Abstract
Natriuretic peptides (NPs) comprise a family of structurally related but genetically distinct hormones that regulate a variety of physiological processes such as cardiac growth, blood pressure, axonal pathfinding and endochondral ossification leading to the formation of vertebrae and long bones. The biological actions of NPs are mediated by natriuretic peptide receptors (NPRs) A, B and C that are located on the cell surface. Mutations in NPR-B have been shown to cause acromesomelic dysplasia-type Maroteaux (AMDM), a growth disorder in humans and severe dwarfism in mice. We hypothesized that missense mutations of NPR-B associated with AMDM primarily affect NPR-B function by the arrest of receptor trafficking at the endoplasmic reticulum (ER), due to conformational change, rather than an impairment of ligand binding, transmission of signal through the membrane or catalytic activity. Twelve missense mutations found in AMDM patients and cn/cn mice were generated by site-directed mutagenesis and transiently overexpressed in HeLa cells. Confocal microscopy revealed that 11 out of 12 mutants were retained in the ER. Determination of the ligand-dependent cGMP response confirmed that ER-retained NPR-B mutants are non-functional. Meanwhile, the only cell surface-targeted NPR-B missense mutant (D176E) displayed greatly reduced enzymatic activity due to impaired ligand binding. Thus, in the majority of cases of AMDM associated with missense NPR-B mutation, disease appears to result from defects in the targeting of the ER receptor to the plasma membrane.
INTRODUCTION
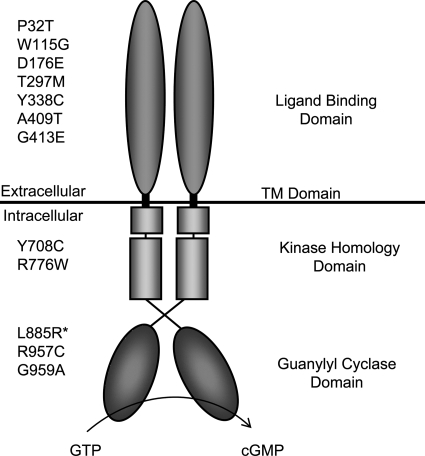
Natriuretic peptides (NPs) comprise a family of structurally related but genetically distinct peptide hormones that play an important role in the regulation of blood pressure, cardiac growth, axonal pathfinding and skeletal growth (1–5). They exert their biological actions by binding to cell surface receptors called natriuretic peptide receptors (NPRs). Three different subtypes have been identified: NPR-A, -B and -C (6). Whereas NPR-C has no cytoplasmic domain, NPR-A and -B harbor an intracellular guanylyl cyclase domain which is thought to mediate their biological function. Both receptors are characterized by a modular structure: an extracellular ligand-binding domain, a transmembrane region and an intracellular ‘kinase homology domain’ (KHD) that regulates guanylyl cyclase activity dependent on its phosphorylation state (Fig. 1) (7). NPR-A is selectively activated by atrial NP (ANP) and B-type NP (BNP), whereas the specific ligand of NPR-B is C-type NP (CNP) (7). NPR-B is expressed in various tissues and cell populations such as the heart, vessels, the brain, the uterus and chondrocytes (6,8,9).
Figure 1.
The homodimeric domain structure of NPR-B showing the position of missense mutations causing AMDM. L885R* has been found in an animal model—the cn/cn mouse (12).
Several studies demonstrated that NPR-B and its ligand CNP play a major role in the regulation of skeletal growth. Mice with the deletion of the NPR-B encoding gene (NPR2) or a loss-of-function mutation (cn/cn) display severe dwarfism (5,10). Homozygous loss-of-function mutation in the human NPR2 gene has been identified in patients with acromesomelic dysplasia-type Maroteaux (AMDM), a rare form of dwarfism that is characterized by reduced body height and shortened limbs (11,12). However, AMDM patients do not show any other manifestations in other organs (9,11). These studies clearly showed that NPR-B is essential in enhancing the development of the skeletal system.
Several genetic models, in vitro studies and mutation in humans provide insight into the prominent role of the NPR-B ligand CNP and the downstream signaling of this receptor in the regulation of skeletal growth. Recent studies have shown that CNP-deficient mice are dwarfed and mice that overexpress CNP have longer bones (13,14). In addition, it has been shown that CNP stimulates or plays a crucial role in growth and size increase of individual chondrocytes in the hypertrophic phase of bone growth. This was shown by studies in which CNP was exogenously administered in vitro to whole-organ cultures of mouse tibia (15). Overexpression of CNP due to a balanced t(2,7) translocation has also been reported in a patient with skeletal overgrowth (16). All of these effects of CNP in skeletal growth are likely to be mediated by NPR-B-dependent signaling. Activation of NPR-B by CNP leads to the production of cGMP with the subsequent activation of cGMP-dependent protein kinase II (cGKII). Consequently, dwarfism has also been observed in mice in which the CNP gene is deleted or rats in which the cGKII gene is mutated (17,18). The substrates of cGKII that promote bone growth are still unknown. A recent report suggests that Sox9, a ‘master’ inhibitor of chondrocyte differentiation, is a possible candidate because cGKII overexpression leads to the inactivation of this factor due to its translocation from the nucleus to the cytoplasm (17).
In human, the NPR2 gene, located on chromosome 9, encodes the NPR-B protein (19). Studies of AMDM patients have identified 28 different causative mutations in the NPR2 gene, including missense, nonsense, frame-shift mutations, insertions and deletions and splice site mutations. AMDM is an autosomal recessive rare genetic disorder characterized by various developmental abnormalities of the skeletal bones and facial anomalies with a prevalence of approximately 1/1 000 000. This disorder exhibits disproportionate effects on the middle and distal segments of the limbs (forearms, forelegs, hands and feet) (11,12).
Endoplasmic reticulum (ER)-associated protein degradation (ERAD) eliminates misfolded or orphan proteins from the ER, thereby preventing toxicity associated with their accumulation. ERAD targets are selected by a quality-control system including molecular chaperone within the ER lumen and are ultimately destroyed by the cytoplasmic ubiquitin–proteasome system (20,21). As NPR-B normally traffics through the ER on its way to the plasma membrane, we hypothesized that in the case of AMDM-associated NPR-B missense mutations, the loss-of-function results from the ER retention of receptor and the prevention of its normal trafficking to the plasma membrane, possibly due to the defective folding/oligomerization of the receptor. To address this issue, we generated all the NPR-B missense mutations found in AMDM patients and cn/cn mice (Fig. 1) and examined their subcellular localization and function in cultured human cells.
