Abstract
Mutations in the mitochondrial helicase Twinkle underlie autosomal dominant progressive external ophthalmoplegia (PEO), as well as recessively inherited infantile-onset spinocerebellar ataxia and rare forms of mitochondrial DNA (mtDNA) depletion syndrome. Familial PEO is typically associated with the occurrence of multiple mtDNA deletions, but the mechanism by which Twinkle dysfunction induces deletion formation has been under debate. Here we looked at the effects of Twinkle adPEO mutations in human cell culture and studied the mtDNA replication in the Deletor mouse model, which expresses a dominant PEO mutation in Twinkle and accumulates multiple mtDNA deletions during life. We show that expression of dominant Twinkle mutations results in the accumulation of mtDNA replication intermediates in cell culture. This indicated severe replication pausing or stalling and caused mtDNA depletion. A strongly enhanced accumulation of replication intermediates was evident also in six-week-old Deletor mice compared with wild-type littermates, even though mtDNA deletions accumulate in a late-onset fashion in this model. In addition, our results in cell culture pointed to a problem of transcription that preceded the mtDNA depletion phenotype and might be of relevance in adPEO pathophysiology. Finally, in vitro assays showed functional defects in the various Twinkle mutants and broadly agreed with the cell culture phenotypes such as the level of mtDNA depletion and the level of accumulation of replication intermediates. On the basis of our results we suggest that mtDNA replication pausing or stalling is the common consequence of Twinkle PEO mutations that predisposes to multiple deletion formation.
INTRODUCTION
Mitochondrial diseases manifest with a broad spectrum of symptoms but frequently involve skeletal muscle and/or the nervous system. Diseases with mitochondrial DNA (mtDNA) mutations are quite frequent, with prevalences of ∼1:11 000 in North-Eastern England (1) and 1:6000 for a single-point mutation A3243G in Finland (2). mtDNA disease can be split into two groups. A first group comprises diseases caused by primary mtDNA mutations that lead to impairment of respiratory function. A second group is often referred to as mtDNA maintenance diseases, as the underlying cause is a dysfunction of the replication and maintenance machinery of mtDNA, encoded by nuclear genes, that leads to a secondary loss or damage of the mitochondrial genome and subsequent mitochondrial dysfunction. These disorders have also been shown to be common in the population, with carrier frequencies for single mutations reaching 1:100 in some populations (3,4). The most severe examples of mtDNA maintenance dysfunction are early-onset, recessively inherited syndromes characterized by severe mtDNA depletion in one or more tissues, such as the Alpers-Huttenlocher syndrome associated with mutations in the mtDNA polymerase gamma catalytic subunit POLG1 (5,6). In contrast, autosomal progressive external ophthalmoplegia (PEO) is late-onset form of mtDNA maintenance disease. It affects mainly the function of skeletal muscles, initiating with weakness of the extra-ocular muscles during adulthood. Patients with autosomal PEO do not typically show loss of mtDNA in tissues analyzed (7–9). Instead, large deletions of mtDNA accumulate in post-mitotic tissues over time and the appearance of a considerable amount of deletions is associated with the onset and progression of symptoms (9).
Several essential proteins for mtDNA maintenance have been described and characterized. The mitochondrial polymerase gamma (PolG) functions as a heterotrimer consisting of a catalytic alpha and two accessory beta subunits in mammals (10,11). Polymerase gamma is assumed to be the replicative polymerase in mitochondria. Mutations in the alpha subunit cause mtDNA depletion syndromes (MDS) like Alpers-Huttenlocher, SANDO (sensory-atactic neuropathy, dysarthria and ophthalmoplegia) or MIRAS (mitochondrial recessive ataxia syndrome) (3,12), but are also associated with recessive and sporadic forms of PEO (13–15). Only one mutation of the beta subunit associated with PEO has been described to date (16). Twinkle, a hexameric DNA helicase of the RecA superfamily (17,18), is thought to be the replicative helicase of mtDNA, as it functions together with PolG and single-stranded DNA binding protein (mtSSB) in a minimal mtDNA replisome (19), while catalytic mutations or RNAi cause mtDNA depletion (20–22) in human and insect cell culture. To date and to our knowledge, 23 mutations in the Twinkle gene (PEO1) have been shown to be associated with PEO, and PEO1 mutations were recently shown to be the most prevalent cause of adPEO in Italy (18,23). A single recessive mutation (Y508C) has been uniquely identified in Finland and associates with infantile onset spinocerebellar ataxia. The same mutation was recently identified in a compound heterozygous configuration with a A318T mutation in two siblings with a severe MDS (24). A second recessive T457I mutation was identified homozygous in a family with MDS (25).
While MDS might be caused by a complete impairment of replication due to a non-functional component of the replication machinery, the precise molecular mechanism(s) for the creation and slow accumulation of mtDNA deletions in PEO is unknown. The Deletor mice, expressing an adPEO mutation, a 13 amino acid duplication in the Twinkle linker region equivalent to human amino acid 352–364, accumulate substantial amounts of mtDNA deletions in the skeletal muscle and show similar histological features as the human patients, but are functionally only mildly affected (26). This mouse clearly proves that impaired Twinkle function is the underlying cause of mtDNA deletions and respiratory chain dysfunction and provides an excellent tool to study disease progression. However, studies focusing in the actual molecular mechanism of mtDNA deletion formation is hindered by the fact that the deletions accumulate only slowly in vivo. In this study, we therefore used cultured human cells to study the effects of different Twinkle PEO mutations on mtDNA replication. We show that PEO mutations impair the helicase activity of Twinkle and lead to mtDNA replication stalling or pausing and reduced mitochondrial transcript levels. Similarly, already in young Deletor mouse tissues, we observed accumulation of replication intermediates, indicative of pausing. Since mtDNA deletions in the Deletor model accumulate only in a late-onset fashion, this could suggest that early problems in mtDNA replication predispose to the accumulation of deletions later in life. We hypothesize that the accumulation of replication intermediates can mediate an enhanced formation of mtDNA deletions allowing for their later accumulation in adult tissues in PEO patients.
RESULTS
Expression of dominant Twinkle PEO mutations in proliferating cells leads to mtDNA depletion
In order to study the effect of Twinkle mutations found in patients suffering from adPEO in a reduced cell model, we established stable HEK293 Flp-In™ T-REx™ cell lines expressing human Twinkle variants containing a C-terminal Myc-His tag. This system has the advantage of allowing controllable expression of transgenes in a proliferating human cell background, and as the endogenous Twinkle gene is unaffected, resembles under conditions of low induction, the heterozygous situation in patients with PEO (22).
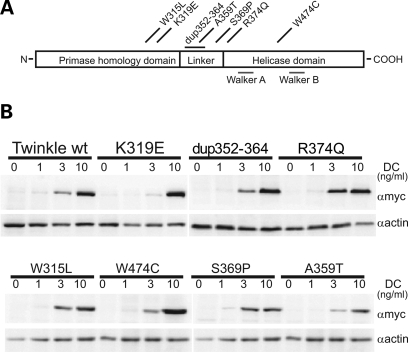
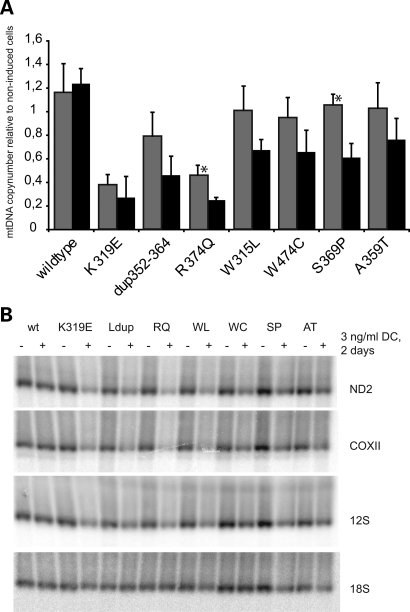
Seven adPEO-related mutations of Twinkle causing mild to severe symptoms were chosen, including several mutations in the linker region of Twinkle (dup352–364, A359T, S369P, R374Q), two mutations located in the N-terminal domain in close proximity to the linker region (W315L, K319E) and one mutation in the helicase domain (W474C) (see Fig. 1A for an overview). All mutations showed expression levels comparable to transgenic wild-type Twinkle-Myc-His, and overexpression could be modulated in a range from 1- to 20-fold over the endogenous wild-type Twinkle (Fig. 1B, and not shown). Overexpression of wild-type Twinkle resulted in a slight but statistically significant copy-number increase, as previously observed (22). In contrast, depletion of mtDNA levels was observed after expression of all adPEO mutations after thee days (Fig. 2A). The level of depletion depended on the mutation. The point mutations K319E located at the end of the N-terminal domain as well as the R374Q mutation in the linker region reduced mtDNA levels to 25% within three days, indicating the complete absence of successful replication. Expression of the 13 amino acid duplication within the linker region also depleted mtDNA levels more than half, while the remaining mutants caused a less severe drop of mtDNA copy-number to 60–76%. None of the mutants caused an increase of mtDNA copy-number when expressed for several days, but for the milder mutations S369P and A359T an initial increase in mtDNA levels after one to two days of expression could sometimes be observed.
