Abstract
Current endeavour focuses on human genetic factors that contribute to susceptibility to or protection from tuberculosis (TB). Monocytes are crucial in containing Mycobacterium tuberculosis infection, and the monocyte chemoattractant protein-1 (MCP-1) cytokine plays a role in their recruitment to the site of infection. The G allele of the MCP-1 promoter polymorphism at position −2581 relative to the ATG transcription start codon has been described to be associated in Mexican and Korean TB patients with increased susceptibility to TB. We genotyped this and additional MCP-1 variants in sample collections comprising more than 2000 cases with pulmonary TB and more than 2300 healthy controls and 332 affected nuclear families from Ghana, West Africa, and more than 1400 TB patients and more than 1500 controls from Russia. In striking contrast to previous reports, MCP-1 −2581G was significantly associated with resistance to TB in cases versus controls [odds ratio (OR) 0.81, corrected P-value (Pcorr) = 0.0012] and nuclear families (OR 0.72, Pcorr = 0.04) and not with disease susceptibility, whereas in the Russian sample no evidence of association was found (P = 0.86). Our and other results do not support an association of MCP-1 −2581 with TB. In the Ghanaian population, eight additional MCP-1 polymorphisms were genotyped. MCP-1 −362C was associated with resistance to TB in the case–control collection (OR 0.83, Pcorr = 0.00017) and in the affected families (OR 0.7, Pcorr = 0.004). Linkage disequilibrium (LD) and logistic regression analyses indicate that, in Ghanaians, the effect results exclusively from the MCP-1 −362 variant, whereas the effect of −2581 may in part be explained by its LD with −362.
INTRODUCTION
Increasing evidence indicates that susceptibility to tuberculosis (TB) in humans largely depends on the individual genetic architecture (1). Among the genetic variants implicated in studies to determine their impact on TB susceptibility, no major risk factor has been identified so far. Observed differences in genotype frequencies between TB cases and controls were often small and have resulted in either weak associations or failure of confirmation in other studies and different ethnicities.
Linkage of TB susceptibility with the 17q chromosomal region (2,3) where monocyte chemoattractant protein-1 (MCP-1) clusters together with CCL7, CCL11, NOS2A, CCL3-5 and CCR7 has previously been observed. It was also reported that a promoter variant of the gene encoding MCP-1 [syn. CCL2 (MIM +158105; 17q11.2–q12)] is associated with increased susceptibility to clinical TB in Mexicans and Koreans (4). The risk to develop pulmonary TB of both Mexican and Korean carriers of the MCP-1 allele G at position −2581 relative to the ATG transcription start codon (rs1024611, alias −2518; http://snpper.chip.org/bio/view-snpset/moao587463/T) was significantly higher compared with individuals carrying the A allele. However, no association of MCP-1 −2581G has been found in the Brazilian multicase families (3) and in a study group of Chinese TB patients from Hong Kong (5).
MCP-1 belongs to the CC chemokines (two adjacent cysteine residues close to the amino terminus) which trigger chemokine receptors on monocytes and other immune cells. The receptor for MCP-1 is the chemokine, CC motif, receptor 2 (monocyte chemotactic protein 1 receptor; CCR2A), a receptor that serves in its isomeric form (CCR2B) in the recognition of both MCP-1 and MCP-3. Different cell types are able to produce MCP-1, whereby it is not clear whether MCP-1 is mainly secreted by infected cells or by uninfected cells in response to inflammatory cytokines or to microbial components (6). Expression of MCP-1 may be found in many disorders characterized by mononuclear cell infiltration. Upon infection with Mycobacterium tuberculosis, MCP-1 is preferentially produced by CD14+ blood monocytes, but also by A549 alveolar epithelial cells (2,7). Microbial components are able to stimulate secretion of cytokines and expression of the CCR2 receptor and to induce, dependent on the presence of MCP-1, migration of monocytes to the site of infection.