RESULTS
Most AMDM-associated NPR-B missense mutant proteins are poorly targeted to the plasma membrane and co-localize well with ER-resident protein calnexin
To test the hypothesis that AMDM pathology associated with NPR-B missense mutation results from defects in protein trafficking, we examined the intracellular localization of NPR-B and patient mutant variants in cultured human cells. To do this, we used site-directed mutagenesis to introduce documented AMDM mutations into wild-type NPR-B cDNA subcloned in a mammalian expression vector that allows the overexpression of the protein of interest as a fusion protein containing a C-terminus HA epitope of the influenza hemagglutinin protein (Fig. 1). The resulting plasmids together with control plasmid encoding HA-tagged wild-type NPR-B were transiently transfected into HeLa cells, and their intracellular localization and co-localization with plasma membrane (H-Ras) and ER (calnexin) marker were examined by confocal immunofluorescence microscopy and by the processing of image data using image analysis software (as described in Materials and Methods).
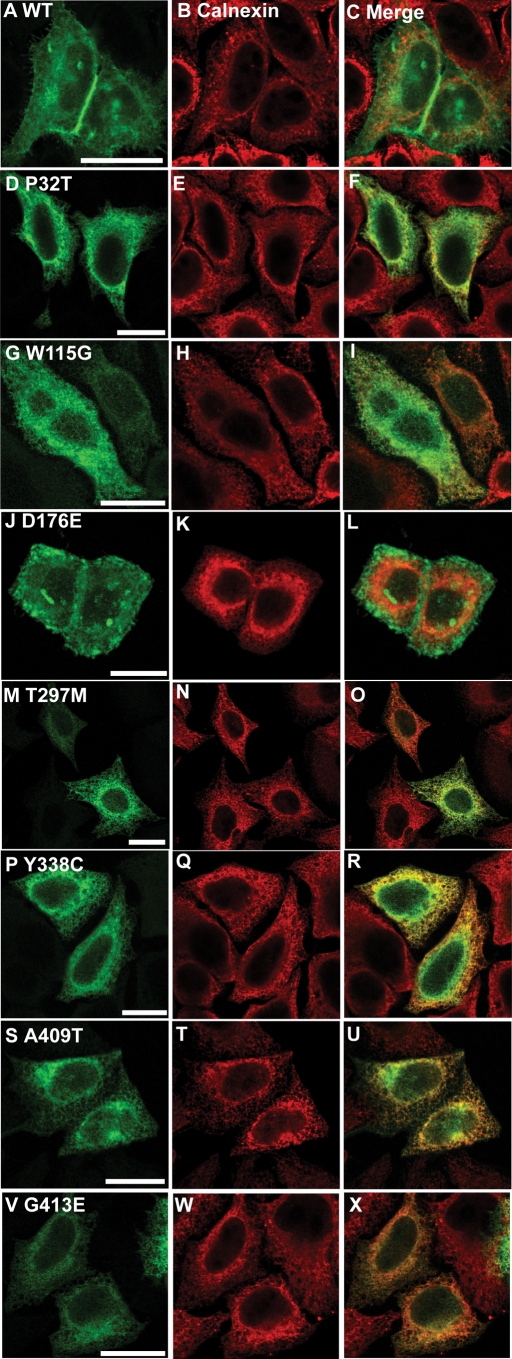
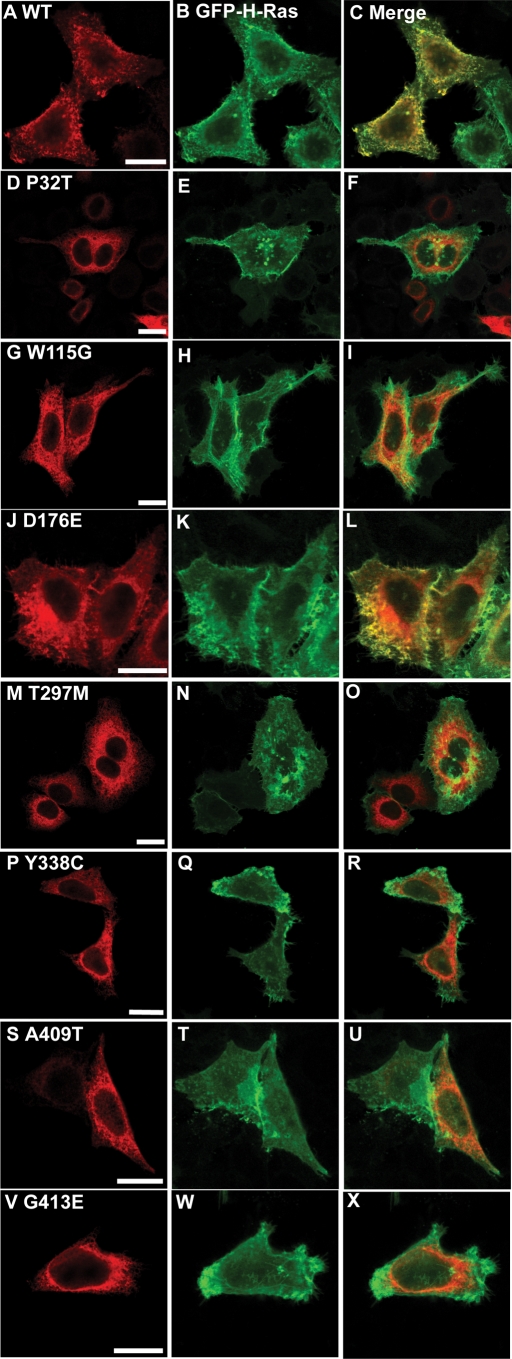
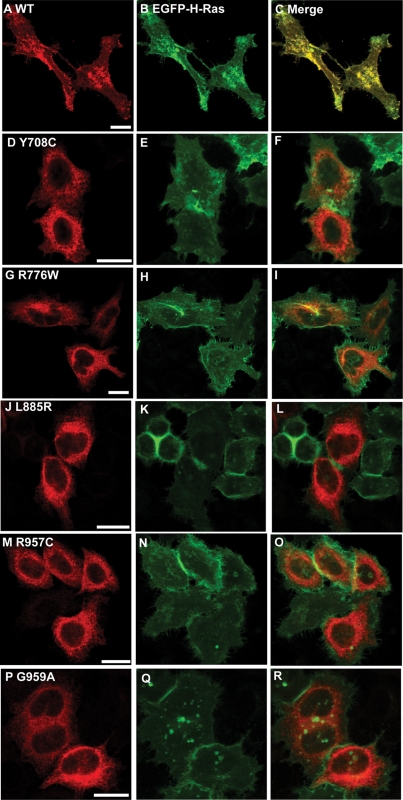
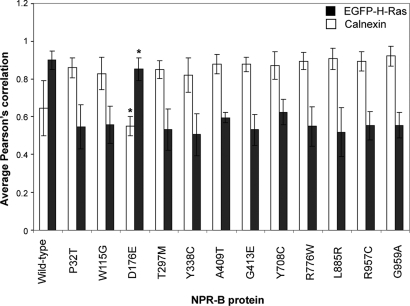
Wild-type NPR-B was observed in a uniform pattern across the surface of expressing cells and showed strong (Pearson correlation 0.90) co-localization with the plasma membrane associated protein H-Ras but not the ER-resident calnexin (Pearson correlation 0.64) (Figs 2A–C, 3A–C, 4A–C, 5A–C and 6). This is consistent with the idea that NPR-B is a plasma membrane associated receptor protein. In contrast, several NPR-B missense mutants in which the extracellular CNP-binding domain is altered [P32T, W115G, T297E, Y338C, A409T, G413E (Fig. 1)] resulted in a protein that was localized in the cytoplasm with a reticular pattern and showed strong co-localization with calnexin (Pearson correlation >0.83) but not with H-Ras (Pearson correlation <0.60) (Figs 2D–I, M–X, 3D–I, M–X and 6). The exception to this picture was the D176E mutant that appeared to undergo normal trafficking to the plasma membrane as shown by stronger co-localization with H-Ras than with calnexin (Pearson correlation of 0.85 and 0.55, respectively) (Figs 2J–L, 3J–L and 6).
Figure 2.
