Abstract
After the recent discovery that common genetic variation in 8q24 influences inherited risk of prostate cancer, we genotyped 2,973 SNPs in up to 7,518 men with and without prostate cancer from five populations. We identified seven risk variants, five of them previously undescribed, spanning 430 kb and each independently predicting risk for prostate cancer (P = 7.9 × 10−19 for the strongest association, and P < 1.5 × 10−4 for five of the variants, after controlling for each of the others). The variants define common genotypes that span a more than fivefold range of susceptibility to cancer in some populations. None of the prostate cancer risk variants aligns to a known gene or alters the coding sequence of an encoded protein.
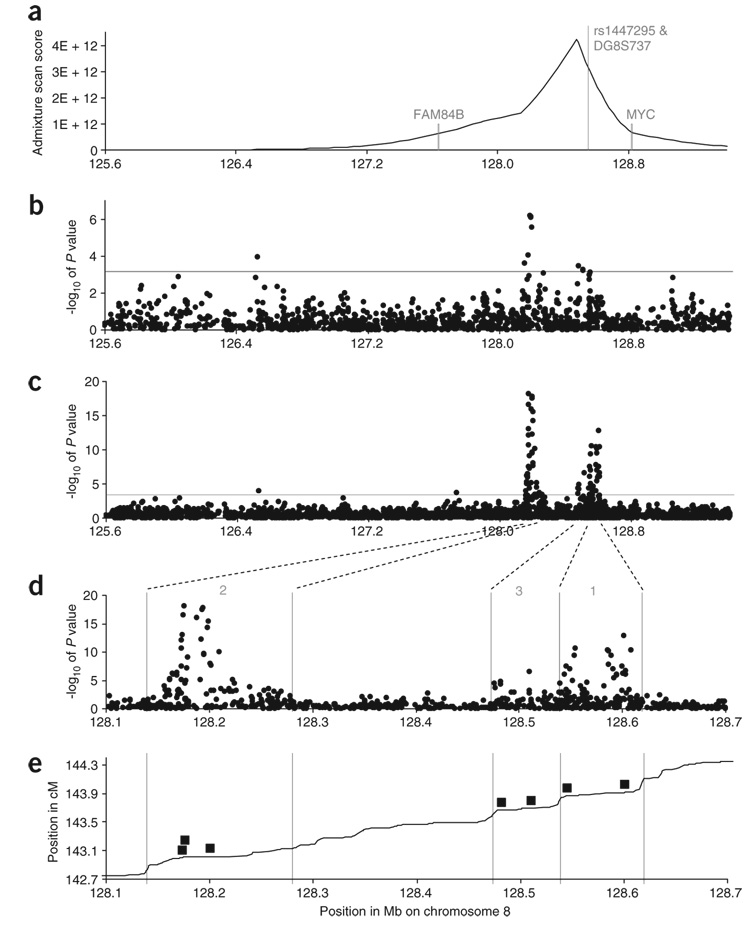
We recently carried out an admixture scan in African Americans with prostate cancer1, highlighting a 3.8-Mb region of chromosome 8 (125.68–129.48 Mb in build 35 of the reference sequence) as containing risk alleles that are highly differentiated in frequency between West Africans and European Americans (Fig. 1a and Supplementary Table 1 online). Independently, another group2 localized the same region via linkage analysis and identified specific variants in a region spanning from 128.54–128.62 Mb (denoted ‘region 1’) that were associated with increased risk of prostate cancer. We replicated the associations after genotyping the same variants in independent samples1. However, our data and analyses indicated that the variants in region 1 are insufficient to explain the magnitude of the admixture signal in African Americans with prostate cancer.
Figure 1.
Results of fine-mapping across the admixture peak, spanning the region 125.6–129.4 Mb defined in ref. 1. (a) The posterior probability distribution for the position of the disease alleles based on our updated admixture mapping scan (Supplementary Table 1). (b) P values from genotyping of 1,521 variants in 1,175 African American affected individuals diagnosed at age <72 years and 837 African American controls, and 465 European American affected individuals and 446 European American controls. This analysis uncovers a ‘region 2’ containing multiple statistically significant associations between 128.14–128.28 Mb, hundreds of kb away from the previously associated ‘region 1’ (128.54–128.62 Mb, identified in ref. 2). (c) Follow-up genotyping of a total of 2,973 alleles in five populations strongly confirms the evidence for association, with the strongest single marker (rs6983561 at 128.176 Mb) significant at P = 7.9 × 10−19. (d) A focused examination of the span 128.1–128.7 Mb uncovers three independent regions of alleles associated with prostate cancer risk, including a new ‘region 3’ of association spanning 128.47–128.54 Mb. (e) We define the boundaries of these regions based on a comparison of genetic and physical maps, which show breakdown of linkage disequilibrium between these regions. The positions of the seven variants (Table 2) we have identified as efficiently capturing the evidence for association are shown in solid boxes. The variants are (from left to right) rs13254738, rs6983561, Broad11934905 (region 2); rs6983267 and rs7000448 (region 3) and DG8S737-8 and rs10090154 (region 1).