The mode of action of MCP-1 in migration and attraction of monocytes is not completely understood. MCP-1 could be released directly from infected cells and, by forming a chemical gradient, escort sensitive monocytes to the site of tissue injury. High and balanced levels of serum MCP-1 could, however, abrogate effective recruitment of monocytes to sites of lesions by depletion of the gradient. An alternative scenario is that chemokines bind to distinct glycosaminoglycans in the bone marrow and pilot monocytes from there via the bloodstream to affected tissues (6). Recent results from studies of genetically modified mice have demonstrated effects of both MCP-1 and CCR2 on the development of T helper cells, with MCP-1 preferably stimulating Th2 immune responses and CCR2 rather triggering Th1 polarization (8).
Several findings, although partly contradictory and possibly dependent on the ethnic background of the population studied, as well as the biological function of MCP-1, strongly suggest an influence of MCP-1 variation on TB susceptibility. Positive linkage of the 17q region (2) and an association of MCP-1 −2581G (rs1024611) with TB susceptibility in Mexican and Korean TB patients (4) have been observed. In contrast, no association of MCP-1 −2581 was found in Brazilian multicase families (3) and in a Chinese patient group from Hong Kong (5).
We have studied the MCP-1 −2581 and eight additional MCP-1 polymorphisms in a sample of more than 2000 sputum/culture-positive pulmonary TB patients and more than 2300 healthy control individuals from Ghana and also assessed the role of MCP-1 −2581 variation in a TB case–control collection (more than 1400 TB patients and more than 1500 controls) from Russia.
RESULTS
Ghanaian study population
A total of nine MCP-1 variants were genotyped in 2010 Ghanaian pulmonary TB cases and 2346 healthy controls. Assuming an approximative TB prevalence of 0.003 in West Africa, a frequency of 0.1 for high-risk alleles and a genotype relative risk of 1.2 (α = 0.05), a detection power of >80% was achieved for additive and multiplicative models with the Ghanaian sample. The frequencies of the nine MCP-1 variants were adjusted for gender, age and ethnicity. Genotype frequencies did not deviate from Hardy–Weinberg equilibria (HWE) beyond that expected at random (P < 0.01).
Alleles and genotypes
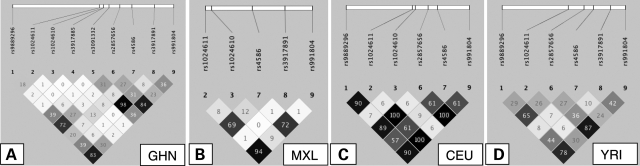
Allele and genotype frequencies of the MCP-1 variants tested in the Ghanaian sample are summarized in Tables 1 and 2 and the respective r2 values of pairwise linkage disequilibria (LDs) are given in Figure 1.
Table 1.
Allelic associations
| MCP-1 allele | Cases (n) (frequency) | Controls (n) (frequency) | OR 95% CI | Pnom | Pcorr |
|---|---|---|---|---|---|
| Ghana | |||||
| −11822G | 2537 (0.63) | 2778 (0.59) | 1 | ||
| −11822A | 1467 (0.37) | 1898 (0.41) | 0.84 (0.77–0.92) | 0.00013 | 0.001 |
| −2581A | 3256 (0.83) | 3692 (0.80) | 1 | ||
| −2581G | 672 (0.17) | 932 (0.20) | 0.81 (0.73–0.91) | 0.0002 | 0.0012 |
| −2138A | 3759 (0.96) | 4408 (0.96) | 1 | ||
| −2138T | 177 (0.04) | 192 (0.04) | 1.09 (0.89–1.35) | 0.41 | |
| −2134T | 3796 (0.96) | 4478 (0.97) | 1 | ||
| −2134G | 140 (0.04) | 122 (0.03) | 1.35 (1.05–1.73) | 0.017 | |
| −1549Aa | 3170 (0.80) | 3600 (0.79) | 1 | ||
| −1549T | 780 (0.20) | 980 (0.21) | 0.89 (0.80–0.99) | 0.038 | |
| −362G | 2266 (0.58) | 2441 (0.53) | 1 | ||
| −362C | 1670 (0.42) | 2161 (0.47) | 0.83 (0.76–0.90) | 0.000019 | 0.00017 |
| +900C | 2906 (0.74) | 3496 (0.76) | 1 | ||
| +900T | 1034 (0.26) | 1122 (0.24) | 1.12 (1.01–1.24) | 0.025 | |
| +3318Ca | 3178 (0.80) | 3621 (0.78) | 1 | ||
| +3318T | 784 (0.20) | 1003 (0.22) | 0.89 (0.80–0.98) | 0.024 | |
| +5356C | 2398 (0.61) | 2634 (0.57) | 1 | ||
| +5356T | 1556 (0.39) | 2014 (0.43) | 0.84 (0.77–0.92) | 0.00013 | 0.00093 |
| Russia | |||||
| −2581A | 2048 (0.71) | 2168 (0.71) | 1 | ||
| −2581G | 832 (0.29) | 890 (0.29) | 0.99 (0.89–1.11) | 0.86 | |
P-values are adjusted for age, gender and ethnicity. Pnom, nominal P-value; Pcorr, P-value after Bonferroni-Holm correction.