Comparison of the intracellular localization of NPR-B wild-type and patient mutant variants (extracellular ligand-binding domain) with the ER. HeLa cells were transiently transfected with plasmids encoding the indicated HA-tagged NPR-B proteins, fixed and stained with antibodies as described in Materials and Methods. (A), (D), (G), (J), (M), (P), (S) and (V) show the distribution of overexpressed HA-tagged NPR-B protein, (B), (E), (H), (K), (N), (Q), (T) and (W) show the distribution of endogenous ER-resident calnexin, whereas (C), (F), (I), (L), (O), (R), (U) and (X) show the extent of co-localization of NPR-B proteins and calnexin. Bars: 20 µm.
Figure 3.
Comparison of the intracellular localization of NPR-B wild-type and patient mutant variants (extracellular ligand-binding domain) with the plasma membrane. HeLa cells were transiently co-transfected with plasmids encoding the indicated HA-tagged NPR-B proteins and EGFP-tagged H-Ras, fixed and stained with antibodies as described in Materials and Methods. (A), (D), (G), (J), (M), (P), (S) and (V) show the distribution of overexpressed HA-tagged NPR-B protein, (B), (E), (H), (K), (N), (Q), (T) and (W) show the distribution of overexpressed EGFP-tagged H-Ras, which is predominantly localized to the plasma membrane, whereas (C), (F), (I), (L), (O), (R), (U) and (X) show the extent of co-localization of NPR-B proteins and EGFP-H-Ras. Bars: 20 µm.
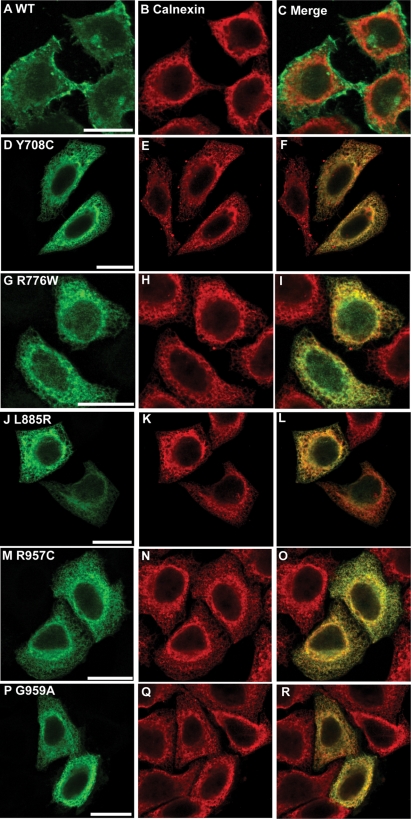
Figure 4.
Comparison of the intracellular localization of NPR-B wild-type and patient mutant variants (intracellular KHD and GCD) with the ER. HeLa cells were transiently transfected with plasmids encoding the indicated HA-tagged NPR-B proteins, fixed and stained with antibodies as described in Materials and Methods. (A), (D), (G), (J), (M) and (P) show the distribution of overexpressed HA-tagged NPR-B protein, (B), (E), (H), (K), (N) and (Q) show the distribution of endogenous ER-resident calnexin, whereas (C), (F), (I), (L), (O) and (R) show the extent of co-localization of NPR-B proteins and calnexin. Bars: 20 µm.
Figure 5.
Comparison of the intracellular localization of NPR-B wild-type and patient mutant variants (intracellular KHD and GCD) with the plasma membrane. HeLa cells were transiently co-transfected with plasmids encoding the indicated HA-tagged NPR-B proteins and EGFP-tagged H-Ras, fixed and stained with antibodies as described in Materials and Methods. (A), (D), (G), (J), (M) and (P) show the distribution of overexpressed HA-tagged NPR-B protein, (B), (E), (H), (K), (N) and (Q) show the distribution of overexpressed EGFP-tagged H-Ras, which is predominantly localized to the plasma membrane, whereas (C), (F), (I), (L), (O) and (R) show the extent of co-localization of NPR-B proteins and EGFP-H-Ras. Bars: 20 µm.
Figure 6.