Figure 1.
Expression of Twinkle adPEO mutations in HEK293 Flp-In™ T-REx™ cells. (A) Schematic presentation of the human Twinkle protein and localization of the adPEO-related mutations investigated in this study. Most mutations are located at the end of the N-terminal domain or in the linker region of Twinkle, only the W474C point mutation lies within the helicase domain. (B) Expression levels of adPEO Twinkle variants. Expression was induced for three days with the indicated amounts of doxycycline (DC) and protein expression was analyzed by western blot analysis against the myc-tagged Twinkle variants. At 1 ng/ml DC no increase in transgene expression could be observed, although mild effects on mtDNA replication were already detectable (data not shown). Maximal expression was reached with 10 ng/ml DC.
Figure 2.
Mutant expression causes rapid mtDNA copynumber and transcript level depletion. (A) While overexpression of wild-type Twinkle transiently increased the relative copynumber of mtDNA per cell, expression of adPEO variants lead to rapid mtDNA depletion. The loss of mtDNA correlated approximately with the severity of the biochemical and replication defects measured in this study. MtDNA levels of cells induced with 3 ng/ml doxycycline for two days (grey bars) or three days (black bars) were quantified by duplex Taqman PCR and normalized to non-induced cells of the same cell line. Four to six independently isolated samples were measured in triplicate with the exception of the two samples indicated with an asterisk that were isolated twice and measured in triplicate. Similar results were obtained by Southern blot analysis (not shown). Error bars show the standard deviation. (B) Also the steady-state levels of mtDNA transcripts were decreased upon Twinkle mutant expression, while the overexpression of wild-type Twinkle and the resulting mtDNA increase did not change transcript levels. RNA from cells grown for two days with or without 3 ng/ml DC was analysed by Northen blot using probes against several mitochondrial transcripts as indicated and 18S rRNA. Note that the decrease in transcripts occurred in advance of mtDNA depletion (Fig. 2A). No effect on protein levels of representative, nuclear encoded, respiration complex subunits were detected by western blot analysis after 3 days (data not shown). Ldup refers to dup352–364.
Interestingly, mtDNA transcript levels were also affected by Twinkle mutations (Fig. 2B). After two days of overexpression, steady-state transcript levels of all analyzed H-strand transcripts were strongly reduced. The reduction of H-strand transcripts was stronger than the observed mtDNA copy-number depletion after two days of expression (Fig. 2A), indicating that the negative effect of Twinkle dysfunction on transcription precedes the depletion of mtDNA and is not due to reduced mtDNA template levels. The reduction in mitochondrial transcript levels appeared to cause a deficiency of mitochondrial respiration indicated by increased acidification of the cell culture medium (not shown).
adPEO mutations affect nucleoid structure
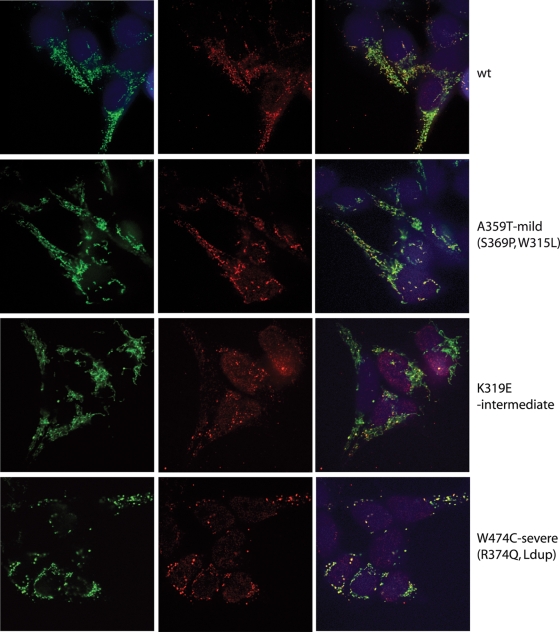
The mtDNA nucleoids in cells overexpressing Twinkle variants for three days were visualized using immunocytochemistry against DNA and the Twinkle protein tag. Twinkle wild-type and W315L (Fig. 3 and Supplementary Material, Fig. S2) co-localized with mtDNA in a pattern of small punctate structures that we have previously found to constitute mtDNA nucleoids (27). The Twinkle variants K319E, R374Q and dup352–364, and A359T to a lesser extent, showed a more uniform mitochondrial matrix staining, but clearly also nucleoid-like spots based on their co-localization with mtDNA. The nucleoids in the K319E, R374Q, W474C and dup352–364 expressing cell lines appeared larger in size and fewer in number compared with wild-type. Finally, the S369P mutation co-localized well with mtDNA similar to W315L. Based on the mtDNA antibody, the number of mtDNA foci seemed normal in cells overexpressing W315L and A359T or even increased for S369P. All the other mutations lead to a reduced number of spots, which could be expected from the copy-number determination of these mutants. Based on the observed effects we have categorized the severity of the nucleoid phenotype as mild for mutants showing no or a limited effect on nucleoid structure and number based on the mtDNA immunofluorescence (A359T, W315L, S369P), moderate (K319E) and severe (R374Q, dup352–364, W474C) and show one example each in Figure 3, A359T, K319E and W474C respectively, while all mutants are shown in Supplementary Material, Fig. S2. Particularly interesting is the observation that especially for the Dup 352–364 and the W474C one can observe a mosaic pattern of cells with some cells being devoid entirely of the strong mtDNA positive foci, whereas other cells of the same culture still showed nucleoid structures. As the cell line used ensures single insertion of the transgene in the genome and similar expression levels in all cells studied, this variability in cell phenotypes could be indicative of an mtDNA segregation defect.
Figure 3.
Some adPEO Twinkle mutants alter the localization of the protein in mtDNA nucleoids. The subcellular localization of the Twinkle variants was determined by immunofluorescence. While wild-type Twinkle co-localized with mtDNA in punctate nucleoids within mitochondria, the disease variants K319E Twinkle variants localized only partially with nucleoids, a proportion of the protein was diffusely localized in the mitochondrial lumen, while the nucleoids often appeared enlarged (similar results were observed with the R374Q and dup 352–364 mutants; Supplementary Material, Fig. S2). The W474C variant caused aggregation of nucleoids without a more diffuse distribution of the protein, while the A359T mutation decreased the Twinkle localization in nucleoids without altering their size and number extensively. The effects of expression of the various mutants were categorized as mild, moderate and severe as indicated in the Figure. Cells were induced with 3 ng/ml doxycycline for 3 days, immunocytochemistry was performed using specific antibodies against the myc-tagged Twinkle variants (green) and DNA (red). Images were taken with the same settings for exposure, laser intensity, etc. and in addition were processed for brightness and contrast in the same manner. All mutants are shown in Supplementary Material, Fig. S2.
