Abstract
Pollen represents an important nitrogen sink in flowers to ensure pollen viability. Since pollen cells are symplasmically isolated during maturation and germination, membrane transporters are required for nitrogen import across the pollen plasma membrane. This study describes the characterization of the ammonium transporter AtAMT1;4, a so far uncharacterized member of the Arabidopsis AMT1 family, which is suggested to be involved in transporting ammonium into pollen. The AtAMT1;4 gene encodes a functional ammonium transporter when heterologously expressed in yeast or when overexpressed in Arabidopsis roots. Concentration-dependent analysis of 15N-labeled ammonium influx into roots of AtAMT1;4-transformed plants allowed characterization of AtAMT1;4 as a high-affinity transporter with a Km of 17 μM. RNA and protein gel blot analysis showed expression of AtAMT1;4 in flowers, and promoter–gene fusions to the green fluorescent protein (GFP) further defined its exclusive expression in pollen grains and pollen tubes. The AtAMT1;4 protein appeared to be localized to the plasma membrane as indicated by protein gel blot analysis of plasma membrane-enriched membrane fractions and by visualization of GFP-tagged AtAMT1;4 protein in pollen grains and pollen tubes. However, no phenotype related to pollen function could be observed in a transposon-tagged line, in which AtAMT1;4 expression is disrupted. These results suggest that AtAMT1;4 mediates ammonium uptake across the plasma membrane of pollen to contribute to nitrogen nutrition of pollen via ammonium uptake or retrieval.
Keywords: Ammonium uptake, AMTs, Germination, Nitrogen transport, Pollen, Pollen tube
Introduction
Pollen development, pollen germination and pollen tube growth are critical stages during sexual reproduction of higher plants. At these stages in particular, pollen represents a significant sink for carbon and nitrogen which must be imported from source organs such as leaves (Schneidereit et al. 2003, Lee and Tegeder 2004). Since there are no plasmodesmal connections to surrounding cells after microspores have been released from the tetrad, pollen grains and pollen tubes develop as symplasmically isolated organs (Scott et al. 1991). This isolation requires the presence of plasma membrane-localized transporters that load nutrients into the pollen. Therefore, pollen has been proposed as an ideal subject to study the role of membrane transport processes in nutrition and development (Schwacke et al. 1999). In fact, a recent genome-wide analysis of membrane transporter genes expressed in the male gametophytes of Arabidopsis revealed 757 transporter genes which are expressed in pollen, and even 16% of those are specifically or at least preferentially expressed relative to sporophytic tissues, revealing the essential roles of membrane transporters in pollen function (Bock et al. 2006). Based on further Arabidopsis transcriptome data (Honys and Twell 2004, Pina et al. 2005, Schmid et al. 2005, Sze et al. 2006), several Lys/His transporters (LHT) are specifically expressed in pollen at certain developmental stages of the flower, suggesting distinct roles of the individual transporters for organic nitrogen supply during pollen development, pollen germination and pollen tube growth. Further amino acid transporters, such as tobacco NsAAP1 and tomato LeProT1, have also been suggested to contribute to nitrogen transport into pollen (Lalanne et al. 1997, Schwacke et al. 1999).
Ammonium is a primary nitrogen source and key metabolite for plant nitrogen metabolism. Membrane transport of ammonium is mediated by ammonium transporters of the ammonium transporter/methylamine permease/rhesus (AMT/MEP/Rh) protein family, which has been found in all domains of life (von Wirén and Merrick 2004). While NH3 has been identified as the substrate being transported by either prokaryote AMT or eukaryote Rh-type ammonium transporters (Khademi et al. 2004, Ripoche et al. 2004, Zheng et al. 2004), plant transporters of the AMT1 subfamily have been described as NH4+ uniporters or NH3/H+ co-transporters (Ludewig et al. 2002, Ludewig et al. 2003, Mayer et al. 2006). All plant AMT proteins investigated so far were localized to the plasma membrane, implicating their roles in ammonium acquisition by plant cells (Ludewig et al. 2002, Sohlenkamp et al. 2002, Simon-Rosin et al. 2003, Ludewig et al. 2003, Loqué et al. 2006, Yuan et al. 2007a). In Arabidopsis, five members of the AMT1 subfamily have been identified (Loqué and von Wirén 2004), and the physiological roles of four of them have been intensively investigated in roots (Kaiser et al. 2002, Loqué et al. 2006, Yuan et al. 2007a). AtAMT1;1, AtAMT1;3 and AtAMT1:5 are highly expressed in nitrogen-deficient rhizodermal cells, including root hairs, and contribute additively to approximately 70–80% of the overall high-affinity ammonium uptake capacity in Arabidopsis roots (Loqué et al. 2006, Yuan et al., 2007a). In contrast, AtAMT1;2, presenting a relatively lower affinity for ammonium than the other members of the subfamily, is localized in endodermal and cortical cells, and appears to be responsible for the uptake of ammonium that has entered the root tissue via the apoplastic route (Yuan et al. 2007a). In contrast to roots, the physiological functions of AMTs in other plant organs remain to be elucidated.
So far, AtAMT1;4 (At4g28700) represents the only member of the AMT1 subfamily in Arabidopsis which has not yet been characterized. Here, we describe the functional expression in yeast and in planta, as well as the tissue-specific expression and membrane localization of AtAMT1;4. Even though a transposon-tagged line in AtAMT1;4 did not show any phenotype, substantial evidence is provided to propose a role for AtAMT1;4 in ammonium uptake or retrieval in pollen.
Results
Heterologous expression of AtAMT1;4 in yeast
Out of the six AMT proteins identified in Arabidopsis, five have been subjected to functional analysis to date. For a functional characterization of AtAMT1;4 (At4g28700), the corresponding gene was amplified by PCR from genomic DNA of the Arabidopsis ecotype Col-0 which was possible due to the absence of introns. The open reading frame (ORF) in AtAMT1;4 is 1,515 bp long and encodes a protein of 504 amino acids with a calculated molecular mass of 53.7 kDa. At the amino acid level, AtAMT1;4 shares a high degree of homology (69.7–74.3% identity) to other members of the AMT1 family in Arabidopsis.
To investigate the functional properties of the AtAMT1;4 protein, the ORF of AtAMT1;4 was cloned into the yeast expression vector p426 and placed under control of the constitutively active yeast hexose transporter promoter HXT7. The resulting construct was used for transformation of the yeast stain 31019b, which is defective in the expression of three endogenous ammonium transporter genes (mep1, mep2 and mep3) (Marini et al. 1997). This strain grows poorly on low ammonium concentrations (<5 mM) as a sole nitrogen source. In contrast to transformants carrying the empty vector, yeast cells expressing either AtAMT1;4 or AtAMT1;1 were able to grow on 1 mM ammonium, indicating that AtAMT1;4, similarly to AtAMT1;1, encodes a functional ammonium transporter (Fig. 1).
Fig. 1.
Growth complementation of the ammonium uptake-defective yeast strain 31019b by AtAMT1;4. The triple mep yeast mutant 31019b was transformed with the empty vector p426, p426-AtAMT1;4 or p426-AtAMT1;1. Transformants were pre-cultured for 24 h on YNB medium supplemented with 5 mM arginine. A 1 ml aliquot of a saturated culture was harvested, and resuspended in 1 ml of water. One- to 5-fold dilutions of a 5 μ l aliquot of yeast cell suspension were spotted on YNB medium supplemented with either 5 mM arginine or 1 mM NH4Cl at pH 6.0.
