Introduction
Control of gene expression can occur locally and over large genomic distances. Regulatory elements are frequently positioned far upstream or downstream of the genes they control and can even influence the expression of genes that lie on separate chromosomes. Most genes exhibit rather unique characteristics with regard to the site and level of expression as well as the timing of transcription throughout development and the cell cycle. This level of specificity is achieved through the combinatorial actions of multiple regulatory elements. In artificial systems to study gene expression, enhancers usually activate linked promoters promiscuously. Hence, in vivo, mechanisms are in place to prevent enhancers from producing unwanted influence on neighboring genes or even genes that are dispersed throughout the genome. This can be achieved in two ways that are not mutually exclusive. In the first, the enhancer may only be able to interact with its target promoter efficiently in the presence of a unique combination of enhancer-binding proteins. Such an enhancer would be expected not to function promiscuously. For example, the locus control region (LCR) of the β-globin locus interacts with embryonic and adult type β-globin promoters at the appropriate time of development. It is thought that the stage-specific transcription factor milieu determines promoter-selective interactions with the LCR. The second way to ensure gene-specific regulation involves enhancer-blocking insulators and barrier elements that reduce the effects of enhancers and block the spreading of repressive chromatin, respectively. Intriguingly, enhancer-blocking insulators resemble enhancers in that they can interact over large distances and between chromosomes. How specific communication among distal regulatory elements is achieved has been the subject of discussion and speculation for many years [1-10]. Substantial recent experimental evidence favors two major models for long-range control of gene expression that might occur alone or in combination, namely looping and tracking/scanning. Specifically, the advent of new techniques has enabled investigators to directly examine long-range interactions between chromosomal sequences in vivo. This has produced strong evidence that genes can be configured into looped structures or chromatin hubs that juxtapose regulatory elements to activate or repress transcription. However, these studies typically provide snapshot images of chromosomal interactions and do not rule out some form of tracking intermediate as a guide to establish gene-specific regulatory chromatin loops. Nevertheless, the detection of loops in vivo at numerous genes has shifted the debate towards the factors that are involved in forming, maintaining and resolving such loops, and how they impact on gene expression. This is the subject of this review.
What are chromatin loops?
A chromatin loop occurs when stretches of genomic sequence that lie on the same chromosome (configured in cis) are in closer physical proximity to each other than to intervening sequences. This simple definition does not consider the degree of proximity required to be functionally meaningful nor does it speak to the length of the intervening sequence, i.e. the size of the loop. Looped structures have been detected at numerous gene loci in a fashion that juxtaposes important genetic elements. Physical interactions have also been observed between elements residing on separate chromosomes and although they do not represent loops in the true sense of the word, these interactions might serve the same functions. Electron micrographs have also impressively demonstrated large chromosomal loops in the so-called lampbrush chromosomes of amphibian and avian oocytes [11], in preparations of metaphase chromosomes [12], and even in interphase chromatin where it appears that large loops are anchored to an insoluble structure often referred to as the nuclear matrix (for review see [13, 14]). The term “looping” has also been used to describe chromosomal segments that protrude outwards from their chromosomal territory (for review see [15, 16]. However, the impact of these loops on the regulation of gene expression is less well understood. Here we specifically discuss loops that spatially configure defined sequences of known gene loci.
Techniques to detect loop formation
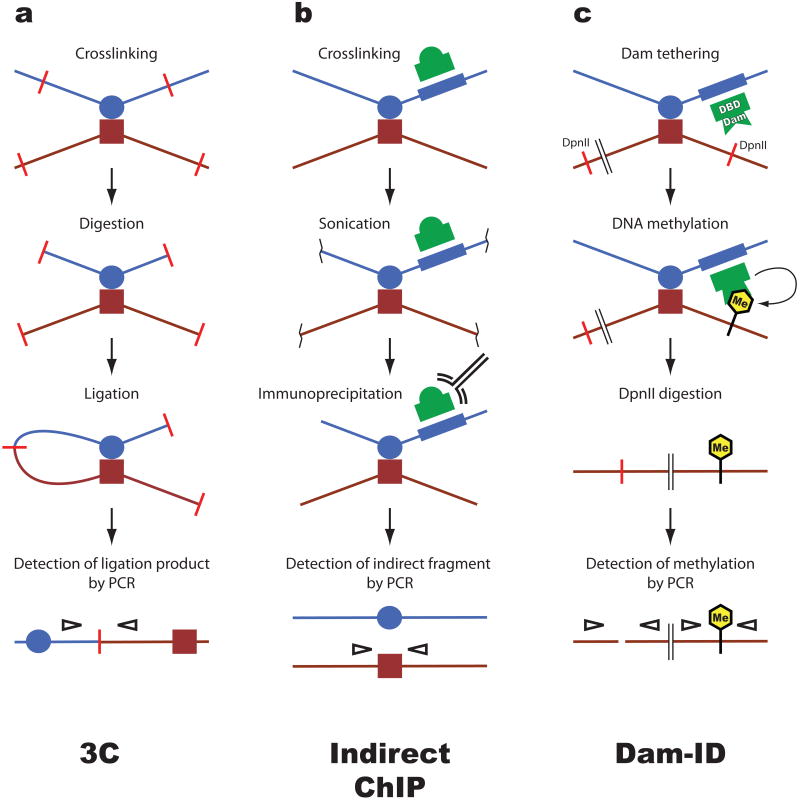
Several approaches have been taken to determine whether distal regulatory elements are juxtaposed to form the base of genomic loops. The most widely used method currently is chromosome conformation capture (3C) in varied form also known as nuclear ligation assay ([17, 18]. This method relies on the assumption that distal genomic sequences that are brought in close vicinity by protein complexes can be chemically crosslinked. Subsequent restriction digestion produces DNA fragments that can be re-ligated to each other. DNA fragments that are close to each other will be crosslinked and hence ligated with a higher efficiency than those that are not. Quantitative PCR using primers spanning the ligated fragments is used to measure the amounts of specific ligation products. The 3C assay requires numerous essential controls [19] since the results are influenced by cross-linking conditions, digestion efficiency, and have a significant potential for PCR artifacts. Large scale versions of the 3C assay (called 4C and 5C) have been devised using dedicated microarray platforms or high throughput sequencing [20-23]. Results obtained by 3C do not easily translate into absolute distances that can be described on a micrometer scale. Instead, by comparing the interaction frequencies of multiple fragments, only relative proximities to each other can be inferred. Another consideration is that 3C requires large numbers of cells and only provides averages of the interactions in question, which hampers the analysis of individual genes in rare cell populations.
The formation of chromatin loops has also been deduced from chromatin immunoprecipitation (ChIP) assays. For example, if an enhancer binding protein is detected by ChIP at a distal promoter that lacks binding sites for this factor, this might result from the enhancer having looped to the promoter (e.g. [24]). An obvious caveat to this interpretation is that said transcription factor might associate directly with the promoter through a protein intermediate. This problem can be addressed by artificially introducing a binding site for a DNA binding protein into a specified genomic location. Expression of a tagged form of the DNA binding protein followed by ChIP might allow detection of this protein at distant sites suggesting the presence of chromatin loops [25]. Combining the ChIP assay with the DNA ligation and the PCR amplification steps of 3C (ChIP-3C or ChIP loop) has been employed to determine whether a transcription factor of interest is present at juxtaposed genomic sites [26]. Of course, the presence of a transcription factor at the base of a loop does not necessarily mean that this factor is required for loop formation.
