinski and Weckwerth 2006). However, many difficulties exist in achieving this goal, e.g. it is technically difficult and extremely time-consuming to merge all the data obtained in different formats by different instruments, and to identify the unknown metabolites (Werner et al. 2008b). Hence, the application of non-targeted metabolomics to hundreds of biological samples is not yet practical. Therefore, we need an alternative methodology for targeted but high-throughput metabolomics.
Generally, MS provides us with information on the molecular masses of compounds and the fragment patterns of compounds, depending on the ionization techniques used. Electron ionization (EI), used in gas chromatography (GC)-MS, gives rise to fragment ions derived by cleavage of a compound. GC-MS spectra are highly reproducible, and standard mass spectrum databases of GC-MS data have already been constructed (Kopka et al. 2005, Schauer et al. 2005). Electrospray ionization (ESI), used in liquid chromatography (LC)-MS, is a soft ionization method (Fenn et al. 1989) and causes much less fragmentation of compounds than GC-EI-MS. LC-ESI-MS is a highly sensitive technique that provides information on the molecular masses of compounds. To obtain structural information based on the fragmentation patterns of compounds, ESI-tandem mass spectrometry (MS/MS) is applied to LC-MS-based methods. In the case of targeted metabolomics, multiple reaction monitoring (MRM) using tandem quadrupole mass spectrometry (TQMS) enables high sensitivity, reproducibility and a broad dynamic range of analysis. During MRM, the first quadrupole (Q) transmits only an ion of specific m/z, which is then fragmented in the collision cell. The second quadrupole is set to transmit a specific product ion from among the fragments. Detection of this product is therefore diagnostic information for compound identification. MRM requires two ions (a precursor and a product ion) to generate a positive result, making it very specific and with very low background, enhancing the sensitivity of detection (Unwin et al. 2005). Because the amount of publicly available information about MRM conditions is currently very limited, we needed to determine the optimal ionization conditions for each compound used in our experiments.
Here, we describe a highly selective and sensitive procedure that utilizes ultra performance liquid chromatography (UPLC)-TQMS for widely targeted metabolomics studies. First we established a method for the automated flow injection analysis (FIA) of a library of authentic compounds. This method enabled us to obtain a huge data set of ESI-MS and ESI-MS/MS spectra, leading to optimization of MRM conditions for approximately 500 authentic compounds. We then applied MRM conditions to the analysis of biological samples using high-throughput UPLC-TQMS to obtain metabolite accumulation patterns. The strategy followed in this study is summarized in Fig. 1. In terms of comprehensiveness and throughput, the analytical system established here is, to our knowledge, the first that can be used to quantify hundreds of metabolites in thousands of biological samples. As a case study, we determined the accumulation patterns of about 100 metabolites in representative plant species of Brassicaceae, Gramineae and Fabaceae. The data sets of the ESI-MS and ESI-MS/MS spectra, MRM conditions and metabolite accumulation patterns are accessible at our website PRIMe (Platform for RIKEN Metabolomics; http://prime.psc.riken.jp/) (Akiyama et al. 2008).
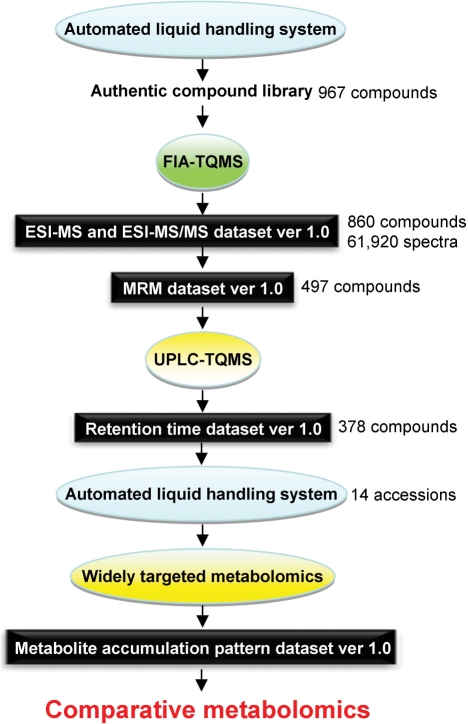
Fig. 1.
Flowchart of the strategy used to conduct a widely targeted metabolomics analysis. Automated FIA-TQMS was applied to a total of 967 authentic compounds to obtain an ESI-MS/MS data set consisting of 61,920 spectra for 860 authentic compounds. The remaining 107 compounds did not show reproducible spectra with FIA-TQMS. The MRM data set consists of optimized analytical conditions for 497 compounds, determined automatically, based on the ESI-MS/MS data set. UPLC-TQMS analysis was applied to these 497 compounds using the optimized UPLC and MRM conditions. Approximately 80% (378) of the compounds were successfully detected, to give a retention time index data set. The conditions that were established, as described above, were used in combination with an ALHS to conduct a widely targeted metabolomics study of 14 accessions representing nine plant species from three families, and the metabolite accumulation patterns were determined. All large-scale data sets mentioned here are freely available at the PRIMe website.