Ectopic expression of AtAMT1; 4 in an Arabidopsis line defective in high-affinity ammonium uptake
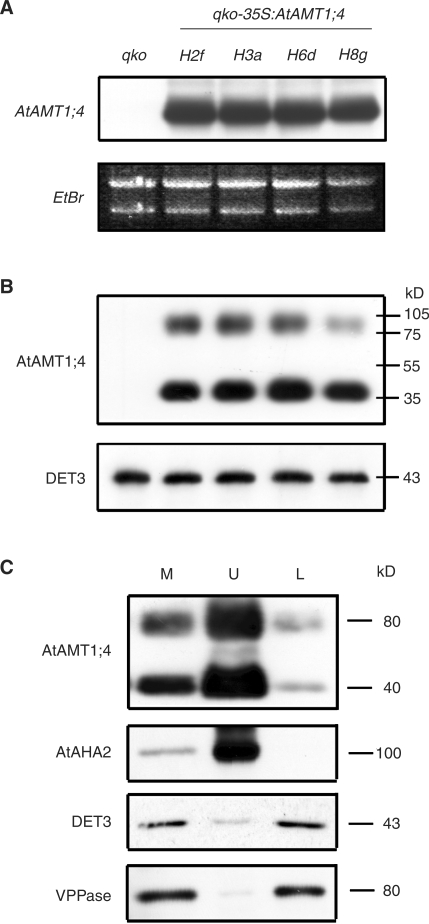
AtAMT1;4 was then constitutively expressed in Arabidopsis plants to examine whether AtAMT1;4 has the potential to contribute to ammonium uptake in roots. To circumvent transport activities from endogenous AtAMTs in Arabidopsis roots, a multiple insertion line (qko) was chosen for transformation, in which the high-affinity ammonium uptake capacity is reduced by approximately 90% due to the genetic disruption of the AtAMT1;1, 1;2, 1;3 and 2;1 genes (Yuan et al. 2007a). Four independent homozygous lines from the T2 generation of CaMV35S:AtAMT1;4 transformants were selected for further investigation. RNA gel blot analysis of root RNA from 6-week-old plants grown in nitrogen-sufficient nutrient solution showed that AtAMT1;4 transcripts were highly expressed in roots of the transgenic lines H2f, H3a, H6d and H8g (Fig. 2A). No hybridization signal was detected in qko, confirming that AtAMT1;4 is not or at least very weakly expressed in roots under these conditions. Furthermore, large amounts of AtAMT1;4 protein were found in the same transgenic lines by protein gel blot analysis of microsomal membrane fractions from roots using a rabbit antibody directed against 15 amino acid residues in the C-terminus of AtAMT1;4. Also no protein signal was detected in qko (Fig. 2B). Two AtAMT1;4-specific bands were observed with an apparent molecular mass of approximately 40 and 80 kDa. Although the predicted molecular weight of the AtAMT1;4 protein is approximately 53.7 kDa, the 40 kDa band is supposed to correspond to the monomer, since lipophilic membrane proteins usually migrate at a lower apparent molecular mass (Sauer and Stadler 1993). The 80 kDa band probably represents the dimer of AtAMT1;4 or a stable complex with another unknown protein.
Fig. 2.
Overexpression of AtAMT1;4 in a multiple insertion line defective in high-affinity ammonium uptake. (A) RNA gel blot analysis of root RNA from a quadruple knock-out line (qko), and qko-35S:AtAMT1;4 transgenic plants (lines H2f, H3a, H6d and H8g) using the ORF of AtAMT1;4 as a probe. Ethidium bromide-stained rRNA served as loading control. Six-week-old plants were cultured hydroponically under continuous supply of 2 mM ammonium nitrate. (B) Protein gel blot analysis of microsomal membrane fractions from roots of the same plants as in (A) using an antibody directed against 15 amino acids from the C-terminus of AtAMT1;4 (anti-AtAMT1;4). Numbers at the right border indicate molecular mass (kDa). Protein levels of DET3 served as a loading control. (C) Microsomal fractions (M) from shoots of qko-35S:AtAMT1;4 transgenic plants (H3a) were separated by aqueous two-phase partitioning into a plasma membrane-enriched upper phase (U) and an endosomal membrane-enriched lower phase (L). Protein gel blot analysis was conducted with antibodies against AtAMT1;4, an Arabidopsis plasma membrane H+-ATPase (AtAHA2), a V-ATPase (DET3) and a VPPase.
To examine the subcellular localization of AtAMT1; 4 in these transgenic lines, shoot microsomal membrane fractions of the line H3a were separated by two-phase partitioning into a plasma membrane-enriched upper phase (U) and a lower phase (L) enriched in endosomal membranes including endoplasmic reticulum, Golgi, chloroplast and tonoplast membranes (Fig. 2C). The purity of these membrane fractions was verified by the abundance of the DET3 protein, a subunit of the vacuolar ATPase (V-ATPase; Schumacher et al. 1999), and, alternatively, by the abundance of the vacuolar pyrophosphatase (VPPase; Takasu et al. 1997), which both were enriched in the lower phase. In contrast, AtAMT1;4 protein accumulated in the upper phase, coinciding with high expression levels of the plasma membrane H+-ATPase AHA2 (DeWitt et al. 1996). This observation strongly indicated a localization of the AtAMT1;4 protein in the plasma membrane.
Analysis of the in planta transport capacity of AtAMT1;4
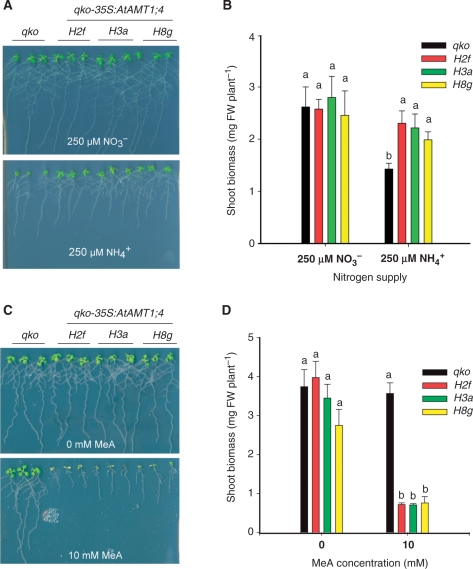
To test the functionality of overexpressed AtAMT1;4 protein in roots, qko and three transgenic lines (H2f, H3a and H8g) were grown on agar medium supplemented with either ammonium or nitrate as the sole nitrogen source. At 250 μM ammonium, qko plants grew poorly because of a low ammonium uptake capacity in roots (Yuan et al. 2007a), while the transgenic lines were able to increase shoot biomass significantly, indicating that AtAMT1;4 is able to mediate ammonium uptake into plant roots (Fig. 3A, B). In contrast, plant growth did not differ between qko and transgenic plants with the supply of 250 μ M nitrate. Besides ammonium, plant AMT1-type transporters have also been shown to permeate the substrate analog methylammonium (MeA) which is toxic to plants (Loqué et al. 2006, Yuan et al. 2007a). When grown on 1 mM nitrate in the presence of 10 mM MeA, the transgenic lines showed a much higher sensitivity to MeA than qko plants (Fig. 3C, D), supporting that AtAMT1;4 is also able to transport MeA.
Fig. 3.
Growth of qko-35S-AtAMT1;4 transgenic lines on ammonium and its toxic analog methylammonium (MeA). (A) Growth of qko plants and transgenic qko-35S:AtAMT1;4 plants (lines H2f, H3a and H8g) on agar containing 250 μ M NH4+ (125 μ M as ammonium succinate) or 250 μ M NO3− (250 μ M as potassium nitrate) for 10 d after pre-culture on half-strength MS medium (containing 5 mM nitrate as sole nitrogen source) for 7 d. (B) Shoot dry weights of the same plants as in (A). Bars indicate means ± SD (n = 6), and significant differences at P < 0.001 within each group are indicated by different letters. (C) Growth of qko plants and transgenic qko-35S:AtAMT1;4 plants (lines H2f, H3a and H8g) on agar containing no or 10 mM MeA in the presence 1 mM nitrate and 1% sucrose for 10 d after pre-culture on half-strength MS medium (containing 5 mM nitrate as sole nitrogen source) for 8 d. (D) Shoot dry weights of the same plants as in (C). Bars indicate means ± SD (n = 4), and significant differences at P < 0.001 within each group are indicated by different letters.