In vitro enzyme activities of mutated Twinkle are impaired
For in vitro assays, wild-type and mutant variants of Twinkle were purified from crude mitochondrial fractions with a single-step procedure using TALON® Co2+ affinity resin (see also Materials and Methods). TALON affinity resin eluates were routinely checked using Coomassie staining (Supplementary Material, Fig. S1), and showed a high level of purification of the Twinkle protein using this single-step procedure. Specific activity of the wild-type Twinkle protein (specified as the concentration of Twinkle that gives 50% unwinding activity at a given substrate concentration) purified in this manner seems very similar to that observed with enzyme purified using a baculovirus expression system (28,29), although a precise comparison is not possible since we have used less DNA substrate and measured at 37°C compared with 32°C used by Korhonen et al. Using approximately 5-fold less M13 substrate we find 50% unwinding activity with ∼25 fmol of purified Twinkle, whereas Korhonen et al. reaches 50% unwinding activity at ∼1.2 pmol of purified Twinkle (28) and at slightly less than half that concentration in a later publication that used an improved protocol to purify the enzyme (29). Given that we used less substrate, our purified Twinkle is five to ten times more active which seems in very good agreement given that we measure the activity at 37°C and not at 32°C. For each repeat of the various in vitro assays, −80°C frozen aliquots were first re-analyzed by Coomassie staining to estimate the protein concentration of each sample using BSA as a standard, to ensure the same concentration of wild-type and mutant proteins were being used.
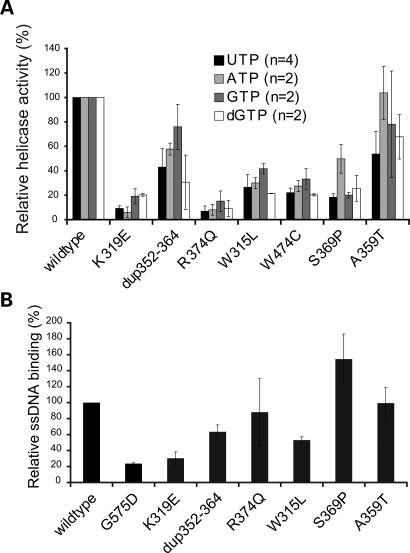
Twinkle is a low-processive helicase in vitro and alone can unwind only short stretches of up to 25 nucleotides. In our hands, the highest activity is achieved when 3 mm UTP is used as an energy source, but also ATP, GTP and dGTP can drive the unwinding reaction. As the natural concentrations of these nucleotides within mitochondria are not known, we measured the helicase activity of the wild-type Twinkle and the various mutants separately with all four nucleotides (Fig. 4A). All mutants had reduced unwinding activity compared with the wild-type protein. The helicase activity of K319E and R374Q was reduced to <20%with all nucleotides, consistent with the strong depletion observed with these mutants in cell culture. However, the Dup 352–364 mutant had ∼70% remaining helicase activity, when GTP was used as an energy source. The relative helicase activity was considerably lower with dGTP and UTP. The W315L and W474C mutations showed an ∼70% reduction in helicase activity irrespective of the nucleotide used. The S369P showed a considerable reduction in helicase activity despite its mild effect on mtDNA copy-number. The mutation with the smallest effect on mitochondrial copy-number, A359T, also had near to normal helicase activities.
Figure 4.
AdPEO mutations affect the unwinding and ssDNA binding activity of Twinkle. (A) Purified protein extracts of the various Twinkle mutations showed clearly reduced maximal unwinding activity in in vitro assays using an M13-based substrate. Most adPEO mutants had less than 25% remaining unwinding activity with UTP, while dup352–364 and A359T were around 50% active compared with wild-type protein (P < 0.05 for dup 352–364 and A359T, P < 0.01 for all others). Other nucleotides (ATP, GTP and dGTP) stimulated the unwinding activity of Twinkle to a lesser extend, but were giving approximately similar decreases for the mutants, only A359T exhibited full activity with ATP. (B) Twinkle ssDNA binding assays were performed essentially as described previously (30) (see also Materials and Methods). We used the G575D mutation previously described by us (22) as an additional control as this mutation was expected to have clearly reduced DNA affinity and indeed showed very poor binding even compared with the various adPEO Twinkle mutants. Of the adPEO mutants, K319E, dup352–364 and W315L showed reduced binding, whereas R374Q and A359T showed near normal ssDNA affinity. The S369P in our hands showed a moderately increased binding. Note that the W474C is not shown because this protein with several repeated isolations gave spurious results. Binding is expressed relative to the binding of the wild-type protein set at 100%.
As the nucleotide concentration of 3 mm is saturating and might conceal any alteration in nucleotide affinity in Twinkle, we measured helicase activities of some mutations with UTP, ATP and GTP over a broader range of nucleotide concentrations. While the unwinding activity was reduced gradually with reduced nucleotide concentrations, no mutant showed a clear change in nucleotide kinetics (not shown).
The binding of Twinkle to single-stranded DNA in vitro was evaluated by electromobility shift assay described recently by Farge et al. (30). (Fig. 4B). The G575D mutation affecting the DNA binding domain of Twinkle (22) was used as a control and gave <25% binding efficiency. Also the adPEO mutations K319E, dup352–364 and W315L had clearly reduced binding affinities, while no clear effect was seen with R374Q and A359T. As the only variant of those investigated, the S369P mutation increased the affinity to ssDNA by 50% compared with wild-type.
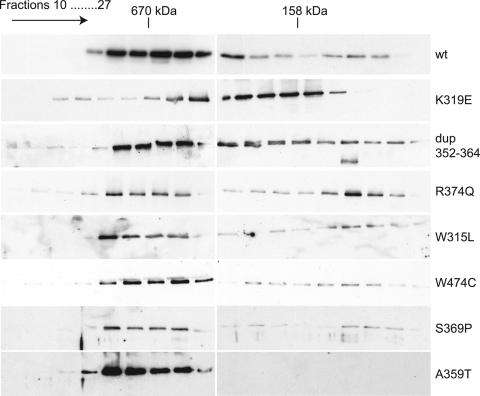
The majority of adPEO mutations are located close to or in the linker region of Twinkle. The homologous region in T7 was shown to be important for subunit interaction and formation of a functional hexamer (31). Therefore, to investigate whether also in Twinkle the reduction in helicase activity and DNA binding are caused by impaired oligomerization, we analyzed the multimeric state of Twinkle with isolated proteins in vitro using a similar chromatographic approach, as described previously (29). As already described by Farge et al. (30), wild-type Twinkle forms hexamers even under high ionic-strength conditions and in the absence of any nucleotide. Using similar conditions, all proteins bearing adPEO mutations formed hexamers, although differences in the degree of oligomerization could readily be detected. Wild-type Twinkle, A359T, W474C, W315L and S369P were mostly eluted at high molecular weight in agreement with normal multimer stability (Fig. 5). The R374Q, K319E and dup352–364 appeared less stable, but in all three cases high molecular weight multimers could still be detected. These results, as far as they involve the analysis of the same mutations, are largely in agreement with Korhonen et al. (29) (see Discussion). Similar results were obtained using glutaraldehyde crosslinking (not shown).
Figure 5.
Some adPEO mutations decrease the stability of Twinkle hexamers. Gel filtration chromatography of purified Twinkle proteins indicated the formation of hexameric complexes in both wild-type and mutant proteins. The same results were obtained in the presence or absence of Mg2+ ions and under salt conditions from 100 to 500 mm NaCl (data not shown). The partial delay and the extensive trailing during the chromatography run indicated the relative instability of the hexamers of K319E, R374Q and dup352–364. Following gel filtration all collected fractions were concentrated and analyzed by western blot analysis as detailed in Materials and Methods.
Twinkle adPEO mutations cause mtDNA replication stalling in cell culture
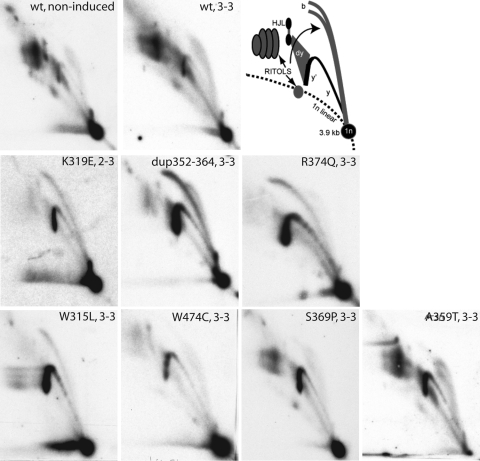
As we described earlier, structural mutations that abolish the helicase activity of Twinkle affected replication of mtDNA in cultured cells and led to strong replication stalling and the accumulation of replication intermediates (22). To test whether the reduced helicase activity of Twinkle adPEO variants impaired mtDNA replication in a similar way, we analyzed the mtDNA replication pattern of cultured cells expressing wild-type and mutant Twinkle variants using two-dimensional DNA gel electrophoresis (Fig. 6). The expression of Myc-His-tagged wild-type Twinkle on top of the endogenous Twinkle levels did not influence the replication pattern or the abundance of replication intermediates except for one species of molecules believed to be part of the termination process in the non-coding region of mtDNA (22). In contrast, a strong effect was visible when Twinkle K319E was expressed in a similar fashion. Low expression levels of Twinkle K319E lead to the appearance of a sharp double-stranded bubble-arc in a fragment containing the non-coding region of mtDNA, while the club-headed double bubble-arc typical for RNA-rich initiation intermediates was reduced. At the same time, the abundance of incompletely digested intermediates containing RNA or single-stranded patches was decreased. With higher expression levels these intermediates vanished completely, with the y- and bubble-arc remaining as the only visible features. This effect could be observed also in other parts of the mitochondrial genome, indicating a general problem with replication fork progression. At full expression levels also the y-shaped intermediates were decreased, indicating an accumulation of replicating molecules shortly after initiation. This change resembled the effect of catalytically inactive Twinkle mutations described by Wanrooij et al. (22).
