Abstract
High salinity and drought have received much attention because they severely affect crop production worldwide. Analysis and comprehension of the plant's response to excessive salt and dehydration will aid in the development of stress-tolerant crop varieties. Signal transduction lies at the basis of the response to these stresses, and numerous signaling pathways have been implicated. Here, we provide further evidence for the involvement of phospholipase D (PLD) in the plant's response to high salinity and dehydration. A tomato (Lycopersicon esculentum) α-class PLD, LePLDα1, is transcriptionally up-regulated and activated in cell suspension cultures treated with salt. Gene silencing revealed that this PLD is indeed involved in the salt-induced phosphatidic acid production, but not exclusively. Genetically modified tomato plants with reduced LePLDα1 protein levels did not reveal altered salt tolerance. In Arabidopsis (Arabidopsis thaliana), both AtPLDα1 and AtPLDδ were found to be activated in response to salt stress. Moreover, pldα1 and pldδ single and double knock-out mutants exhibited enhanced sensitivity to high salinity stress in a plate assay. Furthermore, we show that both PLDs are activated upon dehydration and the knock-out mutants are hypersensitive to hyperosmotic stress, displaying strongly reduced growth.
Keywords: Phospholipase D, High salinity, Drought, Phosphatidic acid, Arabidopsis, Tomato
Introduction
High salinity and hyperosmotic stress are major determinants of crop yield throughout the world (Epstein et al. 1980, Boyer 1982). Hyperosmotic extracellular conditions lead to a loss of turgor that necessitates a response from the plant in order to survive. Plants respond by accumulating intracellular osmolites and reducing water loss, consequently reacquiring their hypertonic state and regaining/maintaining rigidity (Yancey et al. 1982, Fricke 2004, Li et al. 2006). High salinity stress is also detrimental because it disturbs ion homeostasis. High cytosolic sodium concentrations are toxic and plants respond to this condition by removing the sodium from the cytosol, making use of antiporter and co-porter ion channel activity across the plasma and vacuolar membranes. This ion flux causes a loss of homeostasis that has to be actively restored (Kinraide 1998, Zhu 2003, Yamaguchi and Blumwald 2005).
Salt and drought tolerance depend on complex signaling networks, allowing plants to respond rapidly and efficiently to the stress (Zhu 2002). Signal transduction in response to these stresses has become an intensively studied subject because it is believed that a better understanding of this process will lead to the discovery of ways to generate stress-tolerant crops that do not have a fitness penalty (Kasuga et al. 1999, Flowers 2004, Jakab et al. 2005, Yamaguchi and Blumwald 2005). Numerous signal transduction pathways have been demonstrated to be activated in response to high salinity and hyperosmotic stress (Munnik and Meijer 2001, Xiong et al. 2002, Zhu 2002). Our interest focuses especially on lipid signaling events and in particular on phospholipase D (PLD) activity during such processes.
PLD catalyzes the hydrolysis of structural phospholipids, e.g. phosphatidylcholine, producing phosphatidic acid (PA) and a free head group. Twelve PLD genes are present in the genome of the model plant thale cress (Arabidopsis thaliana), whereas just two PLD genes are found in animals and one in yeast. The plant PLD family can be partitioned into six classes, the α-, β-, γ-, δ-, ε- and ζ-PLDs, depending on protein sequence homology and biochemical properties (Qin and Wang 2002, Wang 2005). Roles have been suggested for PLD in numerous processes including vesicular transport, membrane degradation and intracellular signaling. PLD has been reported to be involved in signaling events occurring in response to a multitude of stimuli, e.g. freezing, wounding, plant–pathogen interactions, dehydration and salt stress (Wang 2002, Bargmann and Munnik 2006). PA is believed to act as a second messenger in such signaling events; it is generated rapidly and transiently during various stress responses (Munnik 2001), functioning in signaling cascades by recruiting target proteins to particular membranes and/or influencing their activity (Testerink and Munnik 2005, Wang 2005).
PLD has been linked to high salinity and hyperosmotic stress in several independent studies. Tomato (Lycopersicon esculentum) LePLDα1 gene expression increases in cell suspensions treated with NaCl (Laxalt et al. 2001) and AtPLDδ expression is induced upon both high salinity and dehydration treatments in Arabidopsis (Katagiri et al. 2001, Mane et al. 2007). PLD is activated in response to hyperosmotic stress and dehydration (Frank et al. 2000, Munnik et al. 2000, Katagiri et al. 2001). Arabidopsis plants expressing antisense AtPLDα1 have been shown to have increased sensitivity to drought stress (Sang et al. 2001, Mane et al. 2007). Additionally, Arabidopsis pldα3 knock-out mutants have recently been shown to be hypersensitive to both salt and hyperosmotic stress, and AtPLDα3-overexpressing plants were found to be more resistant to these stresses (Hong et al. 2008). Furthermore, PLD has been suggested to be a negative regulator of the biosynthesis of the osmolite proline in Arabidopsis (Thiery et al. 2004).
This investigation examines the role of PLDα and δ in high salinity as well as water deficit stress, employing LePLDα1-silenced tomato cell suspension cultures and plants, in addition to Arabidopsis pldα1 and pldδ single and double knock-out mutants. We demonstrate that both classes are activated by the different stresses and that lack of either AtPLDα1 or AtPLDδ leads to decreased tolerance on high salt-containing medium and hyperosmotic medium. Notably, the Arabidopsis pldα1/pldδ double mutant is even more sensitive than either of the single knock-out mutants. The results and their implications are discussed.
Results
High salinity-induced PLD activity in LePLDα1-silenced tomato cell suspension cultures
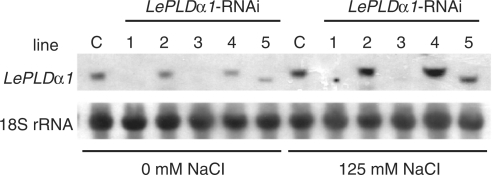
Plant cell suspension cultures are a useful model system for biochemical research because they can be treated with various conditions in parallel, are homogeneous and can be genetically modified relatively easily and quickly. The tomato cell suspension culture Msk8 has been used in several studies to investigate plant responses to both biotic (Felix et al. 1991, Felix et al. 1999) and abiotic stress (Felix et al. 2000). Our laboratory has also used this cell suspension culture in the study of lipid signaling and especially the function of PLD therein (Munnik et al. 2000, van der Luit et al. 2000, Laxalt et al. 2001, Bargmann et al. 2006). LePLDα1 gene expression in Msk8 cultures has been knocked-down using an RNAi (RNA interference) construct targeting the 3′-untranslated region (UTR); the UTR was used to minimize chances of unspecific cross-silencing (see materials and methods). Five independently transformed cell suspension culture lines were obtained as well as an empty vector control line. RNA blot analysis showed that three of the five lines carrying the RNAi construct were in effect completely silenced (Fig. 1). Analysis of the expression of other PLD isoforms (Laxalt et al. 2001) indicated that only LePLDα1 was knocked-down in these lines (data not shown). Although there was an apparently truncated transcript visible in line #5 (Fig. 1), the LePLDα1 protein was absent and PLDα activity was deficient in all silenced lines (data not shown). Salt treatment induced a clear increase in LePLDα1 expression in the empty vector control and the non-silenced cultures (lines #2 and #4), which was lacking in the silenced lines #1 and #3 and truncate in line #5.
Fig. 1.
Silencing and salt-induced expression of LePLDα1 in tomato cell suspension cultures. RNA was extracted from independently transformed Msk8 cultures carrying an LePLDα1-RNAi construct (lines #1–5) or an empty control vector (C) that had been treated with 0 or 125 mM NaCl for 5 h. RNA was separated by gel electrophoresis, blotted and hybridized with a 32P-labeled LePLDα1 probe. An 18S rRNA probe was used as a loading control.