Analysis of the co-localization of NPRB with ER and plasma membrane markers. Images of HeLa cells transiently transfected with wild-type NPRB and indicated AMDM patient mutant variants were analyzed in order to determine the extent of localization of the protein with plasma membrane and ER. Pearson correlations were derived for NPR-B versus calnexin or EGFP-H-Ras signals for at least four individual transfected cells, and the mean together with standard error (bars) are presented. *P > 0.05 versus wild-type, P-values for all other NPR-B mutants co-localization with calnexin and EGFP-H-Ras compared with wild-type were <0.01.
Similar analysis of AMDM patient mutations within the NPR-B intracellular KHD (Y708C and R776) and GCD (L885R, R957C and G959A) resulted in all cases in protein manifesting an intracellular reticular pattern and strong co-localization with the ER marker calnexin (Pearson correlation >0.87) but not the plasma membrane marker H-Ras (Pearson correlation <0.63) (Figs 4D–R, 5D–R and 6).
t-Test analysis of Pearson correlation data for AMDM patient mutant versus wild-type NPR-B, as described in Materials and Methods, confirms that a significant difference (P < 0.0033) exists between the localization of all mutants studied except D176E whose intracellular distribution is similar to wild-type (P > 0.05) (Fig. 7).
Figure 7.
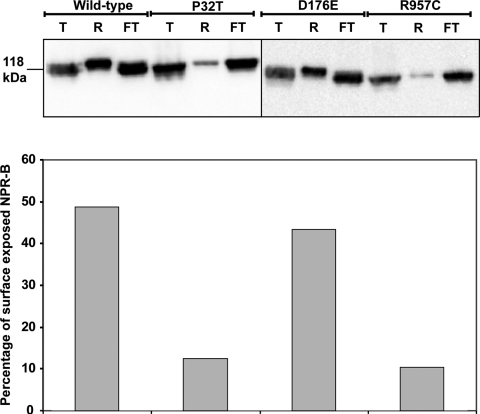
Measurement of cell surface-exposed NPR-B and mutant proteins. HeLa cells were transiently transfected with wild-type NPR-B or AMDM-associated NPR-B mutants, and the cell surface-exposed NPR-B proteins were biotinylated and purified as described in Materials and Methods. Total NPR-B protein together (T) with equivalent amounts of biotinylated/neutravidin-retained (R) and non-biotinylated/neutravidin flow-through (FT) protein fractions were detected by immunoblotting using anti-HA antibody. Differences in the percentage of the total NPR-B that is biotinylated, i.e. surface‐exposed between wild-type and AMDM-associated mutants, are shown in the bar chart. This experiment has been conducted in duplicate with similar results obtained in both instances.
In order to confirm the conclusions of our microscopy study, we surface-biotinylated intact HeLa cells transfected with wild-type NPR-B and AMDM patient mutant NPR-B, purified the biotinylated proteins from cell lysates and quantified the percentage of NPR-B that was surface-exposed by densitometry as described in Materials and Methods. We observe similar overall levels of the expression of each NPR-B protein and that, as expected, a high proportion of wild-type (48.8%) and D176E (43.3%) is present in the neutravidin-retained fraction, indicating that it is biotinylated/targeted to the cell surface, whereas a lower proportion of P32T (12.5%) and R957C (10.4%) is present in this fraction, confirming that these mutants are poorly targeted to the plasma membrane (Fig. 7). Similar cell-surface biotinylation experiments confirm our microscopy observations that other AMDM-associated NPR-B mutants (W115G, T297M, Y338C, A409T, G413E, Y708C, R776W, L885R, G959A) are poorly targeted to the plasma membrane (data not shown).
Together these observations suggest that AMDM pathology in all patients harboring NPR-B missense mutations, with the exception of D176E, results from defects normal in the trafficking of NPR-B to the plasma membrane that lead to its retention within the ER.
NPR-B missense mutant proteins associated with AMDM are non-functional
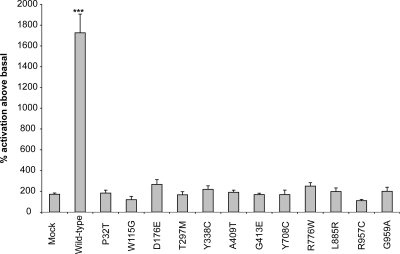
To confirm that ER-retained missense NPR-B mutant protein is non-functional, we measured CNP-dependent NPR-B guanylate cyclase activity, a read-out of receptor activity, in transiently transfected HeLa cells (as described in Materials and Methods). As expected, we observed that CNP stimulation resulted in a 17-fold elevation of guanylate cyclase activity over basal in cells transfected with wild-type NPR-B (Fig. 8). In contrast, CNP stimulation of cells expressing NPR-B missense mutations associated with AMDM, including D176E, which is targeted normally, resulted in 2-fold elevation of guanylate cyclase activity and showed no statistical difference from mock-transfected cells (Fig. 8). This indicates that all NPR-B missense mutations associated with AMDM result in a loss of NPR-B function.
Figure 8.
Measurement of CNP-dependent guanylate cyclase activity of NPR-B wild-type and mutant proteins. HeLa cells transiently transfected with wild-type NPR-B, AMDM-associated NPR-B mutants or no plasmid (mock) were incubated with CNP, and guanylate cyclase activity was measured as described in Materials and Methods. CNP-dependent receptor activity is expressed as a percentage relative to non-stimulated guanylate cyclase activity. Bars are standard error of the mean of experiments conducted in triplicate. ***P < 0.05.
The D176E mutant inhibits ligand binding by NPR-B
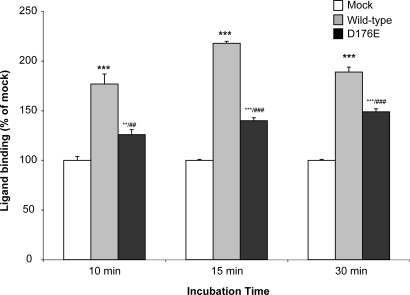
These findings show that, in most cases, the failure of trafficking of mutated NPR-B underlies AMDM. However, the trafficking of the D176E mutant appears normal, indicating that this mechanism is not responsible for AMDM in this instance. The D176E mutation is present in the extracellular ligand-binding domain of NPR-B, raising the possibility that pathology results from the perturbation of CNP binding. To test this possibility, we assayed the ability of plasma membrane-associated D176E to bind CNP (as described in Materials and Methods). We observed that although CNP binding by D176E was significantly reduced compared with wild-type NPR-B at all time-points measured, it was significantly higher than that of mock-transfected cells (Fig. 9). This finding concurs with the idea that AMDM pathology in NPR-B D176E patients results, in part, from defects in CNP binding. Consistent with an important role for D176 in NP-binding sequence analysis confirms that this amino acid is conserved between NPR-B and NPR-A isoforms from different species (data not shown). Furthermore, a previous study of ANP:NPR-A interaction revealed that an adjacent site within bovine NPR-A interacts directly with ANP (22).