To search for additional variants that might also contribute to risk at 8q24, we selected SNPs to capture common genetic variation across the admixture peak based on data from the International HapMap Project (see Methods). We genotyped a total of 1,521 variants (including the alleles of microsatellite DG8S737) in 1,175 African American affected individuals with age at diagnosis <72 years and 837 African American controls (Table 1). We genotyped the same variants in 465 European American cases and 446 European American controls.
Table 1.
Genotyping summary
| Number of polymorphisms tested in various population combinations |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Phase of study | Description | Number of samples (cases/controls) | AA | AA,EA | JA,HA | AA, JA, HA ,EA | JA, HA, LA | AA, JA, HA, LA, EA | Any combination |
| Admixture scan | Admixture association | AA: 1,617/838 Age <72: 1,177/838 |
1,539 | – | – | – | – | – | 1,539 |
| Phase 1 | LD scan focused on admixture peak | AA: 1,175/837 EA: 465/446 |
1,521 | – | – | – | – | – | 1,521 |
| Phase 2 | Gap filling and scanning in all populations between 125.5 and 130.5 Mb | AA: 1,614/837 JA: 722/728 HA: 111/112 LA: 637/633 EA: 1,182/942 |
2,111 | 2,056 | 1,565 | 690 | 275 | 212 | 2,973 |
| Phase 3 | Variants typed in all five populations between 128.1 and 128.7 Mb | AA: 1,614/837 JA: 722/728 HA: 111/112 LA: 637/633 EA: 1,182/942 |
– | – | – | – | – | 186 | 186 |
AA = African American, JA = Japanese American, LA = Latino, HA = Native Hawaiian and EA = European American. For the AA and EA populations, the full set of SNPs was assessed in only a subset of samples: for AA, 666 cases (primarily age at diagnosis <72 years) and 586 controls, and for EA, 465 cases and 446 controls. Summary association statistics for each polymorphism are presented in Supplementary Table 2.
Analysis of these data identified a cluster of genetic variants that we denote ‘region 2’ in a span of linkage disequilibrium from 128.14–128.28 Mb. These variants are hundreds of kilobases away from the region 1 described in ref. 2, and the strongest single-SNP association is significant at P = 6.5 × 10−7 (Fig. 1b and Supplementary Table 2 online). We followed up by genotyping the most associated SNPs in additional cases and controls from five populations: African Americans, Japanese Americans, Native Hawaiians, Latinos and European Americans (for a total sample size of 4,266 individuals with prostate cancer and 3,252 controls) (see Methods and Supplementary Table 3 online). Analysis of the data, correcting for the potentially confounding covariate of genome-wide ancestry proportion and local ancestry proportion in the African American, Native Hawaiian and Latino admixed populations (see Methods and Supplementary Methods online), further strengthened the evidence for association, with the strongest single-SNP association at rs16901979 (P = 1.5 × 10−18). The risk allele at this SNP is more common in West Africans (54%) than in European Americans (3%; frequencies are from HapMap), suggesting that variants in region 2 might contribute to the admixture signal at 8q24 we previously detected in African Americans1.
To clarify the genetic risk for prostate cancer due to variants in regions 1 and 2, and to screen for additional prostate cancer risk variants within the admixture peak, we increased the number of samples and SNPs typed in all five populations (Table 1). In African Americans and European Americans, we increased the number of variants to 2,111. In Japanese Americans and Native Hawaiians, we carried out a new linkage disequilibrium scan across the admixture peak with the goal of capturing all common variation present in HapMap Japanese samples, genotyping 1,565 variants3. In Latinos, we genotyped 275 variants focused on regions of highest interest. To choose SNPs for follow-up genotyping, we not only mined variation in the HapMap database but also used information from an effort to genotype previously uncharacterized genetic variation in the regions of highest interest. To discover new polymorphisms, we sequenced eight African American individuals with prostate cancer and eight African American controls over 282 kb and also sequenced the exonic regions of genes under the admixture peak (Supplementary Methods); we then characterized these variants in samples from the HapMap West African, Japanese and European American populations (genotyping data for 547 newly characterized polymorphisms is provided in Supplementary Table 4 online). Our genotyping in prostate cancer cases and controls successfully tagged a high proportion of common variation in HapMap samples across 8q24 (Supplementary Fig. 1 online).
Analysis of these data further clarified the evidence for association in regions 1 and 2 (Fig. 1c,d) and showed evidence for a third region of association, which we denote ‘region 3’ and define as the linkage disequilibrium span from 128.47–128.54 Mb (Fig. 1d,e). The SNP associations in region 3 were significant at rs7000448 (P = 3.0 × 10−7) and rs6983267 (P = 1.6 × 10−5). The association at rs6983267 was also seen in the Cancer Genetic Markers of Susceptibility (CGEMS) genome-wide prostate cancer scan in European Americans(P = 2.4 × 10−4); combining the two data sets together, the association at rs6983267 was significant at P = 1.0 × 10−7. Both SNPs were highly different in frequency between West Africans and European Americans (98% versus 46% in HapMap for rs6983267), suggesting a possible contribution to the admixture signal in African Americans1. Association scores for the variants in each population separately are given in Supplementary Table 2 and Supplementary Figure 2 online.