aVariants −1549 and +3318 are in almost perfect LD (r2 = 0.98).
Table 2.
Genotype associations
| MCP-1 GT | Cases (n) (frequency) | Controls (n) (frequency) | OR 95% CI | Pnom | Pcorr | OR (trend) 95% CI | Pnom | Pcorr |
|---|---|---|---|---|---|---|---|---|
| Ghana | ||||||||
| −11822GG | 817 (0.41) | 827 (0.35) | 1 | 0.85 (0.77–0.92) | 0.00015 | 0.0012 | ||
| −11822AG | 903 (0.45) | 1124 (0.48) | 0.81 (0.71–0.93) | 0.002 | 0.014 | |||
| −11822AA | 282 (0.14) | 387 (0.17) | 0.73 (0.61–0.88) | 0.001 | 0.007 | |||
| −2581AA | 1355 (0.69) | 1472 (0.64) | 1 | 0.81 (0.73–0.91) | 0.0003 | 0.0018 | ||
| −2581AG | 546 (0.28) | 748 (0.32) | 0.79 (0.69–0.90) | 0.001 | 0.006 | |||
| −2581GG | 63 (0.03) | 92 (0.04) | 0.73 (0.53–1.02) | 0.064 | ||||
| −2138AA | 1796 (0.91) | 2113 (0.92) | 1 | 1.09 (0.89–1.35) | 0.4 | |||
| −2138AT | 167 (0.08) | 182 (0.08) | 1.09 (0.87–1.36) | 0.44 | ||||
| −2138TT | 5 (<0.01) | 5 (<0.01) | 1.27 (0.36–4.41) | 0.71 | ||||
| −2134TT | 1835 (0.93) | 2178 (0.95) | 1 | 1.34 (1.05–1.72) | 0.019 | |||
| −2134TG | 126 (0.06) | 122 (0.05) | 1.22 (0.94–1.58) | 0.13 | ||||
| −2134GG | 7 (<0.01) | 0 | NA | |||||
| −1549AA | 1286 (0.65) | 1410 (0.61) | 1 | 0.89 (0.81–0.99) | 0.039 | |||
| −1549AT | 598 (0.30) | 780 (0.34) | 0.83 (0.73–0.95) | 0.006 | 0.024 | |||
| −1549TT | 91 (0.05) | 103 (0.04) | 0.97 (0.72–1.30) | 0.84 | ||||
| −362GG | 672 (0.34) | 654 (0.28) | 1 | 0.83 (0.76–0.91) | 0.000026 | 0.00023 | ||
| −362CG | 922 (0.47) | 1133 (0.49) | 0.80 (0.69–0.92) | 0.001 | 0.009 | |||
| −362CC | 374 (0.19) | 514 (0.22) | 0.70 (0.59–0.83) | 0.00005 | 0.00045 | |||
| +900CC | 1069 (0.54) | 1326 (0.57) | 1 | 1.12 (1.01–1.24) | 0.025 | |||
| +900CT | 768 (0.39) | 844 (0.37) | 1.14 (1.00–1.29) | 0.045 | ||||
| +900TT | 133 (0.07) | 139 (0.06) | 1.21 (0.94–1.56) | 0.14 | ||||
| +3318CC | 1286 (0.65) | 1412 (0.61) | 1 | 0.89 (0.80–0.98) | 0.024 | |||
| +3318CT | 606 (0.31) | 797 (0.34) | 0.83 (0.72–0.94) | 0.004 | 0.02 | |||
| +3318TT | 89 (0.04) | 103 (0.04) | 0.95 (0.71–1.28) | 0.74 | ||||
| +5356CC | 746 (0.38) | 747 (0.32) | 1 | 0.85 (0.78–0.92) | 0.00016 | 0.0011 | ||
| +5356CT | 906 (0.46) | 1140 (0.49) | 0.80 (0.70–0.92) | 0.001 | 0.008 | |||
| +5356TT | 325 (0.16) | 437 (0.19) | 0.73 (0.61–0.88) | 0.001 | 0.008 | |||
| Russia | ||||||||
| −2581AA | 726 (0.50) | 794 (0.52) | 1 | 0.99 (0.89–1.11) | 0.86 | |||
| −2581AG | 596 (0.41) | 580 (0.38) | 1.12 (0.97–1.31) | 0.13 | ||||
| −2581GG | 118 (0.08) | 155 (0.10) | 0.83 (0.64–1.08) | 0.17 | ||||
GT, genotype; OR (trend), estimates of an additive genetic model.
P-values are adjusted for age, gender and ethnicity. Pnom, nominal P-value; Pcorr, P-value after Bonferroni–Holm correction.