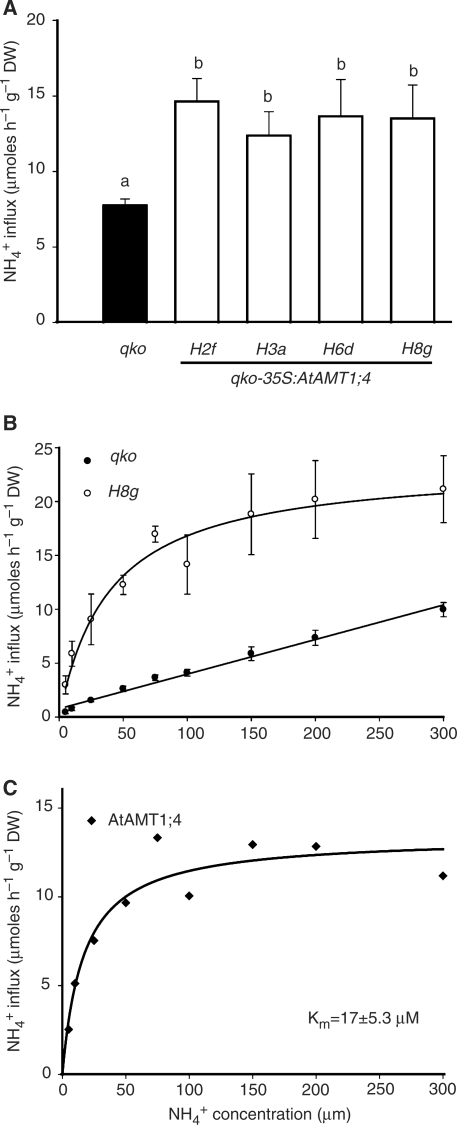
To evaluate the ammonium uptake capacity in roots, a short-term influx study was conducted using 15N-labeled ammonium, and 15N was subsequently determined by stable isotope mass spectrometry. Six-week-old plants grown hydroponically under nitrogen-sufficient conditions were subjected to influx analysis at 200 μ M 15N-labeled ammonium as a measure for high-affinity uptake. All four lines showed an almost 2-fold higher ammonium uptake capacity compared with qko (Fig. 4A). In contrast, no significant difference was observed when the influx analyses were conducted at 5 mM 15N-labeled ammonium supply (Supplementary Fig. S1). Low-affinity ammonium uptake capacity, as calculated by subtraction of the influx value at 200 μ M from that at 5 mM, did not differ among the lines (data not shown). Therefore, these results indicate that overexpression of AtAMT1;4 in qko roots increases the capacity for high-affinity but not for low-affinity ammonium uptake. To investigate the biochemical properties of AtAMT1;4 in more detail, a concentration-dependent influx analysis was performed with qko and the transgenic line H8g (Fig. 4B). Ammonium influx in qko roots followed almost a linear concentration dependency, indicative of a lack of high-affinity uptake as described in Yuan et al. (2007a). However, ammonium influx in H8g roots saturated above approximately 100 μ M. Net ammonium influx mediated by the AtAMT1;4 protein was then determined by subtraction of values of qko from those of H8g, allowing calculation of a Km value of approximately 17 μ M (Fig. 4C).
Fig. 4.
Increased high-affinity ammonium uptake capacity in qko-35S:AtAMT1;4 transgenic plants. (A) Influx of 15N-labeled ammonium into roots of qko and transgenic qko-35S:AtAMT1;4 lines (H2f, H3a, H6d and H8g). 15N-labeled ammonium was supplied at 200 μ M. Bars indicate means ± SD, n = 8–10 plants, and significant differences at P < 0.001 are indicated by different letters. (B) Concentration-dependent influx of 15N-labeled ammonium into roots of qko and the transgenic qko-35S:AtAMT1;4 line H8g. Symbols indicate means ± SD, n = 5–6 plants. (C) 15N-labeled ammonium influx mediated by AtAMT1;4 was calculated by subtracting the value of qko from H8g in (B). Curves were directly fitted to the data for H8g in (B) and those in (C) using the Michaelis–Menten equation. A linear curve was used to fit the data for qko in (B).The caclulated Km value for AtAMT1;4 was 17 ± 5.3 μ M. All plants were cultured hydroponically under continuous supply of 2 mM ammonium nitrate for 6 weeks.
Tissue-specific expression of AtAMT1;4
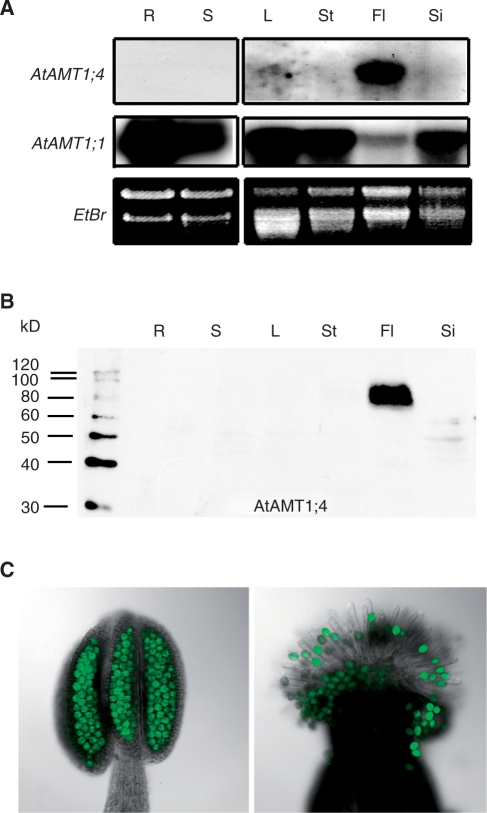
Organ-specific expression of AtAMT1;4 was determined by RNA and protein gel blot analysis (Fig. 5A, B). Total RNA was extracted from roots and shoots of hydroponically grown Arabidopsis plants, or from leaves, stems, flowers and siliques of mature plants grown in soil (Fig. 5A). The ORFs of AtAMT1;4 and AtAMT1;1 were used as probes. While AtAMT1;1 showed a ubiquitous expression pattern with relatively weak expression in flowers, AtAMT1;4 was detected exclusively in flowers. Furthermore, using the AtAMT1;4-specific antibody, AtAMT1;4 protein was also exclusively detected in the microsomal membrane fractions of flower tissue (Fig. 5B). There, the AtAMT1;4 protein appeared at a molecular mass of approximately 80 kDa. In contrast to the overexpressed AtAMT1;4 protein in transgenic roots and shoots of qko (Fig. 2B, C), the putative monomer at 40 kDa was not observed in flowers.
Fig. 5.
Expression of AtAMT1;4 is restricted to pollen. (A) RNA gel blot analysis of RNA from various Arabidopsis tissues using the ORFs of AtAMT1;4 and AtAMT1;1 as probes. Ethidium bromide-stained rRNA served as loading control. The tissues of roots (R) and shoots (S) were harvested from 6-week-old plants grown hydroponically under continuous supply of 2 mM ammonium nitrate, while leaves (L), stems (St), flowers (Fl) and siliques (Si) were obtained from 6-week-old plants grown in soil. (B) Protein gel blot analysis of microsomal membrane fractions from the same tissue as in (A) using anti-AtAMT1;4 antibody. Numbers at the left border indicate molecular mass (kDa). The AtAMT1;4-specific signal was detected at approximately 80 kDa. (C) Fluorescence microscopy of stamen (left) and pollinated stigmata (right) of transgenic plants expressing AtAMT1;4-promoter:ORF:GFP construct.
Analysis of transgenic lines expressing AtAMT1;4-promoter:ORF:GFP or AtAMT1;4-promoter:GFP fusion constructs
To localize AtAMT1;4 expression in more detail, a genomic DNA fragment corresponding to 1,935 bp of the upstream region of AtAMT1;4 including the start codon and the ORF of AtAMT1;4 was fused to the EGFP (enhanced green fluorescent protein) reporter gene and used for transformation of Arabidopsis wild-type plants. Thus, in these transgenic lines, the AtAMT1;4–GFP fusion protein was expressed under control of its native promoter. Within all of six independent T2 homozygous transgenic lines, GFP-dependent fluorescence of the fusion protein was detected exclusively in pollen (Fig. 5C). According to Smyth et al. (1990), green fluorescence in pollen was initiated at flower stage 10 and became more intense with pollen maturity during floral development (Supplementary Fig. S2). AtAMT1;4 promoter activity also appeared to be high during pollen germination on the stigmata of flowers (Fig. 5C).