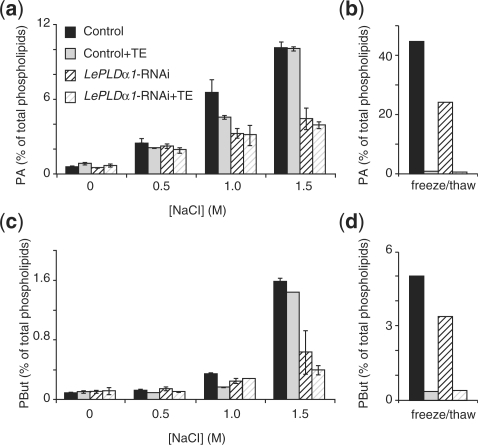
PA increases in response to salt stress have been reported to be produced by both the PLD and phospholipase C/diacylglycerol kinase pathways (PLC/DGK; Munnik et al. 2000, Katagiri et al. 2001). Addition of primary alcohols allows for the distinction of PLD activity from other PA-producing reactions, making use of this enzyme's ability to substitute primary alcohols for water in its transphosphatidylation reaction, whereas non-primary alcohols (e.g. sec- and tert-butanol) cannot be used in such a reaction (Munnik et al. 1995, Dhonukshe et al. 2003). In this case we used n-butanol to visualize PLD activity separately from DGK-generated PA. In the presence of 0.5% n-butanol, high salt treatment of cell suspension cultures led to a dose-dependent accumulation of PA and phosphatidylbutanol (PBut; Fig. 2a, c). A 15 min treatment with 500 mM NaCl led to a clear PA accumultion, while treatment with 1 and 1.5 M NaCl induced a marked increase in both PA and PBut. A manifest reduced induction of PA and PBut formation was evident when LePLDα1-silenced lines were treated with high salt concentrations (e.g. Fig. 2a, c). These results show that LePLDα1 activity is induced in response to high salinity stress in a tomato cell suspension culture. The fact that some salt-induced PLD activity still remained in the silenced cell suspension culture suggests that at least one other PLD is activated under these conditions.
Fig. 2.
Salt-induced LePLDα1 activity in tomato cell suspension cultures. 32Pi-labeled control or LePLDα1-silenced (line 1) cell suspensions were left untreated, snap-frozen and thawed or treated with an equal volume of cell-free medium supplemented with increasing NaCl concentrations for 15 min, either without buffer or buffered with 50 mM Tris–HCl pH 7.5 and 10 mM EGTA (TE). Lipids were extracted, separated by TLC and analyzed by phosphoimaging. PA (a and b) and PBut (c and d) were quantified as a percentage of total radiolabeled lipids and are presented in a histogram (salt treatment n = 2, min and max values indicated; freeze/thaw n = 1).
Reduced PLD activity in the LePLDα1-silenced line could only be detected after severe salt treatments of ≥1 M NaCl. The finding that PLDs are also active upon loss of cell membrane integrity (B.O.R.B. and T.M., unpublished) prompted us to investigate whether PLD activity, after these high salt treatments, might be due to a ‘cell lysis’ effect. As shown in Fig. 2b and d, cells that were lysed by snap-freezing and thawing displayed a marked increase in PLD activity, and this activity could be completely inhibited when cells were buffered in 50 mM Tris–HCl pH 7.5 and 10 mM EGTA (TE). However, when cell suspension cultures were treated with high salt concentrations in the presence of TE, a PLD activation similar to unbuffered samples was observed (Fig. 2a, c). This was true for both control- and LePLDα1-silenced cell suspension cultures. Correspondingly, vitality staining of salt-treated cell suspension cultures with fluorescein diacetate revealed that no loss of plasma membrane integrity had occurred (data not shown). These results suggest that the increase in LePLDα1 activity in response to high salt concentrations is not due to loss of cell membrane integrity.
Silencing LePLDα1 in tomato plants
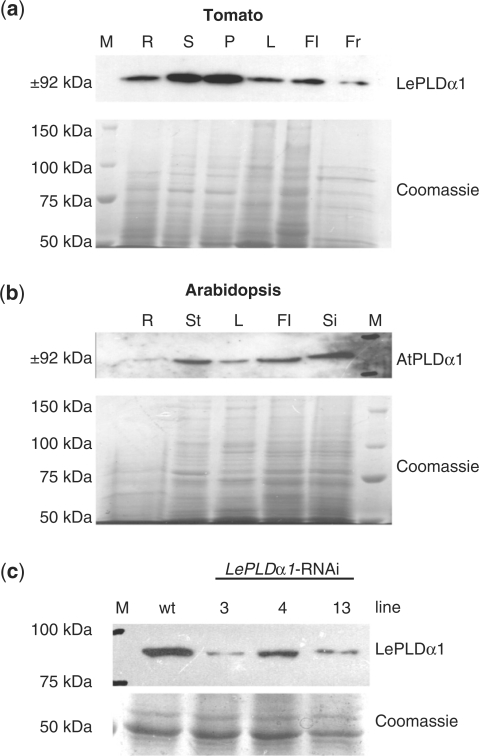
Previously, Laxalt et al. (2001) reported that the LePLDα1 transcript was present in all tested tomato plant organs. Here, expression levels were analyzed using a peptide-specific PLDα1 antibody. As shown in Fig. 3a, LePLDα1 could be detected in roots, stems, petioles, leaves, flowers and fruit of mature tomato plants. This ubiquitous presence of LePLDα1 in tomato mirrors AtPLDα1 expression in Arabidopsis, which could also be detected in all tested organs. AtPLDα1 was detected in roots, leaves, stems, flowers and siliques (Fig. 3b). In order to study LePLDα1 function in the salt tolerance of tomato plants, transgenic lines were generated carrying the same RNAi construct that was used to knock-down LePLDα1 in the Msk8 cell suspension cultures. Several independent transformants were obtained and three lines were selected in which knock-down of LePLDα1 was verified by protein blot analysis of seedlings (Fig. 3c). Compared with wild-type tomato seedlings (GCR161), the LePLDα1-silenced plant lines had reduced LePLDα1 protein levels. Nonetheless, a protein band could still be detected in the different silenced lines (Fig. 3c). LePLDα1-silenced tomato plants developed normally, displaying no obvious growth phenotype.
Fig. 3.
Silencing LePLDα1 in tomato plants. (a) Proteins were extracted from roots (R), stems (S), petioles (P), leaves (L), flowers (Fl) and fruit (Fr) harvested from mature tomato plants. Proteins were separated by SDS–PAGE and blotted or stained with Coomassie brilliant blue as a loading control. A precision protein marker (M) was used to gauge the size of the detected band. (b) Protein blot analysis of AtPLDα1 protein levels was performed on proteins extracted from roots (R), inflorescence stems (St), leaves (L), flowers (Fl) and siliques (Si) of flowering Arabidopsis plants. (c) Protein blot analysis of LePLDα1 protein levels was performed on proteins extracted from 1-week-old wild-type (wt) and LePLDα1-silenced tomato seedlings.