aVariants −1549 and +3318 are in almost perfect LD (r2 = 0.98; Fig. 1).
Figure 1.
Pairwise LD (r2) plots of MCP-1 variants of the Ghanaian study group (GHN) (A), a Mexican population from California (MXL) (B), a population of the US residents of northern and western European ancestry (CEU) (C), the Yoruba population from Nigeria (YRI) (D). MXL, CEU and YRI data are extracted from the HapMap database. Nucleotide positions of rs numbers are given in Table 6. LDs compared between the different populations consisted of the same allelic combinations.
Allele frequencies of SNPs at positions −11822, −2581, −362 and +5356 differed significantly between cases and controls with corrected P-values (Pcorr) of 1.0 × 10−3, 1.2 × 10−3, 1.7 × 10−4 and 9.3 × 10−4, respectively (Table 1). In particular, the promoter allele −2581G, which has previously been described to be associated with susceptibility to TB (4), was significantly more frequent among control individuals [17% cases versus 20% controls; odds ratio (OR) 0.81, confidence interval (CI) 0.73–0.91, nominal P-value (Pnom) = 2 × 10−4, Pcorr = 1.2 × 10−3]. The promoter variant at position −362C, which was in moderate linkage with −2581G (r2 = 0.27), was more strongly associated with protection from TB than was −2581G. With its occurrence of 42% in cases and 47% in controls, an OR of 0.83 (CI 0.76–0.90; Pnom = 1.9 × 10−5, Pcorr = 1.7 × 10−4) was achieved. Two further variants, −11822 and +5356, were identified after imputation. Genotyping of these two variants, which were in linkage of r2 = 0.72 and r2 = 0,84, respectively, with −362 revealed similar associations of −11822A and +5356T with resistance to TB.
Genotype frequencies were calculated in trend tests to compare them in an additive model (Table 2). The strongest association was found for the C variant at position −362 which occurred significantly more frequently among controls than in cases [OR (trend) 0.83, CI 0.76–0.91, Pcorr = 2.3 × 10−4), followed by the T variant at position +5356 associated with resistance (OR 0.85, CI 0.78–0.92, Pcorr = 1.1 × 10−3). MCP-1 −11822A was more frequent among controls (OR 0.85, CI 0.77–0.92, Pcorr = 1.2 × 10−3), and the A variant at position −2581 was more frequent in cases (OR 0.81, CI 0.73–0.91, Pcorr = 1.8 × 10−3). The differences that we observed were not dependent on differing mycobacterial species or genotypes or the age of patients, as, when stratifying cases for the mycobacterial species M. tuberculosis and Mycobacterium africanum or grouping cases and controls according to age (6–30, >30 years), respectively, the findings remained significant (data not shown).