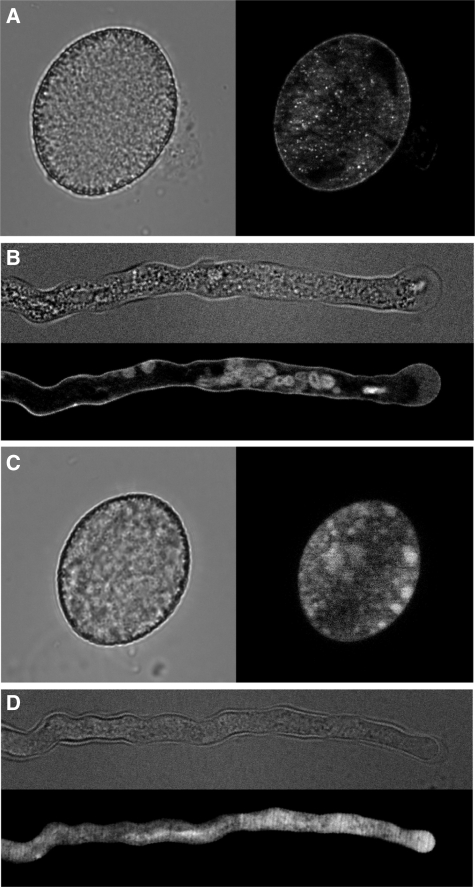
To determine the subcellular localization of the AtAMT1;4 protein in pollen, transgenic plants expressing the AtAMT1;4-promoter:ORF:GFP construct were investigated in parallel using the same promoter sequence as for generation of the AtAMT1;4-promoter:GFP lines. Mature pollen from transgenic plants was germinated in vitro on solid agar medium. By fluorescence imaging, AtAMT1;4-dependent green fluorescence appeared as a thin layer along the border of pollen grains (Fig. 6A) and pollen tubes (Fig. 6B), although some fluorescence was also observed in internal cell compartments. In the transgenic plants expressing the AtAMT1;4-promoter:GFP construct, GFP-derived fluorescence was also found to be expressed only in pollen grains and pollen tubes (Fig. 6C, D), which further confirmed the pollen-specific promoter activity of AtAMT1;4. In contrast to the AtAMT1;4–GFP fusion protein, soluble GFP produced fluorescence throughout the whole body of the pollen grain (Fig. 6C) or pollen tube (Fig. 6D). These observations indicated that the AtAMT1;4 protein localizes to the plasma membrane of pollen grains and pollen tubes.
Fig. 6.
AtAMT1;4 localizes to the plasma membrane of pollen grains and pollen tubes. Fluorescence microscopy of (A) a mature pollen grain and (B) a pollen tube of transgenic plants expressing the AtAMT1;4-promoter:ORF:GFP construct or (C and D) expressing the AtAMT1;4-promoter:GFP construct. GFP fluorescence micrographs in (A–D) were present on a black background, alongside equivalent light micrographs on a gray background. Flowers and pollen grains were harvested from 6-week-old plants grown in soil, and pollen tubes were germinated in vitro.
Analysis of AtAMT1;4 T-DNA and transposon insertion lines
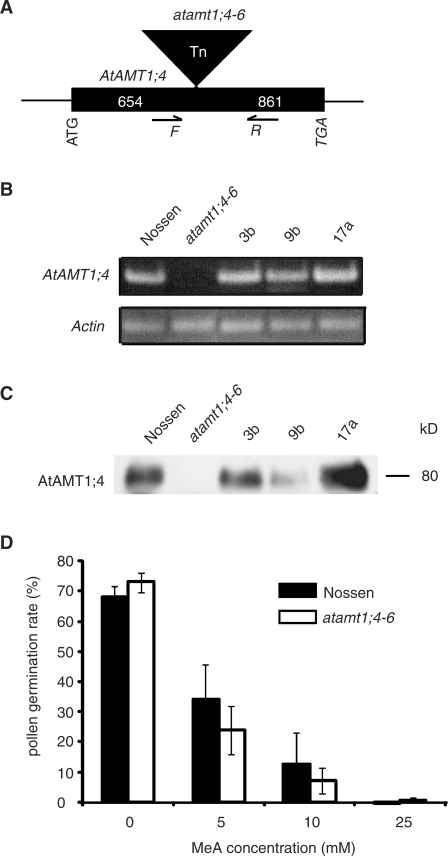
To investigate further the physiological role of AtAMT1;4 in Arabidopsis pollen, six independent insertion lines for the AtAMT1;4 gene were identified by screening of publicly available libraries, and homozygous plants were generated and selected by PCR analysis. The T-DNA insertion sites were verified by sequencing to be upstream of the start codon of the AtAMT1;4 gene in five independent lines, named atamt1;4-1 to atamt1;4-5 (Supplementary Fig. S3). However, RNA gel blot analysis revealed that AtAMT1;4 gene expression was neither absent nor repressed. When compared with their corresponding wild-type Col-0, AtAMT1;4 expression was even higher in the lines atamt1;4-2, -3, -4 and -5, probably resulting from the presence of an additional promoter activity within the T-DNA insertion (Uelker et al. 2008), or from the ectopic expression of AtAMT1;4 in other parts of flowers due to the disruption of pollen-specific elements in the AtAMT1;4 promoter region. With regard to the considerable level of AtAMT1;4 mRNA that could still be detected, only atamt1;4-1 appeared to represent a knock-down line (Supplementary Fig. S3). However, in the line atamt1;4-6, a transposon insertion was identified to be located within the ORF of AtAMT1;4, 654 bp downstream of the start codon (Fig. 7A). As a consequence, AtAMT1;4 transcript abundance was absent, as revealed by RNA gel blot analysis of flower RNA (Fig. 7B) and, in addition, no more AtAMT1;4 protein was detected in microsomal membrane fractions of flower tissue (Fig. 7C). Subsequently, three independent transgenic lines were generated, in which AtAMT1;4 gene expression was conferred to atamt1;4-6 plants under control of its native promoter. The expression of AtAMT1;4 in these lines was evaluated at the transcript and protein levels as shown in Fig. 7B and C.
Fig. 7.
Characterization of the transposon-tagged line atamt1;4-6. (A) The position of the transposon insertion in the ORF of AtAMT1;4 located 654 bp downstream of the start codon. (B) RT–PCR analysis of RNA from flowers of atamt1;4-6, the corresponding wild-type (ecotype Nössen), and three independent lines recomplemented with the ORF of AtAMT1;4 driven by its native promoter (3b, 9b and 17a). Expression of the Actin8 gene served as loading control. The plants were grown in soil for 6 weeks. (C) Protein gel blot analysis of microsomal membrane fractions from the same tissues as in (B) using anti-AtAMT1;4 antibody. Numbers at the right border indicate molecular mass (kDa). The AtAMT1;4-specific signal was detected at approximately 80 kDa. (D) In vitro pollen germination assay of atamt1;4-6 and wild-type Nössen in the medium with the supply of different concentrations of MeA.
The mutant line atamt1;4-6 was then analyzed regarding phenotypic differences in comparison with the corresponding wild-type Nössen and the complemented transgenic lines. However, when grown in nutrient solution or in soil substrate, no phenotypic difference appeared. The mutant plants were fertile and produced normal seeds. Moreover, no significant difference was observed with regard to pollen development, pollen size or pollen abundance (data not shown). In in vitro pollen germination assays, germination rates did not differ between mutant and wild-type pollen (Fig. 7D). Since the AtAMT1;4 protein is able to transport MeA (Fig. 3C, D), in vitro pollen germination in the presence of MeA was analyzed (Fig. 7D). Pollen germination was strongly inhibited by increasing concentrations of MeA in the medium, indicating the acquisition of toxic MeA by pollen during the germination process. However, no significant difference in the pollen germination rate was observed between mutant and wild-type plants.
Discussion
AtAMT1;4 mediates high-affinity ammonium uptake across the plasma membrane
The present study describes the cloning of a new member of the Arabidopsis ammonium transporter family and its functional characterization in yeast and plant roots. First, AtAMT1;4 complemented growth of an ammonium uptake-defective yeast strain using ammonium as the sole nitrogen source (Fig. 1). Secondly, AtAMT1;4 mediated ammonium uptake in roots of transgenic plants overexpressing AtAMT1;4, as assayed by short-term influx studies using 15N-labeled ammonium (Fig. 4A; Supplementary Fig. S1) or by a long-term growth assay on agar supplemented with ammonium as a sole nitrogen source (Fig. 3A, B). In addition, these transgenic plants showed a strongly enhanced growth sensitivity to MeA (Fig. 3C, D), indicating that AtAMT1;4 also permeates this toxic ammonium analog similarly to other members of the AtAMT1 subfamily but unlike AtAMT2 (Gazzarrini et al. 1999, Sohlenkamp et al. 2002, Yuan et al., 2007a).