Although we observed many strong signals of association, it was important to evaluate to what extent these were independent. We performed stepwise logistic regression, incorporating each SNP into the model based on the strength of association, and repeating the analysis of all other SNPs conditional on those already incorporated into the model. We applied this procedure for the variants that had been successfully typed in all five populations in the span 128.1–128.7 Mb (Fig. 2) until none of the remaining ones were statistically significant after correcting for 186 hypotheses tested.
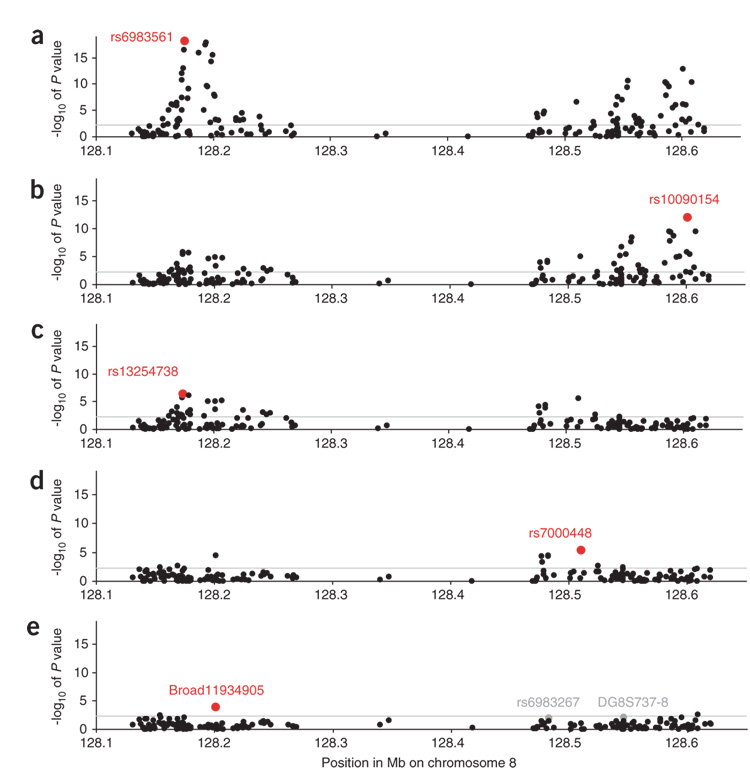
Figure 2.
Case-control association statistics by logistic regression for the 186 alleles for which we collected data in all five populations and are in the region 128.1–128.7 Mb. (a) The strongest association is at marker rs6983561 (P = 7.9 × 10−19; in region 2). (b) Controlling for this SNP as an additional covariate, the next strongest association is rs10090154 (P = 8.8 × 10−13). This SNP is in region 1 (originally identified in ref. 2) and is correlated with their most significantly associated SNP (rs1447295) in all populations (r2 ≥ 0.64) except African Americans, although it appears to capture significant additional association (P = 5.4 × 10−4 after controlling for rs1447295; Supplementary Table 5). (c) Controlling for the top two SNPs, the next strongest association is rs13254738 (P = 3.8 × 10−7), also in region 2. (d) Controlling for the top three SNPs, the next strongest association is rs7000448 in region 3 (P = 5.1 × 10−6). (e) Controlling for the top four SNPs, the next strongest association is Broad11934905 in region 2 (P = 1.5 × 10−4). In gray, we highlight two additional markers, DG8S737-8 and rs6983267, which are of borderline statistical significance after controlling for the top five SNPs (P = 0.008 and P = 0.035 respectively; the latter is significant at 0.0031 in African Americans alone). We believe that the latter two variants are likely to predict additional risk, because they have been independently highlighted in other studies (ref. 2, http://cgems.cancer.gov), so we include them in our modeling of prostate cancer risk.
Notably, this procedure identified five SNPs with independent P values from 7.9 × 10−19 to 1.5 × 10−4 (Fig. 2). After we controlled for the top five SNPs, none crossed the threshold of statistical significance correcting for 186 hypotheses tested (Fig. 2e). Nevertheless, we considered two additional variants that achieve nominal significance after a single hypothesis test controlling for the top five SNPs. The allele DG8S737-8 (region 1; P = 3.1 × 10−8 uncorrected; P = 0.0080 after correcting for the top five alleles) was previously shown to be significantly associated with prostate cancer risk after controlling for other variants in this region2. We also believe that rs6983267 is likely to capture additional risk (P = 2.3 × 10−5 uncorrected; P = 0.035 after correcting for the top five alleles), as it was highly differentiated in frequency between African Americans and European Americans (Table 2) and could potentially contribute to the admixture signal. In African Americans alone, the significance of rs6983267 after controlling for the others was P = 0.0031. Supplementary Table 5 online provides details of the linkage disequilibrium and association patterns among the seven variants we selected for subsequent analysis.
Table 2.