Figure 6.
MtDNA replication is stalling upon Twinkle mutant expression. 2DNAGE analysis of mtDNA replication intermediates from cells expressing Twinkle variants for two to three days (2–3: two days of induction with 3 ng/ml doxycycline; 3–3: three days induction with 3 ng/ml doxycycline). 2DNAGE samples for all panels consisted of purified mtDNA digested with HincII and probed with a radiolabeled cytochrome b gene fragment (nt 14 846–15 357). The detected fragment includes the non-coding region of mtDNA, the cytochrome b, ND6, part of the ND5 gene and intervening tRNA genes (nt 13 636–1006). The upper two panels show a comparison of mtDNA replication in non-induced and Twinkle wild-type overexpressing cells and the interpretation based on earlier 2DNAGE analysis of mtDNA RIs (22). 1n, 3.9 kb non-replicating HincII fragment. b, bubble arcs. MtDNA bubble arcs are usually very sensitive to RNase H due to the presence of patches of RNA–DNA especially on the lagging strand; therefore a second, degraded bubble arc is visible in cells incorporating RNA into the lagging strand. y and y′ indicate ascending and descending parts of the y arc and (dy) indicates double-Y structures typical for the termination region of replication. These will eventually form resolution intermediates resembling Holliday junctions (HJL-Holliday junction like molecules). Other higher molecular weight fragments arise from the failure to cut RNA–DNA hybrids at a restriction site, these as well as the RNA-rich bubble arcs are marked as RITOLS (RNA Incorporation ThroughOut the Lagging Strand). Overexpression of wild-type Twinkle did not change the replication pattern, the only notable difference was a reproducible reduction in one of the HJL RIs as described earlier. In contrast the stronger disease mutations caused accumulation of replication intermediates on the y- and bubble arc (y and b) and a decrease in all RITOLS related features. The milder disease mutations S359P and A369T had only a moderately enhanced descending y-arc (y′) (note that the 1n spot in the A359T panel has run of the gel). The helicase domain mutation W474C was exceptional as no bubble arc, indicating initiation in the non-coding region, was clearly visible, while similar to other strong mutants RITOLS had almost completely disappeared. This was even more clearly visible after induction with 10 ng/ml DC for three days as is illustrated in Supplementary Material, Fig. S3.
A similar although milder replication stalling was induced when expressing R374Q, dup352–364 or W315L Twinkle variants; while molecules on the y- and double-stranded bubble-arc strongly accumulated, the levels of RNA-rich intermediates decreased. Expression of the Twinkle S369P and A359T mutations had the mildest effect, although also they resulted in clear replication stalling and a shift to less RNA-containing intermediates.
The W474C variant, the only helicase domain mutation analyzed had a different effect: also here RNA-rich intermediates vanished and y-shaped molecules accumulated upon expression, but in contrast to the previously described mutants bubble-containing intermediates did not accumulate, but vanished completely (Fig. 6 and Supplementary Material, Fig. S3).
Transgenic animals expressing an adPEO Twinkle variant show impaired replication already at young age
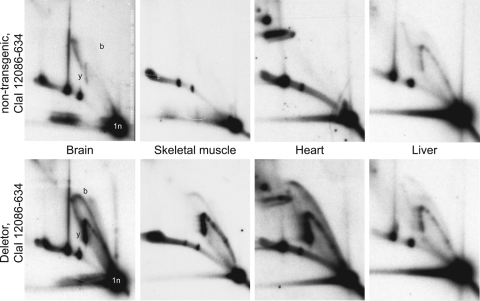
Although the replication stalling that is observed when expressing Twinkle adPEO mutants in cell culture leads to rapid loss of mtDNA, this effect has not been reported in adPEO patients. To determine whether similar phenomena as in cultured cells are found in vivo, we studied the mtDNA replication patterns of the Deletor mice. In Deletor muscle, the first histological signs of disease and detectable amounts of mtDNA deletions appear at the age of 9–12 months. Similarly, in Deletor brain, multiple mtDNA deletions as well as depletion are becoming apparent after approximately one year of age (26 and unpublished data). Since the number of replicating mtDNA molecules is low in mice of old age, we studied the heart, liver, skeletal muscle, kidney and brain of animals of six weeks of age. The expression levels of the transgenic mutant Twinkle compared with the endogenous wild-type Twinkle mRNA were between 13 and 59% depending on the tissue. All these tissues at this age, including brain, showed normal mtDNA copy-number levels. In contrast, the analysis of replication patterns showed a striking accumulation of replication intermediates in mutant animals in all tissues examined except for liver. In the liver, the effect of transgene expression on the accumulation of replication intermediates was rather mild. Figure 7 shows the comparison for brain, muscle, heart and liver mtDNA with the mutant transgene expression level measured to be 39, 59, 45 and 16% of the endogenous transcript level. The results for kidney (13% of endogenous transcript level) were very similar to those observed for brain (not shown). Similar results were obtained for three different Deletor founder lines (26) (not shown), demonstrating that the observed effect was not a consequence of the transgene insertion sites. Also, transgenic overexpression of wild-type Twinkle at much higher levels (20) only showed a slight quantitative change in replication intermediates in all tissues (not shown) except in heart where also qualitative changes were observed (Pohjoismäki et al., manuscript submitted) that were however not seen in the Deletor mouse heart.
Figure 7.
Deletor mice expressing the mouse variant of dup352–364 Twinkle also show mtDNA replication stalling. 2DNAGE analysis of brain, skeletal muscle, heart and liver mtDNA from young Deletor mice expressing the mouse Twinkle mutation equivalent to the human dup352–364 Twinkle variant show, except for liver, a similar strong accumulation of replication intermediates on both y- and bubble arc as in cultured cells. MtDNA was isolated from 6-week-old transgenic mice and non-transgenic siblings and probed against the OH-containing ClaI fragment (nts 12086–634). A similar replication stalling as observed in brain was also observed in kidney (data not shown).
DISCUSSION
Mutations in the mtDNA helicase Twinkle are one of the major causes of adPEO (18,23). Understanding how Twinkle mutations cause mitochondrial dysfunction and multiple mtDNA deletions in post-mitotic tissues in patients is one of the primary goals in studying the effects of Twinkle mutants using a combination of in vitro assays, cell culture and animal models. Using these approaches we show here that the Twinkle adPEO mutations cause a defect in mtDNA replication both in human cell culture and in a transgenic mouse model (26), resulting in the accumulation of replication intermediates, indicative of replication pausing/stalling. In addition, we suggest that replication stalling can also result in a mitochondrial transcription defect. Although the defects of several of the mutations we studied are largely in agreement with recent studies using both in vitro assays and insect cell culture (21,29), we here extend these findings by studying additional mutants in human cell culture, and show that the cell culture findings are relevant for the disease pathogenesis, as they can be replicated in vivo, in the Deletor mice.