AtAMT1;4-mediated ammonium uptake into roots presumed a localization of the corresponding protein in the plasma membrane. Indeed, a large majority of the AtAMT1;4 protein expressed in transgenic qko plants partitioned into a plasma membrane-enriched protein fraction (Fig. 2C). Furthermore, using a GFP fusion protein, AtAMT1;4 was shown to be localized to the plasma membrane of pollen grains and pollen tubes (Fig. 6A, B). Thus, plasma membrane localization represents a common feature of all members of plant AMT proteins that have been investigated so far (Sohlenkamp et al. 2002, Ludewig et al. 2003, Simon-Rosin et al. 2003, Loqué et al. 2006, Yuan et al. 2007a), supporting their role in ammonium acquisition by plant cells. Interestingly, in addition to the monomer observed in both roots and shoots, the overexpressed AtAMT1;4 protein formed a putative dimer or a stable complex with another unknown protein of similar size (Fig. 2B, C). A similar feature was also observed in the case of AtAMT1;2, but not for AtAMT1;1 or AtAMT1;3 (Yuan et al. 2007a). This complex probably represents the functional form in plants, since the endogenous AtAMT1;4 protein in pollen was detected exclusively as a dimer (Fig. 5B). However, other members of the AMT/MEP/Rh family have been demonstrated to form trimeric complexes (Khademi et al. 2004, Zheng et al. 2004, Andrade et al. 2005), and homology modeling also suggested a trimeric structure for AtAMT1;1 (Loqué et al. 2007). Considering a high degree of structural conservation within the AMT family, the AtAMT1;4 protein might also form a trimer, but the trimeric complex may be sensitive to SDS and hence become undetectable on SDS–PAGE.
Short-term influx studies revealed that AtAMT1;4 mediates ammonium uptake in the high-affinity range but not in the low-affinity range (Fig. 4A; Supplementary Fig. S1). Taking advantage of the genetic background of qko, in which high-affinity uptake is reduced by >90%, the kinetic properties of AtAMT1;4 were investigated and a Km value for ammonium of 17 μ M was determined in nitrogen-sufficient roots (Fig. 4B, C). Although influx analysis was conducted with an overexpressed protein, the measured Km value most probably also reflects the natural substrate affinity in pollen. In comparison, the Km values of AtAMT1;1 and AtAMT1;3 were determined to be approximately 50–60 μ M, while that of AtAMT1;2 was > 230 μ M in nitrogen-deficient roots (Yuan et al. 2007a). The residual high-affinity uptake capacity in nitrogen-deficient qko roots with a Km of 4.5 μ M was supposed to reflect the activity of AtAMT1;5. Thus, AtAMT1;4 together with AtAMT1;5 represent those ammonium transporters in Arabidopsis plants which exhibit the highest affinity for ammonium.
AtAMT1;4 is expressed specifically in pollen
A major finding of the present study was the localization of AtAMT1;4 exclusively to pollen, which was demonstrated by several independent approaches. First, RNA and protein gel blot analysis showed that the AtAMT1;4 transcript is only detected in flowers but not in other tissues such as roots, leaves, stems or siliques (Fig. 5A, B). Secondly, a more detailed expression in floral tissues was obtained using transgenic plants expressing either an AtAMT1;4-promoter:GFP or an AtAMT1;4-promoter:ORF:GFP construct, and showed the corresponding promoter activity to be localized only to pollen grains and pollen tubes (Figs. 5C, 6). This pollen-specific expression was further confirmed by microarray data from the AtGenExpress project, in which AtAMT1;4 was reported to be specifically expressed in the tissue of stamens (ATGE_36) or mature pollen (ATGE_73), but not in other floral parts, such as sepals, petals, carpels and pedicels, or in any other Arabidopsis tissue (Schmid et al. 2005).
Transcriptome analysis of pollen at four different developmental stages revealed that many membrane transporter genes are developmentally regulated in pollen, most probably reflecting their distinct physiological roles in pollen (Bock et al. 2006, Sze et al. 2006). The genes expressed in early stages (microspores or bicellular pollen) are most likely to be important for pollen development, while those expressed at later stages (tricellular or mature pollen) seem to be more essential for post-pollination events such as pollen tube growth. By these transcriptome analyses, AtAMT1;4 was assigned to a group of pollen-specific and pollen-preferential genes whose expression peaked during early stages, but was low or even undetectable during later stages (Bock et al. 2006). In our study, AtAMT1;4 was found to be constitutively expressed throughout these stages, with a high promoter activity from flower stage 10 to 14, corresponding to the period of microspore development to pollen maturity. The promoter activity of AtAMT1;4 at later developmental stages did not correlate with reported transcript levels, which might be due to a slow turnover of GFP in vivo. Moreover, AtAMT1;4 was also expressed in pollen tubes after their germination in vitro (Figs. 5B, 6; Supplementary Fig. S2). In the case of the monosaccharide transporter gene AtSTP2, which was clustered in the same group as AtAMT1;4 in the above-mentioned transcriptome studies, mRNA and protein were not detected by in situ hybridization and immunohistochemistry in mature pollen or pollen tubes, respectively, whereas promoter-dependent β-glucuronidase (GUS) activity was detectable, which might be supported by the high stability of the soluble GUS protein (Truernit et al., 1999). However, our approach with the AtAMT1;4–GFP fusion protein confirmed expression in mature pollen and pollen tubes (Fig. 6A, B), which does not agree with the transcriptome-based characterization (Bock et al. 2006). It has been shown for several membrane transporters that their mRNA in developing pollen was not translated before hydration and germination of pollen on the stylar tissue (Stadler et al. 1999, Schneidereit et al. 2003, Schneider et al. 2006). It is thus recommended to verify the occurrence of the endogenous AtAMT1;4 protein at certain developmental stages in pollen by an immunohistochemical approach using the AtAMT1;4-specific antibody.
The possible physiological role of AtAMT1;4 in pollen
Nitrogen presents a major limiting factor not only during vegetative plant growth but also during reproductive plant growth, because transport of nitrogen to the developing flowers has a significant influence on flower set, pollen and embryo development, as well as seed production (Lee and Tegeder 2004). It has also been shown that the nitrogen status of pollen donor plants affects kernel set and seed yield in maize (Zhang et al. 2007). Nitrogen import into pollen appears essential for protein synthesis not only during the development of pollen grains in the anthers or during pollen germination but in particular during the rapid pollen tube growth in the pistil. Because of the symplasmic isolation of pollen from the surrounding tissue, nitrogen sources present in the fluid of the anther locule and exudates from the stylar canal need to be taken up via transporters across the plasma membrane of the pollen grains and the pollen tubes, respectively. In general, amino acids are supposed to be the major nitrogen form being imported into pollen, which was supported by studies on the amino acid transporters LeProT1 or NsAAP1 in pollen grains and pollen tubes (Lalanne et al. 1997, Schwacke et al. 1999). Moreover, there are even further transporter homologs in Arabidopsis found to be specifically or at least preferentially expressed in pollen, e.g. several AtLHT genes and AtProT1 (Bock et al. 2006, Sze et al. 2006, Foster et al. 2008). Whether ammonium, a major nitrogen form being continuously generated in metabolically active tissue, is present in the apoplasmic fluid around pollen still remains to be shown. However, we show here that ammonium can be taken up by pollen, because pollen germination in vitro was sensitive to the toxic ammonium analog MeA (Fig. 7D). In addition, proteome studies indicated the presence of glutamine synthetase in the pollen tubes of rice and white pine (Fernando et al. 2005, Dai et al. 2006), and pollen-specific gene expression of AtGLN1;5 (At1g48470) was reported in mature Arabidopsis pollen as well as in growing pollen tubes on the basis of transcriptome data (Honys and Twell 2004, Wang et al. 2008). This suggests that in pollen ammonium can be assimilated into amino acids. The importance of functional ammonium assimilation in pollen has been further supported by antisense NADH-GOGAT alfalfa plants, which were found to be male sterile due to inviable pollen (Schoenbeck et al. 2000). Besides uptake across the plasma membrane, large amounts of cytoplasmic ammonium in pollen may also derive from an internal breakdown of amino acids, particularly in fast growing pollen tubes. Since ammonium efflux across the plasma membrane has been demonstrated in roots and leaves (Wang et al. 1993, Husted et al. 2002), ammonium leakage from pollen or pollen tubes should be expected. In this case, an ammonium retrieval process might be required for pollen nutrition, which would be mediated by high-affinity ammonium transporters, such as AtAMT1;4. Together with other amino acid transporters, AtAMT1;4 may thus contribute to nitrogen nutrition during pollen development, pollen germination and pollen tube growth.