Independent contribution of seven alleles to 8q24 association
| Marker, region and position | African Americansa,b (1,614/837) | Japanese Americans (722/728) | Native Hawaiiansa (111/112) | Latinosa (637/633) | European Americansb (1,182/942) | PHetc | Pooled OR 95% c.i.)d (unadjusted for other markers) | Pooled ORAdj (95% c.i.)e (adjusted for other markers) |
|---|---|---|---|---|---|---|---|---|
| rs13254738 | 1.24 | 1.57 | 1.46 | 1.25 | 1.11 | |||
| Region 2 | (1.09–1.42) | (1.33–1.83) | (1.00–2.12) | (1.07–1.46) | (0.97–1.26) | 0.02 | 1.26 | 1.18 |
| 128173525 | 58% | 62% | 50% | 49% | 33% | (1.18–1.36) | (1.09–1.27) | |
| rs6983561 | 1.34 | 1.78 | 3.17 | 1.99 | 1.16 | |||
| Region 2 | (1.18–1.53) | (1.47–2.15) | (1.87–5.36) | (1.34–2.96) | (0.86–1.58) | 0.001 | 1.51 | 1.42 |
| 128176062 | 40% | 16% | 12% | 3% | 4% | (1.37–1.67) | (1.28–1.58) | |
| Broad11934905f | 2.45 | – | – | – | – | |||
| Region 2 | (1.65–3.62) | – | – | – | – | – | 2.45 | 2.24 |
| 128200991 | 2% | <1% | 0% | <1% | 0% | (1.65–3.62) | (1.43–3.21) | |
| rs6983267 | 1.43 | 1.22 | 1.29 | 1.05 | 1.13 | |||
| Region 3 | (1.17–1.75) | (1.05–1.42) | (0.88–1.89) | (0.89–1.24) | (0.99–1.28) | 0.17 | 1.18 | 1.14 |
| 128482487 | 84% | 32% | 28% | 62% | 51% | (1.09–1.27) | (1.03–1.26) | |
| rs7000448g | 1.33 | 1.23 | 1.38 | 1.29 | 1.14 | |||
| Region 3 | (1.12–1.58) | (1.04–1.46) | (0.89–2.14) | (1.07–1.56) | (0.93–1.40) | 0.83 | 1.26 | 1.19 |
| 128510352 | 61% | 24% | 22% | 29% | 37% | (1.15–1.38) | (1.08–1.32) | |
| DG8S737–8g | 1.25 | 1.48 | 2.55 | 1.46 | 1.45 | |||
| Region 1 | (1.06–1.49) | (1.16–1.88) | (1.33–4.89) | (1.05–2.02) | (0.96–2.19) | 0.27 | 1.39 | 1.23 |
| 128545681 | 16% | 16% | 15% | 6% | 5% | (1.23–1.57) | (1.08–1.40) | |
| rs10090154 | 1.11 | 1.49 | 2.54 | 1.98 | 1.44 | |||
| Region 1 | (0.94–1.32) | (1.23–1.81) | (1.61–4.02) | (1.49–2.61) | (1.17–1.76) | 0.0005 | 1.43 | 1.32 |
| 128601319 | 16% | 15% | 17% | 7% | 9% | (1.30–1.58) | (1.17–1.50) |
Each cell of the table gives odds ratios (and 95% confidence intervals) for allele dosage effects along with the risk allele frequency in controls. Odds ratios in this table do not correct for local ancestry estimates in African Africans, Latinos and Native Hawaiians, as we know local ancestry is correlated to some of these alleles. P values establishing a contribution of these alleles to disease risk, above and beyond the admixture association, are given in (Figure 2 and Supplementary Table 2
Adjusted for genome-wide European ancestry.
OR adjusted for study.
P value testing for heterogeneity of allelic effects across all populations.
OR adjusted for population, study and genome-wide European ancestry (African Africans, Latinos and Native Hawaiians).
OR adjusted for population, study and genome-wide European ancestry (African Africans, Latinos and Native Hawaiians) and all other markers in the same region (i.e. region 1, 2 or 3). Within a region, individuals missing data for any marker were excluded from analysis.
Analysis of Broad11934905 is presented for African Americans only, as this is the only population in which the risk variant has an appreciable frequency.
A smaller number of subjects were genotyped for rs7000448 (2,422 affected individuals and 2,311 controls) and the microsatellite (3,036 cases and 2,208 controls).
The evidence for association at the seven variants is summarized in Table 2. (Supplementary Table 6 online presents the same analysis restricted to the prospectively collected Multiethnic Cohort (MEC)). Mutual adjustment for risk variants is consistent with each independently contributing to risk, although the odds ratios were heterogeneous across populations for rs13254738, rs6983561 and rs10090154, suggesting that we may not yet have genotyped the true causal variants2 or that gene-gene or gene-environment interactions may be different across populations. For each variant, we did not find any evidence for a departure from multiplicative effects per allele or epistatic interaction of the risk alleles within or across regions (Supplementary Table 7 online). In African Americans, these seven variants were sufficient to account for our previously described signal of admixture association (Supplementary Table 7).