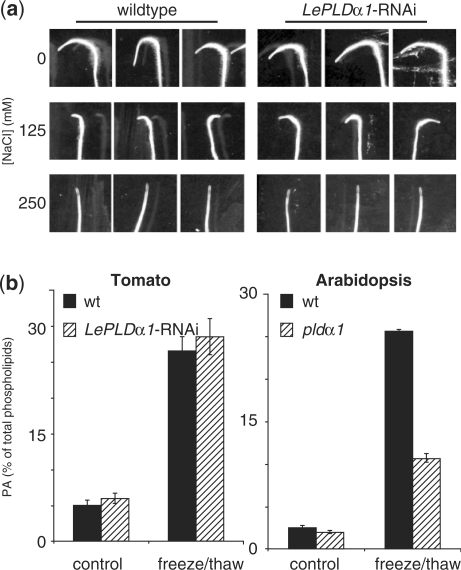
Salt tolerance can easily be assayed by transferring 1-week-old seedlings to agar plates supplemented with NaCl and assessing their root growth (Wu et al. 1996). Accordingly, seedlings were transferred to new plates containing 0, 125 or 250 mM NaCl, and rotated 180° to be able to monitor the new growth. As shown in Fig. 4a, primary root growth after 24 h was greatly reduced on plates with 125 mM NaCl compared with control plates, and was completely abolished when seedlings were transferred to plates containing 250 mM NaCl. LePLDα1-silenced plant lines had the same magnitude of growth reduction on plates containing 125 mM NaCl as the wild-type control lines and also exhibited complete growth inhibition on 250 mM NaCl. These results show that, although it was able to reduce LePLDα1 protein levels, the introduction of a LePLDα1-RNAi construct did not lead to altered salt tolerance in tomato seedlings.
Fig. 4.
Salt tolerance and lysis-induced PLD activity in LePLDα1-silenced tomato plants. (a) Tomato seeds from wild-type and LePLDα1-silenced plant lines were sown on agar plates and grown vertically in a growth chamber. After 1 week, seedlings were transferred to fresh plates supplemented with 0, 125 or 250 mM NaCl, rotated 180° and placed back in the growth chamber. Plates were scanned after 24 h. A representative silenced line is shown (line #3). (b) Leaf discs were excised from fully expanded leaves from wild-type and LePLDα1-silenced (line #13) tomato plants as well as wild-type and pldα1 knock-out Arabidopsis plants, and labeled overnight with 32Pi. Leaf discs were either left untreated or snap-frozen and thawed for 15 min. Lipids were extracted, separated by TLC and analyzed by phosphoimaging. PA was quantified as a percentage of total phospholipids and is presented in histograms ± SD (n = 3).
PLDα1 deficiency leads to reduced cell lysis-induced PLD activity in tomato cell suspension cultures and Arabidopsis knock-out lines (Figs. 2 and 4b right panel). However, when phospholipid analysis was performed on LePLDα1-silenced plants, no consistent reduction in cell lysis-induced PLD activity could be observed in LePLDα1-silenced plant lines compared with the wild type (Fig. 4b, left panel). This result suggest that the reduction of LePLDα1 protein levels seen in the LePLDα1-silenced tomato plant lines is not sufficient to cause an observable decrease in PLD activity upon snap-freezing and thawing of leaf discs.
Salt and water deficit tolerance in Arabidopsis requires both AtPLDα1 and AtPLDδ
The lack of a detectable phenotype in silenced tomato plants prompted us to continue our research into PLD involvement in high salinity tolerance in Arabidopsis, where knock-out mutants are available for multiple PLD isoforms. Whereas gene silencing can be incomplete, non-specific and inconsistent, T-DNA insertional mutagenisis gives a higher degree of confidence that the gene of interest is no longer functional and that no other genes are affected. AtPLDα1 and AtPLDδ are the two predominant PLD isoforms present in Arabidopsis and, for both, silencing has been linked to altered responses to water stress (Katagiri et al. 2001, Sang et al. 2001). T-DNA insertion lines were obtained for both genes (SALK_067533 and SALK_023247), and a pldα1/pldδ double mutant was generated by crossing the two. T-DNA insertion was verified by PCR, and knock-out was confirmed on the protein and transcript level for AtPLDα1 and AtPLDδ, respectively (data not shown). Both the individual and double mutants developed normally, exhibiting no obvious phenotype when grown under standard greenhouse or growth chamber conditions.
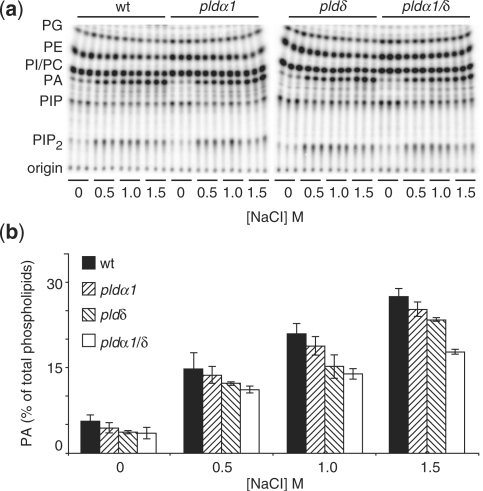
When leaf discs from wild-type (Col-0), pldα1, pldδ and pldα1/pldδ Arabidopsis plants were labeled and lipids were extracted and analyzed, no difference in basal composition levels could be detected (Fig. 5). Treatment of leaf discs with high salt concentrations caused up to a 5-fold increase in PA levels within 15 min in wild-type plants. However, when the pld mutants were treated with high salt concentrations, a consistently lower PA response was found compared with the wild type: wild type > pldα1 > pldδδ> pldα1/pldδ double mutants (Fig. 5b). This outcome demonstrates that AtPLDα1 and AtPLDδ are activated in unison during the response to high salt exposure in Arabidopsis.
Fig. 5.
Salt-induced PLD activity in Arabidopsis T-DNA insertion lines. Leaf discs were excised from fully expanded leaves from control and pld knock-out plant lines and labeled overnight with 32Pi. Leaf discs were treated with increasing NaCl concentrations for 15 min. Lipids were extracted, separated by alkaline TLC (a) and analyzed by phosphoimaging (b). PA was quantified as a percentage of total radiolabeled lipids and is presented in a histogram ± SD (n = 3).
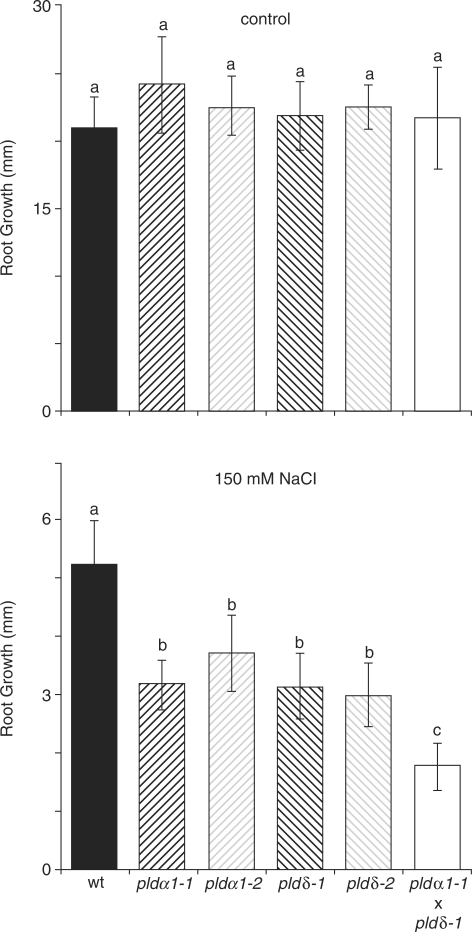
As with LePLDα1-silenced tomato plants, root growth of pld knock-out mutants and wild-type Arabidopsis seedlings was assayed under high salinity conditions (Fig. 6). When 4-day-old seedlings were transferred to control plates (supplemented with 0 mM NaCl), no reduced growth compared with the wild type was seen in pldα1, pldδ or pldα1/pldδ mutant seedlings. However, transfer to plates containing 150 mM NaCl caused a significant reduction in root growth in mutant vs. wild-type seedlings (Fig. 6). Statistical analysis revealed that pldα1 and pldδδ mutants grouped in a class of their own, displaying approximately 60% of the growth seen in the wild type after 4 d. Interestingly, pldα1/pldδ double mutants exhibited significantly less growth than both the wild type and the single mutants, which was about 40% of that measured in the wild type (Fig. 6). Independent T-DNA insertions, pldα1-2 (SALK_053785) and pldδ-2 (SALK_023808), were included to verify that the observed effect was not due to secondary insertions. These results show that AtPLDα1 and AtPLDδ are both required for the salt-induced PA production and salt tolerance in Arabidopsis, and that their combined deletion gives rise to plants that are even more sensitive than the single mutants alone.