Oligomerization
As Twinkle PEO is a dominantly inherited disease, the mutations are heterozygous and the mutant protein coexists with the wild-type protein. Therefore, adPEO Twinkle variants are assumed to be dominant negative and to impair the function of wild-type Twinkle by interaction, which requires co-hexamerization. We show here that the Twinkle mutants K319E, dup352–364 and R374Q resulted in a moderate to severe decrease of hexamer formation under all tested conditions, although some ability to form multimers was maintained. This is in apparent contrast with a recent study (29) who found that several mutations including the R374Q studied caused the helicase to be completely monomeric. In seeming agreement with our results, the Drosophila R341Q mutant, analogous to R374Q in humans, expressed and isolated from Drosophila Schneider S2 cells retained the ability to form multimers using glycerol-gradient sedimentation (21), although the resolution of these experiments makes it difficult to assess to what extent hexamers were stable.
The apparent discrepancy of identical mutations showing different multimerization abilities can be explained by the different origin of the proteins in our study and that of Korhonen et al. (29). In the latter study, human Twinkle variants were expressed in the cytosol of insect cells using baculovirus which also ensures that the isolated mutant proteins are free from any wild-type Twinkle. However, it was shown clearly that co-expression with the wild-type form increased the ability to form hexamers of otherwise monomeric mutant protein, such as the R374Q mutant (29). In contrast, the proteins used in our study are expressed in the mitochondria of human cells that do contain functional wild-type Twinkle. Although the mutant form is overexpressed to a great extent, a small proportion of the extracted Twinkle hexamers does contain one or more subunits of the wild-type Twinkle. This might be especially the case for the R374Q variant that in the studied cells was expressed at normal levels, but consistently gave low yields when isolated. The diffuse mitochondrial signal in immunocytochemistry supports the possibility of increased destabilization of this variant that might lead to the selective purification of heteromultimers containing wild-type Twinkle. The same argument can be applied to the expression of the R341Q Drosophila Twinkle protein (21), as previously discussed (29). A second difference that might apply to this discussion is that the localization of Twinkle variants in mitochondria allows for potential post-translational modification that may influence and regulate the activity of Twinkle in vivo. Mitochondrial chaperones that could assist in protein stability and folding could also influence the ability of mutants to form multimers. In conclusion, based on the present and previous studies (21,29), the lost ability of some of the mutant Twinkle PEO variants to form multimers in their pure form does not reflect the in vivo ability of the protein to interact with wild-type Twinkle, the situation found in heterozygous PEO patients. Clearly, the presence of wild-type Twinkle rescues a quarternary structural defect of several mutant Twinkle variants to the extent that a heteromultimeric complex can be formed that is still able to perform its function to some degree in mtDNA replication. The lost ability to form homomultimers for some of the mutants does suggest that either heteromultimer formation can be less efficient, perhaps even to the extent that there is a preferential formation of wild-type-only multimers, or that heteromultimers form that are, for example, less processive or that are defective in the coupling of nucleotide hydrolysis with DNA unwinding. It is again important to note here that the nucleotide binding and hydrolysis pocket of ring helicases is formed at the interface of subunits (see 32, for a review). Any disturbance by mutation, however subtle, could be anticipated to affect the hydrolysis cycle and its mechanistic coupling to DNA unwinding, thus resulting in a less-effective enzyme.
Helicase activity and DNA binding
As described earlier, Twinkle has affinity both to single-stranded and double-stranded DNA (30). We measured the affinity of purified protein to a single-stranded DNA substrate and found reduced binding for the mutations K319E, dup352–364 and W315L, in agreement with their reduced helicase activity (all three) and hexamerization (K319E and dup352–364). Surprisingly, in our hands the S369P variant had a clearly increased affinity to ssDNA in contrast to the results of Korhonen et al. (29), who reported a decreased binding affinity for the same variant. As we found an increased amount of mtDNA nucleoids in cells expressing this variant in contrast to other variants showing less nucleoids, the tight binding of S369P Twinkle to mtDNA might influence the segregation of nucleoids. The observed differences in binding activity compared with Korhonen et al. could be explained as described above for hexamerization. The unwinding ability of all mutations in vitro was diminished compared with wild-type Twinkle. Depending on the oligonucleotide used as energy source the range of residual activity ranged from <10 to 76% remaining activity, but all mutations had <55% maximal activity with UTP.
mtDNA replication in human cells expressing Twinkle mutants and in Deletor mice
As all available in vitro assays for Twinkle function have the disadvantage of working with a relatively inactive enzyme, we studied the effect of adPEO variants on mtDNA replication directly under physiological conditions. When expressed approximately in a one-to-one ratio with wild-type Twinkle, the ‘strong’ mutations showed a slowing-down of mtDNA replication rate and an accumulation of replication intermediates (not shown). Under these conditions, comparable to the situation in patients, mtDNA depletion is not detected, indicating that the slow replication is still sufficient to keep mtDNA copy-numbers normal. When mutant levels are increased, the stalling is increased and results in reduced mtDNA levels. This might explain why only the relatively mild A359T mutation has been seen as homozygous in patients: a severely deleterious Twinkle variant would lead to mtDNA loss already early in the development. We used the exaggerated situation of a mutant to wild-type ratio of ca. 4:1 (22) to study the immediate effect on the replication machinery in detail. All mutations localized in or close to the linker domain, except for the A359T, caused relatively severe replication stalling over the whole length of mtDNA. The observed stalling/pausing phenotypes with most of the adPEO Twinkle mutants are entirely consistent with our previous analysis of catalytic and structural mutations in the Twinkle protein (22); all of the Twinkle mutants except for the W474C mutant (see later) showed more or less severe accumulation of what we previously showed to be mostly double-stranded DNA bubble and y-shaped replication intermediates, while at the same time there was a strong reduction in the RNA-containing RITOLS intermediates. Overexpression of wild-type Twinkle, even at full induction, did not substantially modify the types of replication intermediates observed, such as RITOLS, though there did appear to be a small increase in y-shaped replication intermediates (for a more extensive comparison of the effects of wild-type and mutant proteins on replication intermediates, we have also included a Supplementary Material, Fig. S3 comparing effects at different levels of induction). It should be pointed out again here that overexpression of all mutants resulted in a more or less severe drop in mtDNA levels, while no such drop was observed for the wild-type protein even at full induction. For a discussion of the implications of the stalling/pausing phenotype of Twinkle mutants with reference to the mechanisms of mtDNA replication we refer to our previous paper (22). The W474C mutation localized in the helicase domain of Twinkle showed a slightly different effect on replication. The stalling effect was limited to the non-coding region, while replication in other parts of the mtDNA seemed to be normal (not shown). Analysis of an OH-containing fragment showed that only y-shaped replication intermediates accumulated, while bubble intermediates, indicative of replication initiation, were reduced or absent. This suggests a specific effect of this mutation on the initiation in the non-coding region of mtDNA, while elongation elsewhere seemed to be less affected.
Analysis of various tissues of six-week-old Deletor mice showed strong accumulation of replication intermediates compared with non-transgenic littermates. This was a surprising finding, since multiple mtDNA deletions are only detectable in animals older than approximately nine months of age (26), thus suggesting that perhaps the critical events that in later live result in the typical molecular phenotype of PEO occur at a much earlier age when mtDNA replication is still frequent. Similarly, depletion of brain mtDNA is apparent in older mice (26) but not in the 6-week-old animals, yet replication intermediates clearly accumulated at this stage already. Finally, multiple mtDNA deletions accumulate in muscle, heart and brain but not in liver and kidney (26 and unpublished data), yet 6-week-old kidney also showed clear signs of replication stalling similar to what was observed in brain. These differences could be explained by variable degrees of self-regenerating capacity or apoptosis resistance of the various tissues, but remain still to be clarified.
Transcription
The expression of Twinkle mutations in human cells impaired not only replication, but also the transcription of mtDNA. Already after 2 days of expression the steady-state levels of H-strand transcripts were depleted, an effect that clearly preceded the depletion of mtDNA. The impairment of RNA synthesis might be due to several mechanisms. Firstly, as replication is severely slowed down, an increasing number of mtDNA molecules are in the state of replication. The increased presence of replisomes on the mtDNA could cause a collision of replication and transcription machineries and abort successful transcription. Secondly, a change in mtDNA topology was observed upon the expression of mutant Twinkle. The proportion of relaxed molecules increased, while supercoiled mtDNA molecules were less abundant (unpublished data). At this point, however, the exact cause and effect relationship is unclear. It remains to be determined whether in adPEO patients a defect in the mitochondrial transcription precedes the accumulation of multiple deletions and could in fact contribute to the pathophysiology of adPEO by inducing an oxidative phosphorylation deficiency.