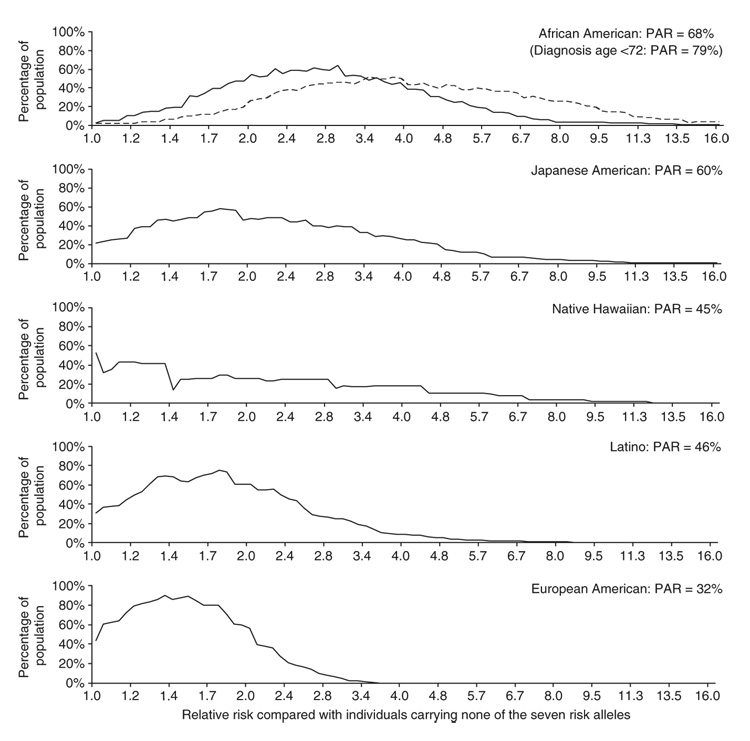
We used these results to build a quantitative model of prostate cancer risk associated with different genotypes. To estimate the distribution of risk relative to noncarriers of any allele in each of the populations, we used the empirically observed distribution of genotypes at the seven variants in control samples from that population. There are combinations of these risk alleles that span a more than fivefold range of risk in many populations, with both extremes of risk common (>5% frequency) in some populations (Fig. 3). The population attributable risk (PAR)—the expected reduction in prostate cancer incidence if the risk alleles did not exist in the population—is 68% in African Americans, 60% in Japanese Americans, 45% in Native Hawaiians, 46% in Latinos and 32% in European Americans.
Figure 3.
The distributions of relative risks for prostate cancer in each of the populations, compared with the baseline relative risk (relative risk = 1) for individuals who do not carry any of the risk alleles at the seven markers we identified. For each population, we included the alleles into population-specific risk models in a stepwise fashion, in the order rs6983561, rs13254738, rs10090154, rs6983267, DG8S737-8, rs7000448, Broad11934905 (the last SNP was included for African Americans only, as it is appreciably polymorphic only in this population). Based on the assumption of independent multiplicative effects for each marker, we calculated the relative risks for all possible genotype combinations for these seven risk alleles. The estimated proportion of individuals in each population is plotted (based on the frequencies in the controls). For the purpose of curve smoothing, at each point we plot the percentage of the population in a relative risk range within a factor of a square root of 2 above and below the value on the x-axis (in other words, a range of 0.71–1.41 around that value). Genotypes at the extremes of risk are assigned to the lowest or highest category. For African Americans, we also plot the distribution of risk for cases diagnosed at age <72 years, because of our previous observation of significantly strong genetic risk at 8q24 in this group1. Population attributable risk (PAR) is the expected reduction in prostate cancer incidence in the populations if the risk alleles at each polymorphism did not exist.
Finally, we tested for association of the seven risk alleles in the three regions with specific phenotypes of prostate cancer: age at diagnosis, family history of prostate cancer in a first-degree relative, stage at diagnosis and tumor grade (Supplementary Table 8 online).When we considered all populations together, associations with all variants except rs6983267 and rs7000448 were nonsignificantly greater among younger affected individuals (that is, less than the median age of 68 years). For African Americans, the effects of rs13254738 and rs6983561 were significantly greater for those diagnosed at younger ages (P = 0.02; Supplementary Table 8), consistent with the stronger admixture signal that we observed previously among younger African American cases1. The effect of rs6983561 was modestly greater among those without a first-degree family history (P = 0.04) and among those with high-stage disease (P = 0.02). We also detected an association of DG8S737-8 with tumor grade (Gleason score >7, P = 0.04) providing some support for the previous finding that the genetic variants at 8q24 are associated with cellular differentiation in prostate cancer tumors2. Findings from these stratified analyses will need to be replicated in other large studies.
What could explain the presence of independent sets of alleles at 8q24 that together contribute to prostate cancer risk in multiple populations but that do not lie in known genes? It is possible that there are multiple unknown prostate cancer susceptibility genes in 8q24, which by chance occur within a few hundred kilobases. More likely, the variants converge on a common biological mechanism, and these regions may independently influence the regulation of the same nearby cancer-causing gene (for example, the protooncogene MYC). Somatic amplifications at 8q are one of the most common acquired events in prostate tumors4,5, and a speculative model is that risk alleles make the entire region (including cancer-related genes like MYC) prone to amplification.
These results are also notable from the point of view of gene mapping, demonstrating the great power that comes from mapping in multiethnic populations. Region 1 was originally localized in populations of European descent2; we were alerted to regions 2 and 3 by an admixture and fine-mapping scan in African Americans and the Japanese provided the strongest statistical signals of association of any single population. In the initial identification2 of risk variants in region 1, the PAR measured in European-derived populations was 8%. With the consideration of the seven variants we describe here, it increases to 32%. The effect of the locus on prostate cancer is even higher in non-European populations, with the PAR as high as 68% in African Americans (Fig. 3). The difference in PAR at this locus may contribute to the higher incidence rate of prostate cancer in African Americans than in European Americans, although further studies will be necessary to prove that genetic factors account for the epidemiological differences. The large effects of the alleles in multiple populations demonstrate the importance of 8q24 in prostate cancer. It should now be a priority to further elucidate the contribution of genetic variation at 8q24 to risk and to understand the biological mechanism by which these variants lead to cancer.