Fig. 6.
Reduced salt tolerance in Arabidopsis pld mutants. Seeds from wild-type, pldα1-1, pldα1-2, pldδ-1, pldδ-2 and pldα1-1/pldδ-1 knock-out mutant lines were sown on agar plates and grown vertically in a growth chamber. After 4 d, seedlings were transferred to fresh plates supplemented with 0 or 150 mM NaCl. Primary root growth was measured 4 d after transfer and is represented in histograms ± SD. Data were analyzed for significance by one-way ANOVA (Tukey post hoc, α = 0.001, n = 18–20).
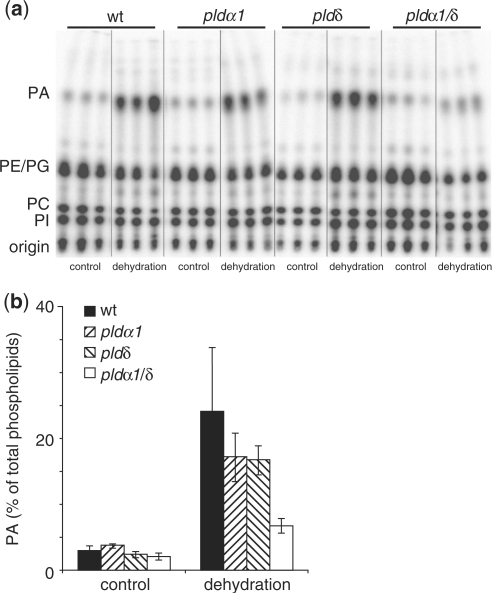
Earlier studies have also demonstrated PLD activation in response to hyperosmotic stresses other than high salinity, both in cell suspension cultures (Munnik et al. 2000) and in plants (Frank et al. 2000, Katagiri et al. 2001, Hong et al. 2008). Katagiri and co-workers (2001) went on to show that the activity in dehydrated leaf discs of Arabidopsis plants expressing an antisense AtPLDδ construct was reduced compared with wild-type plants, implicating this PLD isoform as one of the PLDs activated under hyperosmotic conditions. When leaf discs from the pld single and double knock-outs were tested for dehydration-induced PLD activity, a clear difference could be noted between the wild type, pldα1, pldδ and pldα1/pldδ mutants (Fig. 7). PA production in the single mutants was inferior to that in the wild type and lower still in the double mutant. The single mutants showed, on average, 70% of the PA level observed in the wild type, whereas the double mutant displayed only 30% (Fig. 7). This finding shows that both AtPLDα1 and AtPLDδ are activated upon drought stress in Arabidopsis plants, and that they together are responsible for the bulk of the PA formed in response to this stress.
Fig. 7.
Dehydration-induced PLD activity in Arabidopsis T-DNA insertion lines. Leaf discs were excised from fully expanded leaves from control and pld knock-out plant lines and labeled overnight with 32Pi. Leaf discs were treated by removing them from the labeling solution and placing them on filtration paper for 2 h. Lipids were extracted, separated by ethyl acetate TLC (a) and analyzed by phosphoimaging (b). PA was quantified as a percentage of total radiolabeled lipids and is presented in a histogram ± SD (n = 3).
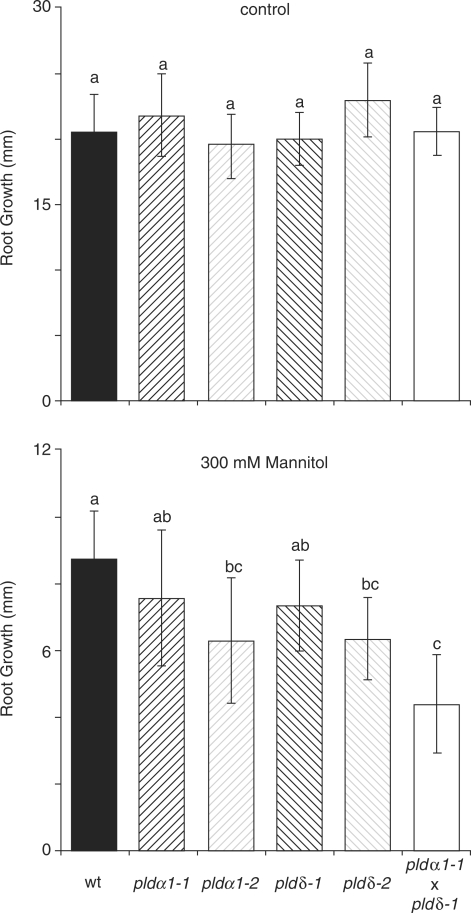
Although AtPLDδ activity in response to dehydration has been demonstrated before (Katagiri et al. 2001), a phenotype in the hyperosmotic stress response for the Arabidopsis plants expressing an antisense AtPLDδ construct used in the above-mentioned study was not found. On the other hand, plants expressing an antisense AtPLDα1 construct have been shown to be hypersensitive to drought (Sang et al. 2001, Mane et al. 2007). We opted to use a more quantitative assay for hyperosmotic stress sensitivity in order to be able to distinguish better between the wild type, the pldα1 and pldδ single mutants and the pldα1/pldδ double mutant. Four-day-old seedlings were transferred to plates supplemented with 0 and 300 mM mannitol, and root growth was monitored after 4 d (Fig. 8). As with the salt tolerance assay, the different plant lines showed altered responses to hyperosmotic stress. Whereas the single mutants displayed an ∼30% reduction in root growth, the double mutant exhibited an ∼50% reduction. These results establish that AtPLDα1 and AtPLDδ are required for an adequate hyperosmotic stress response, and that the absence of both leads to a further increased sensitivity.
Fig. 8.
Increased sensitivity to hyperosmotic stress in Arabidopsis pld mutants. Seeds from wild-type, pldα1-1, pldα1-2, pldδ-1, pldδ-2 and pldα1-1/pldδ-1 knock-out mutant lines were sown on agar plates and grown vertically in a growth chamber. After 4 d, seedlings were transferred to fresh plates supplemented with or without 300 mM mannitol. Primary root growth was measured 4 d after transfer and is represented in histograms ± SD. Data were analyzed for significance by one-way ANOVA (Tukey post hoc, α = 0.001, n = 15–16).
Taken together, the findings gathered in the assays for PLD activity and stress tolerance in response to high salinity and hyperosmotic stress in the Arabidopsis knock-out mutants demonstrate a concerted activity of both AtPLDα1 and AtPLDδ that is essential for endurance upon exposure to such environments.
Discussion
LePLDα1 and salt treatment of tomato cell suspension cultures and plants
LePLDα1 gene expression is induced in Msk8 tomato cell suspension cultures upon exposure to 125 mM salt (Fig. 1). This recaptures findings of Laxalt and co-workers (2001), who studied changes in LePLDα1 expression in Msk8 cultures in response to various stresses. These authors found an induction of expression after a 1 h treatment with 250 mM NaCl. Yet no induction was seen in this period when cells were treated with 400 mM sorbitol, suggesting this response was specific to salt and not due to hyperosmotic stress (Laxalt et al. 2001). Increased PLD activity in response to high salinity has also been previously demonstrated in Msk8 cells (Munnik et al. 2000). The generation of cell suspension cultures lacking LePLDα1 by the use of an RNAi construct (Fig. 1) made it possible to assess whether this PLD contributed to the induced activity. Whereas basal PA and PBut levels were not affected in the silenced lines, the PLD activity induced with salt treatment was markedly less (Fig. 2), indicating that this isoform is indeed responsible for, at least part of, the observed PLD activity. The remaining activity could be attributed to either incomplete silencing or the activity of yet another PLD in response to this treatment. The latter seems more likely, as the silencing seen on the transcript level (Fig. 1) and on the protein level (data not shown) was effectively complete.