Haplotypes
Haplotypes and LD were reconstructed with the UNPHASED and HaploView software, respectively (Table 3, Fig. 1). The global and adjusted (age, gender, ethnicity) P-value of haplotype comparisons (Pg) was 2.6 × 10−5. The P-value after permutation analyses to correct for multiple testing (10 000 permutations) was 1 × 10−4. MCP-1 −2581G/−362C was strongly associated with resistance to TB. Inclusion in haplotype reconstruction of the two other variants that we found associated with protection, −11822A and +5356T (Tables 1 and 2), did not affect the association (Pg = 2.4 × 10−4).
Table 3.
Haplotype associations
| Haplotype |
Cases (n) (frequency) | Controls (n) (frequency) | OR 95% CI | P-value | |
|---|---|---|---|---|---|
| −2581 | −362 | ||||
| A | G | 2214 (0.58) | 2405 (0.53) | 1.21 (1.11–1.32) | 0.000014 |
| A | C | 976 (0.25) | 1227 (0.27) | 0.92 (0.83–1.01) | 0.088 |
| G | C | 658 (0.17) | 906 (0.20) | 0.82 (0.74–0.92) | 0.0003 |
ORs and P-values refer to comparisons of one haplotype to the others combined.
Interaction analysis of MCP-1 −2581 and −362
We tested whether the effect of MCP-1 −362 was independent of that of MCP-1 −2581 in a logistic regression model. A likelihood ratio (LR) test was applied to evaluate whether a given variant improved the models as given in Table 4. Focusing on the main effects of the −362 and −2581 variants in model 3 did not refine the predictive value of the phenotype than did model 2 with the −362 variant only (model 3 versus model 2; P = 0.15). Thus, the −362 variant was verified to exclusively explain the observed association with resistance to TB, however, implying that the association of the −2581 variant was not independent of that of MCP-1 −362. To analyse synergistic effects between the −362 and −2581 variants, interaction terms were tested in logistic regression calculations. When comparing model 4 (interaction terms of −2581 and −362) with model 3 (individual variants), no improvement in the potential to predict the disease status was observed (P = 0.16).
Table 4.
Model comparisons of main effects including MCP1 −2581 and −362
| Model | Model terms | Model comparisons | LR test, P-value |
|---|---|---|---|
| 0 | |||
| 1 | MCP1 −2581 | 1 versus 0 | 0.0006 |
| 2 | MCP1 −362 | 2 versus 0 | 0.00003 |
| 3 | MCP1 −2581 + MCP1 −362 | 3 versus 2 | 0.15 |
| 4 | MCP1−2581 × MCP1 −362 | 4 versus 3 | 0.16 |
To test whether the further variants that gave significant associations, MCP-1 −11822 and +5356, were associated with the TB phenotype independently of MCP-1 −362, each variant was added in turn to the regression model. None of these variants improved the model that predicts the phenotype (data not shown).
Transmission disequilibrium test
The results of the transmission disequilibrium tests (TDT; MCP-1 −2581 and MCP-1 –362) are given in Table 5. The TDT is robust against biases caused by ethnic stratification. TDT revealed less than expected at random transmission of the −362G allele to TB patients (OR 0.7, P = 0.003), indicating the protective effect of this variant and supporting our finding in the Ghanaian case–control sample. Permutation analyses (10 000 permutations) to correct for multiple testing confirmed the results with Pcorr = 0.04 (MCP-1 −2581) and Pcorr = 0.004 (MCP-1 −362).
Table 5.