To elucidate further the physiological role of AtAMT1;4 in pollen, the insertion line atamt1;4-6, in which AtAMT1;4 gene expression is completely disrupted, was subjected to phenotypic analysis (Fig. 7). However, there was no visible difference between the mutant line and its corresponding wild type in terms of pollen function. One explanation might be a compensation by other AtAMTs, such as by AtAMT1;1, which is also expressed in mature pollen according to microarray data (Honys and Twell 2004). Additionally, a compensation for the lack of AtAMT1;4 expression might be conferred by amino acid transporters, which may transport organic nitrogen forms into pollen. Considering nitrogen uptake by pollen as an essential prerequisite for pollen function, it is not surprising if this function could be conferred by several independent membrane proteins. For example, expression in pollen of five monosaccharide transporter genes (AtSTP2, 4, 6, 9 and 11) indicates that the import of carbon into pollen is of major importance for pollen survival (Truernit et al. 1999, Schneidereit et al. 2003, Scholz-Starke et al. 2003, Schneidereit et al. 2005). However, so far, no pollen-specific phenotype has been observed in any single knock-out line of these genes. Therefore, the generation of double or multiple knock-out lines might be required before a pollen-specific role for AtAMT1;4 can be observed, similar to the situation in roots, where AtAMTs display non-overlapping but additive functions in nitrogen nutrition (Yuan et al. 2007a).
Materials and Methods
AtAMT1;4 cloning and expression in yeast
The ORF of AtAMT1;4 was amplified by PCR from genomic DNA (Arabidopsis thaliana Col-0) using the specific primers AMT1;4-HindIII, CCCAAGCTTATGGCGTCGGCTCTCTCTTG; and AMT1;4-XhoI, CCGCTCGAGTCAAACACTACATTGGGAT. The amplified fragment was cloned into the pGEM-T Easy vector (Promega, Madison, WI, USA) and DNA sequences were verified. The HindIII–XhoI fragment was subcloned into the yeast expression vector p426-HXT7 (kindly provided by Eckhard Boles, University of Düsseldorf, Germany) resulting in the plasmid, which was used for the transformation of the triple mep deletion yeast strain 31019b (Marini et al. 1997). Growth complementation assays were performed on solid YNB medium supplemented with 3% glucose, and 5 mM arginine or 1 mM ammonium chloride as nitrogen sources, and buffered at pH 6.0 by 50 mM MES-Tris.
Generation of transgenic Arabidopsis plants overexpressing AtAMT1;4
Using the NotI restriction site, the DNA fragment containing the ORF of AtAMT1;4 was released from pGEM-T, and then subcloned into the plant transformation vector pGreen-Hyg which was modified based on pGreen0029 between the cauliflower mosaic virus (CaMV) 35S promoter and terminator sequence. A multiple knock-out line (qko) defective in the expression of AtAMT1;1, 1;2, 1;3 and 2;1 (Yuan et al. 2007a) was transformed by Agrobacterium, and transformants were selected via hygromycin resistance. Homozygous T2 lines were further selected by segregation analysis.
Generation of transgenic Arabidopsis plants expressing AtAMT1;4-promoter:GFP and AtAMT1;4-promoter:ORF:GFP constructs
The 1,935 bp promoter region upstream of the start codon of AtAMT1;4 was amplified by PCR from genomic DNA (A. thaliana Col-0) using the specific primers AMT1;4-pro-BamHI, CGGATCCCCAGACGTTTTGTGAGATGGTAAGAATGTG; and AMT1;4-pro-NcoI, GCCATGGTGTTGCAAAGATTAAGAGAGATTTTGTGAG, and then cloned into the PCR-Blunt II-TOPO vector (Invitrogen, San Diego, CA, USA). The BamHI–NcoI fragment was subcloned into the plant transformation vector pBI101 (Clontech, Palo Alto, CA, USA) upstream of the EGFP coding sequence, resulting in the plasmid AtAMT1;4-promoter:GFP. The fragment of the AtAMT1;4 promoter was inserted in front of the AtAMT1;4 ORF sequence in pGEM-T, and then the AtAMT1;4 promoter:ORF fragment was subcloned into the binary vector pTKan derived from pPZP212 (kindly provided by Karin Schumacher, ZMBP, Tübingen, Germany) at the Bsp120I restriction site. The ORF of AtAMT1;4 was amplified without a stop codon by PCR, ligated to the EGFP coding sequence, and this fragment was introduced into the plasmid AtAMT1;4-promoter:ORF-pTKan using the PstI restriction site and yielding the plasmid AtAMT1;4-promoter:ORF:GFP. Arabidopsis Col-0 transformed with both of these constructs by Agrobacterium were selected via kanamycin resistance, and homozygous T2 lines were further selected by segregation analysis. The fluorescence of GFP in the transgenic plants was observed by an inverted fluorescence microscope equipped with an Apotome (Zeiss Axiovert 200M, Jena, Germany) and by a confocal laser scanning microscope (Olympus, Tokyo, Japan) as described by Loqué et al. (2005, 2006).
Plant culture
Arabidopsis thaliana plants were grown hydroponically as described by Loqué et al. (2006) under non-sterile conditions for 6 weeks (±2 d) in a growth cabinet under the following conditions: 10/14 h light/dark at a temperature of 22°C/18°C and a light intensity of 280 μmol m−2 s−1 at 70% humidity. The nutrient solution contained 1 mM KH2PO4, 1 mM MgSO4, 250 μM K2SO4, 250 μM CaCl2, 2 mM NH4NO3, 100 μM Na-Fe-EDTA, 50 μ M KCl, 50 μM H3BO3, 5 μ M MnSO4, 1 μM ZnSO4, 1 μM CuSO4 and 1 μM NaMoO4, pH adjusted to 6.0 with KOH. In plate growth tests of transgenic lines, Arabidopsis seeds were surface-sterilized and plated onto half-strength Murashige and Skoog (MS) medium (containing 5 mM nitrate as the sole nitrogen source) solidified with Difco agar. The plants were pre-cultured for 7 d and transferred to vertical plates containing half-strength MS medium supplemented with different nitrogen sources at the indicated concentrations. Plants were grown under axenic conditions in a growth chamber under the above-mentioned conditions except that the light intensity was 120 μmol photons m−2 s−1. In addition, the plants were grown in a pre-fertilized peat-based substrate (www.einheitserde.de) in a climate-controlled greenhouse (16/8 h light/dark, temperature 22°C/18°C).
RNA extraction and gel blot analysis
Total RNA was extracted from various tissues by the phenol–guanidine extraction method. Northern blot analyses were performed as described by Loqué et al. (2005). Total RNA (20–30 μ g per lane) was separated by formaldehyde agarose gel electrophoresis, and RNA was transferred to a nylon membrane. DNA fragments carrying the ORFs of AtAMT1;1 or AtAMT1;4 were labeled with 32P as probes for RNA hybridization.