An alternative approach to assay genomic interactions in vivo involves the fusion of a DNA binding protein to the bacterial DNA methylase Dam [27]. Targeting of this fusion protein to a specific location in the genome results in methylation of the N6 position of adenines in cis but also at other genomic sites that are in close proximity. Since endogenous methylation of adenines does not occur in most eukaryotes, methylated sequences can be attributed to chromatin looping. In this fashion long-range interactions have been detected in yeast and Drosophila [28, 29]. This method also has the potential for unbiased high-throughout analysis. A method based on nucleic acid hybridization-mediated targeting of a biotinylation reaction to primary transcripts (RNA-TRAP) allowed the detection of long-range genomic interactions [30]. While elegant in principle, this method has not found widespread use perhaps in part due to its dependence on active gene transcription.
Finally, high resolution 3D FISH (fluorescent in situ hybridization) with probes against primary transcripts or DNA can detect the juxtaposition of distinct genomic regions (e.g. [31, 32]) thus directly visualizing long-range genomic interactions. However, the current resolution limits of fluorescence microscopy preclude reliable distinction between looped and linear conformation of regulatory sites that are spaced closely within a single gene locus. However, if two cis-configured elements are being actively transcribed, primary transcript FISH can produce sufficiently strong signals to measure distances between them as close as 10 to 20 kb [32]. A potential limitation of FISH-based methods is that juxtaposition or even co-localization does not necessarily mean that two loci are functionally interacting.
Where are the loops?
A rapidly growing number of studies support the general hypothesis that distal regulatory elements are configured in a manner that brings them into close proximity with their target promoters. Moreover, enhancer-blocking insulator elements have been found to cluster in the nuclear space. At the mammalian β-globin locus, long-range interactions between a distal powerful enhancer called locus control region (LCR) and the active globin promoters have been described by two independent methods, 3C and RNA-TRAP [30, 33]. This lends strong and direct support to looping models that were established many years ago but, although based on substantial amounts of indirect evidence, were not definitive ([34], for reviews see [1, 2, 35]).
Additional examples of long-range interactions that have been observed within mammalian gene loci include the α-globin gene cluster in erythroid cells [36], the TH2, IFNγ, and MHC loci in T cells [37-39], The IgH and Igκ loci in B-cells [40, 41], HoxB1 [23], and at the imprinted gene clusters Dlx5, Dlx6 and H19-Igf2 [25, 26, 42, 43]. Promoter-enhancer interactions have been observed at the prolactin [18] and androgen receptor-dependent PSA genes [44].
Interactions among regulatory elements residing on different chromosomes have been observed at the IFNγ and TH2 cytokine loci [45], among olfactory receptor genes [46] and select estrogen-regulated genes [47].
In Drosophila, higher-order interactions have been described at the homeotic genes of the bithorax complex, including contacts between widely-spaced polycomb response elements (PREs) [29, 32, 48-50], and among insulator elements surrounding the heat shock genes [51] and the gypsy transposon [52]. The assembly of multiple simultaneous interactions that leads to the clustering of several activating elements has been termed active chromatin hub (ACH) [33]. At developmentally controlled genes, enhancer/promoter interactions form according to the developmental stage of gene expression. For example, 3C experiments showed that in murine erythroid cells containing a transgenic human β-globin locus, the LCR interacts with embryonic/fetal globin genes and adult globin in primitive and definitive erythroid cells, respectively [53]. Moreover, the frequency of interactions appears to increase upon cell maturation correlating with the onset of gene transcription.
In concert, these studies show that looped chromatin configurations represent a general feature of chromatin organization in eukaryotes that serve diverse genomic functions including transcription activation, repression, and DNA recombination.
Inhibitory chromatin loops
It is easy to envision how chromatin looping might also serve to inhibit gene expression. Therefore, it is not surprising that genes that are temporarily or permanently repressed have also been associated with specific chromatin loops. At the imprinted H19-Igf2 locus differentially methylated regions (DMR) control parent of origin-specific gene expression such that H19 and Igf2 are only expressed from the maternal and paternal alleles, respectively. Using 3C and ChIP, it was shown that these regions interact in an allele-specific manner to form distinct chromatin loops [25, 42, 43], (for review see [9]). Enhancer-promoter interactions correlate positively with gene activity. Moreover, on the maternal chromosome the transcriptionally silent Igf2 gene is placed within an inactive chromatin loop that prevents its promoter from interacting with the downstream enhancer. This chromatin configuration leaves the active H19 gene and the enhancer free to interact. Deletion of either of two DMRs that reside at the base of the loop leads to activation of maternal Igf2 expression ([54, 55], suggesting that this interaction contributes causally to maternal Igf2 silencing. Additional analysis suggests that the imprinting control region (ICR) that bears maternal allele-specific insulator activity interacts physically with the silent maternal Igf2 promoter and enhancer to block their function ([43]; see also below). A corollary of this finding is that enhancer-blocking insulators can form chromatin loops with positively acting regulatory elements to negate their activities.
Looped structures were also observed at the Dlx5-Dlx6 locus in mice where the methyl-binding protein MeCP2 is required for the formation of a distinct inhibitory chromosome configuration [26]. A direct involvement of MeCP2 in looped chromatin organization was supported by the ChIP-loop assay.
Recent work showed the presence of a higher-order chromatin structure at the Kit cytokine receptor gene [56]. Concurrent with repression of Kit expression during terminal erythroid differentiation, transcription factor GATA-1 triggers a dynamic structural transition of distinct chromatin loops. Specifically, an interaction among GATA elements that reside downstream of the transcriptional start site increases at the expense of an upstream enhancer-promoter chromatin loop.
Interactions have also been described among enhancer blockers, including CTCF (C-binding factor) binding sites [57], and the scs and scs' elements flanking the heat shock genes and the gypsy insulators in Drosophila ([51, 52]. Moreover, PREs that are important for the maintenance of repressed chromatin states can cluster in nuclear space ([50], see below). Together, the studies outlined above indicate that genes are organized into distinct configurations when active and repressed. Chromatin loops might therefore be established to promote or inhibit gene expression. Whether higher order folding is a cause or a consequence of transcriptional regulation is largely an open question.
How are loops formed?
The precise mechanism of loop formation is not understood. However, several transcription factors have been implicated in this process. The Drosophila GAGA factor that is encoded by the Trithorax-like gene (Trl) activates the expression of many genes by antagonizing the repression exerted by heterochromatin. Evidence based on the use of specially designed reporter constructs in yeast and in vitro transcription assays supports the idea that GAGA can contribute to the formation of chromatin loops ([58, 59]. Loop formation depends on the POZ/BTB domain involved in multimerization of GAGA. This domain is also found in the mod(mdg4) protein that contributes to clustering of the gypsy insulator elements. It is noteworthy that GAGA can also trigger enhancer-dependent activation of transcription even when the enhancer and the promoter are located on a separate plasmid, indicating that GAGA can promote physical proximity among DNA sequences located in trans, and that a tracking mechanism is not required for enhancer activity under these circumstances [58].