Figure 9.
Binding of CNP to HeLa cells transiently transfected with wild-type NPR-B. HeLa cells cultured in six-well plates were incubated with 125I-labeled CNP for 10, 15 or 30 min. After washing, cells were disrupted and radioactivity of each well was determined. Compared with cells transfected with the empty vector (Mock—white bars), CNP displayed the highest affinity to HeLa cells overexpressing wild-type NPR-B (gray bars) and a modest one to those expressing NPR-B D176E (black bars). Experiments were carried out five times and values are expressed as percentage of binding achieved by mock-transfected cells. Error bars are standard error of the mean. **P < 0.01 versus Mock; ***P < 0.001 versus Mock; ##P < 0.01 versus NPR-B wild-type; ###P < 0.001 versus NPR-B wild-type.
DISCUSSION
Correct folding of nascent polypeptide chains within the ER is critical for function, assembly into multi-subunit complexes and trafficking through the exocytic pathway for secretory and cell-surface proteins. This process appears inefficient, and a substantial proportion of nascent polypeptides are rejected by the stringent ER quality-control machinery and targeted for degradation by the ubiquitin/proteasome systems (20,21). For instance, pulse-chase studies showed that 50–70% of wild-type CFTR is inefficiently folded and is degraded intracellularly (23,24) by the ubiquiting/proteasome systems (25,26). Therefore, it is not surprising that the smallest alteration to polypeptide primary structure (i.e. missense point mutation) can result in the complete loss of function due to retention in the ER and degradation by the proteasome system with inherent pathological consequences.
The group of human genetic diseases associated with ER retention and degradation have been named ERAD (endoplasmic reticulum-associated protein degradation) diseases and include cystic fibrosis and emphysema (20,21). In view of the fact that ∼30% of all cellular proteins contain a predicted ER-targeting signal, we reasoned that many more ERAD diseases remain undiscovered. We therefore exploited bioinformatic algorithms and databases of disease genes to identify more than 30 potential diseases where molecular defects consistent with protein misfolding were evident from the position and nature of disease-associated allele, but significantly, where the molecular etiology was unknown (20) (B. Ali, unpublished data). We validated this approach by the analysis of ROR2 gene, an orphan receptor tyrosine kinase with alleles associated with Robinow and brachydactyly-type B syndromes. Robinow syndrome is a recessive disorder found in several parts of the Middle East, including Oman and Saudi Arabia, and caused by loss-of-function mutations in ROR2 gene. Indeed, we determined that such alleles were responsible for the misfolding of the ROR2 protein, confirming ER retention and therefore loss-of-function as the underlying mechanism in Robinow syndrome (20,27).
AMDM-causing mutations were among the ERAD candidates identified by our bioinformatics approach mainly due to their nature as missense mutations in a receptor that normally traffics through the ER. For example, a number of AMDM-causing mutations (P32T, W115G, Y338C, G413E, Y708C, R776W and R957C) cause structurally significant alterations in the NPR-B protein and therefore are likely to result in at least partial misfolding and oligomerization of the nascent NPR-B polypeptide and retention by the ER quality-control system. Our fluorescence microscopy (Figs 2–6) and surface biotinylation (Fig. 7) data demonstrating that 11 out of the 12 AMDM-missense-causing mutations result in NPR-B being poorly targeted to the plasma membrane and instead show strong co-localization with the ER-resident calnexin strongly support this hypothesis. Taken together, the data on ROR2 and NPR2 strongly support our hypothesis that ERAD is contributing to the mechanisms of many more human genetic diseases that have been described in the literature.
Our fluorescence microscopy (Figs 2–6), surface biotinylation (Fig. 7), the measurement of NPR-B-associated guanylate cyclase activity (Fig. 8) and ligand-binding assay (Fig. 9) indicate that the D176E mutant causes AMDM by a different mechanism compared with the other mutants. We argue that the disease mechanism underlying AMDM patients carrying this mutation is the reduction of peptide-binding ability due to the replacement of aspartic acid with glutamic acid in this position. Aspartic acid at position 176 is located at the peptide-binding site of the receptor and therefore it is plausible that any alteration will affect the peptide binding and loss of NPR-B-signaling pathway as has been suggested previously (11).
Finally, further studies are needed to fully document the mechanisms of possible ERAD diseases such as those caused by the mutation of ROR2 and NPR2. Elucidation of the mechanisms of disease is important for disease management and perhaps developing new therapies. It has been recently shown that manipulating the ER quality control might be a good drug target for genetic diseases (28,29). The realization that some of those ER-retained proteins might be biologically functional if they escaped from this organelle is encouraging to search for ways to manipulate ERAD to gain therapeutic advantage.