Excessive salt concentrations were needed to make out the induction of PLD activity and the disparity between control and silenced lines, possibly due to the short incubation time (15 min) used in these experiments (Fig. 2). Although the treatment was severe, the observed PLD activity was not due to cell lysis, as demonstrated by the enduring activity seen when the treatment was carried out in the presence of TE buffer (Fig. 2). Cell lysis-induced PLD activity is completely lost in this buffer, as demonstrated by snap-freezing and thawing of cells (Fig. 2).
The fact that LePLDα1 gene expression and enzymatic activity were induced by salt treatment of cell suspension cultures provided an incentive to study the role of LePLDα1 during high salinity stress in tomato plants. The LePLDα1 protein was detected in all organs of mature tomato plants as well as in the seedling stage (Fig. 3). Introduction of the LePLDα1-RNAi construct in tomato plants again resulted in gene silencing; however, it did not lead to an absolute attenuation of LePLDα1 protein levels (Fig. 3) as seen in cell suspension cultures (data not shown). This incomplete silencing could explain the lack of an effect on salt tolerance (Fig. 4a) and the cell lysis-induced PLD activity, which can be seen in an Arabidopsis pldα1 knock-out mutant (Fig. 4b). Silencing in planta might also not be as consistent as in the cell suspension cultures, perhaps due to the less efficient 35S-driven expression of the silencing construct in certain cells and tissues (van Leeuwen et al. 2001). Alternatively, the effects could be explained by the action of another PLD isoform.
AtPLDα1 and AtPLDδ are required for high salinity and hyperosmotic stress tolerance in Arabidopsis
Extending our research to include Arabidopsis conferred the opportunity to analyze actual null mutants rather than looking at plant material expressing silencing constructs. This gave the assurance of examining the complete knock-out of a gene and no possibility of non-specific or secondary effects of a silencing construct. Furthermore, this extension made it possible to incorporate analysis of another PLD isoform that has been implicated in hyperosmotic stress responses, i.e. AtPLDδ (Katagiri et al. 2001). Notably, a mutant lacking both AtPLDα1 and AtPLDδ could be generated and scrutinized along with the two single knock-out mutants.
Analysis of PLD activity in Arabidopsis leaf discs treated with high salt concentrations demonstrated that both AtPLDα1 and AtPLDδ are responsible for PA production in reaction to this treatment (Fig. 5). Yet, even in the double knock-out mutant, there is still a considerable accumulation of PA, indicating that there is one (or more) other route(s) of PA synthesis active under these conditions. Again, severe salt treatments were required to bring out the difference between the wild type and mutants in this assay. Although these are distant from physiological salt concentrations encountered in nature, it is reasonable to believe that these PLD isoforms are activated by high salinity, especially in light of the fact that the knock-out mutants have a growth defect under physiological salt concentrations of 150 mM NaCl (Fig. 6). Parallel to the differences in activity discernible between the wild type and the mutants, the double knock-out mutant fared worse on plates supplemented with salt than the wild type or either of the single mutants. These results demonstrate that PLDs are activated in response to salt treatment in plants and that the lack of these PLDs leads to enhanced salt sensitivity, pointing to the importance of these enzymes in the high salinity response.
Previously, activation of AtPLDδ by dehydration in leaf discs has been demonstrated by Shinozaki's lab using antisense AtPLDδ lines (Katagiri et al. 2001). However, this study did not find a phenotype for these lines under hyperosmotic stress conditions. Conversely, antisense AtPLDα1 lines have been reported to be hypersensitive to drought (Sang et al. 2001, Mane et al. 2007) but AtPLDα1 has not been shown to be activated by hyperosmotic stress. Here, we demonstrate that both PLD isoforms in Arabidopsis are activated by dehydration (Fig. 7) and that both are required for an adequate response to hyperosmotic stress (Fig. 8). The double pldα1/pldδ mutant showed a hypersensitivity that is more acute than either of the single mutants alone. As in the PA response to high salt concentrations, a residual PA production could be noted in the double knock-out mutant, once more implicating other routes of PA synthesis.
Recently, Wang and co-workers (Hong et al. 2008) discovered a novel PLD that is involved in salt stress, i.e. AtPLDα3. They showed that this isoform is activated upon salt and hyperosmotic stress, and is required for wild-type growth under these conditions. It is not unlikely that this is the PLD isoform that is responsible for the observed residual PA production in the pldα1/pldδ double knock-out mutant (Figs. 5, 7). It will be interesting to find out how salt and hyperosmotic stress sensitivity is affected in a pldα1/pldα3/ pldδ triple mutant.
PLD function in high salinity and water deficit stress responses
This study has made apparent that two separate PLD isoforms, PLDα1 and PLDδ, are concomitantly activated by high salinity and hyperosmotic stress in planta (Figs. 5, 7). Furthermore, the importance of their involvement in the response to these stresses is emphasized by the fact that both are required for wild-type growth under these conditions (Figs. 6, 8). That these two PLDs are not redundant during these responses is demonstrated by the finding that the single knock-out mutants, by themselves, have an effect on PA production as well as stress tolerance and that the double knock-out mutant displays an effect that verges upon additive. There is, however, a remaining accumulation of PA in the pldα1/pldδ double mutant (Figs. 5, 7), suggesting that another route of PA production is active under these conditions. Theoretically, this could be accounted for by the activity of another PLD or the concerted action of PLC and DGK. The latter is also known to be activated by salt treatment and hyperosmotic stress (Munnik et al. 2000, DeWald et al. 2001, Katagiri et al. 2001). We have noted a remnant of PBut production in the salt-treated and dehydrated double mutant in the presence of n-butanol (data not shown), indicating that at least one other PLD is active under these circumstances. A good candidate would be the newly implicated AtPLDα3, which has been shown to be involved in the response to both of these stresses (Hong et al. 2008).
The discovery that the single and double mutants are hypersensitive to growth on 300 mM mannitol as well as 150 mM NaCl (Figs. 6, 8) leads one to question whether the effect seen upon high salt treatment is just due to the hyperosmotic stress or whether sodium toxicity also plays a role in the inferior root growth. Several findings argue that PLDs also play a role in the response to sodium toxicity alone. Laxalt and co-workers (2001) found that LePLDα1 (and LePLDα2) expression is up-regulated in response to salt treatment but not upon hyperosmotic treatment with 400 mM sorbitol. In addition, PLD-derived PA has been proposed to regulate the activity of vacuolar H+-ATPases upon high salt treatment (Zhang et al. 2006). These proton pumps help maintain the proton gradient that drives Na+/H+ antiporter activity.
The premise that PLDs play a role in the hyperosmotic stress response is supported by several other independent lines of research. PA has been found to bind Snf-related protein kinases (SnRKs) (Testerink et al. 2004), and these signaling components have been shown to be activated in response to hypersomotic stress and high salinity (Munnik et al. 1999, Mikołajczyk et al. 2000, Hrabak et al., 2003, Boudsocq et al. 2004, Kelner et al., 2004, Boudosq and Laurière 2005, Burza et al. 2006). In addition, several papers have pointed to a role for PA and PLD in stomatal aperture (Jacob et al. 1999, Zhang et al. 2004, Mishra et al. 2006, Distéfano et al. 2007). Lastly, PLD has also been implicated in Arabidopsis proline biosynthesis (Thiery et al. 2004). Together, these findings argue a strong case for PLD function in hyperosmotic and salinity stress signaling. Yet, it must also be taken into account that these stresses induce significant membrane remodeling and rearrangements, and that PLD activity could be physically involved in such membrane alterations (Gigon et al. 2004). In conclusion, PLDs are key players in the plant response to hyperosmotic stress and high salinity, and their important role warrants further investigation of how, when and where which isoforms are involved. Future analysis will require (i) exploration of the mechanism by which these enzymes are activated; (ii) scrutiny of the activation temporally and spatially, throughout the plant and within the cell, i.e. by looking at reporter constructs, green fluorescent protein-tagged PLDs and fluorescent PA biosensors (as in Vermeer et al. 2006, van Leeuwen et al. 2007, Kusuno et al. 2008, Vermeer at al. 2008); (iii) study of the involvement of other PLD isoforms (e.g. PLDα3); and (iv) performing genome-wide transcriptional profiling of the mutants upon exposure to salt and drought stress.