Results
Establishment of a 96-well-formatted authentic compound library
In metabolomics, the metabolites detected in biological samples are generally identified by reference to data from authentic compounds. To obtain the reference data for a wide range of compounds, we constructed an authentic compound library as part of our PRIMe project (Akiyama et al. 2008). The library consists of >1,000 commercially available compounds which we selected from the metabolite databases KEGG (Kanehisa and Goto 2000), AraCyc (Mueller et al. 2003) and KNApSAcK (Shinbo et al. 2006). In this study, 967 compounds were weighed and dissolved in water, methanol, ethanol, acetone or chloroform, depending on the solubility of each compound. The concentrations of the stock solutions were adjusted to 250 μM and the solutions were stored in 10 ml glass vials. Then the stock solutions were transferred from the vials to 96-well plates using an automated liquid handling system (ALHS, see Supplementary Fig. S1), because this plate format is standard for automated robotics in high-throughput analyses. The ALHS can detect the liquid level in the plate using electrical potential and pressure, and pressure monitoring also enables anti-droplet control for organic solvents (Palandra et al. 2007). All solutions except those in chloroform, which is difficult to use in LC-MS analysis, were successfully transferred.
Optimization of MRM conditions by FIA
In GC-MS analysis, the GC retention index and standardized EI-MS spectra are essential for the identification of compounds. More than 100,000 spectra obtained by EI-MS have been stored in commercial databases, such as the NIST library (http://www.nist.gov). However, in the case of ESI and atmospheric pressure chemical ionization (APCI), which are applied to LC- and capillary electrophoresis-MS, the ionization conditions have not been standardized, and only a few hundred to a thousand MS and MS/MS spectra obtained by ESI and APCI are accessible in databases, including MassBank (http://www.massbank.jp/) (Horai et al. 2008), The Human Metabolome Database (HMDB, http://www.hmdb.ca/), METLIN (http://masspec.scripps.edu/index.php) and NIST2008 (http://www.nist.gov).
In this study, we optimized the MRM conditions for polar metabolites, which can be easily applied to LC-MS analysis. To establish a large-scale ESI-MS and ESI-MS/MS data library, authentic compounds were analyzed by automated FIA using TQMS (Fig. 2A). During the automated FIA, the ESI-MS (Fig. 2B) and ESI-MS/MS (Fig. 2C) ionization patterns were measured after two injections as follows. In the first injection, the MS conditions were optimized in quadrupole 1 (Q1), which transmits only an ion of specific m/z, using fast polarity switching with six levels of cone voltage (CV). In the second injection, the second MS conditions were optimized in quadrupole 2 (Q2), which transmits a specific product ion generated by fragmentation of the transmitted ion in the collision cell, using fast polarity switching with six levels of collision voltage. The experiments were performed in triplicate for each of 967 compounds (Fig. 2D). As a result of these analyses the MRM conditions were successfully optimized for 497 compounds, based on reproducibility in conditions for both the selected precursor ion and the production in the triplicate experiments. During this procedure, >60,000 ESI-MS and ESI-MS/MS spectra were acquired and deposited in the publicly available database PRIMe. The number of spectra deposited is much greater than the numbers in NIST2008 and MassBank. NIST2008 is a commercially available database which consists of 14,802 ESI-MS spectra (as of September 2008) of 5,308 precursor ions (3,898 positive ions and 1,410 negative ions) obtained by ion trap MS. MassBank is a web-searchable database (http://www.massbank.jp) of ESI-MS and ESI-MS/MS spectra obtained by using multiple instruments [TQMS, time-of-flight (TOF) MS, QTOF MS and ion trap MS], consisting of 13,125 spectra (as of September 2008).
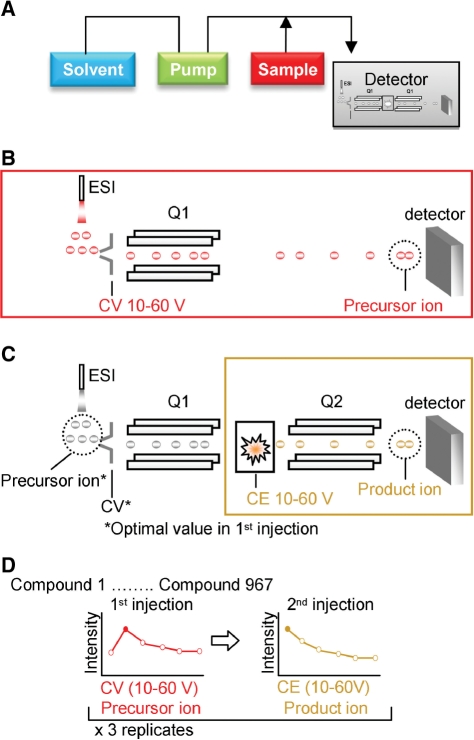
Fig. 2.
Strategy for the optimization of MRM conditions for authentic compounds. (A) Schematic diagram of automated FIA using TQMS for high-throughput acquisition of ESI-MS and ESI-MS/MS spectra. FIA was applied to the authentic compound library using samples (shown in red) in a 96-well format. (B) In the first injection, the precursor ion was detected within a range of five atomic mass units of the specified molecular masses of each compound. The optimal CV, which determines ionization in ESI-MS, was identified from among six levels in the range 10–60 eV, in the positive and negative polarity. (C) In the second injection, the optimal product ion and CE were examined using the conditions determined for the first injection. (D) Triplicate experiments were used to determine optimal MRM conditions for 967 compounds. Q1, quadrupole 1; Q2, quadrupole 2.