Preparation of membrane fractions and protein gel blot analysis
Total microsomal membrane fractions were extracted from Arabidopsis roots, shoots and flowers, and fractionated by aqueous two-phase partitioning as described by Yuan et al. (2007b). Protein concentrations were determined using the Bradford Protein Assay (Bio-Rad, Munich, Germany) and bovine serum albumin served as the standard. A polyclonal antibody was raised against the peptide representing a sequence of the C-terminus of AtAMT1;4 (n-MVRRVGGDNDPNVGV-c) (Biotrend, Köln, Germany). The antiserum from rabbits was further affinity purified using a nitrocellulose membrane for binding of the corresponding peptide. For protein gel blot analysis, proteins (5–10 μg per lane) were electrophoresed on SDS–polyacrylamide gels, transferred to a polyvinylidene fluoride membrane, and blots were developed using an ECL Advance Western Blotting Detection Kit (Amersham, Little Chalfont, UK). Primary antibodies and secondary antibody (peroxidase-conjugated anti-rabbit IgG, Amersham) were diluted in blocking solution at the following combinations: anti-AtAMT1;4 at 1 : 200 with secondary antibody at 1 : 10,000; anti-AtAHA2 (DeWitt, et al. 1996) at 1 : 20,000, anti-VPPase (Takasu et al. 1997) at 1 : 2,000, and anti-DET3 (Schumacher et al., 1999) at 1 : 20,000 with secondary antibody at 1 : 100,000. Rainbow marker (Amersham) was used as molecular weight marker. Protein blots of DET3, a subunit of the V-ATPase, were used as a control for equal loading.
[15N]Ammonium uptake analysis
Ammonium influx measurements in Arabidopsis roots were conducted after rinsing the roots of hydroponically grown plants in 1 mM CaSO4 solution for 1 min, followed by an incubation during 6 min in nutrient solution containing different concentrations of 15N-labeled NH4+ (95 atom% 15N) as the sole nitrogen source, and a final wash in 1 mM CaSO4 solution. Roots were harvested and stored at –70°C before freeze-drying. Samples were ground and approximately 1.6 mg of powder was used for 15N determination by isotope mass spectrometry (Thermo-Finnigan, Bremen, Germany).
Isolation of insertion lines for AtAMT1;4
Information about T-DNA and transposon mutants for the AtAMT1;4 gene was obtained from the SIGnAL database (Alonso et al. 2003). The atamt1;4-1 (SAIL_912_E05), atamt1;4-2 (SAIL_761_F02) and atamt1;4-3 (SAIL_557_C08) lines were obtained from the Syngenta SAIL collection (Torrey Mesa Research Institute, San Diego, CA, USA). The atamt1;4-4 (194B10) and atamt1;4-5 (359E07) lines were obtained from the GABI-KAT collection (MPIZ, Köln, Germany) (Rosso et al. 2003). The atamt1;4-6 (53-2537-1) line was obtained from RIKEN BioResource Center (Tsukuba, Japan). The genotypes of plants were determined by PCR using left or right border-specific primers and gene-specific primers, and the locations of the insertions were verified by sequencing of the PCR products.
Pollen germination
Mature pollen was harvested from flowers at stage 13. Pollen was germinated on a liquid or solid agar medium containing 0.6% agar, 1.65 mM boric acid, 2 mM CaCl2, 17% sucrose (adjusted to pH 7.0 with 1 M NaOH), and incubated for 12–24 h at room temperature at 100% humidity.
Supplementary data
Supplementary data are available at PCP online.
Funding
Financial support from the Deutsche Forschungsgemeinschaft (WI1728/4) and the European International Association for the promotion of cooperation with scientists from the New Independent States of the former Soviet Union (INTAS) initiative is gratefully acknowledged.
Supplementary Material
Acknowledgments
We thank Elke Dachtler and Susanne Reiner, University of Hohenheim, for skillful technical support. We are grateful to Dr. Masayoshi Maeshima, University of Nagoya, for the VPPase antibody, and to Dr. Karin Schumacher, ZMBP Tübingen, for kindly providing the AHA2 and DET3 antibodies, and pPTKan plant transformation vector. We also thank Torrey Mesa Research Institute (San Diego, CA, USA) for SAIL lines, MPIZ (Köln, Germany) for GABI-KAT lines, and RIKEN BioResource Center (Tsukuba, Japan) for the transposon-tagged line.
Glossary
Abbreviations:
- AMT/MEP/Rh
ammonium transporter/methylamine permease/rhesus
- CaMV
cauliflower mosaic virus
- GFP
green fluorescent protein
- GUS
β-glucuronidase
- MeA
methylammonium
- MS
Murashige and Skoog
- ORF
open reading frame
- V-ATPase
vauolar ATPase
- VPPase
vacuolar pyrophosphatase.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shin P, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Andrade SL, Dickmanns A, Ficner R, Einsle O. Crystal structure of the archaeal ammonium transporter Amt-1 from Archaeoglobus fulgidus. Proc. Natl Acad. Sci. USA. 2005;102:14994–14999. doi: 10.1073/pnas.0506254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock KW, Honys D, Ward JM, Padmanaban S, Nawrocki EP, Hirschi KD, et al. Integrating membrane transport with male gametophyte development and function through transcriptomics. Plant Physiol. 2006;140:1151–1168. doi: 10.1104/pp.105.074708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S, Li L, Chen T, Chong K, Xue Y, Wang T. Proteomic analyses of Oryza sativa mature pollen reveal novel proteins associated with pollen germination and tube growth. Proteomics. 2006;6:2504–2529. doi: 10.1002/pmic.200401351. [DOI] [PubMed] [Google Scholar]
- DeWitt ND, Hong B, Sussman MR, Harper JF. Targeting of two Arabidopsis H(+)-ATPase isoforms to the plasma membrane. Plant Physiol. 1996;112:833–844. doi: 10.1104/pp.112.2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando DD. Characterization of pollen tube development in Pinus strobus (Eastern white pine) through proteomic analysis of differentially expressed proteins. Proteomics. 2005;5:4917–4926. doi: 10.1002/pmic.200500009. [DOI] [PubMed] [Google Scholar]
- Foster J, Lee YH, Tegeder M. Distinct expression of members of the LHT amino acid transporter family in flowers indicates specific roles in plant reproduction. Sexual Plant Reprod. 2008;21:143–152. [Google Scholar]
- Gazzarrini S, Lejay L, Gojon A, Ninnemann O, Frommer WB, von Wirén N. Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into Arabidopsis roots. Plant Cell. 1999;11:937–948. doi: 10.1105/tpc.11.5.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Twell D. Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 2004;5:R85. doi: 10.1186/gb-2004-5-11-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husted S, Mattsson M, Mollers C, Wallbraun M, Schjoerring JK. Photorespiratory NH4+ production in leaves of wild-type and glutamine synthetase 2 antisense oilseed rape. Plant Physiol. 2002;130:989–998. doi: 10.1104/pp.006759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser BN, Rawat SR, Siddiqi MY, Masle J, Glass AD. Functional analysis of an Arabidopsis T-DNA ‘knockout’ of the high-affinity NH4+ transporter AtAMT1;1. Plant Physiol. 2002;130:1263–1275. doi: 10.1104/pp.102.010843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khademi S, O’Connell J., III, Remis J, Robles-Colmenares Y, Miercke LJ, Stroud RM. Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 Å. Science. 2004;305:1587–1594. doi: 10.1126/science.1101952. [DOI] [PubMed] [Google Scholar]
- Lalanne E, Mathieu C, Roche O, Vedel F, De Pape R. Structure and expression of a Nicotiana sylvestris putative amino acid transporter gene in mature and in vivo germinating pollen. Plant Mol. Biol. 1997;35:855–864. doi: 10.1023/a:1005812419151. [DOI] [PubMed] [Google Scholar]
- Lee YH, Tegeder M. Selective expression of a novel high-affinity transport system for acidic and neutral amino acids in the tapetum cells of Arabidopsis flowers. Plant J. 2004;40:60–74. doi: 10.1111/j.1365-313X.2004.02186.x. [DOI] [PubMed] [Google Scholar]
- Loqué D, Lalonde S, Loger LL, von Wirén N, Frommer WB. A cytosolic trans-activation domain essential for ammonium uptake. Nature. 2007;446:195–198. doi: 10.1038/nature05579. [DOI] [PubMed] [Google Scholar]