Can we understand the functional consequences of replication stalling in proliferating cells versus post-mitotic tissues?
In proliferating cells mtDNA replication needs to keep pace with the mitochondrial and cellular proliferation rate. Thus, if mtDNA replication significantly pauses or stalls without immediately affecting cell proliferation one can expect a drop in mtDNA copy-number and this is exactly what is observed when we overexpress Twinkle mutants (this study and 22), POLG1 mutants (22) and, for example, wild-type TFAM (33) or when we knock-down Twinkle expression (20). It is unclear at the moment what happens with stalled replication forks during cell division, but expression of Twinkle PEO mutants in cultured cells does not seem to result in mtDNA deletions (none could be detected, neither by Southern blot analysis nor by long range PCR, unpublished data), which is in line with our previous observations with cultured adPEO patient cells (unpublished data). One might expect that a failure to complete replication would result in a failure to divide nucleoids resulting in enlarged nucleoid structures and a mosaic cell distribution of nucleoids. This is indeed one of the phenotypes of some of our Twinkle variants in cell culture. In contrast, in post-mitotic tissues the situation is different as mtDNA replication is no longer coupled to the cell cycle. Presumably in this case it is the balance between mitochondrial (DNA) turnover and replication that determines whether there will be a net loss of mtDNA copy-number. How this different requirement for mtDNA replication might cause multiple deletions to accumulate remains a matter of speculation but we can envisage several scenarios. First, the persistent presence of stalled/paused replication forks could elicit an mtDNA repair response similar to SOS responses resulting in the recruitment of specific repair proteins. We recently discussed a scenario of how enhanced rates of mtDNA repair could result in enhanced rates of deletion formation (34). A second scenario that cannot be excluded at this point would be frequent focal depletion of mtDNA followed by reamplification. Especially in a situation where replication is frequently paused due to a partially defective replication machinery, this could favour the amplification of mtDNA deletions that naturally occur at low frequencies. Finally, and in line with the previous suggestion, replication stalling might induce an increased rate of mitochondrial (DNA) turnover, for example, via mitophagy (see also 26), combined with an increased rate of replication in order to maintain normal mtDNA steady-state levels. The observation that replication intermediates abnormally accumulate in tissues of young mice but multiple deletions and depletion are only observed in much older animals could suggest that one or several mechanisms to efficiently deal with mtDNA replication problems are in place and that the later onset mtDNA defects are a direct consequence of an age-associated deterioration of these repair mechanisms. We are currently pursuing the various possibilities suggested here.
MATERIALS AND METHODS
Cloning of expression constructs
The full-length cDNA of Twinkle variants were originally cloned in the pcDNA3.1(-)/Myc-His A (Invitrogen), as previously described (18,35). All constructs were re-cloned in the pcDNA5/FRT/TO vector (Invitrogen) taking advantage of two PmeI restriction sites flanking the multiple cloning sites of the original pcDNA3 vector and the target vector. The resulting fusion proteins contained the sequence of the respective proteins followed by an Myc-His tag. All resulting plasmid constructs were confirmed by DNA sequencing.
Creation and maintenance of stable transfected inducible expression cell lines
Stable cell lines expressing various Twinkle mutants upon induction were created as described (22) using the Flp-In™ T-Rex™ 293 host cell line (Invitrogen), an HEK293 variant containing a Flip recombination site at a transcriptionally active locus. The resulting cells were grown in DMEM medium (Sigma) supplemented with 10% FCS (Sigma), 1 mm l-glutamine, 50 µg/ml uridine (Sigma), 150 µg/ml Hygromycin and 15 µg/ml Blasticidin in a 37°C incubator at 8.5% CO2. To test cell lines for mosaic expression of transgenes, which is a common problem with inducible cell lines that are generated by allowing for random integration, we routinely tested all cell lines using immunofluorescence (as described below). All cell lines tested in this manner showed that >99% of cells were positive for expression and that the protein was correctly localized in mitochondria. Even at maximal induction levels there was no evidence of mistargeting.
To induce expression the indicated amount of doxycycline (DC) (Sigma) was added to the growth medium, and cells were processed for further analyses. With longer than 2 day induction, medium was refreshed every 2 days.
Mouse tissue collection
All animal procedures were performed according to protocols approved by the ethical boards for animal experimentation of the National Public Health Institute and Helsinki University (agreement number STU575A/2004) and all experiments were done in accordance with good practice of handling laboratory animals and of genetically modified organisms. The generation and the phenotypes of the Deletor and the wild-type Twinkle overexpressor mice have been previously described (20,26). The mice were housed in a humidity- and temperature-controlled environment with free access to chow and water. Mouse tissue samples were carefully dissected and immediately processed for DNA extraction.
Western blots
Cell lysates were prepared and analyzed for protein expression by immunoblotting after SDS–PAGE (35). A primary monoclonal c-myc (Roche Molecular Biochemicals) antibody was used for detection of recombinant proteins. Detection of actin (polyclonal antibody from Novus Biologicals) was used as a loading control. Peroxidase-coupled secondary antibody horse-anti-mouse or goat-anti-rabbit was obtained from Vector Laboratories. Enhanced Chemiluminescence detection was done essentially as described (35).
Southern blots
The mtDNA levels per cell and the conformation of mtDNA were analyzed by one-dimensional agarose gels and Southern blotting. Total DNA was extracted from cells by proteinase K digest and Phenol–Chloroform extraction. mtDNA levels were analyzed by separating 2 µg of HindIII digested total DNA on a 0.6% agarose gel in TBE. For conformational studies, 1 µg of DNA was digested with BglII that does not cut mtDNA, and separated over a 0.4% agarose gel in TBE without ethidium bromide. All gels were blotted and hybridized with 32P-labelled DNA probes containing the D-loop region (nts 16341–151) or part of the cytochrome b sequence (nts 14846–15357) of human mtDNA. The signal was quantified by phosphorimager and for mtDNA copy-number determination normalized against the signal for 18S rDNA.
Northern blots
RNA for northern blot analysis was extracted with Trizol (Sigma) using the manufacturer's recommendations. 4 µg of total RNA per sample was run on a 1% agarose MOPS/formaldehyde gel, blotted and hybridized using standard techniques (36). Blots were probed with ca. 500 bp double-stranded DNA probes radioactively labelled by random-primed labelling. Exposures were performed using a phosphorimager (Storm 840, Molecular Dynamics).
Quantitative PCR
The copy-number of mtDNA per cell was determined essentially as described (22). Total cellular DNA was extracted by Proteinase K digest and isopropanol precipitation, and copy-numbers of cytochrome b (mtDNA) and APP (nuclear DNA) were determined in a Taqman assay on an Abiprism 7000 (Applied Biosciences, Foster City, CA, USA) using plasmids containing the amplicons as standards.
Immunocytochemistry
Immunofluorescent detection was done as described previously (37) using a monoclonal anti-DNA antibody AC-30-10 (PROGEN, Shingle Springs, CA, USA) and a monoclonal c-myc antibody (Roche Molecular Biochemicals) as primary antibodies. Secondary antibodies were anti-mouse IgG-Alexa-Fluor®488 (Invitrogen, myc) and anti-mouse IgM-Alexa Fluor®568 (DNA). Image acquisition using confocal microscopy was done as described (37).
Protein extraction, helicase, ssDNA binding and oligomerization assays
In vitro assays for determination of helicase activities were performed with highly enriched Twinkle preparations derived from 293 Flp-In™ T-REx™ cells. Cells induced for 36 h were disrupted after short cytochalasin treatment (38); mitochondria were isolated using differential centrifugation and sucrose gradient purification and then lyzed and sonicated in high salt buffer (50 mm KH2PO4, pH 7.4, 1 m NaCl, 0.05% Triton X-100, 10 mm Imidazol, 7 mm β-mercaptoethanol). The lysate was incubated with TALON Co2+ affinity resin (Clontech) for 1 h at 4°C, the resin was washed twice each with high and low salt buffer (25 mm Tris pH 7.6, 200 mm NaCl, 100 mm l-arginine, pH 7.6, 40 mm Imidazol, 10% glycerol, 7 mm β-mercaptoethanol) and His-tagged proteins binding to the resin were isolated with elution buffer containing 25 mm Tris pH 7.6, 200 mm NaCl, 100 mm l-arginine, pH 7.6, 200 mm Imidazol, 50% glycerol. Protein extracts were aliquoted, shock frozen in liquid nitrogen and stored at –80°C. The concentration of Twinkle in the eluate was judged by SDS–PAGE and Coomassie Brilliant blue staining using BSA as a reference. Typically 100 mg mitochondrial wet weight yielded 2–4 µg of Twinkle protein.