Optimization of UPLC for widely targeted metabolomics
For high-throughput analyses of biological samples, we chose UPLC-TQMS. In a novel analytical UPLC platform, smaller particles (<2 μm; called sub-2-μm particles) packed in the column have markedly improved flow speed, resolution and sensitivity compared with conventional LC particles (5 μm) (Citova et al. 2007). We determined the best gradient conditions for UPLC using acetonitrile and water with 0.1% formic acid, which are representative of solvents used in LC-MS analysis. Since we were already familiar with the analysis of glucosinolates by UPLC-QTOFMS (Hirai et al. 2007), we optimized the UPLC gradient program with these solvents using 12 selected compounds of methionine-derived glucosinolates with different side chains. We chose the gradient program by which these glucosinolates were distributed over a wide range of retention times (0.3–1.5 min) among the MRM data recording times (0–2.1 min) (see Materials and Methods). The 497 compounds for which MRM conditions were determined were analyzed by UPLC-TQMS under optimized gradient conditions. A total of 390 compounds were successfully detected from the 497 independent injections before column conditioning steps (2.2–3.0 min) in the gradient cycle. Quantification limits were determined based on signal-to-noise ratios (S/N ratios), and 378 compounds showed quantitative S/N ratios >10.
High-throughput analysis of metabolite accumulation patterns
As a case study of high-throughput, widely targeted metabolomics, we chose to analyze dry matter such as mature seeds due to their ease of handling compared with fresh samples such as leaves. Almost all of the steps of sample preparation after sample quenching and metabolite extraction were successfully automated by using the ALHS (Supplementary Fig. S1). In this case, the MS conditions [CV, collision energy (CE) and polarity, see Fig. 2] were set to detect five compounds with similar MRM conditions in each run of 3 min per cycle, i.e. 390 compounds could be detected in 78 runs. The combination of automated sample preparation and widely targeted UPLC-TQMS analysis was applied to seeds and seed coats of 14 accessions derived from nine species of Brassicaceae, Fabaceae and Gramineae (see Materials and Methods for a complete list of the accessions and species analyzed). A data matrix of metabolite accumulation patterns in the 14 samples was acquired by reference to the database of authentic compounds described above.
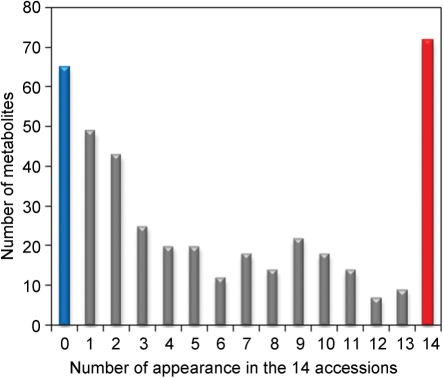
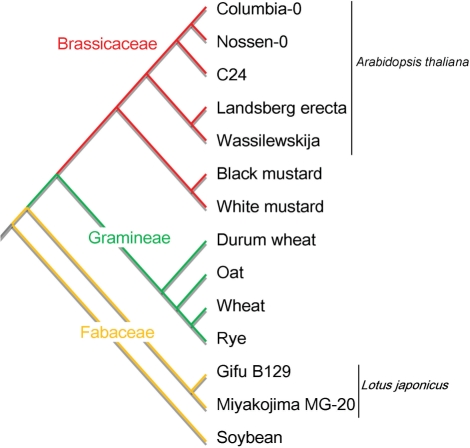
The number of detected metabolites was determined for each of the 14 plant samples (Fig. 3). A total of 343 metabolites were detected in at least one of the samples, and 18% of the 378 compounds detectable in UPLC-TQMS analysis (see Fig. 1) were identified in all of the plants analyzed (Fig. 3). For an overview, the detected metabolites and detectable authentic compounds were superimposed on a map of metabolic pathways using the KEGG Atlas (Okuda et al. 2008) (Supplementary Fig. S2). They were broadly distributed across the metabolic map. To evaluate the ability of this analytical method to detect and quantify metabolites in biological samples, we then classified the 14 plant accessions based on accumulation patterns of detected metabolites whose S/N ratios were >30. Classification was conducted by hierarchical clustering analysis and visualized as a dendrogram (Fig. 4). Fourteen plant accessions were classified as Brassicaceae, Gramineae or Fabaceae, indicating that this analysis could be used to determine metabolite accumulation patterns specific to plant families.
Fig. 3.
Histogram showing the frequencies of metabolites detected in 14 plant accessions by widely targeted metabolomics. The vertical axis indicates the number of metabolites that were detected in a given number of plant accessions, as shown on the horizontal axis. The red bar indicates the number of metabolites detected in all accessions, while the blue bar indicates the number of compounds that were detected in none of the 14 accessions.
Fig. 4.
Hierarchical clustering of plant accessions and species based on their metabolite accumulation patterns. The dendrogram shows the cluster relationships among the 14 accessions representing nine species from the families Brassicaceae, Gramineae and Fabaceae (see Materials and Methods for further details). The families are colored red, green and yellow.