By 3C it was shown that a combination of transcription factors including EKLF, GATA-1 and its cofactor FOG-1, as well as the SCL complex component Ldb-1/NLI is needed for juxtaposing the β-globin LCR with the active globin promoter ([60-62]. This suggests that select combinations of transcription factors mediate gene specific long-range interactions. However, not all transcription factors required for globin gene expression appear to be involved in loop formation. For example, erythroid cells from mice lacking transcription factor p45 NF-E2 display essentially normal LCR-promoter interactions despite some reduction in globin expression [63]. Compensation by other factors remains a distinct possibility. It is worth noting that similar to its loop-promoting function at the globin locus, GATA-1 also triggers loop formation at the repressed Kit locus involving GATA sites downstream of the transcription start site (TSS) ([56]). It is possible that the location of the loop with respect to regulatory elements and the TSS determines transcriptional outcomes.
At the cytokine locus in TH2 cells, the full extent of spatial interactions among the promoters and the LCR required transcription factor STAT6. Nevertheless, a core configuration of the TH2 cytokine locus remained intact in the absence of STAT6 [37], indicating the existence of multiple stages in chromatin folding. Similarly, SATB1 (special AT-rich sequence binding protein 1) is required for the complete folding of the chromatin fiber at this locus [64]. Notably, the SATB1 ChIP-loop assay and the unbiased 3C assay produced very similar findings suggesting that SATB1 contributes to the anchoring of most if not all measurable chromatin loops at the TH2 locus.
In several of the cases discussed above it remains uncertain whether the effects of transcriptional regulators on chromatin looping are direct. Attempts to show a more direct involvement of nuclear factors in loop formation have been made at the β-globin locus. Conditional restoration of EKLF (erythroid Krüppel-like factor) [60] or GATA-1 [61] in erythroid cells null for either factor triggered LCR-β-globin gene interactions in the presence of the protein translation inhibitor cycloheximide. Another approach to address this problem involves the deletion or mutation of critical cis-acting elements. Removal of DNase1 hypersensitive site (HS) 3 of the LCR in combination with deletion of the β-globin promoter in the context of a transgenic human β-globin locus resulted in widespread reduction in ACH formation whereas neither deletion by itself had dramatic effects [65]. Also, deletion of a critical portion of the TH2 LCR reduced some but not all LCR - promoter interactions at the TH2 cytokine locus [66].
Thus, multiple interactions contribute to the stable assembly of a regulatory hub consistent with a requirement of several transcription factors for this process. The deletions used in these studies were too large to implicate single transcription factors. However, these studies could be developed further by generation of point mutations at critical transcription factor binding sites. Although extremely labor intensive, this approach could help discerning direct from indirect roles of DNA binding proteins during chromatin loop formation. Nevertheless, in the end one has to concede that neither the use of conditional transcription factors with cycloheximide nor the generation of DNA point mutations can completely exclude indirect effects since changes in transcription factor occupancy can influence the binding of neighboring factors.
The above studies inferred transcription factor requirements through loss of function approaches, leaving unresolved the question as to the minimal requirements of transcription factors sufficient for looping. Erythroid cells that lack GATA-1 but maintain expression of other known erythroid transcription factors including EKLF and NF-E2, fail to establish the LCR-β-globin promoter interaction [61]. This suggests that EKLF, NF-E2 and presumably other erythroid expressed transcription factors are insufficient to form this interaction. Similarly, erythroid cells that lack EKLF but retain the remaining complement of erythroid transcription factors including GATA-1, fail to set up normal LCR-β-globin promoter communication [60]. Nevertheless, substantial folding of the chromatin fiber at the β-globin locus persists in EKLF-null erythroid cells when compared to non-erythroid cells. This indicates that the remaining erythroid factors are capable of forming higher order structures but in the absence of EKLF these structures are insufficiently developed for globin gene expression. At the α-globin locus, occupancy of erythroid transcription factors including GATA-1, EKLF, and SCL occurs in immature cells prior to loop formation indicating that these proteins are insufficient for the assembly of all relevant chromosome interactions at least at the levels at which they are expressed at this stage of maturation [36].
Finally, forced expression of GATA-3 in fibroblasts in combination with calcium ionophore treatment, but not GATA-3 or ionophore alone can trigger chromosomal interactions at the TH2 locus but are insufficient to stimulate cytokine gene expression [37].
In concert these studies provide evidence for the general concept that chromosomal interactions are set up by transcription factors in distinct steps prior to the onset of gene transcription. Such interactions in the absence of ongoing transcription have been called “poised” and “pre-poised” and their functional significance remains to be determined [37].
Are there unifying models for activating chromatin loops?
Based on the available evidence it is reasonable to assume that most if not all distal enhancers contact promoters by some kind of looping mechanism and that DNA-binding proteins contribute to this process. Nevertheless, it remains unresolved how these proteins interact with each other to form or stabilize chromatin loops. The simplest of all possibilities is that interactions are established by proteins that can form dimers. This requires the presence of binding sites for such factors at both anchors of the loop. For example, GATA-1 can homodimerize [67] and GATA sites are found at both the globin promoters and the LCR thus fulfilling those basic criteria. However, a direct role of GATA-1 homodimerization in loop formation has not been evaluated. Of course, non-DNA-binding transcription co-factors might mediate interactions among transcription factors. For example, the hematopoietic-specific GATA-1 cofactor FOG-1 is required for GATA-1-induced chromatin loop formation [56, 61]. Since each FOG-1 molecule contains four potential GATA-1 binding surfaces, it could mediate interactions among multiple sites bound by GATA-1. However, truncated forms of FOG-1 that retain a single GATA-1 binding surface are biologically active in erythroid cells calling this simple model into question [68]. It is also conceivable that more general features of active genes might be involved in loop formation. This includes general transcription factors or cofactors, RNA polymerase II, histone modifications or proteins that bind to them, and remodeled nucleosomes, all of which could theoretically contribute at various levels to mediating enhancer-promoter interactions. For example, general cofactors such as CBP and the P220/PBP component of mediator were required for interchromosomal clustering of estrogen-responsive genes [47]. In fact, it has been proposed that association with nuclear compartments enriched for RNA polymerase 2 (pol2), so-called transcription factories, generally mediates the juxtaposition of distal chromosomal fragments and is thus a key determinant of the nuclear organization of the genome [69]. At first glance such a model seems attractive since the number of active genes exceeds the number of detectable transcription factories, implying that active genes need to share such factories. Enhancers and promoters but not intervening sequences are recruited towards these sites thus forming chromatin loops. Although there is ample evidence that active genes can localize to pol2 factories, such a model has difficulty in explaining many observations regarding how the chromatin fiber is folded. As detailed below, numerous results favor the existence of gene-specific chromosomal interactions that cannot be simply attributed to clustering around pol2 factories and that are established by dedicated transcription factors.