METHODS
Human subjects
This study was approved by ethical review boards at the University of Southern California, the University of Hawaii, the Massachusetts Institutes of Technology and Harvard Medical School, and informed consent forms were approved at each clinical site. All individuals who participated provided informed consent. The majority of affected individuals and controls for this study came from the Multiethnic Cohort (MEC)6, a large prospective cohort that was established between 1993 and 1996 and comprises mainly African Americans, Japanese American, Native Hawaiians, Latinos and European Americans living in Hawaii and California (mainly Los Angeles county). A total of 2,788 prostate cancer cases in the MEC were identified by record linkage to the California Cancer Registry, the Los Angeles County Cancer Surveillance Program and the Hawaii Cancer Registry. We chose a total of 2,613 male controls from within the MEC (frequency-matched to the affected individuals based on age within each population). In addition to genotypes from the MEC samples, we also included genotypes from men with and without prostate cancer from six other studies: 761 African American cases and 143 African American controls, and 717 European American cases and 496 European American controls. The non-MEC African American samples came from the same six studies that we included in a previous admixture scan1: the Los Angeles County Men’s Health Study, the Bay Area Men’s Health Study7 the Study of Early Onset Prostate Cancer8, Genomics Collaborative, the Flint Men’s Health Study9 and the University of Michigan Prostate Cancer Genetics Project10. The non-MEC European American samples came from two of these studies: the Los Angeles County Men’s Health Study and the Bay Area Men’s Health Study7.
SNP choices for targeted association scan in African Americans and European Americans
Because the risk alleles at 8q24 were likely to be under the admixture peak and highly differentiated in frequency between West Africans and European Americans, we chose SNPs for the follow-up scan by assigning them an ad hoc priority score that was the product of four factors (A × B × C × D). The factors were A, the number of SNPs of >5% minor allele frequency in the International Haplotype Map11 with which they were in linkage disequilibrium (LD) at r2 > 0.8 in the combined European American (CEU) and West African (YRI) populations; B, 10 to the power of the LOD score given in our recently published admixture scan across the peak1; C, an increased factor of 2 if the allele had a frequency differential across populations consistent with explaining a substantial part of the admixture signal, and D, a down-weighting factor of 5 for SNPs in r2 > 0.8 linkage disequilibrium with other SNPs already at higher priority on the list. We also subsequently genotyped additional SNPs to fill in gaps across the peak, to more densely tag the 160-kb region surrounding MYC, to capture variation in the genes across the admixture peak (which we resequenced) and to study new variants we identified by sequencing. All SNPs were genotyped in both African and European Americans.
SNP choices for targeted association scan in Japanese Americans and Native Hawaiians
We genotyped the Japanese Americans and Native Hawaiians at SNPs chosen to comprehensively capture variation in the East Asian HapMap samples from 125.7–130.5 Mb. We used the Tagger program3 to select SNPs that were correlated at r2 > 0.8 with all SNPs of >5% minor allele frequency in the combined Japanese and Chinese HapMap samples. We increased tag SNP coverage to r2 ≥ 0.97 centered on 128.15 Mb–129.2 Mb, the span of highest interest containing regions 1–3.
Characterization of new SNPs by resequencing and genotyping
To supplement the database from which we could choose SNPs for genotyping, we carried out bidirectional, PCR-based resequencing in eight African American individuals with prostate cancer diagnosed at age <72 years and in eight controls, attempting genotyping over the spans 128141270–128267120 Mb (much of region 2) and 128510300–128617550 Mb (much of regions 1 and 3), with 79.5% of bases covered at high quality (Supplementary Methods). We also sequenced the coding regions of 12 genic or potential potential coding sequences under the admixture peak: MYC, FAM84B, TRIB1, TMEM75, C8ORF36, MTSS1, ZNF572, SQLE, KIAA0196, AK093407, DQ515896 and DQ515898. We followed up by genotyping the discovered variants and other previously uncharacterized SNPs in regions of high interest in European American (n = 54),West African (n = 57) and Japanese (n = 39) samples from HapMap. Our polymorphism discovery and characterization effort is described in more detail in the Supplementary Methods. Supplementary Table 4 provides the genotypes for the newly characterized variants, which we have submitted to the dbSNP database (see URL below).
Filling in gaps in genotyping
We carried out additional genotyping in the regions of highest interest (regions 1–3) as well as near SNPs that had shown evidence of suggestive or significant association in earlier rounds of genotyping for the study. We chose tagging SNPs to capture as much variation as possible in the combined West African, European American and Japanese populations, mining the variation data in HapMap and Supplementary Table 4.