MATERIALS AND METHODS
Generation of NPR-B missense mutants in PRK5 vector
For origin of vector(s) (30). The missense mutations (Fig. 1) were generated by Stratagene’s QuickChange Site-directed Mutagenesis Kit according to the manufacturer’s instructions using pRK5-NPR-B WT as template and the following primers with the changed nucleotide in boldface:
Npr2-P32T-FCTGGCGGTGGTGCTGACAGAACACAACCTGAGC
Npr2-P32T-R GCTCAGGTTGTGTTCTGTCAGCACCACCGCCAG
Npr2-W115G-F CGCTTTGCCTCGCACGGGCACCTTCCCCTCCTG
Npr2-W115G-R CAGGAGGGGAAGGTGCCCGTGCGAGGCAAAGCG
Npr2-D176E-F CTGGATGCTCGCACAGAGGACCGGCCCCACTAC
Npr2-D176E-R GTAGTGGGGCCGGTCCTCTGTGCGAGCATCCAG
Npr2-T297M-F ACTGTCCTGGTGATCATGTACCGAGAGCCCCCG
Npr2-T297M-R CGGGGGCTCTCGGTACATGATCACCAGGACAGT
Npr2-Y338C-F ATTGCGGGCTGCTTCTGTGATGGAATCCTGCTC
Npr2-Y338C-R GAGCAGGATTCCATCACAGAAGCAGCCCGCAAT
Npr2-A409T-F GACTTTCAGCCTGCAACCCATTACTCAGGAGCT
Npr2-A409T-R AGCTCCTGAGTAATGGGTTGCAGGCTGAAAGTC
Npr2-G413E-F GCAGCCCATTACTCAGAAGCTGAGAAGCAGATT
Npr2-G413E-R AATCTGCTTCTCAGCTTCTGAGTAATGGGCTGC
Npr2-Y708C-F CAGAAGGCGGATGTCTGTAGCTTTGCCATCATTC
Npr2-Y708C-R GAATGATGGCAAAGCTACAGACATCCGCCTTCTG
Npr2-R776W-F CAGGACCCAACAGAATGGCCAGACTTTGGGCAA
Npr2-R776W-R TTGCCCAAAGTCTGGCCATTCTGTTGGGTCCTG
Npr2-L885R-F ATGCAGGTGGTGACACGTCTTAATGACCTTTAT
Npr2-L885R-R ATAAAGGTCATTAAGACGTGTCACCACCTGCAT
Npr2-R957C-F GACCAGCTAAGGTTATGCATAGGCGTCCATACTG
Npr2-R957C-R CAGTATGGACGCCTATGCATAACCTTAGCTGGTC
Npr2-G959A-F CTAAGGTTACGCATAGCCGTCCATACTGGGCC
Npr2-G959A-R GGCCCAGTATGGACGGCTATGCGTAACCTTAG
All generation of the desired mutations was confirmed by direct DNA sequencing using the purified plasmids (www.macrogen.com).
Construction of the HA-tagged NPR-B constructs
The HA-tagged NPR-B was made by PCR using the following primers: forward primer: cggaattcccaccATGGCACTGCCATCCCTGC; reverse primer: gcaagcttTTAAGCGTAATCTGGAACATCGTATGGGTACAGGAGTCCAGGAGGTCC.
The forward primer included an EcoRI (lower case bold letters) site, whereas the reverse primer included a HindIII site (lower case bold letters). The reverse complement sequence of the HA tag (italicized capitals) was included in the reverse primer just before the stop codon to insert the tag at the C-end of the protein. The wild-type and mutants NPR2 cDNAs were amplified from pRK5-NPR-B constructs using Stratagene PfuTaq polymerase. The amplified cDNAs were digested with EcoRI and HindIII enzymes and subcloned into pcDNA3.1 vector digested with the same enzymes using T4 DNA ligase. The subcloning of the cDNAs was confirmed by direct DNA sequencing. The HA-tagged NPR-B constructs were generated due to the failure of the commercially available polyclonal antibodies to detect HeLa cells that exogenously expressed NPR-B.
Cell culture and transfection
HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% fetal calf serum, 2 mm l-glutamine and 100 U/ml penicillin/streptomycin at 37°C with 10% CO2. For transfection, cells were grown in a 24-well tissue culture plates. Transfection was performed using the liposomal transfection reagent FuGENE 6 (Roche Biochemicals), according to the manufacturer’s instructions. A mixture of 1 µg of NPR-B wild-type or mutant plasmid DNA and 4 µl of FuGENE 6 in 94 µl of OPTIMEM I medium was applied to each well of the cultured cells. Twenty-four hours after transfection, the cells were fixed and processed for microscopy.
Immunochemistry
Antibodies were purchased from the following sources: mouse anti-HA-Tag monoclonal antibody (dilution 1:200 for immunofluorescence and 1:1000 for western blotting; Cell Signaling Technology), rabbit anti-calnexin polyclonal antibody (dilution 1:500; StressGen Biotechnologies), Alexa Fluor 568-goat anti-mouse IgG (dilution 1:200; Molecular Probes), Alexa Fluor 488-goat anti-rabbit IgG (dilution 1:200; Molecular Probes) and HRP-conjugated to goat anti-mouse IgG (dilution 1:5000; Dako). For immunofluorescence, cover slip-grown HeLa cells were washed with phosphate-buffered saline (PBS), fixed in 3% paraformaldehyde in PBS for 15 min at room temperature, washed in PBS three times, quenched with 50 mm NH4Cl in PBS for 10 min and incubated in permeabilization/blocking solution (0.05% saponin, 0.5% BSA and 1× PBS) for 15 min. The fixed cells were then incubated at room temperature for 1 h with either mouse monoclonal anti-HA antibody alone or co-stained with both mouse monoclonal anti-HA antibody and rabbit polyclonal anti-calnexin antibody. After washing with PBS, the cells were incubated with the appropriate secondary antibodies for 1 h at room temperature, washed several times with PBS and mounted in immunofluor medium (ICN Biomedicals). Data were acquired as 8 bit gray-scale images using Leica DM-IRBE confocal microscope controlled by Leica TCS-NT software associated with the microscope. For presentation, images were pseudocolored as either red (NPR-B) or green (calnexin and EGFP-H-Ras), contrast-enhanced and overlayed using Adobe Photoshop® (Adobe Inc.). All images presented are single sections in the z-plane. For co-localization analysis, raw 8 bit red-green overlay data for NPR-B were imported into Volocity 4.3.2 image analysis software and Pearson correlations were determined using the co-localization function available in this software. Briefly, this function plots the red and green intensity values (0–255 gray-scale values) for individual pixels derived from the area of transfected cells and determines the correlation of these intensities. According to this measurement, perfect co-localization corresponds to a value of 1. The statistical significance of differences in the pattern of co-localization data between populations of cells (n ≥ 4) expressing wild-type or AMDM mutant NPR-B proteins was determined by pairwise Student’s t-testing of Pearson correlation data for each mutant compared with H-Ras and calnexin and the corresponding wild-type data.