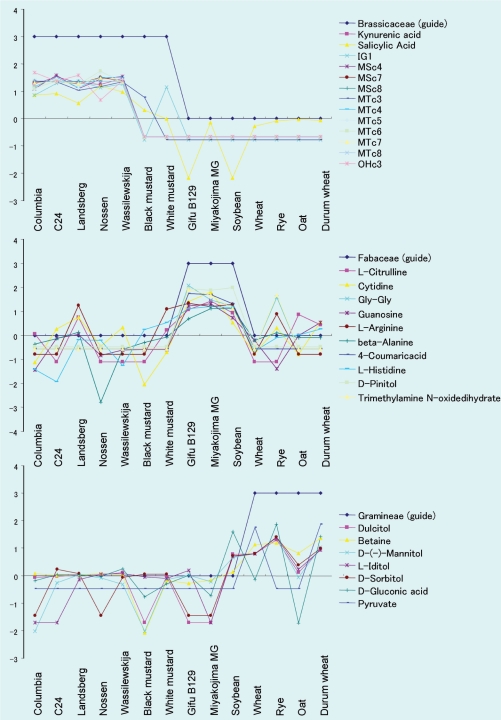
The data generated from actual detected metabolites were combined with fictitious, model metabolite data representing three families, and metabolites were classified in a batch-learning self-organizing map (BL-SOM) (Fig. 5, see Materials and Methods). This analysis was used to search for family-specific metabolites, and classified the actual and fictitious metabolites into 48 classes according to their accumulation patterns in the 14 plant accessions (data not shown). The fictitious metabolites were classified into one of three classes (Fig. 5). Glucosinolates, which are well-known Brassicaceae-specific metabolites, were correctly classified with the model data for Brassicaceae-specific metabolites (Fig. 5, top panel), indicating that this analysis can be used in at least some cases to predict family-specific metabolites.
Fig. 5.
Prediction of family-specific metabolites by BL-SOM analysis. Three fictitious model data sets, Brassicaceae (guide), Gramineae (guide) and Fabaceae (guide), were combined with actual metabolite accumulation data, and metabolites were classified by BL-SOM based on the accumulation patterns in 14 plant accessions. The actual metabolites that were grouped with the respective model data are shown as putative family-specific metabolites of Brassicaceae (upper), Gramineae (middle) and Fabaceae (lower). Vertical values represent normalized peak area values in each accession (see Materials and Methods). IG1, indol-3-ylmethyl glucosinolate; MSc4, 4-methylsulfinylbutyl glucosinolate; MSc7, 7-methylsulfinylheptyl glucosinolate; MSc8, 8-methylsulfinyloctyl glucosinolate; MTc3, 3-methylthiopropyl glucosinolate; MTc4, 4-methylthiobutyl glucosinolate; MTc5, 5-methylthiopentyl glucosinolate; MTc6, 6-methylthiohexyl glucosinolate; MTc7, 7-methylthioheptyl glucosinolate; MTc8, 8-methylthiooctyl glucosinolate; OHc3, 3-hydroxypropyl glucosinolate.
Data storage at the PRIMe website
In order to create a powerful research infrastructure for practical metabolomics, a large-scale database, consisting of ESI-MS and ESI-MS/MS spectra, MRM conditions, UPLC-TQMS retention time indices and metabolite accumulation patterns in representative plant species, was created. The database can be used for widely targeted metabolomics and is available to the public at the PRIMe website. The download page is linked to the top page of PRIMe (http://prime.psc.riken.jp/).
Discussion
Comparison between metabolomics and transcriptomics
Here we introduce an automated high-throughput methodology for widely targeted metabolomics as an alternative to non-targeted metabolomics, which includes time-consuming steps, and therefore cannot be applied to large numbers of biological samples. The difference between non-targeted metabolomics and widely targeted metabolomics is analogous to the difference between a whole genome array and an expressed sequence tag-based custom array in transcriptomics. Needless to say, the custom array often works effectively for many applications. Likewise, widely targeted analyses based on available authentic compounds can provide the information needed to elucidate whole biological processes (DellaPenna and Last 2008, Lu et al. 2008a, Lu et al. 2008b).
Significance of the large-scale ESI-MS and ESI-MS/MS spectrum data sets
We have developed an automated procedure to determine optimal MRM conditions using FIA-TQMS. This means that if thousands of authentic compounds become available in the future, we can easily determine the optimal MRM conditions of each compound, and thus can quantify their levels in biological samples. Once the MRM conditions have been determined in our analytical system, they can be applied to other LC-MS-based analytical systems with different instruments to obtain the MS/MS spectra in those systems. This is necessary because ESI-MS and ESI-MS/MS spectra are highly instrument dependent and difficult to standardize (Lu et al. 2008a). The concept of MRM can be applied to metabolites only when the corresponding authentic compounds are available and reference data can be obtained. In the case of non-targeted metabolomics, the identification of unknown peaks is the most time-consuming step, because no pre-existing reference data are available. Recently, Matsuda et al. performed non-targeted metabolomics of Arabidopsis using UPLC-QTOFMS, and gathered ESI-MS/MS spectra of about 500 unidentified metabolites which they referred to as MS/MS spectral tags (MS2Ts) (Matsuda et al. 2008). These can be used as a reference data set for comparison with unidentified metabolites, in the same way that expressed sequence tags are used in transcriptomics. We intend to establish a methodology for ultra-widely targeted metabolomics using UPLC-TQMS by combining the non-targeted MS2Ts with the large-scale reference data set of authentic compounds. In this case, metabolites detected in biological samples will be given either compound names or metabolite IDs (not identified as specific compounds), by reference to the combined data sets.
The advantage of MRM also resides in its potential to distinguish between metabolites of the same molecular mass, which give the same retention times in chromatography. For example, in LC-MS analysis the peaks corresponding to leucine and isoleucine cannot be separated either in LC chromatograms or in MS chromatograms. However, if MRM is performed, leucine and isoleucine can be separated by monitoring their respective product ions (Gu et al. 2007).