Materials and Methods
Plant material
Tomato plants (L. esculentum cv. GCR161) were grown on soil under a 13.5 h light/10.5 h dark regime in the greenhouse. Transgenic tomato plant lines were generated with the empty vector and LePLDα1-RNAi construct in Agrobacterium tumefaciens strain EHA105 carrying the pJIC.SaRep plasmid. For the LePLDα1-RNAi construct, an inverted repeat specific for LePLDα1 was generated targeting the gene's 3′-UTR. PCR amplification of the LePLDα1 cDNA, cloned previously by Laxalt et al. (2001), was performed with the following oligonucleotides: 1, 5′-CGGGATCCCCATCGTCAGTCAATTAAAGCATCTC-3′(reverse) with a BamHI and a ClaI restriction site; 2, 5′-CCGGAATTCCCCCGACACCAAGG-3′ (forward) with an EcoRI restriction site; and 3, 5′-CCGGAATTCCATCCAGAAAGTGAGG-3′ (forward) with an EcoRI restriction site. The PCR products resulting from primer combinations 1–2 and 1–3 were ligated in a 1–2/3–1 orientation into the pGreen1K binary vector which was modified to contain the 35S-Tnos cassette from pMON999. Tomato plants were transformed as specified by van Roekel et al. (1993). Arabidopsis thaliana var. Col-0 T-DNA insertion lines were obtained from the SALK Institute. Homozygous lines for pldα1-1 (SALK_067533), pldα1-2 (SALK_053785), pldδ-1 (SALK_023247) and pldδ-2 (SALK_023808) were generated and checked by PCR: SALK_067533F, 5′-GACGATGAATACATTATCATTGG-3′; SALK_067533R, 5′-GTCCAAAGGTACATAACAAC-3′; SALK_053785F, 5′-CAAGGCTGCAAAGTTTCTCTG-3′; SALK_053785R, 5′-ATTAAGTGCAGGGCATTGATG-3′; SALK_023247/0238 08F, 5′-TGTACTCGGTGCTTCGGGAAA-3′; SALK_023247/02380 8R, 5′- TCGAGAAACAATGGTGCGACA-3′; SALK_LeftBorderA, 5′-TGGTTCACGTAGTGGGCCATCG-3′. SALK_LeftBorderA was used in combination with SALK_053785F and SALK_ 023247/023808R in compliance with the direction of T-DNA insertion. For routine plant growth, seeds were sown on soil and vernalized at 4°C for 2 d. Plants were grown in a growth chamber under a 12 h light/12 h dark regime at 21°C and 70% humidity.
Plant cell suspension cultures
Suspension-cultured cells (L. esculentum cv. Mill. line Msk8; Felix et al. 1991) were grown at 24°C in the dark, shaking at 125 r.p.m. in MS medium supplemented with 3% (w/v) sucrose, 5.4 μM naphthaleneacetic acid, 1 μM 6-benzyladenine and vitamins (pH was adjusted to 5.7 with 1 M KOH) as described by Felix et al. (1991), and used 4–6 d after weekly subculturing. Cell suspension culture transformation with the LePLDα1-RNAi construct was achieved as described by Bargmann et al. (2006).
RNA blot analysis
Total RNA from tomato cell suspension cultures was isolated using the Trizol-LS reagent method (Gibco, Gaithersburg, MD, USA). A 10 μg aliquot of RNA was separated by denaturing 1.4% (w/v) formaldehyde–agarose gel electrophoresis, transferred onto Hybond-XL nylon membranes (Amersham Pharmacia, Buckinghamshire, UK), and hybridized with 32Pi-labeled probes in modified Church solution at 65°C. Membranes were washed three times for 15 min with wash buffer [1× SSC, 0.1% (w/v) SDS] and the probe signal was visualized by autoradiography.
Protein blot analysis
Protein extraction buffer [9.5 M urea, 0.1 M Tris–HCl pH 6.8, 2% (w/v) SDS and 2% (v/v) β-mercaptoethanol] was added to an equal volume of ground leaf tissue, vortexed and centrifuged in an Eppendorf centrifuge for 10 min at 1,000×g. Four times sample buffer [8% (w/v) SDS, 40% (v/v) glycerol, 20% (v/v) β-mercaptoethanol, 240 mM Tris–HCl pH 6.8 and 0.08% (w/v) bromophenol blue] was added to the supernatant and samples were loaded onto a 10% SDS–polyacrylamide gel, blotted on nitrocellulose and incubated overnight in 10 ml of PBST [0.8% (w/v) NaCl, 0.02% (w/v) KCl, 0.144% (w/v) Na2HPO4, 0.02% (w/v) KH2PO4 and 0.05% (v/v) Tween-20] with 5% (w/v) powdered milk and affinity-purified polyclonal anti-LePLDα1 antibody (rabbit; Eurogentech, Liege, Belgium). Antibodies were generated using the final 12 amino acids of the LePLDα1 protein: N-TKSDYLPPNLTT-C. The blot was washed three times with PBST, incubated for 1 h in 10 ml PBST with 5% (w/v) powdered milk and horseradish peroxidase-coupled goat anti-rabbit IgG antibody (Pierce, Rockford, IL, USA) and washed three more times in PBST. The peroxidase activity was detected by enhanced chemiluminescence (Amersham). A duplicate gel was stained with Coomassie brilliant blue [0.25% (w/v) Coomassie brilliant blue, 30% (v/v) methanol and 10% (v/v) acetic acid] as a loading control.
In vivo phospholipid analysis
Cell suspension cultures were labeled by incubation of 100 μl aliquots with 100 μCi of carrier-free PO43– in growth medium. Treatments were performed in the presence of 0.5% (v/v) n-butanol, and treatments were stopped and lipids extracted as described before (van der Luit et al. 2000). Leaf discs (Ø 5 mm) were labeled by floating them overnight on 100 μl of 10 mM MES buffer pH 5.7 (KOH) supplemented with 100 μCi of carrier-free PO43– in a 2 ml microcentrifuge tube (Frank et al. 2000). Treatments were stopped by addition of perchloric acid to a final concentration of 5% (w/v). Leaf discs were then transferred to a new tube containing 375 μl of CHCl3/MeOH/HCl [50 : 100 : 1 (v/v)] where lipids were extracted during vigorous shaking for 10 min. A two-phase system was induced by the addition of 375 μl of CHCl3 and 200 μl of 0.9% (w/v) NaCl. The remainder of the extraction was performed as described before (van der Luit et al. 2000).
For quantitative analysis, lipids were separated on silica thin-layer chromatography (TLC) plates using the organic upper phase of an ethyl acetate mixture: ethyl acetate/iso-octane/formic acid/water [12 : 2 : 3 : 10 (by vol.); Munnik et al. 1998] or using an alkaline solvent system: CHCl3/MeOH/25%NH4OH/H2O [90 : 70 : 4 : 16 (by vol.); Munnik et al. 1994] when indicated. Phospholipids were visualized and quantified by phosphoimaging (Molecular Dynamics, Sunnyvale, CA, USA).