Association with pol2 factories correlates tightly with the activity of a gene [31]. Therefore, this model does not explain the presence of chromatin loops at genes prior to their activation (i.e. the “poised” configurations) or at repressed genes (e.g. [37]). Further insights into this question came from 3C experiments of the Kit cytokine receptor locus where it was shown that a distal enhancer is in proximity with the active promoter [56]. Notably, the enhancer does not contact most sequences within the transcribed region despite the presence of high levels of pol2. In addition, treatment with the transcription elongation blocker DRB led to a loss of pol2 from the Kit gene but did not reduce the enhancer-promoter interaction. This strongly suggests that the enhancer-promoter contacts can be established by gene-specific transcription factors in a manner independent of elongating pol2. Another example where loop formation at an active gene can be uncoupled from pol2 recruitment is the β-globin locus. Deletion of the β-globin promoter abrogates β-globin transcription but has minimal effects on the formation of the ACH [65]. More generally, transcription of α- and β-globin genes can occur outside of pol2 factories, and inactive genes can be close to them [31] suggesting that gene activity and, by inference, enhancer promoter-communication does not strictly depend on clustering around these active foci. Together these observations invoke a more intricate model to explain the complex folding patterns found at many gene loci. Thus, it is likely that gene-specific chromosomal interactions are established by transcription factors and their co-regulators independent of pol2 factories. Moreover, we speculate that contacts among genes are layered such that intragenic interactions are closer than those resulting from aggregation around pol2 factories. In this manner the activity of looped enhancers would be restricted to the dedicated promoters thus avoiding unwanted effects on other genes. This would also explain the finding that erythroid-specific genes that share transcriptional regulators are no more likely to form contacts with each other than with house keeping genes that are activated by unrelated mechanisms [20]. Very recent reports support such a model showing that inhibition of transcription with concomitant loss of pol2 association did not fundamentally influence the folding of the β-globin locus [70, 71]. Improvements in biochemical (3C) or microscopy (3D FISH) technologies might lead to more precise estimates of the absolute distances of both intragenic and intergenic chromosomal interactions and perhaps allow discrimination between interactions that function in a gene-specific manner and those that do not.
Are there unifying models for repressive chromatin loops?
Similar to gene activation, pathways that control gene repression might ultimately converge on a common set of rules. Polycomb response elements (PREs) are sequences that mediate repression of nearby genes in a heritable manner. Polycomb proteins that bind to PREs are part of a complex that maintains gene repression throughout numerous cell divisions during development. Polycomb response elements associate with each other and with repressed promoters leading to looped chromosomal structures [50]. The presence of such folded structures might exist to bock nearby enhancers from communicating with the repressed promoters. Moreover, polycomb proteins form recognizable foci, called polycomb group (PcG) bodies in the nucleus [72]. How association with PcG bodies contributes to silencing is unclear. However, components of the RNAi machinery are found in a subset of these foci and are also required for the long distance interaction among certain PREs [73]. Thus, PcG bodies represent subnuclear sites that promote repression by employing the RNAi machinery, or through the enrichment of co-repressor complexes and protein sumoylation [74] or other mechanisms. Regardless of the route leading to repression, PRE recruitment to PcG bodies contributes to loop formation among elements in cis and also fosters interactions in trans.
Enhancer-blocking insulators provide another example of elements that can cluster to form chromatin loops with the effect of preventing enhancers from inappropriately activating promoters (for review see [9, 10]. Insulators inhibit enhancer can activity only when placed between the enhancer and the promoter and thus distinguish themselves from silencer elements (for review see [8]). Work from Drosophila and vertebrates suggests that the function of enhancer blockers somewhat parallels that observed with PREs, including their ability to associate with one another in cis and trans as well as the tendency for enrichment in defined nuclear compartments. For example, the Su(Hw) insulator that is found at numerous locations across the Drosophila genome including the gypsy retrotransposon, is associated with loop formation as determined by FISH [52]. Moreover, proteins associated with Su(Hw) including CP190, Mod(mdg4) and dTopors mediate clustering of Su(Hw) insulators leading to the formation of “insulator bodies” [75-77]. These insulator bodies are recruited to the nuclear periphery through anchoring with lamin proteins, components of the nuclear skeleton [75, 77]. However, recent experiments with mutant forms of Mod(mdg4) revealed that sequestration to subnuclear sites enriched for insulator proteins can be uncoupled from insulator activity [78]. This study raises questions as to the true character and function of insulator bodies and their role in partitioning the genome. Another tantalizing set of observations suggests that the similarity among PREs and Su(Hw) insulators extends to the involvement of components of the RNAi machinery ([73, 79].
Similar to the gypsy insulators, the scs and scs' sequences, the first known insulators that flank the Drosophila 87A7 hsp70 heat shock locus, contact each other as shown by both FISH and 3C experiments [51]. The interaction depends on Zw5 and BEAF that bind to scs and scs', respectively. Zw5 and BEAF interact in solution and likely play a direct role in loop formation.
The theme of both looping and tethering to defined nuclear compartments extends to vertebrate insulators (for review see [8]). Sequences bound by CTCF not only share the ability to associate in vivo both in cis and in trans [80, 81] but also their tendency for enrichment at defined nuclear compartments. Specifically, CTCF binding sites in the chicken HS4 insulator were required to target associated DNA sequences to the outer rim of the nucleolus [57]. This targeting, which might be mediated by the CTCF-binding protein nucleophosmin, is thought to contribute to organizing associated genes into looped domains. Whether clustering and nucleolar targeting occurred simultaneously or independently remains unanswered.
CTCF is also required for the formation of some of the looped structures found at the murine β-globin locus. However, loss of CTCF binding and the resulting loss of looped structures had no measurable effect on LCR-promoter contacts or β-globin gene expression, nor did it lead to activation of the adjacent silent olfactory receptor genes [80]. This suggests that not all long-range interactions are of functional significance. Alternatively, clustering of CTCF-bound sites at the β-globin locus might prevent them from contacting other regions in the genome and produce unwanted effects on gene expression. As mentioned above, interchromosomal interactions between CTCF binding sites have been reported [81].
It is worth noting that certain boundary/barrier elements also appear to cluster at distinct nuclear compartments. In S. cerevisiae, boundary activity was associated with proteins that interact with the nuclear pore complex in the nuclear periphery [82]. In S. pombe, inverted repeat elements that serve as boundaries at the silent mating type loci associate with the nuclear periphery and also to some extent with nucleoli [83]. Binding sites for the general transcription factor TFIIIC are required for barrier activity and mediate the association of DNA with nuclear structures referred to as TFIIIC bodies. Thus, both barrier and enhancer blocking elements share this particular property of being enriched at specific nuclear sites.
Together, the above studies suggest that both looped chromatin organization and distinct subnuclear localization either alone or in combination contribute to enhancer-blocking activity. This might be accomplished in two ways. In the first, loop formation might prevent the insulated enhancers and promoters from interacting productively perhaps by separating them physically or by interfering with some kind of tracking or spreading mechanism. In the second, the nuclear periphery and especially the insulator bodies might be enriched for repressive protein complexes.
If shared mechanisms underlie the function of inhibitory elements, a key question becomes how long-range interactions are restricted such that a consistent organization of the genome is achieved. What promotes the correct interactions and what prevents inappropriate contacts among insulator elements that belong to the same class or between insulators and promoters/enhancers? It is likely that multiple proteins function in specific combinations to set up long-range interactions. Another unanswered mechanistic question is how loop formation leads to loss of enhancer activity. In other words, what prevents the enhancer from looping beyond the insulator element to regulate a distal promoter? One of the hypotheses addressing this problem invoked the formation of specific inhibitory chromatin loops between the insulator and either the enhancer, the promoter, or both. An example for such interactions was provided recently from studies of the imprinted Igf2-H19 locus. Specifically, it was shown by 3C that the H19 imprinting control region (ICR) insulator, when active, interacts with both the blocked promoter and enhancer [43]. While elegant and provocative, this observation begs the question as to how the insulator “knows” to which regulatory element it should loop. Also, if the insulator can associate with key regulatory elements, why does this require that the insulator be positioned between the promoter and enhancer? Additional models such as tracking mechanisms are required to accommodate all of these observations.