Large-scale acquisition of metabolite accumulation patterns in plants
In the case of transcriptomics, thousands of microarray data were systematically acquired using the same analytical platforms, i.e. Affymetrix GeneChip, in AtGenExpress (Schmid et al. 2005, Kilian et al. 2007, Goda et al. 2008) and NASCArrays (Craigon et al. 2004). These data sets are available to the public and are used to develop web-based tools for gene co-expression analyses (Steinhauser et al. 2004, Zimmermann et al. 2004, Toufighi et al. 2005, Zimmermann et al. 2005, Obayashi et al. 2007, Horan et al. 2008, Srinivasasainagendra et al. 2008). The availability of these tools has accelerated the functional identification of unknown genes (Saito et al. 2008). High-throughput analysis of widely targeted metabolites will enable the systematic acquisition of metabolite accumulation pattern data for large numbers of biological samples from a single species (e.g. in screening mutants) as well as from a wide range of species. In this study, an interspecies comparison of metabolite accumulation patterns was successfully conducted (Figs. 4, 5), indicating that our methodology has paved the way for comparative metabolomics. Certain taxon-specific metabolites and/or metabolite accumulation patterns are highly relevant to the usefulness of plants to humans (e.g. as resources of phytochemicals) and therefore influence the commercial values of crops (Surh 2003). Comparative metabolomics will be a key approach to understanding diverse plant metabolisms and to developing value-added crops.
By systematically acquiring the metabolite accumulation patterns in large numbers of samples, such as mutant collections and natural variants/ecotypes within a single species, we can carry out large-scale reverse genetics and quantitative trait locus analyses aimed at the elucidation of regulatory mechanisms in plant metabolism (Keurentjes et al. 2006, Kuromori et al. 2006, Loudet et al. 2007). The metabolite accumulation pattern is the final phenotype, resulting from gene expression and protein function in metabolic pathways. Thus, the acquisition of a large-scale data set of metabolite accumulation patterns is essential to our work in understanding plant metabolism, which cannot be fully elucidated using the other omics approaches.
High-throughput UPLC-TQMS analysis for widely targeted metabolomics
In this study, five metabolites were simultaneously monitored in each 3 min run, and thus a total of about 400 metabolites in a single sample were quantified in 240 min. This level of throughput may seem relatively low. However, unlike non-targeted metabolomics, the metabolites to be measured are already identified and directly monitored in this process, without the necessity for signal separation from other unnecessary signals and noises. Moreover, the number of metabolites monitored in a single run could be increased by up to about 100 compounds by adjusting the MRM monitoring times based on the specific retention times. Thus, we expect that the actual throughput will be 1,000 biological samples per week for the quantification of a few hundred metabolites, depending on the number of detectable compounds.
By employing our prototype method, based on UPLC-single quadrupole MS, we have already analyzed the contents of soluble amino acids and glucosinolates in the seeds of about 3,000 transposon-tagged lines (Kuromori et al. 2006) and about 400 natural variants/ecotypes of Arabidopsis (to be published elsewhere). In the novel method introduced here, the sensitivity of detection and selectivity of compounds, as well as the rate of throughput, have been greatly improved. We intend to re-analyze the Arabidopsis lines mentioned above for the accumulation patterns of a few hundred metabolites, by using UPLC-TQMS. This will provide us with a large data set that will help us to understand plant metabolism, and it will be used as a model data set for other bioinformatics studies.
Future prospects—systems analysis and comparative metabolomics
Metabolomics is a young field, and thus the necessary analytical technologies and informatics are still developing in response to the specialized demands of this new area. In order to develop metabolomics effectively as a robust tool for basic biology, some technical difficulties, such as metabolite identification, will need to be overcome.
Over the last decade, many studies have investigated the ‘metabolomes’ of biological samples from plants, mammals and bacteria. In some cases, metabolic fingerprints without metabolite identification were used for clinical diagnosis or for a description of metabolic status. In other cases, targeted metabolomics for hundreds of identified metabolites possessing similar chemical properties were conducted to understand a particular biological event fully. As above, developing ‘metabolic descriptions’ and ‘targeted approaches’ is still a major trend in metabolomics, partly due to technical limitations. In contrast, the widely targeted metabolomics approach established in this study covers a broad range of metabolites encompassing plant primary and secondary metabolism. Such wide coverage is necessary to understand fully the plant metabolic system as a whole. For example, any perturbation in primary metabolism almost certainly affects many secondary metabolic pathways, and vice versa (Kaimoyo et al. 2008).
In addition, the high-throughput measurement advances realized in this study are essential to metabolomics-based biology. Scant metabolome data require other types of biological data and/or a priori knowledge, such as gene expression patterns or kinetic parameters of enzymes, to make sense of the biological information embedded in metabolomic data. On the other hand, metabolome data in the order of thousands or tens of thousands of data points would allow the statistical treatment of data sets and, for example, create algorithms describing relationships among metabolites. Such analyses could be conducted independently from genome information, and thus it may be possible to extrapolate the results to many species. Furthermore, unlike genes, a given metabolite (compound) is identical in all organisms (i.e. there is no ‘orthologous’ metabolite); thus, metabolite accumulation patterns can be directly compared among different species.
Our final goal is to understand complex plant metabolic systems which are fine-tuned to sense and react to both developmental and environmental changes. The high-throughput and widely targeted metabolomics methodologies established here will enable us to acquire a huge metabolome data set and to generate data-driven hypotheses about plant metabolism. They will also provide the basis for comparative metabolomics that can be used to elucidate plant-specific but widely diverse metabolic phenomena.