Helicase assays were performed as described (22) with a radioactively end-labelled 60 nt oligonucleotide hybridized to M13 (+) ssDNA, forming a 20 nt double-stranded stretch with a 40 nts 5′ end overhang. The reaction conditions were essentially as described previously (22), but the reaction mix did not contain other DNA besides the substrate which was used at a concentration of ∼3 fmol. UTP, ATP, GTP or dGTP were added at 3 mm, if not indicated otherwise. Short-term oligomerization of isolated Twinkle variants in vitro was tested by crosslinking the protein with low concentrations of glutaraldehyde followed by separation on an SDS–PAGE and western blotting. Ca. 20 ng of Twinkle protein was diluted in 20 µl of oligomerization buffer (25 mm Tris–HCl pH 7.6, 100 mm l-arginine, pH 7.6, 1 mm DTT, 10% glycerol) containing either 50 mm NaCl (helicase conditions) or 400 mm NaCl (high salt conditions). Glutaraldehyde measuring 0.02% was added and the reaction mix incubated for 5 min at room temperature. The crosslinking was stopped by adding 10 µl of SDS-containing sample buffer and directly heat-denatured for 10 min at 95°C. The denatured sample was separated on a 3–8% Tris–Acetate gel (NuPAGE, Invitrogen) and Twinkle proteins were detected by regular western blotting.
The stability of Twinkle hexamers was analyzed by gel filtration. 100 ng of purified twinkle were separated over a Sephadex 200 10/300GL column (GE healthcare) in high salt buffer (25 mm Tris–HCl pH 7.6, 400 mm NaCl, 100 mm l-arginine pH 7.6, 5 mm EDTA, 10% glycerol), proteins from 0.5 ml fractions were precipitated with deoxycholate and TCA and analyzed by western blot using the anti-myc antibody.
Binding of Twinkle wild-type and mutant protein to a ssDNA oligonucleotide was essentially performed as described previously (30), but with the exception that we used the following oligo nucleotide primer: CT TCC TGG CTT GCT TTG GCT GAG CCA AAA in stead of an oligodT primer; reactions were done in 20 µl with a protein concentration of 50–100 ng as judged by Coomassie staining. Binding was quantified by phosphorimaging and is expressed as the level of binding relative to the wild-type protein.
Brewer-Fangman 2D neutral/neutral agarose electrophoresis
Mitochondrial nucleic acids were extracted using cytochalasine (Sigma-Aldrich) as described (38). Purified mtDNA was digested with HincII (human) or ClaI (mouse). The fragments were separated by 2DNAGE as described (39,40) and the gels were blotted and hybridized with a 32P-labelled DNA probe for human mtDNA nts 14846–15357 or mouse mtDNA nts 16356–136.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Academy of Finland [110689, 103213 and CoE funding to J.N.S.]; Sigrid Juselius Foundation [to A.S. and J.N.S.]; the University of Helsinki [to A.S.]; the EU sixth Framework Programme for Research, Priority 1 ‘Life sciences, genomics and biotechnology for health [LSHM-CT-2004-503116 to J.N.S. and S.G.]; and the Tampere University Hospital Medical Research Fund [9G072, 9H079 to J.N.S., 9H102 to H.M.C.]. Funding to Pay the Open Access Charge was provided by the Academy of Finland.
Supplementary Material
ACKNOWLEDGEMENTS
We are very grateful for suggestions and many discussions we have had with the Ian Holt laboratory (Cambridge, UK) and with Howy Jacobs and Jaakko Pohjoismäki.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Schaefer A.M., Blakely E.L., He L., Whittaker R.G., Taylor R.W., Chinnery P.F., Turnbull D.M. Prevalence of mitochondrial DNA disease in adults. Ann. Neurol. 2008;63:35–39. doi: 10.1002/ana.21217. [DOI] [PubMed] [Google Scholar]
- 2.Majamaa K., Moilanen J.S., Uimonen S., Remes A.M., Salmela P.I., Karppa M., Majamaa-Voltti K.A., Rusanen H., Sorri M., Peuhkurinen K.J., et al. Epidemiology of A3243G, the mutation for mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes: prevalence of the mutation in an adult population. Am. J. Hum. Genet. 1998;63:447–454. doi: 10.1086/301959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hakonen A.H., Heiskanen S., Juvonen V., Lappalainen I., Luoma P.T., Rantamaki M., Goethem G.V., Lofgren A., Hackman P., Paetau A., et al. Mitochondrial DNA polymerase W748S mutation: a common cause of autosomal recessive ataxia with ancient European origin. Am. J. Hum. Genet. 2005;77:430–441. doi: 10.1086/444548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winterthun S., Ferrari G., He L., Taylor R.W., Zeviani M., Turnbull D.M., Engelsen B.A., Moen G., Bindoff L.A. Autosomal recessive mitochondrial ataxic syndrome due to mitochondrial polymerase gamma mutations. Neurology. 2005;64:1204–1208. doi: 10.1212/01.WNL.0000156516.77696.5A. [DOI] [PubMed] [Google Scholar]
- 5.Naviaux R.K., Nguyen K.V. POLG mutations associated with Alpers’ syndrome and mitochondrial DNA depletion. Ann. Neurol. 2004;55:706–712. doi: 10.1002/ana.20079. [DOI] [PubMed] [Google Scholar]
- 6.Chinnery P.F., Zeviani M. 155th ENMC workshop: Polymerase gamma and disorders of mitochondrial DNA synthesis, 21–23 September 2007, Naarden, The Netherlands. Neuromuscul. Disord. 18:259–267. doi: 10.1016/j.nmd.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Zeviani M., Servidei S., Gellera C., Bertini E., DiMauro S., DiDonato S. An autosomal dominant disorder with multiple deletions of mitochondrial DNA starting at the D-loop region. Nature. 1989;339:309–311. doi: 10.1038/339309a0. [DOI] [PubMed] [Google Scholar]
- 8.Suomalainen A., Majander A., Haltia M., Somer H., Lonnqvist J., Savontaus M.-L., Peltonen L. Multiple deletions of mitochondrial DNA in several tissues of a patient with severe retarded depression and familial progressive external opthalmoplegia. J. Clin. Invest. 1992;90:61–66. doi: 10.1172/JCI115856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suomalainen A., Majander A., Wallin M., Setala K., Kontula K., Leinonen H., Salmi T., Paetau A., Haltia M., Valanne L., et al. Autosomal dominant progressive external ophthalmoplegia with multiple deletions of mtDNA: clinical, biochemical, and molecular genetic features of the 10q-linked disease. Neurology. 1997;48:1244–1253. doi: 10.1212/wnl.48.5.1244. [DOI] [PubMed] [Google Scholar]
- 10.Yakubovskaya E., Chen Z., Carrodeguas J.A., Kisker C., Bogenhagen D.F. Functional human mitochondrial DNA polymerase gamma forms a heterotrimer. J. Biol. Chem. 2006;281:374–382. doi: 10.1074/jbc.M509730200. [DOI] [PubMed] [Google Scholar]
- 11.Yakubovskaya E., Lukin M., Chen Z., Berriman J., Wall J.S., Kobayashi R., Kisker C., Bogenhagen D.F. The EM structure of human DNA polymerase gamma reveals a localized contact between the catalytic and accessory subunits. Embo J. 2007;26:4283–4291. doi: 10.1038/sj.emboj.7601843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Goethem G., Martin J.J., Dermaut B., Lofgren A., Wibail A., Ververken D., Tack P., Dehaene I., Van Zandijcke M., Moonen M., et al. Recessive POLG mutations presenting with sensory and ataxic neuropathy in compound heterozygote patients with progressive external ophthalmoplegia. Neuromuscul. Disord. 2003;13:133–142. doi: 10.1016/s0960-8966(02)00216-x. [DOI] [PubMed] [Google Scholar]