A role for chromatin loops in the transcription cycle
Chromatin loops do not only form to establish long-range enhancer-promoter or boundary element interactions. Studies in yeast indicate that the promoters and termination sites of genes as small as 1 kb are in close physical proximity [84, 85]. This seems to be a widespread phenomenon in yeast and requires elongating RNA polymerase, suggesting a model in which an initial “pioneer” round of transcription establishes contacts between the promoter and termination sites. Of note, both the basal transcription factor TFIIB and RNA processing factors are required for loop formation [86]. Specifically, Ssu72, a member of the 3′ RNA processing complex CPF, dephosphorylates serine-5 of the pol2 C-terminal domain (CTD), converting it from an elongation- to an initiation-competent form. Furthermore, Ssu72 interacts with TFIIB [87]. In actively transcribed genes, both Ssu72 and TFIIB occupy both the promoter and the terminator, and both are required for their juxtaposition [85]. Therefore, it is attractive to speculate that loop formation coordinates recycling of RNA polymerase 2 by converting it from the elongation form into the initiation form and delivering it back to the promoter. A point mutation in TFIIB was identified that abrogates loop formation and lowers TFIIB association with the terminator but not the promoter, suggesting that looping by TFIIB can be uncoupled from its role in transcription initiation. However, this mutation did not affect transcript levels of any of the genes examined, raising the question as to the requirement for loop formation for the transcription cycle [86]. Perhaps efficient transfer of pol2 from the 3′ to the 5′ end of transcribed gene is only required for the most highly expressed genes. Another possibility is that the looping mechanism may have evolved to discourage illegitimate transcription of irrelevant sequences. The promoter-terminator loop requires an ordered gene structure including a poly(A) signal. If RNA polymerase II initiates a transcript lacking such a sequence it will not be efficiently recycled back to the start site.
Provocative recent studies further suggest that the phenomenon of promoter/terminator juxtaposition appears to be a general mechanism during gene expression. First, initiation and termination sites are juxtaposed at the mitochondrial rDNA genes, and loop formation requires the termination factor mTERF that associates with both initiation and termination sites [88]. Second, both episomal and integrated proviral HIV constructs exhibit interactions between the promoter and poly(A) sequences located at the 5′ and 3′ LTRs, respectively [89]. Similar to the findings in yeast, loop formation required ongoing transcription. Importantly, substitution of the HIV promoter with another viral promoter (i.e. CMV) or substitution of the natural poly(A) signal with a synthetic one did not abrogate loop formation. This resonates not only with the theme of juxtaposing transcriptional and 3′ processing elements but underscores the general nature of linkage of termination with initiation as a feature of actively transcribed genes. A caveat is that at the human and mouse BRCA1 loci, the promoter was found to interact with the terminator, but only in the repressed state [90]. Following induction of BRCA1 transcription, the promoter-terminator loop dissolved. Interestingly, even in the un-induced state, the BRCA1 promoter-terminator loop but not smaller interspersed loops between the promoter and intronic sequences, still required basal level transcription suggesting that the loop is maintained either by a direct mechanism involving elongating RNA polymerase 2 or indirectly through an unstable protein or RNA intermediate. Together, these findings suggest that promoter-terminator loops might contribute to both gene activation and repression.
General considerations about chromatin loops
Despite the identification of proteins capable of mediating chromatin loops it remains unclear how sequence elements “find” each other to set up specific interactions. One possibility is that loops result from random collisions that are subsequently stabilized by protein-protein interactions. As mentioned above, such a model does not easily reconcile all of the observations regarding the function of enhancers and insulators, e.g. the directionality of enhancer blockers. Another possibility is that tracking/scanning might function as a conduit to establish a loop. In this scenario, proteins anchored to an enhancer might move along the chromatin fiber, thus bringing along associated DNA until a stable interaction is formed with promoter-bound factors. Work on the HNF4α gene locus supports the existence of tracking intermediates, which are predicted by the tracking/scanning but not the random-collision model [24]. Time course ChIP experiments detected the presence of three enhancer-binding proteins, HNF-1, HNF-3 and C/EBP along with the histone modifying enzymes CBP, P/CAF and Brg1 in the intervening region between the enhancer and the promoter. The distribution of these factors progressed directionally towards the promoter and reached the promoter at a time point coinciding with the onset of transcription. Pol2 was initially not found at the enhancer but concurrent with activation pol2 was detected at the enhancer presumably as a result of contacts between the promoter and enhancer. Although no 3C experiments were performed in this study, these results suggest that tracking precedes looping at the HNF4a locus. An enhancer-blocking insulator element might function by blocking such a spreading mechanism. Another open question addresses the energy requirements for reconfiguring chromatin fibers in such a fashion. It is possible that ATPases such as Brg1 provide the necessary energy [24]. Additional support for the tracking model derives from experiments with an episomal construct containing the HS2 enhancer of the β-globin LCR and the embryonic globin gene promoter [91]. In this system pol2 is present at both the promoter and HS2. It was shown that insertion of the chicken HS4 insulator between the HS2 and the promoter prevented the spreading of RNA pol2 and histone acetylation from the enhancer toward the promoter. This provides another example for a model in which tracking, in this case of RNA pol2, might precede loop formation. So far there is no direct evidence that looping of the β-globin LCR to the more distant adult type globin genes also follows tracking and pol2 and active histone modifications are low or undetectable in chromatin between the LCR and the active adult β-globin promoters [92, 93]. However, since the model predicts that tracking is a transient state, protein complexes that travel through intervening regions might escape detection.
Another variation of an integrated tracking/looping model is based on a study of the PSA gene at which the ligand-bound androgen receptor is involved in promoting contacts between the enhancer and proximal promoter [44]. Notably, in contrast to the scenario at the HNF4a gene, RNA pol2 but not co-activator proteins were detected by ChIP at intervening regions, suggesting a mechanism of pol2 tracking that does not involve the enhancer-bound proteins.
In concert, the above studies suggest that while tracking is likely a widespread means to establish communication between regulatory elements, the mechanisms by which this is accomplished seem to vary. Whether similar rules apply to the interactions formed among distant insulators remains unresolved. Insulators and enhancers seem to share important properties with regard to chromatin organization, and insulators can physically interact with enhancer and promoter sequences. Therefore, a speculative scenario of how insulator-enhancer loops could be formed invokes an initial tracking mechanism where spreading of pol2 or transcription factors initiates from an enhancer but is blocked by an insulator. As a result, proteins arrested at the insulator might contribute to the formation of stable loops that render the enhancer inactive. Conceptually, tracking as a prerequisite for looping might be analogous to the above-mentioned situation in yeast where juxtaposition of promoter and terminator sites requires an initial round of transcription.
Since both enhancers and insulators can function quite promiscuously in experimental systems, the question as to how specificity is established in vivo remains unclear (see above). In their native environment, unique combinations of transcription factors likely convey a degree of specificity favoring desirable over inappropriate long-range interactions. However, the question arises whether a mechanism exists that actively constrains unwanted interactions. To our knowledge no such mechanism has been described in metazoan cells. A provocative study in yeast identified several chromatin-associated proteins (SIN4, SPT2, SPT10 and HTA1-HTB1) that limit the distance from which an upstream activating sequence (UAS) can stimulate transcription [94]. Moreover, by 3C it was shown for one of the model genes under the influence of these factors that the UAS loops to its target promoter. In light of the generally close proximity between UAS and promoter elements and the comparatively small size of most genes, it is likely that this particular mechanism is most useful to yeast cells and other organisms with compact genomes. However, this illustrates that, in principle, dedicated nuclear factors can restrict aberrant loop formation.