Materials and Methods
Acquisition of ESI-MS and ESI-MS/MS data
All the solutions of authentic compounds (250 μM) were transferred from 10 ml vials to 1.2 ml glass inserts in 96-well plates (Webseal system, GL Sciences, Tokyo, Japan) and diluted to 50 pmol μl–1 with H2O using an ALHS (Microlab STARplus, Hamilton, Reno, NV, USA). The solutions (20 μl aliquots containing 1 nmol of compound) were analyzed by the flow injection method using a CTC-PAL injection system (AMR, Tokyo, Japan) and a Waters GI Pump solvent system (Waters, Milford, MA, USA) consisting of: 0.05 ml min–1 flow using a micro splitter (GL Sciences), 80% acetonitrile (0.1% formic acid) and 20% water (0.1% formic acid), with a 2.5 min cycle in the isocratic mode. The ionized authentic compounds were detected by TQMS (TQD, Waters) according to the following conditions: capillary voltage +3.0 keV or –2.8 keV, CV 10–60 eV (six levels), source temperature 120°C, desolvation temperature 350°C, cone gas flow 50 l h–1, desolvation gas flow 600 l h–1 and CE 10–60 eV (six levels). A total of 61,920 spectra of 860 compounds were obtained. The optimal MRM conditions, including positive/negative polarity (e.g. [M]+, [M + H]+, [M]–, [M – H]–), m/z of precursor ion and product ion, and optimal CV and CE were determined automatically for 497 of these compounds using the software QuanOptimize (Waters).
UPLC-TQMS analysis
The UPLC (Waters) conditions were manually optimized based on the separation patterns of 12 methionine-derived glucosinolates and were as follows: flow rate 0.24 ml min–1; solvents A, 0.1% formic acid in water and B, 0.1% formic acid in acetonitrile; gradient program of B (0 min, 0%; 0.25 min, 0%; 0.4 min, 9%; 0.8 min, 17%; 1.9 min, 100%; 2.1 min, 100%; 2.11 min, 0%); 3 min cycles with a temperature of 38°C. The TQMS detection conditions were the same as those for FIA-TQMS, except that the source temperature was 130°C.
Automated liquid handling for sample preparation
The plant tissue samples were frozen in liquid nitrogen, quenched using a Mixer Mill (MM300, Retsch, Hann, Germany) as previously reported (Hirai et al. 2007), and the extraction buffer (80% MeOH) was added. The resulting extracts consisted of 400 μl with 20 mg ml–1 (fresh weight) of tissue. The extracts were transferred and treated using an ALHS as follows (Supplementary Fig. S1): 350 μl of each plant extract were transferred to a 96-well plate, and the solutions were dried under N2 gas using a 96-well format spray instrument (40°C, 25 min and 30°C, 20 min). The dried samples were dissolved in 350 μl of H2O using a vortex system (1,300 r.p.m., 6 min), and then filtered through a 96-well filter [Captiva 96-well Filter Plate (pore size 0.45 μm, polyvinylidene fluoride), Varian, CA, USA], using a vacuum manifold with the following program: 30 s, 50 Hpa; 20 s, 100 Hpa; 20 s, 200 Hpa; 30 s, 300 Hpa; and 60 s, 300 Hpa. Plate handling was carried out automatically using robot arms (iSWAP, Hamilton).
Plant materials
In this study, we selected 14 accessions representing nine plant species from three families, and analyzed whole seeds or seed coats. The samples tested were as follows: seeds of the three Brassicaceae species Arabidopsis thaliana (accessions Columbia-0, C24, Landsberg erecta, Wassilewskija and Nossen-0), white mustard (Sinapis alba) and black mustard (Brassica nigra); seed coats of the four Gramineae species wheat (Triticum aestivium), oat (Avena sativa), rye (Secale cereale) and durum wheat (Triticum durum); and seeds of the two Fabaceae species Lotus japonicus (accessions B129 gifu and MG20 miyakojima) and soybean (Glycine max). Arabidopsis seeds were obtained from the Arabidopsis Biological Resource Center (Ohio State University, USA). Seeds of white and black mustards were purchased from Mikasa-Engei (Tokyo, Japan). Seed coats of Gramineae species and soybean seeds were purchased from Cuoca Planning Co., Ltd. (Tokushima, Japan). Lotus japonicus accessions were a gift from Dr. Toshio Aoki (Nihon University, Japan) (Kawaguchi et al. 2001).
Analysis of metabolite accumulation patterns
Quantitative data for metabolite accumulation in the seeds or seed coats of the 14 accessions or species were obtained by UPLC-TQMS. Peaks that showed S/N ratios >30 were selected as the detected metabolites to be used in further analyses. Areas under the selected peaks were converted into logarithms (base 2) after missing values, which appeared when a metabolite was not detected in a sample, were replaced with 0.1. Data were normalized by z-score transformation using the software TM4 MEV (Chu et al. 2008). The resulting data matrix was analyzed using hierarchical clustering based on the Euclidean distance and visualized by MEGA4 (Tamura et al. 2007) as a dendrogram. The family-specific metabolites were identified by BL-SOM analysis of the matrix in combination with a model data set consisting of one hypothetical metabolite specific to each of the families Brassicaceae, Gramineae and Fabaceae.
Supplementary data
Supplementary data are available at PCP online.
Funding
The Japan Science and Technology Agency (CREST grant).
Supplementary Material
Acknowledgments
We thank Ms. Etsuko Suzuki (Nihon Waters KK), Mr. Takehiro Nozawa (Nihon Waters KK) and Mr. Hiroshi Hike (AMR Inc.) for technical support with the FIA-TQMS analysis. We also thank Mr. Kazuto Hashimoto (GL Sciences Inc.) and Mr. Masahito Shimoya (GL Sciences Inc.) for technical support with the ALHS. We wish to thank Dr. Aoki Toshio (Department of Applied Biological Sciences, Nihon University) for the kind gift of Lotus japonicus (accessions B129 gifu and MG20 miyakojima). We are grateful to Dr. Fumio Matsuda for useful comments on our widely targeted metabolomics approach.