Transmission disequilibrium test
| MCP-1 allele | Cases [n (%)] (frequency) | Controls [n (%)] (frequency) | OR 95% CI | Pnom | Pcorr |
|---|---|---|---|---|---|
| −2581A | 558 (85.1) | 527 (80.3) | 1 | ||
| −2581G | 98 (14.9) | 129 (19.6) | 0.72 (0.53–0.98) | 0.04 | 0.04 |
| −362G | 400 (60.6) | 342 (51.9) | 1 | ||
| −362C | 260 (39.4) | 318 (48.1) | 0.70 (0.55–0.89) | 0.003 | 0.004 |
TDT performed with 207 complete trios (affected child and both parents) and 125 pairs of an affected child with a single parent. Pnom, nominal P-value; Pcorr, corrected P-value after 10 000 permutations.
Russian study population
The MCP-1 −2581 (rs1024611) polymorphism was genotyped in 1440 TB patients and 1529 controls from Russia. Given that we observed a minor allele frequency of 0.29, this sample provided 90% power to detect an allele OR of 1.2 at P = 0.05, assuming a multiplicative model. We did not find an association with TB susceptibility (OR 0.99, P = 0.86). Since HapMap CEU data suggested that the MCP-1 −2581 and −362 SNPs are in perfect LD in populations of European descent (r2 = 1.0, Fig. 1), we did not genotype the MCP-1 −362 variant in the Russian study group.
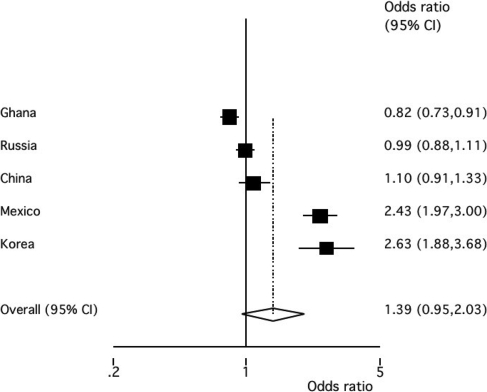
Meta-analysis of five study populations
The meta-analysis of the association between MCP-1 −2581A>G variant and susceptibility to TB in five case–control studies of five ethnicities (Ghanaians, Russians, Chinese, Mexicans, Koreans) showed differences in associations between studies as indicated by a significant test for heterogeneity (P < 0.001). As heterogeneity occurred, a random effect model had to be applied. The DerSimonian and Laird algorithm showed that MCP-1 G was not significantly associated with susceptibility to TB (pooled OR 1.39, CI 0.95–2.03, P = 0.09).
DISCUSSION
The functional significance of the MCP-1 chemokine in attracting monocytes to the site of infectious lesions and its presumed role in the pathogenesis of TB and the granulomatous response suggested that variation of the MCP-1 gene might participate in conferring susceptibility to or protection from TB. This hypothesis prompted us to study several MCP-1 polymorphisms in two case–control sample collections from Ghana and Russia, both of considerable size.
In contrast to the earlier report that suggested an association of MCP-1 −2581G with susceptibility to TB in Mexicans and Koreans (4), we found an opposite association of MCP-1 −2581G, namely with resistance to TB in the Ghanaian population and no effect at all on TB in the Russian sample. Taking into consideration our results and the fact that no association of the MCP-1 −2581 polymorphism with TB has been observed in the Brazilian and Chinese populations (3,5), we believe that, at present, the overall data do not sufficiently support any major involvement of that variant in TB susceptibility or protection. This is supported by the meta-analysis that we performed applying the random effect model of the DerSimonian and Laird algorithm (Fig. 2). The comparison of the five pooled case–control studies did not show a significant contribution of MCP-1 −2581 to TB susceptibility or to protection from disease (P = 0.09). However, as a considerable degree of heterogeneity was observed (P < 0.001), it may be assumed that variability exists in the LD patterns in the different populations that were reviewed in the meta-analysis.
Figure 2.
Meta-analysis of five case–control studies analysing the allelic association between the MCP-1 variant 2581A>G and susceptibility to TB. Black squares represent the ORs of each study/ethnicity. The pooled OR (P = 0.09) is indicated by the diamond.