Cell surface biotinylation of NPR-B
HeLa cells were seeded onto 10 cm diameter dishes and transiently transfected with plasmids encoding wild-type NPR-B or the mutant NPR-B. Twenty-four hours later, cells were surface-labeled with biotin and the biotinylated proteins were purified using the Pierce Cell Surface Protein Isolation Kit (Perbio, Northumberland, UK) in accordance with the manufacturer's recommended protocol. The following fractions for each transfection—total equivalent to 20% of total transfected cell pool, biotinylated/neutravidin-retained equivalent to 25% of retained material and non-biotinylated/neutravidin flow-through equivalent to 25% of flow-through material—were loaded onto 8% SDS–PAGE gels transfered to PVDF membrane, and NPR-B was detected using mouse anti-HA at 1:1000. The percentage of surface-exposed NPR-B in each case was calculated by densitometric measurement of neutravidin-retained and flow-through fractions. Specifically, the value for neutravidin-retained NPR-B was divided by the total retained and flow-through value. Experiments were conducted in duplicate with similar results obtained in both cases. Presented data derive from one of these experiments.
CNP-dependent cGMP response
Transiently transfected HeLa cells were seeded onto six-well plates and grown to confluency. After washing twice with PBS, the stimulation mixture containing CNP (CNP-22 Calbiochem) in a final concentration of 100 nm, 1 mm IBMX and 0.5% BSA dissolved in serum-free DMEM was added. After incubation for 20 min at 37°C, culture plates were immediately placed on ice. Cells were disrupted by sonication, lysates were stored at –20°C until cGMP measurement as described previously (30). Experiments were conducted in triplicate, and the significance of differences between transfected versus mock-transfected cells was determined using Student’s t-test.
Determination of CNP binding
HeLa cells transiently transfected with the empty vector (mock), wild-type NPR-B or the mutant NPR-B D176E were seeded onto six-well plates. After reaching a density of 70%, cells were washed twice with serum-free DMEM. To prepare radiolabelled ligand 125I-labelled C-type NP was dissolved in 5% acetic acid and diluted to a concentration of 1 µm with DMEM supplemented with 1% BSA. Cells were incubated with the ligand for 10, 15 or 30 min at 37°C. After washing twice with PBS, cells were disrupted by adding 1 M NaOH/1% Triton-X100. Lysates were neutralized with 1 M HCl. Total radioactivity of each well was determined. Experiments were conducted in triplicate and the significance of differences between transfected versus mock-transfected cells and between cells transfected with different NPR-B proteins were determined using Student’s t-test.
Conflict of Interest statement. None declared.
FUNDING
This project was funded by the Faculty of Medicine and Health Sciences Research Office (UAE University) grant number NP/07/09. We would also like to thank the Wellcome Trust for funding the work at Imperial College.
REFERENCES
- 1.Kishimoto I., Rossi K., Garbers D.L. A genetic model provides evidence that the receptor for atrial natriuretic peptide (guanylyl cyclase-A) inhibits cardiac ventricular myocyte hypertrophy. Pro. Natl. Acad. Sci. USA. 2001;98:2703–2706. doi: 10.1073/pnas.051625598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Langenickel T.H., Buttgereit J., Pagel-Langenickel I., Lindner M., Monti J., Beuerlein K., Al-Saadi N., Plehm R., Popova E., Tank J., et al. Cardiac hypertrophy in transgenic rats expressing a dominant-negative mutant of the natriuretic peptide receptor B. Proc. Natl. Acad. Sci. USA. 2006;103:4735–4740. doi: 10.1073/pnas.0510019103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopez M.J., Wong S.K., Kishimoto I., Dubois S., Mach V., Friesen J., Garbers D.L., Beuve A. Salt-resistant hypertension in mice lacking the guanylyl cyclase-A receptor for atrial natriuretic peptide. Nature. 1995;378:65–68. doi: 10.1038/378065a0. [DOI] [PubMed] [Google Scholar]
- 4.Schmidt H., Stonkute A., Juttner R., Schaffer S., Buttgereit J., Feil R., Hofmann F., Rathjen F.G. The receptor guanylyl cyclase Npr2 is essential for sensory axon bifurcation within the spinal cord. J. Cell Biol. 2007;179:331–340. doi: 10.1083/jcb.200707176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tamura N., Doolittle L.K., Hammer R.E., Shelton J.M., Richardson J.A., Garbers D.L. Critical roles of the guanylyl cyclase B receptor in endochondral ossification and development of female reproductive organs. Proc. Natl. Acad. Sci. USA. 2004;101:17300–17305. doi: 10.1073/pnas.0407894101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Potter L.R., Abbey-Hosch S., Dickey D.M. Natriuretic peptides, their receptors, and cyclic guanosine monophosphate-dependent signaling functions. Endocr. Rev. 2006;27:47–72. doi: 10.1210/er.2005-0014. [DOI] [PubMed] [Google Scholar]
- 7.Potter L.R., Hunter T. Identification and characterization of the phosphorylation sites of the guanylyl cyclase-linked natriuretic peptide receptors A and B. Methods. 1999;19:506–520. doi: 10.1006/meth.1999.0893. [DOI] [PubMed] [Google Scholar]
- 8.Silberbach M., Roberts Natriuretic peptide signalling: molecular and cellular pathways to growth regulation. Cell Signal. 2001;13:221–231. doi: 10.1016/s0898-6568(01)00139-5. [DOI] [PubMed] [Google Scholar]
- 9.Pagel-Langenickel I., Buttgereit J., Bader M., Langenickel T.H. Natriuretic peptide receptor B signaling in the cardiovascular system: protection from cardiac hypertrophy. J. Mol. Med. 2007;85:797–810. doi: 10.1007/s00109-007-0183-4. [DOI] [PubMed] [Google Scholar]