Future perspective
Many important questions regarding the three-dimensional organization of the genome remain: Can illegitimate gene looping contribute to human disease? A recent study uncovered a rare form of α-thalassemia that is caused by a single gain-of-function nucleotide polymorphism that creates a promoter-like element between the α-globin genes and distal upstream regulatory elements [95]. This leads not only to transcription initiation from this newly formed promoter but also reduces α-globin expression. This is likely a consequence of the enhancer elements looping to the aberrant promoter at the expense of forming contacts with the natural promoters.
How is chromatin folding regulated throughout the cell cycle? Are chromatin loops maintained in some form throughout S-phase and mitosis? The observation that PREs are sequence modules that impart epigenetic inheritance together with their ability to aggregate raises the possibility that higher order chromatin folding might be preserved from one cell generation to the next. This might facilitate reestablishing proper gene expression following cell division.
A major challenge for the future will be to study the three-dimensional configuration of the genome in different cell lineages at distinct stages of differentiation. The development of high-throughput 4C and 5C methods has moved this goal a significant step closer to reality [96]. This line of investigation will conceptually intersect and overlap with continued studies on sub-nuclear compartmentalization and localization of nuclear functions.
Additional important questions for future investigation are a) whether contacts between enhancers and promoters are a cause or consequence of gene activation, and b) whether interactions among insulator elements or between insulators and enhancers might be sufficient to limit enhancer function. The first question was addressed in a study showing that forced loop formation was rate-limiting for transcriptional activation in the context of a transfected reporter gene construct [58]. In this construct an enhancer element was placed at a distance from a promoter-reporter gene such that it was incapable of activating transcription. Forced juxtaposition of the enhancer with the promoter through transcription factor-induced looping led to activation of the promoter. The second question was addressed (again in the context of episomal vectors) by showing that forced loop formation that places an enhancer and promoter on separate loops can abrogate enhancer activity [97]. Together, these observations indicate that forced reorganization of the chromatin fiber can be sufficient to elicit positive and negative changes in gene expression. To better evaluate the roles of looping in gene regulation, the field awaits the directed manipulation of chromatin loops at endogenous gene loci. If loops can be reconfigured to alter promoter-enhancer interactions, one might envision this to become a tool for influencing gene expression for therapeutic purposes.
Figure 1. Methods to detect chromatin loops.
Several assays have been developed to detect proximity between two distant genomic sequences, for example an enhancer (blue circle) and a promoter (red square). a. Chromosome conformation capture (3C). This widely used assay relies on the assumption that sequences which interact with a high frequency can be crosslinked more efficiently than sequences that interact less frequently. Crosslinked chromatin is digested with a restriction enzyme that creates sticky ends (red bars). Ligation generates unique products which do not occur in genomic DNA and which can be detected by PCR (empty arrowheads). The amount of PCR product correlates with the proximity between the genomic fragments in vivo. b. Indirect chromatin immunoprecipitation (ChIP). This technique exploits sequence-specific DNA-binding proteins (green dome) as anchors to recover indirectly associated sequences. Chromatin is crosslinked and fragmented by sonication. DNA associated with the protein is recovered by immunoprecipitation. Sequences that are indirectly associated with the cognate sequence of the DNA-binding protein are recovered in this process and can be detected by PCR. c. DNA adenine methyltransferase identification (Dam ID). This method takes advantage of the fact that adenine methylation, which does not naturally occur in eukaryotes, inhibits DNA cleavage by the DpnII restriction enzyme. The bacterial Dam methylase is fused to a DNA-binding domain (DBD) to tether it to a sequence of interest (blue box). Tethered Dam methylates DNA in vivo, after which genomic DNA is digested with DpnII. PCR is used to amplify sequences in a fashion that distinguishes undigested i.e. methylated from unmethylated sites of interest.
Acknowledgments
We thank Job Dekker, Susan Janicki and members of the lab for critically reading this manuscript. We apologize to those whose work could not be discussed here due to space limitations. G.A.B is supported by NIH grants DK54937, DK58044 and DK075731. S.K. is supported by the University of Pennsylvania School of Medicine and the Benjamin and Mary Siddons Measey Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Engel JD, Tanimoto K. Looping, linking, and chromatin activity: new insights into beta-globin locus regulation. Cell. 2000;100:499–502. doi: 10.1016/s0092-8674(00)80686-8. [DOI] [PubMed] [Google Scholar]
- 2.Bulger M, Groudine M. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 1999;13:2465–77. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- 3.Martin DI, Fiering S, Groudine M. Regulation of beta-globin gene expression: straightening out the locus. Curr Opin Genet Dev. 1996;6:488–95. doi: 10.1016/s0959-437x(96)80072-4. [DOI] [PubMed] [Google Scholar]
- 4.Blackwood EM, Kadonaga JT. Going the distance: a current view of enhancer action. Science. 1998;281:60–3. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- 5.Chambeyron S, Bickmore WA. Does looping and clustering in the nucleus regulate gene expression? Curr Opin Cell Biol. 2004;16:256–62. doi: 10.1016/j.ceb.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 6.Fraser P. Transcriptional control thrown for a loop. Curr Opin Genet Dev. 2006;16:490–5. doi: 10.1016/j.gde.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Dean A. On a chromosome far, far away: LCRs and gene expression. Trends Genet. 2006;22:38–45. doi: 10.1016/j.tig.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7:703–13. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 9.Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr Opin Genet Dev. 2007;17:400–7. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorman ER, Bushey AM, Corces VG. The role of insulator elements in large-scale chromatin structure in interphase. Semin Cell Dev Biol. 2007;18:682–90. doi: 10.1016/j.semcdb.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morgan GT. Lampbrush chromosomes and associated bodies: new insights into principles of nuclear structure and function. Chromosome Res. 2002;10:177–200. doi: 10.1023/a:1015227020652. [DOI] [PubMed] [Google Scholar]
- 12.Marsden MP, Laemmli UK. Metaphase chromosome structure: evidence for a radial loop model. Cell. 1979;17:849–58. doi: 10.1016/0092-8674(79)90325-8. [DOI] [PubMed] [Google Scholar]
- 13.Heng HH, Krawetz SA, Lu W, Bremer S, Liu G, Ye CJ. Re-defining the chromatin loop domain. Cytogenet Cell Genet. 2001;93:155–61. doi: 10.1159/000056977. [DOI] [PubMed] [Google Scholar]
- 14.Martelli AM, Falcieri E, Zweyer M, Bortul R, Tabellini G, Cappellini A, Cocco L, Manzoli L. The controversial nuclear matrix: a balanced point of view. Histol Histopathol. 2002;17:1193–205. doi: 10.14670/HH-17.1193. [DOI] [PubMed] [Google Scholar]
- 15.Cremer T, Cremer M, Dietzel S, Muller S, Solovei I, Fakan S. Chromosome territories--a functional nuclear landscape. Curr Opin Cell Biol. 2006;18:307–16. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 16.Kosak ST, Groudine M. Form follows function: The genomic organization of cellular differentiation. Genes Dev. 2004;18:1371–84. doi: 10.1101/gad.1209304. [DOI] [PubMed] [Google Scholar]