We found another polymorphism, MCP-1 −362, associated with protection from TB both in the Ghanaian case–control sample and in nuclear families. As the −2581 and −362 variants exhibited a moderate LD (r2 = 0.27) in our study population, we tried to define which of the two variants caused the strongest effect and found in regression analyses that the −362CC genotype, and not the −2581 variant, exclusively explained the observed association with resistance to TB.
When reconstructing haplotypes comprising the variants −2581 and −362, none of the three haplotypes was associated with protection stronger than was the −362CC genotype alone. This was confirmed by interaction analyses indicating that, conditional on the −362 variant, no other variant improved the model predicting the phenotype. Our case–control results were corroborated by those of the modified TDT of nuclear families.
In order to compare our findings in the Ghanaian and Russian populations with previously published data, we analysed LDs between MCP-1 polymorphisms. Pairwise MCP-1 LDs of more than 4300 Ghanaian individuals participating in the present study (GHN), populations of Mexican ancestry from California (MXL), US Caucasian residents of northern and western European ancestry (CEU) and of the Nigerian Yoruba ethnicity (YRI) are depicted in Figure 1. The LD analysis provided an important insight. When comparing the linkage plot of our GHN study population to the plots of Caucasians and Mexicans (MXL) using preliminary data from the HapMap Phase 3 release, clear differences of LD patterns became evident. Comparisons of LDs in these ethnicities indicate that, in Ghanaians, the alleles at positions −2581 and −362 are in moderate LD only with each other (r2 = 0.27) as they are in Yoruba (r2 = 0.26), whereas they are in perfect LD in Caucasians (r2 = 1.0). MXL data on MCP-1 −362 are not available so far. As in all populations, except our GHN and the YRI population, for which data are available in the HapMap database, −2581 is in strong LD with +5356 (data not shown), a corresponding LD between the three variants −2581, −362 and +5356 may plausibly be inferred for the MXL population. Notably, when plotting LD patterns with all available HapMap MCP-1 variants, the resulting LD plots appear to differ among ethnicities even to a still higher degree (data not shown in Fig. 1).
The LD pattern that is shown in Figure 1A suggests that, in Ghanaians, the association of MCP-1 −362C is independent of that of −2581G, which is also evident from our logistic regression analysis. In contrast, given the perfect LD in the CEU population as evident from the HapMap data, it is reasonable to expect that the −362 and −2581 alleles will be in perfect LD in the Russian population as well, and −362 would show no association with TB susceptibility/resistance, should we genotype this variant in the Russian sample.
Several factors might explain the disparity of associations that was observed in different studies in various populations. First, our evidence of an association of −362C and resistance to TB in the Ghanaian population (Pcorr = 0.00017 in the case–control data set and Pcorr = 0.004 in the nuclear families) does not completely exclude an association by chance, and, therefore, replication in a large statistically powerful sample collection is important. Alternatively, the association of MCP-1 −362 might reflect the presence of a genuine association owing to LD with another, putatively causal polymorphism in the adjacent MCP-1 gene region. Different LD patterns between such a causal, yet unidentified variant and the −362 SNP in the Ghanaian population, on the one hand, and the Russian and other populations (Chinese, Brazilian), on the other hand, might explain the lack of association that we and others observed in non-African populations. A meta-analysis of findings of the MCP-1 −362 variant is currently not feasible, as TB association data are not available for other than the Ghanaian study population. This is in agreement with the distinct LD patterns that exist in African and non-African populations. Further analyses of other polymorphisms in the MCP-1 gene region are needed to address this question in more detail. Finally, we cannot exclude that environmental variance, including different M. tuberculosis complex genotypes occurring in different parts of the world, might influence the effects of genetic variation on susceptibility to TB. These factors might also have contributed to the discrepant results of different studies, including the original study of Mexicans and Koreans.