Genotyping for the study
Most of the genotyping for this study was carried out on DNA obtained by whole-genome amplification12 (the only samples that were not whole genome amplified were from the European Americans that were not part of the MEC). We used the Illumina BeadStudio technology13 to genotype one panel of 1,536 SNPs in the African American and European American samples as well as a second panel of 1,536 SNPs in the Japanese Americans and Native Hawaiians. The remaining SNP genotyping was carried out with the Sequenom MassArray iPLEX or hME technologies14, as well as the ABI TaqMan technology15 (four columns in Supplementary Table 2 identify the genotyping technology used for each polymorphism). Microsatellite DG8S737 was genotyped using the ABI3730 DNA Analyzer1. A substantial number of SNPs were genotyped with two technologies (Supplementary Table 2), often with duplicate genotyping of the same samples, providing opportunity for error-checking.
Genotyping error rate assessment
Duplicate samples were included in all genotyping runs. For the African American, European American, Japanese American and Native Hawaiian Illumina-based linkage disequilibrium scans, the discrepancy for genotypes obtained in duplicate using the same genotyping assay was <0.1%. We also compared TaqMan genotyping at the University of Southern California with Sequenom and Illumina genotyping at the Broad Institute for SNP-sample combinations genotyped at both sites. Of genotypes obtained in duplicate, the discrepancy rate was 0.3%.
Genotyping database and quality control procedures
To maintain a high level of data quality in the midst of many rounds of genotyping (often using different technologies), we uploaded all data into a database before extracting data for analysis. During each upload, we checked each SNP, sample and genotyping plate and discarded any that showed evidence of a substantial error rate (for example, a SNP with a high discrepancy rate among duplicate samples). We also removed from the data set any variants that failed a Hardy-Weinberg equilibrium test in control samples from nonadmixed populations (European Americans or Japanese Americans). Finally, before allowing data from a polymorphism into the database, we checked that in control samples, its frequency was consistent with that seen in samples from the same population that were already in the database. These multiple data quality checks, which we implemented before allowing data into the database, greatly improved the reliability of the data used for analysis.
Outlier removal and estimates of ancestry in admixed samples
We identified samples whose genetic ancestry was not concordant with self-identified ancestry by genotyping 40 ancestry informative markers genome-wide in non-African American samples and removing from analysis samples that appeared to be outliers based on principal components analysis (Supplementary Methods, Supplementary Table 9 and Supplementary Fig. 3 online). For case-control analysis in the African Americans, Native Hawaiians and Latinos—which are all admixed populations—we needed to adjust for their proportion of genome-wide and local ancestry, in order to test whether alleles were associated above and beyond the confounders of ancestry association. For the African Americans, we obtained local and global estimates of ancestry for use in the logistic regression analysis by applying ANCESTRYMAP software16 to the admixture scanning data we had previously reported1 (Supplementary Table 1). In the Native Hawaiians and Latinos, we used principal components analysis on genome-wide data sets and markers chosen from the vicinity of 8q24 (Supplementary Methods).
Tests for statistical significance of alleles
We used unconditional logistic regression for the main tests of association (Fig. 2), providing a systematic framework to test for allelic association while controlling for covariates such as local and genome-wide ancestry in the three admixed populations (and also allowing us to control for other SNPs in the analysis)17. For testing in the Latinos, Native Hawaiians and African Americans, we controlled for genome-wide and local estimates of European ancestry, which could contribute to false-positive allelic associations (by population stratification or by admixture association, respectively).
Calculations of odds ratios and 95% confidence intervals
Odds ratios (OR) and 95% confidence intervals for Table 2 were calculated using unconditional logistic regression. We first examined associations with each marker separately in population-stratified analyses, controlling for genome-wide but not local estimates of ancestry in the African Americans, Native Hawaiians and Latinos. We examined heterogeneity of effects by including interaction terms between population and each SNP in multivariate models. For each marker, we estimated pooled odds ratios after adjusting for population and study. To assess independence of associations, pooled ORs were estimated by adjusting for these covariates and all other risk alleles within each region (1–3).
Tests for dominant/recessive effects and interaction among prostate cancer–causing genetic variants
For each risk variant, we tested whether the risk was consistent with a simple allele dosage effect by comparing the fit of the data allowing different risks for each genotype, with the fit assuming a multiplicative increase in risk per copy (we used a χ2 test to evaluate significance). Statistical interaction (epistasis) between candidate loci was examined by including genotypes as score variables in logistic regression models. Interactions with age at diagnosis (<68 versus ≥68 years) and family history of prostate cancer in first-degree relatives (yes or no) were also examined. A likelihood ratio test was used to assess statistical significance. Associations by stage (localized versus regional or distant) and grade (Gleason score ≤7 versus >7) were examined by logistic regression in case-only analyses.
Population attributable risk (PAR) calculation
We let i = 1,…,729 index the 3 × 3 × 3 × 3 × 3 × 3 = 729 possible genotype combinations (2,187 for African Americans including Broad11934905) and let kj represent the number of copies of each risk allele j = 1,2,3. We also let Pi denote the proportion of controls in a given population with allele combination i, and let Ri = exp[β1k1 + β2k2 + β3k3] denote the relative risk (odds ratio) for combination i. The PAR for each population is then PAR = (∑PiRi – 1) / ∑PiRi.
URLs
CGEMS: http://cgems.cancer.gov. dbSNP: http://www.ncbi.nlm.nih.gov/projects/SNP. HapMap: http://www.hapmap.org.