- Loqué D, Ludewig U, Yuan L, von Wirén N. Tonoplast intrinsic proteins AtTIP2;1 and AtTIP2;3 facilitate NH3 transport into the vacuole. Plant Physiol. 2005;137:671–680. doi: 10.1104/pp.104.051268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loqué D, von Wirén N. Regulatory levels for the transport of ammonium in plant roots. J. Exp. Bot. 2004;55:1293–1305. doi: 10.1093/jxb/erh147. [DOI] [PubMed] [Google Scholar]
- Loqué D, Yuan L, Kojima S, Gojon A, Wirth J, Gazzarrini S, et al. Additive contribution of AMT1;1 and AMT1;3 to high-affinity ammonium uptake across the plasma membrane of nitrogen-deficient Arabidopsis roots. Plant J. 2006;48:522–534. doi: 10.1111/j.1365-313X.2006.02887.x. [DOI] [PubMed] [Google Scholar]
- Ludewig U, von Wirén N, Frommer WB. Uniport of NH4+ by the root hair plasma membrane ammonium transporter LeAMT1;1. J. Biol. Chem. 2002;277:13548–13555. doi: 10.1074/jbc.M200739200. [DOI] [PubMed] [Google Scholar]
- Ludewig U, Wilken S, Wu B, Jost W, Obrdlik P, El Bakkoury M, et al. Homo- and hetero-oligomerization of ammonium transporter-1 NH4+ uniporters. J. Biol. Chem. 2003;278:45603–45610. doi: 10.1074/jbc.M307424200. [DOI] [PubMed] [Google Scholar]
- Marini AM, Soussi-Boudekou S, Vissers S, André B. A family of ammonium transporters in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997;17:4282–4293. doi: 10.1128/mcb.17.8.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M, Dynowski M, Ludewig U. Ammonium ion transport by the AMT/Rh homologue LeAMT1;1. Biochem. J. 2006;396:431–437. doi: 10.1042/BJ20060051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina C, Pinto F, Feijo JA, Becker JD. Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol. 2005;138:744–756. doi: 10.1104/pp.104.057935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoche P, Bertrand O, Gane P, Birkenmeier C, Colin Y, Cartron JP. Human Rhesus-associated glycoprotein mediates facilitated transport of NH3 into red blood cells. Proc. Natl Acad. Sci. USA. 2004;101:17222–17227. doi: 10.1073/pnas.0403704101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 2003;53:247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a. [DOI] [PubMed] [Google Scholar]
- Sauer N, Stadler R. A sink-specific H+/monosaccharide co-transporter from Nicotiana tabacum: cloning and heterologous expression in baker's yeast. Plant J. 1993;4:601–610. doi: 10.1046/j.1365-313x.1993.04040601.x. [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, et al. A gene expression map of Arabidopsis thaliana development. Nature Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Schneider S, Schneidereit A, Konrad KR, Hajirezaei MR, Gramann M, Hedrich R, et al. Arabidopsis INOSITOL TRANSPORTER4 mediates high-affinity H+ symport of myoinositol across the plasma membrane. Plant Physiol. 2006;141:565–577. doi: 10.1104/pp.106.077123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidereit A, Scholz-Starke J, Buttner M. Functional characterization and expression analyses of the glucose-specific AtSTP9 monosaccharide transporter in pollen of Arabidopsis. Plant Physiol. 2003;133:182–190. doi: 10.1104/pp.103.026674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidereit A, Scholz-Starke J, Sauer N, Büttner M. AtSTP11, a pollen tube-specific monosaccharide transporter in Arabidopsis. Planta. 2005;221:48–55. doi: 10.1007/s00425-004-1420-5. [DOI] [PubMed] [Google Scholar]
- Schoenbeck MA, Temple SJ, Trepp GB, Blumenthal JM, Samac DA, Gantt JS, et al. Decreased NADH glutamate synthase activity in nodules and flowers of alfalfa (Medicago sativa L.) transformed with an antisense glutamate synthase transgene. J. Exp. Bot. 2000;51:29–39. [PubMed] [Google Scholar]
- Scholz-Starke J, Buttner M, Sauer N. AtSTP6, a new pollen-specific H+-monosaccharide symporter from Arabidopsis. Plant Physiol. 2003;131:70–77. doi: 10.1104/pp.012666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher K, Vafeados D, McCarthy M, Sze H, Wilkins T, Chory J. The Arabidopsis det3 mutant reveals a central role for the vacuolar H(+)-ATPase in plant growth and development. Genes Dev. 1999;13:3259–3270. doi: 10.1101/gad.13.24.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R, Grallath S, Breitkreuz KE, Stransky E, Stransky H, Frommer WB, et al. LeProT1, a transporter for proline, glycine betaine, and gamma-amino butyric acid in tomato pollen. Plant Cell. 1999;11:377–392. doi: 10.1105/tpc.11.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R, Hodge R, Paul W, Soufleri I, Draper J. The molecular biology of anther differentiation. Plant Sci. 1991;80:167–191. doi: 10.1007/BF00039494. [DOI] [PubMed] [Google Scholar]
- Simon-Rosin U, Wood C, Udvardi MK. Molecular and cellular characterisation of LjAMT2;1, an ammonium transporter from the model legume Lotus japonicus. Plant Mol. Biol. 2003;51:99–108. doi: 10.1023/a:1020710222298. [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowmanm JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohlenkamp C, Wood CC, Roeb GW, Udvardi MK. Characterization of Arabidopsis AtAMT2, a high-affinity ammonium transporter of the plasma membrane. Plant Physiol. 2002;130:1788–1796. doi: 10.1104/pp.008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler R, Truernit E, Gahrtz M, Sauer N. The AtSUC1 sucrose carrier may represent the osmotic driving force for anther dehiscence and pollen tube growth in Arabidopsis. Plant J. 1999;19:269–278. doi: 10.1046/j.1365-313x.1999.00527.x. [DOI] [PubMed] [Google Scholar]
- Sze H, Frietsch S, Li X, Bock KW, Harper JF. Genomic and molecular analyses of transporters in the male gametophyte. In: Malhó R, editor. The Pollen Tube: A Cellular and Molecular Perspective. Heidelberg: Springer Berlin; pp. 71–93. [Google Scholar]
- Takasu A, Nakanishi Y, Yamauchi T, Maeshima M. Analysis of the substrate binding site and carboxyl terminal region of vacuolar H+-pyrophosphatase of mung bean with peptide antibodies. J. Biochem. 1997;122:883–889. doi: 10.1093/oxfordjournals.jbchem.a021837. [DOI] [PubMed] [Google Scholar]
- Truernit E, Stadler R, Baier K, Sauer N. A male gametophyte-specific monosaccharide transporter in Arabidopsis. Plant J. 1999;17:191–201. doi: 10.1046/j.1365-313x.1999.00372.x. [DOI] [PubMed] [Google Scholar]
- Uelker B, Peiter E, Dixon DP, Moffat C, Capper R, Bouch N, et al. Getting the most out of publicly available T-DNA insertion lines. Plant J. 2008 doi: 10.1111/j.1365-313X.2008.03608.x. 10.1111/j.1365-313X.2008.03608.x. [DOI] [PubMed] [Google Scholar]
- Wirén N, Merrick M. Regulation and function of ammonium carriers in bacteria, fungi and plants. Trends Curr. Genet. 2004;9:95–120. [Google Scholar]
- Wang MY, Siddiqi MY, Ruth TJ, Glass A. Ammonium uptake by rice roots (I. Fluxes and subcellular distribution of 13NH4+). Plant Physiol. 1993;103:1249–1258. doi: 10.1104/pp.103.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang W.-Z, Song L.-F, Zou J.-J, Su Z, Wu W.-H. Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol. 2008;148:1201–1211. doi: 10.1104/pp.108.126375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Loqué D, Kojima S, Rauch S, Ishiyama K, Inoue E, et al. The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. Plant Cell. 2007a;19:2636–2652. doi: 10.1105/tpc.107.052134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Loqué D, Ye F, Frommer WB, von Wirén N. Nitrogen-dependent posttranscriptional regulation of the ammonium transporter AtAMT1;1. Plant Physiol. 2007b;143:732–744. doi: 10.1104/pp.106.093237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Bittman S, Hunt DE, Schaber MM. Nitrogen status of pollen donor affects kernel set and yield components in corn. J. Plant Nutr. 2007;30:1205–1212. [Google Scholar]
- Zheng L, Kostrewa D, Berneche S, Winkler FK, Li XD. The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli. Proc. Natl Acad. Sci.USA. 2004;101:17090–17095. doi: 10.1073/pnas.0406475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.