Plate root growth assay
Seeds were sterilized, placed on MS agar plates (2.2 g l–1 Murashige & Skoog salts, 1% sucrose, 0.5 g l–1 MES, pH 5.7 with KOH) and vernalized for 2 d at 4°C. Plates were placed vertically in growth chambers under a 18 h light/6 h dark regime at 21°C and 70% humidity. Seedlings (4 d after germination in the case of Arabidopsis and 7 d after gemination in the case of tomato) were transferred to fresh plates supplemented with NaCl or mannitol.
Funding
The EU ROSt project (QLK5-CT-2002-00841 to D.B.); the Netherlands Organization for Scientific Research (NWO; 700.56.429 to C.T); NWO (813.06.0039, 863.04.004, 864.05.001); the European Union (COST Action FA0605); Royal Netherlands Academy of Arts and Sciences (KNAW).
Acknowledgements
The authors wish to thank Kenneth Birnbaum (New York University, New York, USA) for helpful advice and the use of growth chambers.
Glossary
Abbreviations:
- DGK
diacylglycerol kinase
- PA
phosphatidic acid
- PBut
phosphatidylbutanol
- PLC
phospholipase C
- PLD
phospholipase D
- RNAi
RNA interference
- TLC
thin-layer chromatography
- UTR
untranslated region
References
- Bargmann BOR, Munnik T. The role of phospholipase D in plant stress responses. Curr. Opin. Plant Biol. 2006;9:515–522. doi: 10.1016/j.pbi.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Bargmann BOR, Laxalt AM, ter Riet B, Schouten E, van Leeuwen W, Dekker HL, et al. LePLDbeta1 activation and relocalization in suspension-cultured tomato cells treated with xylanase. Plant J. 2006;45:58–68. doi: 10.1111/j.1365-313X.2005.02631.x. [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Barbier-Brygoo H, Lauriere C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 2004;279:41758–41766. doi: 10.1074/jbc.M405259200. [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Laurière C. Osmotic signaling in plants: multiple pathways mediated by emerging kinase families. Plant Physiol. 2005;138:1185–1194. doi: 10.1104/pp.105.061275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burza AM, Pekala I, Sikora J, Siedlecki P, Małagocki P, Bucholc M, et al. Nicotiana tabacum osmotic stress-activated kinase is regulated by phosphorylation on Ser-154 and Ser-158 in the kinase activation loop. J. Biol. Chem. 2006;281:34299–34311. doi: 10.1074/jbc.M601977200. [DOI] [PubMed] [Google Scholar]
- Boyer J.S. Plant productivity and environment. Science. 1982;218:443–448. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- DeWald DB, Torabinejad J, Jones CA, Shope JC, Cangelosi AR, Thompson JE, et al. Rapid accumulation of phosphatidylinositol 4,5-bisphosphate and inositol 1,4,5-trisphosphate correlates with calcium mobilization in salt-stressed arabidopsis. Plant Physiol. 2001;126:759–769. doi: 10.1104/pp.126.2.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distéfano AM, García-Mata C, Lamattina L, Laxalt AM. Nitric oxide-induced phosphatidic acid accumulation: a role for phospholipases C and D in stomatal closure. Plant Cell Environ. 2007;31:187–194. doi: 10.1111/j.1365-3040.2007.01756.x. [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, Laxalt AM, Goedhart J, Gadella TW, Munnik T. Phospholipase D activation correlates with microtubule reorganization in living plant cells. Plant Cell. 2003;15:2666–2679. doi: 10.1105/tpc.014977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein E, Norlyn JD, Rush DW, Kingsbury RW, Kelley DB, Cunningham GA, et al. Saline culture of crops: a genetic approach. Science. 1980;210::399–404. doi: 10.1126/science.210.4468.399. [DOI] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999;18:265–276. doi: 10.1046/j.1365-313x.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- Felix G, Grosskopf DG, Regenass M, Boller T. Rapid changes of protein phosphorylation are involved in transduction of the elicitor signal in plant cells. Proc. Natl Acad. Sci. USA. 1991;88:8831–8834. doi: 10.1073/pnas.88.19.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G, Regenass M, Boller T. Sensing of osmotic pressure changes in tomato cells. Plant Physiol. 2000;124:1169–1180. doi: 10.1104/pp.124.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers T.J. Improving crop salt tolerance. J. Exp. Bot. 2004;55:307–319. doi: 10.1093/jxb/erh003. [DOI] [PubMed] [Google Scholar]
- Frank W, Munnik T, Kerkmann K, Salamini F, Bartels D. Water deficit triggers phospholipase D activity in the resurrection plant Craterostigma plantagineum. Plant Cell. 2000;12:111–124. doi: 10.1105/tpc.12.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke W. Rapid and tissue-specific accumulation of solutes in the growth zone of barley leaves in response to salinity. Planta. 2004;219:515–25. doi: 10.1007/s00425-004-1263-0. [DOI] [PubMed] [Google Scholar]
- Gigon A, Matos AR, Laffray D, Zuily-Fodil Y, Pham-Ti AT. Effect of drought stress on lipid metabolism in the leaves of Arabidopsis thaliana (ecotype Columbia). Ann. Bot. 2004;94:345–351. doi: 10.1093/aob/mch150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Pan X, Welti R, Wang X. Phospholipase Dalpha3 is involved in the hyperosmotic response in Arabidopsis. Plant Cell. 2008;20:803–816. doi: 10.1105/tpc.107.056390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrabak EM, Chan CW, Gribskov M, Harper JF, Choi JH, Halford N, et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003;132:666–680. doi: 10.1104/pp.102.011999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob T, Ritchie S, Assmann SM, Gilroy S. Abscisic acid signal transduction in guard cells is mediated by phospholipase D activity. Proc. Natl Acad. Sci. USA. 1999;96:12192–12197. doi: 10.1073/pnas.96.21.12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakab G, Ton J, Flors V, Zimmerli L, Metraux JP, Mauch-Mani B. Enhancing Arabidopsis salt and drought stress tolerance by chemical priming for its abscisic acid responses. Plant Physiol. 2005;139:267–274. doi: 10.1104/pp.105.065698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 1999;17:287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- Katagiri T, Takahashi S, Shinozaki K. Involvement of a novel Arabidopsis phospholipase D, AtPLDdelta, in dehydration-inducible accumulation of phosphatidic acid in stress signalling. Plant J. 2001;26:595–605. doi: 10.1046/j.1365-313x.2001.01060.x. [DOI] [PubMed] [Google Scholar]
- Kelner A, Pekala I, Kaczanowski S, Muszynska G, Hardie DG, Dobrowolska G. Biochemical characterization of the tobacco 42-kD protein kinase activated by osmotic stress. Plant Physiol. 2004;136:3255–3265. doi: 10.1104/pp.104.046151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinraide T.B. Three mechanisms for the calcium alleviation of mineral toxicities. Plant Physiol. 1998;118:513–520. doi: 10.1104/pp.118.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano H, Testerink C, Vermeer JEM, Tsuge T, Oka A, Shimada H, et al. The Arabidopsis phosphatidylinositol phosphate 5-kinase PIP5K3 is a key regulator for root hair tip growth. Plant Cell. 2008;20:367–380. doi: 10.1105/tpc.107.056119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxalt AM, ter Riet B, Verdonk JC, Parigi L, Tameling WI, Vossen J, et al. Characterization of five tomato phospholipase D cDNAs: rapid and specific expression of LePLDbeta1 on elicitation with xylanase. Plant J. 2001;26:237–247. doi: 10.1046/j.1365-313x.2001.01023.x. [DOI] [PubMed] [Google Scholar]