Supplementary Material
Note: Supplementary information is available on the Nature Genetics website.
ACKNOWLEDGMENTS
We thank the men with and without prostate cancer who volunteered to participate in this study. We thank E. Lander, A. Price, J. Seidman and E. Ziv for critical comments and researchers from the NCI Cancer Genetic Markers of Susceptibility Study (CGEMS) Project, whose online data we cited to provide confirmation of the association at rs6983267. Genotyping and personnel were supported by NIH grants CA63464 to B.E.H., C.A.H., D.A., M.L.F. and D.R. and by funds from the Harvard Medical School Department of Genetics (D.R.). M.L.F. was supported by a Department of Defense Health Disparity Training-Prostate Scholar Award (17-02-1-0246) and Dana-Farber/Harvard Partners Cancer Care Prostate SPORE. N.P. was supported by NIH career transition award HG02758. S.C.G. is a Hospital for Sick Children and March of Dimes Fellow of the Pediatric Scientist Development Program, and is also supported by NICHD grant HD00850. C.A.H., D.O.S., M.C.P., L.L.M., L.N.K. and B.E.H. were supported by CA54281, E.M.J. and S.A.I. were supported by California Cancer Research Program Grants 99-00527V-10182 and 99-00524V-10258; S.A.I. was also supported by NIH grant CA84979. The Flint Men’s Health Study was supported by the University of Michigan SPORE in Prostate Cancer (CA69568), the University of Michigan Department of Urology and the University of Michigan Comprehensive Cancer Center. K.A.C. was supported by grants CA69568 and CA79596, and I.O.G. and A.S.W. were supported by CA67044. D.A. is a Charles E. Culpeper Scholar of the Rockefeller Brothers Fund and a Burroughs Wellcome Fund Clinical Scholar in Translational Research. D.R. is supported by a Burroughs-Wellcome Career Development Award in the Biomedical Sciences.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
Published online at http://www.nature.com/naturegenetics
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Freedman ML, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc. Natl. Acad. Sci. USA. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amundadottir LT, et al. A common variant associated with prostate cancer in European and African populations. Nat. Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 3.de Bakker PI, et al. Efficiency and power in genetic association studies. Nat. Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 4.van Duin M, et al. High-resolution array comparative genomic hybridization of chromosome arm 8q: evaluation of genetic progression markers for prostate cancer. Genes Chromosom. Cancer. 2005;44:438–449. doi: 10.1002/gcc.20259. [DOI] [PubMed] [Google Scholar]
- 5.Visakorpi T, et al. Genetic changes in primary and recurrent prostate cancer by comparative genomic hybridization. Cancer Res. 1995;55:342–347. [PubMed] [Google Scholar]
- 6.Kolonel LN, et al. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am. J. Epidemiol. 2000;151:346–357. doi: 10.1093/oxfordjournals.aje.a010213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.John EM, Schwartz GG, Koo J, Van Den Berg D, Ingles S. Sun exposure, vitamin D receptor gene polymorphisms, and risk of advanced prostate cancer. Cancer Res. 2005;65:5470–5479. doi: 10.1158/0008-5472.CAN-04-3134. [DOI] [PubMed] [Google Scholar]
- 8.Oakley-Girvan I, et al. Risk of early-onset prostate cancer in relation to germline polymorphisms of the vitamin D receptor. Cancer Epidemiol. Biomarkers Prev. 2004;13:1325–1330. [PubMed] [Google Scholar]
- 9.Cooney KA, et al. Age-specific distribution of serum prostate-specific antigen in a community-based study of African-American men. Urology. 2001;57:91–96. doi: 10.1016/s0090-4295(00)00873-6. [DOI] [PubMed] [Google Scholar]
- 10.Cooney KA, et al. Prostate cancer susceptibility locus on chromosome 1q: a confirmatory study. J. Natl. Cancer Inst. 1997;89:955–959. doi: 10.1093/jnci/89.13.955. [DOI] [PubMed] [Google Scholar]
- 11.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hosono S, et al. Unbiased whole-genome amplification directly from clinical samples. Genome Res. 2003;13:954–964. doi: 10.1101/gr.816903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan JB, et al. Highly parallel SNP genotyping. Cold Spring Harb. Symp. Quant. Biol. 2003;68:69–78. doi: 10.1101/sqb.2003.68.69. [DOI] [PubMed] [Google Scholar]
- 14.Tang K, et al. Chip-based genotyping by mass spectrometry. Proc. Natl. Acad. Sci. USA. 1999;96:10016–10020. doi: 10.1073/pnas.96.18.10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee LG, Connell CR, Bloch W. Allelic discrimination by nick-translation PCR with fluorogenic probes. Nucleic Acids Res. 1993;21:3761–3766. doi: 10.1093/nar/21.16.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patterson N, et al. Methods for high-density admixture mapping of disease genes. Am. J. Hum. Genet. 2004;74:979–1000. doi: 10.1086/420871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breslow NE, Day NE. Statistical Methods in Cancer Research, Vol 1: the Analysis of Case-Control Studies. Lyon: International Agency on Cancer Research; 1980. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary information is available on the Nature Genetics website.