- 13.Van Goethem G., Dermaut B., Lofgren A., Martin J.J., Van Broeckhoven C. Mutation of POLG is associated with progressive external ophthalmoplegia characterized by mtDNA deletions. Nat. Genet. 2001;28:211–212. doi: 10.1038/90034. [DOI] [PubMed] [Google Scholar]
- 14.Lamantea E., Tiranti V., Bordoni A., Toscano A., Bono F., Servidei S., Papadimitriou A., Spelbrink H., Silvestri L., Casari G., et al. Mutations of mitochondrial DNA polymerase gammaA are a frequent cause of autosomal dominant or recessive progressive external ophthalmoplegia. Ann. Neurol. 2002;52:211–219. doi: 10.1002/ana.10278. [DOI] [PubMed] [Google Scholar]
- 15.Agostino A., Valletta L., Chinnery P.F., Ferrari G., Carrara F., Taylor R.W., Schaefer A.M., Turnbull D.M., Tiranti V., Zeviani M. Mutations of ANT1, Twinkle, and POLG1 in sporadic progressive external ophthalmoplegia (PEO) Neurology. 2003;60:1354–1356. doi: 10.1212/01.wnl.0000056088.09408.3c. [DOI] [PubMed] [Google Scholar]
- 16.Longley M.J., Clark S., Yu Wai Man C., Hudson G., Durham S.E., Taylor R.W., Nightingale S., Turnbull D.M., Copeland W.C., Chinnery P.F. Mutant POLG2 disrupts DNA polymerase gamma subunits and causes progressive external ophthalmoplegia. Am. J. Hum. Genet. 2006;78:1026–1034. doi: 10.1086/504303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leipe D.D., Aravind L., Grishin N.V., Koonin E.V. The bacterial replicative helicase DnaB evolved from a RecA duplication. Genome Res. 2000;10:5–16. [PubMed] [Google Scholar]
- 18.Spelbrink J.N., Li F.Y., Tiranti V., Nikali K., Yuan Q.P., Tariq M., Wanrooij S., Garrido N., Comi G., Morandi L., et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat. Genet. 2001;28:223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- 19.Korhonen J.A., Pham X.H., Pellegrini M., Falkenberg M. Reconstitution of a minimal mtDNA replisome in vitro. Embo J. 2004;23:2423–2429. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tyynismaa H., Sembongi H., Bokori-Brown M., Granycome C., Ashley N., Poulton J., Jalanko A., Spelbrink J.N., Holt I.J., Suomalainen A. Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Hum. Mol. Genet. 2004;13:3219–3227. doi: 10.1093/hmg/ddh342. [DOI] [PubMed] [Google Scholar]
- 21.Matsushima Y., Kaguni L.S. Differential phenotypes of active site and human autosomal dominant progressive external ophthalmoplegia mutations in Drosophila mitochondrial DNA helicase expressed in Schneider cells. J. Biol. Chem. 2007;282:9436–9444. doi: 10.1074/jbc.M610550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wanrooij S., Goffart S., Pohjoismaki J.L., Yasukawa T., Spelbrink J.N. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 2007;35:3238–3251. doi: 10.1093/nar/gkm215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Virgilio R., Ronchi D., Hadjigeorgiou G.M., Bordoni A., Saladino F., Moggio M., Adobbati L., Kafetsouli D., Tsironi E., Previtali S., et al. Novel Twinkle (PEO1) gene mutations in mendelian progressive external ophthalmoplegia. J. Neurol. 2008 doi: 10.1007/s00415-008-0926-3. First published on June 30, 2008, 10.1007/s00415–008–0926–3. [DOI] [PubMed] [Google Scholar]
- 24.Hakonen A.H., Isohanni P., Paetau A., Herva R., Suomalainen A., Lonnqvist T. Recessive Twinkle mutations in early onset encephalopathy with mtDNA depletion. Brain. 2007;130:3032–3040. doi: 10.1093/brain/awm242. [DOI] [PubMed] [Google Scholar]
- 25.Sarzi E., Goffart S., Serre V., Chretien D., Slama A., Munnich A., Spelbrink J.N., Rotig A. Twinkle helicase (PEO1) gene mutation causes mitochondrial DNA depletion. Ann. Neurol. 2007;62:579–587. doi: 10.1002/ana.21207. [DOI] [PubMed] [Google Scholar]
- 26.Tyynismaa H., Mjosund K.P., Wanrooij S., Lappalainen I., Ylikallio E., Jalanko A., Spelbrink J.N., Paetau A., Suomalainen A. Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice. Proc. Natl. Acad. Sci. USA. 2005;102:17687–17692. doi: 10.1073/pnas.0505551102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garrido N., Griparic L., Jokitalo E., Wartiovaara J., Van Der Bliek A.M., Spelbrink J.N. Composition and dynamics of human mitochondrial nucleoids. Mol. Biol. Cell. 2003;14:1583–1596. doi: 10.1091/mbc.E02-07-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korhonen J.A., Gaspari M., Falkenberg M. TWINKLE Has 5′ ->3′ DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J. Biol. Chem. 2003;278:48627–48632. doi: 10.1074/jbc.M306981200. [DOI] [PubMed] [Google Scholar]
- 29.Korhonen J.A., Pande V., Holmlund T., Farge G., Pham X.H., Nilsson L., Falkenberg M. Structure-function defects of the TWINKLE linker region in progressive external ophthalmoplegia. J. Mol. Biol. 2008;377:691–705. doi: 10.1016/j.jmb.2008.01.035. [DOI] [PubMed] [Google Scholar]
- 30.Farge G., Holmlund T., Khvorostova J., Rofougaran R., Hofer A., Falkenberg M. The N-terminal domain of TWINKLE contributes to single-stranded DNA binding and DNA helicase activities. Nucleic Acids Res. 2008;36:393–403. doi: 10.1093/nar/gkm1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo S., Tabor S., Richardson C.C. The linker region between the helicase and primase domains of the bacteriophage T7 gene 4 protein is critical for hexamer formation. J. Biol. Chem. 1999;274:30303–30309. doi: 10.1074/jbc.274.42.30303. [DOI] [PubMed] [Google Scholar]
- 32.Donmez I., Patel S.S. Mechanisms of a ring shaped helicase. Nucleic Acids Res. 2006;34:4216–4224. doi: 10.1093/nar/gkl508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pohjoismaki J.L., Wanrooij S., Hyvarinen A.K., Goffart S., Holt I.J., Spelbrink J.N., Jacobs H.T. Alterations to the expression level of mitochondrial transcription factor A, TFAM, modify the mode of mitochondrial DNA replication in cultured human cells. Nucleic Acids Res. 2006;34:5815–5828. doi: 10.1093/nar/gkl703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krishnan K.J., Reeve A.K., Samuels D.C., Chinnery P.F., Blackwood J.K., Taylor R.W., Wanrooij S., Spelbrink J.N., Lightowlers R.N., Turnbull D.M. What causes mitochondrial DNA deletions in human cells? Nat Genet. 2008;40:275–279. doi: 10.1038/ng.f.94. [DOI] [PubMed] [Google Scholar]
- 35.Spelbrink J.N., Toivonen J.M., Hakkaart G.A., Kurkela J.M., Cooper H.M., Lehtinen S.K., Lecrenier N., Back J.W., Speijer D., Foury F., et al. In vivo functional analysis of the human mitochondrial DNA polymerase POLG expressed in cultured human cells. J. Biol. Chem. 2000;275:24818–24828. doi: 10.1074/jbc.M000559200. [DOI] [PubMed] [Google Scholar]
- 36.Church G.M., Gilbert W. Genomic sequencing. Proc. Natl. Acad. Sci. USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cooper H.M., Spelbrink J.N. The human Sirt3 protein deacetylase is exclusively mitochondrial. Biochem J. 2008;411:279–285. doi: 10.1042/BJ20071624. [DOI] [PubMed] [Google Scholar]
- 38.Yasukawa T., Yang M.Y., Jacobs H.T., Holt I.J. A bidirectional origin of replication maps to the major noncoding region of human mitochondrial DNA. Mol. Cell. 2005;18:651–662. doi: 10.1016/j.molcel.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 39.Friedman K.L., Brewer B.J. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 1995;262:613–627. doi: 10.1016/0076-6879(95)62048-6. [DOI] [PubMed] [Google Scholar]
- 40.Brewer B.J., Fangman W.L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.