- Li S, Assmann SM, Albert R. Predicting essential components of signal transduction networks: a dynamic model of guard cell abscisic acid signaling. PLoS Biol. 2006;4:e312. doi: 10.1371/journal.pbio.0040312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mane SP, Vasquez-Robinet C, Sioson AA, Heath LS, Grene R. Early PLDalpha-mediated events in response to progressive drought stress in Arabidopsis: a transcriptome analysis. J. Exp. Bot. 2007;58:241–252. doi: 10.1093/jxb/erl262. [DOI] [PubMed] [Google Scholar]
- Mikołajczyk M, Awotunde OS, Muszyńska G, Klessig DF, Dobrowolska G. Osmotic stress induces rapid activation of a salicylic acid-induced protein kinase and a homolog of protein kinase ASK1 in tobacco cells. Plant Cell. 2000;12:165–178. [PMC free article] [PubMed] [Google Scholar]
- Mishra G, Zhang W, Deng F, Zhao J, Wang X. A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science. 2006;312:264–266. doi: 10.1126/science.1123769. [DOI] [PubMed] [Google Scholar]
- Munnik T. Phosphatidic acid: an emerging plant lipid second messenger. Trends Plant Sci. 2001;6:227–233. doi: 10.1016/s1360-1385(01)01918-5. [DOI] [PubMed] [Google Scholar]
- Munnik T, Arisz SA, De Vrije T, Musgrave A. G protein activation stimulates phospholipase D signaling in plants. Plant Cell. 1995;7:2197–2210. doi: 10.1105/tpc.7.12.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T, Irvine RF, Musgrave A. Rapid turnover of phosphatidylinositol 3-phosphate in the green alga Chlamydomonas eugametos: signs of a phosphatidylinositide 3-kinase signalling pathway in lower plants? Biochem. J. 1994;298:269–73. doi: 10.1042/bj2980269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T, Ligterink W, Meskiene II, Calderini O, Beyerly J, Musgrave A, et al. Distinct osmo-sensing protein kinase pathways are involved in signalling moderate and severe hyper-osmotic stress. Plant J. 1999;20:381–388. doi: 10.1046/j.1365-313x.1999.00610.x. [DOI] [PubMed] [Google Scholar]
- Munnik T, Meijer H.J. Osmotic stress activates distinct lipid and MAPK signalling pathways in plants. FEBS Lett. 2001;498:172–178. doi: 10.1016/s0014-5793(01)02492-9. [DOI] [PubMed] [Google Scholar]
- Munnik T, Meijer HJ, ter Riet B, Hirt H, Frank W, Bartels D, et al. Hyperosmotic stress stimulates phospholipase D activity and elevates the levels of phosphatidic acid and diacylglycerol pyrophosphate. Plant J. 2000;22:147–154. doi: 10.1046/j.1365-313x.2000.00725.x. [DOI] [PubMed] [Google Scholar]
- Munnik T, van Himbergen JAJ, ter Riet B, Braun FJ, Irvine RF, van den Ende H, et al. Detailed analysis of the turnover of polyphosphoinositides and phosphatidic acid upon activation of phospholipases C and D in Chlamydomonas cells treated with non-permeabilizing concentrations of mastoparan. Planta. 1998;207:133–145. [Google Scholar]
- Qin C, Wang X. The Arabidopsis phospholipase D family. Characterization of a calcium-independent and phosphatidylcholine-selective PLD zeta 1 with distinct regulatory domains. Plant Physiol. 2002;128:1057–1068. doi: 10.1104/pp.010928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Y, Zheng S, Li W, Huang B, Wang X. Regulation of plant water loss by manipulating the expression of phospholipase Dalpha. Plant J. 2001;28:135–144. doi: 10.1046/j.1365-313x.2001.01138.x. [DOI] [PubMed] [Google Scholar]
- Testerink C, Dekker HL, Lim ZY, Johns MK, Holmes AB, Koster CG, et al. Isolation and identification of phosphatidic acid targets from plants. Plant J. 2004;39:527–36. doi: 10.1111/j.1365-313X.2004.02152.x. [DOI] [PubMed] [Google Scholar]
- Testerink C, Munnik T. Phosphatidic acid: a multifunctional stress signaling lipid in plants. Trends Plant Sci. 2005;10:368–375. doi: 10.1016/j.tplants.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Thiery L, Leprince AS, Lefebvre D, Ghars MA, Debarbieux E, Savouré A. Phospholipase D is a negative regulator of proline biosynthesis in Arabidopsis thaliana. J. Biol. Chem. 2004;279:14812–14818. doi: 10.1074/jbc.M308456200. [DOI] [PubMed] [Google Scholar]
- van der Luit AH, Piatti T, van Doorn A, Musgrave A, Felix G, Boller T, et al. Elicitation of suspension-cultured tomato cells triggers the formation of phosphatidic acid and diacylglycerol pyrophosphate. Plant Physiol. 2000;123:1507–1516. doi: 10.1104/pp.123.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen W, Ruttink T, Borst-Vrenssen AW, van der Plas LH, van der Krol AR. Characterization of position-induced spatial and temporal regulation of transgene promoter activity in plants. J. Exp. Bot. 2001;52:949–995. doi: 10.1093/jexbot/52.358.949. [DOI] [PubMed] [Google Scholar]
- van Leeuwen W, Vermeer JEM, Gadella T.W.J., Jr, Munnik T. Visualisation of phosphatidylinositol 4,5-bisphosphate in the plasma membrane of suspension-cultured tobacco BY-2 cells and whole Arabidopsis seedlings. Plant J. 2007;52:1014–1026. doi: 10.1111/j.1365-313X.2007.03292.x. [DOI] [PubMed] [Google Scholar]
- van Roekel JSC, Damm B, Melchers LS, Hoekema A. Factors influencing transformation frequency of tomato (Lycopersicon esculentum). Plant Cell Rep. 1993;12:644–647. doi: 10.1007/BF00232816. [DOI] [PubMed] [Google Scholar]
- Vermeer JEM, Thole JM, Goedhart J, Nielsen E, Munnik T, Gadella T.W.J., Jr Visualisation of PtdIns4P dynamics in living plant cells. Plant J. 2008 doi: 10.1111/j.1365-313X.2008.03679.x. in press. [DOI] [PubMed] [Google Scholar]
- Vermeer JEM, van Leeuwen W, Tobeña-Santamaria R, Laxalt AM, Jones DR, Divecha N, et al. Visualisation of PtdIns3P dynamics in living plant cells. Plant J. 2006;47:687–700. doi: 10.1111/j.1365-313X.2006.02830.x. [DOI] [PubMed] [Google Scholar]
- Wang X. Phospholipase D in hormonal and stress signaling. Curr. Opin. Plant Biol. 2002;5::408–414. doi: 10.1016/s1369-5266(02)00283-2. [DOI] [PubMed] [Google Scholar]
- Wang X. Regulatory functions of phospholipase D and phosphatidic acid in plant growth, development and stress responses. Plant Phys. 2005;139:566–573. doi: 10.1104/pp.105.068809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SJ, Ding L, Zhu J.K. SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell. 1996;8:617–627. doi: 10.1105/tpc.8.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu J.K. Cell signaling during cold, drought, and salt stress. Plant Cell. 2002;14 Suppl:S165–S183. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Blumwald E. Developing salt-tolerant crop plants: challenges and opportunities. Trends Plant Sci. 2005;10:615–620. doi: 10.1016/j.tplants.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero G.N. Living with water stress: evolution of osmolyte systems. Science. 1982;217:1214–1222. doi: 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]
- Zhang W, Qin C, Zhao J, Wang X. Phospholipase D alpha 1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc. Natl Acad. Sci. USA. 2004;101:9508–9513. doi: 10.1073/pnas.0402112101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang L, Liu Y, Zhang Q, Wei Q, Zhang W. Nitric oxide enhances salt tolerance in maize seedlings through increasing activities of proton-pump and Na(+)/H(+) antiport in the tonoplast. Planta. 2006;224:545–555. doi: 10.1007/s00425-006-0242-z. [DOI] [PubMed] [Google Scholar]
- Zhu J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.K. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant. Biol. 2003;6:441–445. doi: 10.1016/s1369-5266(03)00085-2. [DOI] [PubMed] [Google Scholar]