Glossary
Abbreviations:
- ALHS
automated liquid handling system
- APCI
atmospheric pressure chemical ionization
- CE
collision energy
- CV
cone voltage
- EI
electron ionization
- ESI
electrospray ionization
- FIA
flow injection analysis
- GC
gas chromatography
- LC
liquid chromatography
- MRM
multiple reaction monitoring
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- MS2T
MS/MS spectral tag
- NMR
nuclear magnetic resonance
- Q
quadrupole
- S/N ratio
signal-to-noise ratio
- TOF
time-of-flight
- TQMS
tandem quadrupole mass spectrometry
- UPLC
ultra performance liquid chromatography.
References
- Akiyama K, Chikayama E, Yuasa H, Shimada Y, Tohge T, Shinozaki K, et al. PRIMe: a web site that assembles tools for metabolomics and transcriptomics. In Silico Biol. 2008;8:0027. [PubMed] [Google Scholar]
- Chu VT, Gottardo R, Raftery AE, Bumgarner RE, Yeung KY. MeV+R: using MeV as a graphical user interface for bioconductor applications in microarray analysis. Genome Biol. 2008;9:R118. doi: 10.1186/gb-2008-9-7-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citova I, Havlikova L, Urbanek L, Solichova D, Novakova L, Solich P. Comparison of a novel ultra-performance liquid chromatographic method for determination of retinol and alpha-tocopherol in human serum with conventional HPLC using monolithic and particulate columns. Anal. Bioanal. Chem. 2007;388:675–681. doi: 10.1007/s00216-007-1237-8. [DOI] [PubMed] [Google Scholar]
- Craigon DJ, James N, Okyere J, Higgins J, Jotham J, May S. NASCArrays: a repository for microarray data generated by NASC’s transcriptomics service. Nucleic Acids Res. 2004;32:D575–D577. doi: 10.1093/nar/gkh133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DellaPenna D, Last RL. Genome-enabled approaches shed new light on plant metabolism. Science. 2008;320:479–481. doi: 10.1126/science.1153715. [DOI] [PubMed] [Google Scholar]
- Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- Fiehn O. Metabolomics—the link between genotypes and phenotypes. Plant Mol. Biol. 2002;48:155–171. [PubMed] [Google Scholar]
- Fiehn O, Kloska S, Altmann T. Integrated studies on plant biology using multiparallel techniques. Curr. Opin. Biotechnol. 2001;12:82–86. doi: 10.1016/s0958-1669(00)00165-8. [DOI] [PubMed] [Google Scholar]
- Glinski M, Weckwerth W. The role of mass spectrometry in plant systems biology. Mass Spectrom. Rev. 2006;25:173–214. doi: 10.1002/mas.20063. [DOI] [PubMed] [Google Scholar]
- Goda H, Sasaki E, Akiyama K, Maruyama-Nakashita A, Nakabayashi K, Li W, et al. The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J. 2008;55:526–542. doi: 10.1111/j.0960-7412.2008.03510.x. [DOI] [PubMed] [Google Scholar]
- Gu L, Jones AD, Last RL. LC-MS/MS assay for protein amino acids and metabolically related compounds for large-scale screening of metabolic phenotypes. Anal. Chem. 2007;79:8067–8075. doi: 10.1021/ac070938b. [DOI] [PubMed] [Google Scholar]
- Hirai MY, Sugiyama K, Sawada Y, Tohge T, Obayashi T, Suzuki A, et al. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl Acad. Sci. USA. 2007;104:6478–6483. doi: 10.1073/pnas.0611629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horai H, Arita M, Nishioka T. 1st International Conference on BioMedical Engineering and Informatics. 2008. Comparison of ESI-MS spectra in MassBank database. [Google Scholar]
- Horan K, Jang C, Bailey-Serres J, Mittler R, Shelton C, Harper JF, et al. Annotating genes of known and unknown function by large-scale coexpression analysis. Plant Physiol. 2008;147:41–57. doi: 10.1104/pp.108.117366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaimoyo E, Farag MA, Sumner LW, Wasmann C, Cuello JL, VanEtten H. Sub-lethal levels of electric current elicit the biosynthesis of plant secondary metabolites. Biotechnol. Prog. 2008;24:377–384. doi: 10.1021/bp0703329. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi M, Motomura T, Imaizumi-Anraku H, Akao S, Kawasaki S. Providing the basis for genomics in Lotus japonicus: the accessions Miyakojima and Gifu are appropriate crossing partners for genetic analyses. Mol. Genet. Genomics. 2001;266:157–166. doi: 10.1007/s004380100540. [DOI] [PubMed] [Google Scholar]
- Keurentjes JJB, Fu JY, de Vos CHR, Lommen A, Hall RD, Bino RJ, et al. The genetics of plant metabolism. Nat. Genet. 2006;38:842–849. doi: 10.1038/ng1815. [DOI] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, et al. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007;50:347–363. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmuller E, et al. GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics. 2005;21:1635–1638. doi: 10.1093/bioinformatics/bti236. [DOI] [PubMed] [Google Scholar]
- Kuromori T, Wada T, Kamiya A, Yuguchi M, Yokouchi T, Imura Y, et al. A trial of phenome analysis using 4000 Ds-insertional mutants in gene-coding regions of Arabidopsis. Plant J. 2006;47:640–651. doi: 10.1111/j.1365-313X.2006.02808.x. [DOI] [PubMed] [Google Scholar]