- 10.Tsuji T., Kunieda T. A loss-of-function mutation in natriuretic peptide receptor 2 (Npr2) gene is responsible for disproportionate dwarfism in cn/cn mouse. J. Biol. Chem. 2005;280:14288–14292. doi: 10.1074/jbc.C500024200. [DOI] [PubMed] [Google Scholar]
- 11.Bartels C.F., Bukulmez H., Padayatti P., Rhee D.K., van Ravenswaaij-Arts C., Pauli R.M., Mundlos S., Chitayat D., Shih L.Y., Al-Gazali L.I., et al. Mutations in the transmembrane natriuretic peptide receptor NPR-B impair skeletal growth and cause acromesomelic dysplasia, type Maroteaux. Am. J. Hum. Genet. 2004;75:27–34. doi: 10.1086/422013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olney R.C., Bukulmez H., Bartels C.F., Prickett T.C., Espiner E.A., Potter L.R., Warman M.L. Heterozygous mutations in natriuretic peptide receptor-B (NPR2) are associated with short stature. J. Clin. Endocrinol. Metab. 2006;91:1229–1232. doi: 10.1210/jc.2005-1949. [DOI] [PubMed] [Google Scholar]
- 13.Komatsu Y., Chusho H., Tamura N., Yasoda A., Miyazawa T., Suda M., Miura M., Ogawa Y., Nakao K. Significance of C-type natriuretic peptide (CNP) in endochondral ossification: analysis of CNP knockout mice. J. Bone Miner. Metab. 2002;20:331–336. doi: 10.1007/s007740200048. [DOI] [PubMed] [Google Scholar]
- 14.Yasoda A., Komatsu Y., Chusho H., Miyazawa T., Ozasa A., Miura M., Kurihara T., Rogi T., Tanaka S., Suda M., Tamura N., Ogawa Y., Nakao K. Overexpression of CNP in chondrocytes rescues achondroplasia through a MAPK-dependent pathway. Nat. Med. 2004;10:80–86. doi: 10.1038/nm971. [DOI] [PubMed] [Google Scholar]
- 15.Yasoda A., Ogawa Y., Suda M., Tamura N., Mori K., Sakuma Y., Chusho H., Shiota K., Tanaka K., Nakao K. Natriuretic peptide regulation of endochondral ossification. Evidence for possible roles of the C-type natriuretic peptide/guanylyl cyclase-B pathway. J. Biol. Chem. 1998;273:11695–11700. doi: 10.1074/jbc.273.19.11695. [DOI] [PubMed] [Google Scholar]
- 16.Bocciardi R., Giorda R., Buttgereit J., Gimelli S., Divizia M.T., Beri S., Garofalo S., Tavella S., Lerone M., Zuffardi O., et al. Overexpression of the C-type natriuretic peptide (CNP) is associated with overgrowth and bone anomalies in an individual with balanced t(2;7) translocation. Hum. Mutat. 2007;28:724–731. doi: 10.1002/humu.20511. [DOI] [PubMed] [Google Scholar]
- 17.Chikuda H., Kugimiya F., Hoshi K., Ikeda T., Ogasawara T., Shimoaka T., Kawano H., Kamekura S., Tsuchida A., Yokoi N., et al. Cyclic GMP-dependent protein kinase II is a molecular switch from proliferation to hypertrophic differentiation of chondrocytes. Genes Dev. 2004;18:2418–2429. doi: 10.1101/gad.1224204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfeifer A., Aszodi A., Seidler U., Ruth P., Hofmann F., Fassler R. Intestinal secretory defects and dwarfism in mice lacking cGMP-dependent protein kinase II. Science. 1996;274,:2082–2086. doi: 10.1126/science.274.5295.2082. [DOI] [PubMed] [Google Scholar]
- 19.Rehemudula D., Nakayama T., Soma M., Takahashi Y., Uwabo J., Sato M., Izumi Y., Kanmatsuse K., Ozawa Y. Structure of the type B human natriuretic peptide receptor gene and association of a novel microsatellite polymorphism with essential hypertension. Circ. Res. 1999;84:605–610. doi: 10.1161/01.res.84.5.605. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y., Bellamy W.P., Seabra M.C., Field M.C., Ali B.R. ER-associated protein degradation is a common mechanism underpinning numerous monogenic diseases including Robinow syndrome. Hum. Mol. Genet. 2005;14:2559–2569. doi: 10.1093/hmg/ddi259. [DOI] [PubMed] [Google Scholar]
- 21.McCracken A.A., Brodsky J.L. Evolving questions and paradigm shifts in endoplasmic-reticulum-associated degradation (ERAD) Bioessays. 2003;25:868–877. doi: 10.1002/bies.10320. [DOI] [PubMed] [Google Scholar]
- 22.McNicoll N., Gagnon J., Rondeau J.J., Ong H., De Lean A. Localization by photoaffinity labeling of natriuretic peptide receptor-A binding domain. Biochemistry. 1996;35:12950–12956. doi: 10.1021/bi960818q. [DOI] [PubMed] [Google Scholar]
- 23.Lukacs G.L., Mohamed A., Kartner N., Chang X.B., Riordan J.R., Grinstein S. Conformational maturation of CFTR but not its mutant counterpart (delta F508) occurs in the endoplasmic reticulum and requires ATP. Embo. J. 1994;13:6076–6086. doi: 10.1002/j.1460-2075.1994.tb06954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ward C.L., Kopito R.R. Intracellular turnover of cystic fibrosis transmembrane conductance regulator. Inefficient processing and rapid degradation of wild-type and mutant proteins. J. Biol. Chem. 1994;269:25710–25718. [PubMed] [Google Scholar]
- 25.Jensen T.J., Loo M.A., Pind S., Williams D.B., Goldberg A.L., Riordan J.R. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- 26.Ward C.L., Omura S., Kopito R.R. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 27.Ali B.R., Jeffery S., Patel N., Tinworth L.E., Meguid N., Patton M.A., Afzal A.R. Novel Robinow syndrome causing mutations in the proximal region of the frizzled-like domain of ROR2 are retained in the endoplasmic reticulum. Hum. Genet. 2007;122:389–395. doi: 10.1007/s00439-007-0409-0. [DOI] [PubMed] [Google Scholar]
- 28.Cohen F.E., Kelly J.W. Therapeutic approaches to protein-misfolding diseases. Nature. 2003;426:905–399. doi: 10.1038/nature02265. [DOI] [PubMed] [Google Scholar]
- 29.Romisch K. A cure for traffic jams: small molecule chaperones in the endoplasmic reticulum. Traffic. 2004;5:815–820. doi: 10.1111/j.1600-0854.2004.00231.x. [DOI] [PubMed] [Google Scholar]
- 30.Langenickel T.H., Pagel I., Buttgereit J., Tenner K., Lindner M., Dietz R., Willenbrock R., Bader M. Rat corin gene: molecular cloning and reduced expression in experimental heart failure. Am. J. Physiol. Heart Circ. Physiol. 2004;287:H1516–1521. doi: 10.1152/ajpheart.00947.2003. [DOI] [PubMed] [Google Scholar]