- 17.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–11. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 18.Cullen KE, Kladde MP, Seyfred MA. Interaction between transcription regulatory regions of prolactin chromatin. Science. 1993;261:203–6. doi: 10.1126/science.8327891. [DOI] [PubMed] [Google Scholar]
- 19.Dekker J. The three ‘C’ s of chromosome conformation capture: controls, controls, controls. Nat Methods. 2006;3:17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- 20.Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006 doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 21.Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, Green RD, Dekker J. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–7. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 23.Wurtele H, Chartrand P. Genome-wide scanning of HoxB1-associated loci in mouse ES cells using an open-ended Chromosome Conformation Capture methodology. Chromosome Res. 2006;14:477–95. doi: 10.1007/s10577-006-1075-0. [DOI] [PubMed] [Google Scholar]
- 24.Hatzis P, Talianidis I. Dynamics of Enhancer-Promoter Communication during Differentiation-Induced Gene Activation. Mol Cell. 2002;10:1467–77. doi: 10.1016/s1097-2765(02)00786-4. [DOI] [PubMed] [Google Scholar]
- 25.Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet. 2004;36:889–93. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 26.Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- 27.van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat Biotechnol. 2000;18:424–8. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- 28.Lebrun E, Fourel G, Defossez PA, Gilson E. A methyltransferase targeting assay reveals silencer-telomere interactions in budding yeast. Mol Cell Biol. 2003;23:1498–508. doi: 10.1128/MCB.23.5.1498-1508.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cleard F, Moshkin Y, Karch F, Maeda RK. Probing long-distance regulatory interactions in the Drosophila melanogaster bithorax complex using Dam identification. Nat Genet. 2006;38:931–5. doi: 10.1038/ng1833. [DOI] [PubMed] [Google Scholar]
- 30.Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32:623–6. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 31.Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–71. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 32.Ronshaugen M, Levine M. Visualization of trans-homolog enhancer-promoter interactions at the Abd-B Hox locus in the Drosophila embryo. Dev Cell. 2004;7:925–32. doi: 10.1016/j.devcel.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 33.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–65. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 34.Gallarda JL, Foley KP, Yang Z, Engel JD. The β-globin stage selector element factor is erythroid-specific promoter/enhancer binding protein NF-E4. Genes Dev. 1989;3:1845–1859. doi: 10.1101/gad.3.12a.1845. [DOI] [PubMed] [Google Scholar]
- 35.Li Q, Peterson KR, Fang X, Stamatoyannopoulos G. Locus control regions. Blood. 2002;100:3077–86. doi: 10.1182/blood-2002-04-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. Embo J. 2007;26:2041–51. doi: 10.1038/sj.emboj.7601654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol. 2004;5:1017–27. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- 38.Eivazova ER, Aune TM. Dynamic alterations in the conformation of the Ifng gene region during T helper cell differentiation. Proc Natl Acad Sci U S A. 2004;101:251–6. doi: 10.1073/pnas.0303919101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar PP, Bischof O, Purbey PK, Notani D, Urlaub H, Dejean A, Galande S. Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nat Cell Biol. 2007;9:45–56. doi: 10.1038/ncb1516. [DOI] [PubMed] [Google Scholar]
- 40.Sayegh C, Jhunjhunwala S, Riblet R, Murre C. Visualization of looping involving the immunoglobulin heavy-chain locus in developing B cells. Genes Dev. 2005;19:322–7. doi: 10.1101/gad.1254305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Z, Garrard WT. Long-range interactions between three transcriptional enhancers, active Vkappa gene promoters, and a 3′ boundary sequence spanning 46 kilobases. Mol Cell Biol. 2005;25:3220–31. doi: 10.1128/MCB.25.8.3220-3231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci U S A. 2006;103:10684–9. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoon YS, Jeong S, Rong Q, Park KY, Chung JH, Pfeifer K. Analysis of the H19ICR insulator. Mol Cell Biol. 2007;27:3499–510. doi: 10.1128/MCB.02170-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell. 2005;19:631–42. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 45.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–45. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 46.Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–13. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 47.Nunez E, Kwon YS, Hutt KR, Hu Q, Cardamone MD, Ohgi KA, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG, Fu XD. Nuclear receptor-enhanced transcription requires motor- and LSD1-dependent gene networking in interchromatin granules. Cell. 2008;132:996–1010. doi: 10.1016/j.cell.2008.01.051. [DOI] [PubMed] [Google Scholar]
- 48.Comet I, Savitskaya E, Schuettengruber B, Negre N, Lavrov S, Parshikov A, Juge F, Gracheva E, Georgiev P, Cavalli G. PRE-mediated bypass of two Su(Hw) insulators targets PcG proteins to a downstream promoter. Dev Cell. 2006;11:117–24. doi: 10.1016/j.devcel.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 49.Bantignies F, Grimaud C, Lavrov S, Gabut M, Cavalli G. Inheritance of Polycomb-dependent chromosomal interactions in Drosophila. Genes Dev. 2003;17:2406–20. doi: 10.1101/gad.269503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lanzuolo C, Roure V, Dekker J, Bantignies F, Orlando V. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat Cell Biol. 2007;9:1167–74. doi: 10.1038/ncb1637. [DOI] [PubMed] [Google Scholar]
- 51.Blanton J, Gaszner M, Schedl P. Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev. 2003;17:664–75. doi: 10.1101/gad.1052003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Byrd K, Corces VG. Visualization of chromatin domains created by the gypsy insulator of Drosophila. J Cell Biol. 2003;162:565–74. doi: 10.1083/jcb.200305013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W. The beta-globin nuclear compartment in development and erythroid differentiation. Nat Genet. 2003;35:190–4. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- 54.Thorvaldsen JL, Duran KL, Bartolomei MS. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12:3693–702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Constancia M, Dean W, Lopes S, Moore T, Kelsey G, Reik W. Deletion of a silencer element in Igf2 results in loss of imprinting independent of H19. Nat Genet. 2000;26:203–6. doi: 10.1038/79930. [DOI] [PubMed] [Google Scholar]
- 56.Jing H, Vakoc CR, Ying L, Mandat S, Wang H, Zheng X, Blobel GA. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol Cell. 2008;29:232–42. doi: 10.1016/j.molcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell. 2004;13:291–8. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 58.Mahmoudi T, Katsani KR, Verrijzer CP. GAGA can mediate enhancer function in trans by linking two separate DNA molecules. Embo J. 2002;21:1775–81. doi: 10.1093/emboj/21.7.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Petrascheck M, Escher D, Mahmoudi T, Verrijzer CP, Schaffner W, Barberis A. DNA looping induced by a transcriptional enhancer in vivo. Nucleic Acids Res. 2005;33:3743–50. doi: 10.1093/nar/gki689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 2004;18:2485–90. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among Distant Regulatory Elements at the beta-Globin Locus Requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–62. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 62.Song SH, Hou C, Dean A. A positive role for NLI/Ldb1 in long-range beta-globin locus control region function. Mol Cell. 2007;28:810–22. doi: 10.1016/j.molcel.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kooren J, Palstra RJ, Klous P, Splinter E, von Lindern M, Grosveld F, de Laat W. Beta-globin active chromatin Hub formation in differentiating erythroid cells and in p45 NF-E2 knock-out mice. J Biol Chem. 2007;282:16544–52. doi: 10.1074/jbc.M701159200. [DOI] [PubMed] [Google Scholar]