- Loudet O, Saliba-Colombani V, Camilleri C, Calenge F, Gaudon V, Koprivova A, et al. Natural variation for sulfate content in Arabidopsis thaliana is highly controlled by APR2. Nat. Genet. 2007;39:896–900. doi: 10.1038/ng2050. [DOI] [PubMed] [Google Scholar]
- Lu W, Bennett BD, Rabinowitz JD. Analytical strategies for LC-MS-based targeted metabolomics. J. Chromatogr. B. 2008a;871:236–242. doi: 10.1016/j.jchromb.2008.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Savage LJ, Ajjawi I, Imre KM, Yoder DW, Benning C, et al. New connections across pathways and cellular processes: industrialized mutant screening reveals novel associations between diverse phenotypes in Arabidopsis. Plant Physiol. 2008b;146:1482–1500. doi: 10.1104/pp.107.115220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F, Yonekura-Sakakibara K, Niida R, Kuromori T, Shinozaki K, Saito K. Plant J. 2008. MS/MS spectral tag (MS2T)-based annotation of non-targeted profile of plant secondary metabolites. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller LA, Zhang P, Rhee SY. AraCyc: a biochemical pathway database for Arabidopsis. Plant Physiol. 2003;132:453–460. doi: 10.1104/pp.102.017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK, Connelly J, Lindon JC, Holmes E. Metabolomics: a platform for studying drug toxicity and gene function. Nat. Rev. Drug Discov. 2002;1:153–161. doi: 10.1038/nrd728. [DOI] [PubMed] [Google Scholar]
- Obayashi T, Kinoshita K, Nakai K, Shibaoka M, Hayashi S, Saeki M, et al. ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 2007;35:D863–D869. doi: 10.1093/nar/gkl783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda S, Yamada T, Hamajima M, Itoh M, Katayama T, Bork P, et al. KEGG atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res. 2008;36:W423–W426. doi: 10.1093/nar/gkn282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palandra J, Weller D, Hudson G, Li J, Osgood S, Hudson E, et al. Flexible automated approach for quantitative liquid handling of complex biological samples. Anal. Chem. 2007;79:8010–8015. doi: 10.1021/ac070618s. [DOI] [PubMed] [Google Scholar]
- Saito K, Hirai MY, Yonekura-Sakakibara K. Decoding genes with coexpression networks and metabolomics—‘majority report by precogs’. Trends Plant Sci. 2008;13:36–43. doi: 10.1016/j.tplants.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Schauer N, Steinhauser D, Strelkov S, Schomburg D, Allison G, Moritz T, et al. GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett. 2005;579:1332–1337. doi: 10.1016/j.febslet.2005.01.029. [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, et al. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Shinbo Y, Nakamura Y, Altaf-Ul-Amin M, Asahi H, Kurokawa K, Arita M, et al. KNApSAcK: a comprehensive species–metabolite relationship database. In: Saito K, Dixon RA, Willmitzer L, editors. Agriculture and Forestry. Heidelberg: Springer-Verlag; 2006. pp. 165–181. [Google Scholar]
- Srinivasasainagendra V, Page GP, Mehta T, Coulibaly I, Loraine AE. CressExpress: a tool for large-scale mining of expression data from Arabidopsis. Plant Physiol. 2008;147:1004–1016. doi: 10.1104/pp.107.115535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauser D, Usadel B, Luedemann A, Thimm O, Kopka J. CSB.DB: a comprehensive systems-biology database. Bioinformatics. 2004;20:3647–3651. doi: 10.1093/bioinformatics/bth398. [DOI] [PubMed] [Google Scholar]
- Sumner LW, Mendes P, Dixon RA. Plant metabolomics: large-scale phytochemistry in the functional genomics era. Phytochemistry. 2003;62:817–836. doi: 10.1016/s0031-9422(02)00708-2. [DOI] [PubMed] [Google Scholar]
- Surh YJ. Cancer chemoprevention with dietary phytochemicals. Nat. Rev. Cancer. 2003;3:768–780. doi: 10.1038/nrc1189. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. The Botany Array Resource: e-Northerns, expression angling, and promoter analyses. Plant J. 2005;43:153–163. doi: 10.1111/j.1365-313X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- Unwin RD, Griffiths JR, Leverentz MK, Grallert A, Hagan IM, Whetton AD. Multiple reaction monitoring to identify sites of protein phosphorylation with high sensitivity. Mol. Cell Proteomics. 2005;4:1134–1144. doi: 10.1074/mcp.M500113-MCP200. [DOI] [PubMed] [Google Scholar]
- Werner E, Croixmarie V, Umbdenstock T, Ezan E, Chaminade P, Tabet JC, et al. Mass spectrometry-based metabolomics: accelerating the characterization of discriminating signals by combining statistical correlations and ultrahigh resolution. Anal. Chem. 2008a;80:4918–4932. doi: 10.1021/ac800094p. [DOI] [PubMed] [Google Scholar]
- Werner E, Heilier JF, Ducruix C, Ezan E, Junot C, Tabet JC. Mass spectrometry for the identification of the discriminating signals from metabolomics: current status and future trends. J. Chromatogr. B. 2008b;871:143–163. doi: 10.1016/j.jchromb.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hennig L, Gruissem W. Gene-expression analysis and network discovery using Genevestigator. Trends Plant Sci. 2005;10:407–409. doi: 10.1016/j.tplants.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.