- 64.Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet. 2006;38:1278–88. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- 65.Patrinos GP, de Krom M, de Boer E, Langeveld A, Imam AM, Strouboulis J, de Laat W, Grosveld FG. Multiple interactions between regulatory regions are required to stabilize an active chromatin hub. Genes Dev. 2004;18:1495–509. doi: 10.1101/gad.289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee GR, Spilianakis CG, Flavell RA. Hypersensitive site 7 of the TH2 locus control region is essential for expressing TH2 cytokine genes and for long-range intrachromosomal interactions. Nat Immunol. 2005;6:42–8. doi: 10.1038/ni1148. [DOI] [PubMed] [Google Scholar]
- 67.Crossley M, Merika M, Orkin SH. Self-association of the erythroid transcription factor GATA-1 mediated by its zinc finger domains. Mol Cell Biol. 1995;15:2448–2456. doi: 10.1128/mcb.15.5.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cantor AB, Katz SG, Orkin SH. Distinct domains of the GATA-1 cofactor FOG-1 differentially influence erythroid versus megakaryocytic maturation. Mol Cell Biol. 2002;22:4268–79. doi: 10.1128/MCB.22.12.4268-4279.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marenduzzo D, Faro-Trindade I, Cook PR. What are the molecular ties that maintain genomic loops? Trends Genet. 2007;23:126–33. doi: 10.1016/j.tig.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 70.Mitchell JA, Fraser P. Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes Dev. 2008;22:20–5. doi: 10.1101/gad.454008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Palstra RJ, Simonis M, Klous P, Brasset E, Eijkelkamp B, de Laat W. Maintenance of Long-Range DNA Interactions after Inhibition of Ongoing RNA Polymerase II Transcription. PLoS ONE. 2008;3:e1661. doi: 10.1371/journal.pone.0001661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Buchenau P, Hodgson J, Strutt H, Arndt-Jovin DJ. The distribution of polycomb-group proteins during cell division and development in Drosophila embryos: impact on models for silencing. J Cell Biol. 1998;141:469–81. doi: 10.1083/jcb.141.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grimaud C, Bantignies F, Pal-Bhadra M, Ghana P, Bhadra U, Cavalli G. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell. 2006;124:957–71. doi: 10.1016/j.cell.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 74.Kagey MH, Melhuish TA, Wotton D. The polycomb protein Pc2 is a SUMO E3. Cell. 2003;113:127–37. doi: 10.1016/s0092-8674(03)00159-4. [DOI] [PubMed] [Google Scholar]
- 75.Gerasimova TI, Byrd K, Corces VG. A chromatin insulator determines the nuclear localization of DNA. Mol Cell. 2000;6:1025–35. doi: 10.1016/s1097-2765(00)00101-5. [DOI] [PubMed] [Google Scholar]
- 76.Pai CY, Lei EP, Ghosh D, Corces VG. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol Cell. 2004;16:737–48. doi: 10.1016/j.molcel.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 77.Capelson M, Corces VG. The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol Cell. 2005;20:105–16. doi: 10.1016/j.molcel.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 78.Golovnin A, Melnikova L, Volkov I, Kostuchenko M, Galkin AV, Georgiev P. ‘Insulator bodies’ are aggregates of proteins but not of insulators. EMBO Rep. 2008;9:440–5. doi: 10.1038/embor.2008.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lei EP, Corces VG. RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat Genet. 2006;38:936–41. doi: 10.1038/ng1850. [DOI] [PubMed] [Google Scholar]
- 80.Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–54. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–72. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 82.Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell. 2002;109:551–62. doi: 10.1016/s0092-8674(02)00756-0. [DOI] [PubMed] [Google Scholar]
- 83.Noma K, Cam HP, Maraia RJ, Grewal SI. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125:859–72. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 84.O'Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, Proudfoot NJ. Gene loops juxtapose promoters and terminators in yeast. Nat Genet. 2004;36:1014–8. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- 85.Ansari A, Hampsey M. A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes Dev. 2005;19:2969–78. doi: 10.1101/gad.1362305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Singh BN, Hampsey M. A transcription-independent role for TFIIB in gene looping. Mol Cell. 2007;27:806–16. doi: 10.1016/j.molcel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 87.Sun ZW, Hampsey M. Synthetic enhancement of a TFIIB defect by a mutation in SSU72, an essential yeast gene encoding a novel protein that affects transcription start site selection in vivo. Mol Cell Biol. 1996;16:1557–66. doi: 10.1128/mcb.16.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Martin M, Cho J, Cesare AJ, Griffith JD, Attardi G. Termination factor-mediated DNA loop between termination and initiation sites drives mitochondrial rRNA synthesis. Cell. 2005;123:1227–40. doi: 10.1016/j.cell.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 89.Perkins KJ, Lusic M, Mitar I, Giacca M, Proudfoot NJ. Transcription-dependent gene looping of the HIV-1 provirus is dictated by recognition of pre-mRNA processing signals. Mol Cell. 2008;29:56–68. doi: 10.1016/j.molcel.2007.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tan-Wong SM, French JD, Proudfoot NJ, Brown MA. Dynamic interactions between the promoter and terminator regions of the mammalian BRCA1 gene. Proc Natl Acad Sci U S A. 2008;105:5160–5. doi: 10.1073/pnas.0801048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhao H, Dean A. An insulator blocks spreading of histone acetylation and interferes with RNA polymerase II transfer between an enhancer and gene. Nucleic Acids Res. 2004;32:4903–19. doi: 10.1093/nar/gkh832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Forsberg EC, Downs KM, Christensen HM, Im H, Nuzzi PA, Bresnick EH. Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc Natl Acad Sci U S A. 2000;97:14494–9. doi: 10.1073/pnas.97.26.14494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bulger M, Schubeler D, Bender MA, Hamilton J, Farrell CM, Hardison RC, Groudine M. A complex chromatin landscape revealed by patterns of nuclease sensitivity and histone modification within the mouse beta-globin locus. Mol Cell Biol. 2003;23:5234–44. doi: 10.1128/MCB.23.15.5234-5244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dobi KC, Winston F. Analysis of transcriptional activation at a distance in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:5575–86. doi: 10.1128/MCB.00459-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.De Gobbi M, Viprakasit V, Hughes JR, Fisher C, Buckle VJ, Ayyub H, Gibbons RJ, Vernimmen D, Yoshinaga Y, de Jong P, Cheng JF, Rubin EM, Wood WG, Bowden D, Higgs DR. A regulatory SNP causes a human genetic disease by creating a new transcriptional promoter. Science. 2006;312:1215–7. doi: 10.1126/science.1126431. [DOI] [PubMed] [Google Scholar]
- 96.Dekker J. Gene regulation in the third dimension. Science. 2008;319:1793–4. doi: 10.1126/science.1152850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ameres SL, Drueppel L, Pfleiderer K, Schmidt A, Hillen W, Berens C. Inducible DNA-loop formation blocks transcriptional activation by an SV40 enhancer. Embo J. 2005;24:358–67. doi: 10.1038/sj.emboj.7600531. [DOI] [PMC free article] [PubMed] [Google Scholar]