In conclusion, we have identified a genetic variant, MCP-1 −362C, that contributes to protection from pulmonary TB. It remains, however, to be elucidated by re-sequencing and fine mapping whether this variant is truly causal or is only part of a haplotype that comprises the causative genetic variant. This again will require meticulous analyses of LD patterns in different ethnicities. Once unquestionably identified, functional studies will help to determine in more depth the role of MCP-1 variants in their contribution to relative resistance to TB.
MATERIALS AND METHODS
Ghanaian and Russian study groups
Participants were consecutively enrolled in Ghana, West Africa, between September 2001 and July 2004 at Korle Bu Teaching Hospital in Accra, Komfo Anokye Teaching Hospital in Kumasi, plus 15 additional hospitals and polyclinics in Accra and Kumasi and at regional district hospitals. The case group consisted of 2010 HIV-negative individuals with newly diagnosed smear-/culture-positive pulmonary TB. Out of the total of 2346 control individuals, 1211 were unrelated personal contacts of cases and 1135 were community members from neighbouring houses or working contacts of cases. Cases and controls belonged to the ethnic groups of Akan (Ashanti, Fante, Akuapem), Ga-Adangbe, Ewe, all in the south of Ghana, and several other ethnic groups from northern Ghana. The proportions of ethnicities among patients and controls did not differ significantly. The male-to-female ratio in the total study group 1 was 0.58, and the mean age of participants was 33 years without gender differences.
Phenotyping of patients included the medical histories and documentation of major symptoms on structured questionnaires, physical examination, HIV-1/2 testing (Capillus, Trinity Biotech, Bray, Co Wicklow, Ireland), posterior–anterior chest X-rays, Ziehl–Neelsen staining of two independent sputum smears and culturing of M. tuberculosis on solid Loewenstein–Jensen medium with subsequent determination of mycobacterial species, fine-typing of genotypes by spoligotyping, IS6110 fingerprinting and determination of drug resistance as described previously (9–12). Cases were HIV-negative and had characteristic radiological lesions of pulmonary TB. All patients were treated in the framework of the DOTS (Directly Observed Treatment Short-Course strategy) programme organized by the Ghanaian National Tuberculosis Programme.
A number of cases that were identified during the enrolment procedure were excluded for one or several of the following reasons: history of TB or of previous antimycobacterial treatment, HIV positivity, lost to follow-up or refusal after primary enrolment, age not consistent with the age of 6–60 as specified in the study protocol, predisposition to immunosuppression (alcoholism, drug use, diabetes, overt generalized disease) and inappropriateness for matching with controls for sex and age.
Characterization of controls included a medical history and clinical examination, posterior–anterior chest X-ray and a tuberculin skin test (Tuberculin Test PPD Mérieux, bioMérieux, Nürtingen, Germany). A total of 2217 individuals were PPD-positive and 129 individuals were PPD-negative. The controls had no radiological signs of actual or past pulmonary TB and no history of specific antimycobacterial treatment. In order to verify results in transmission disequilibrium tests, genotypes of 332 affected individuals and their parents (trios) or children with single-parents were studied. The family sample was only in part independent from the case–control sample. Further details of the recruitment procedure and the composition of the study group including the distribution of ethnicities have been described previously (12–15).
The study protocol was approved by the Committee on Human Research, Publications and Ethics, School of Medical Sciences, Kwame Nkrumah University of Science and Technology, Kumasi, Ghana, and the Ethics Committee of the Ghana Health Service, Accra, Ghana. Venous blood samples were taken only after a detailed explanation of the aims of the study, and consent was obtained by signature, or thumbprint in case of illiteracy.
One of the SNPs associated with protection in the Ghanaian sample was typed in an independent sample of 1440 TB patients and 1529 controls from Russia. Russian TB patients and controls were collected at St Petersburg. Patients [mean age 43.8 (17–86), 26.2% female] had been diagnosed by the local health care service using information provided on TB contact, clinical symptoms, chest X-rays and sputum smears. Diagnoses were confirmed by sputum culture of M. tuberculosis on solid and/or liquid culture medium. Clinical information, as well as age, ethnicity, socio-economic status and the concurrent medical history were recorded on structured questionnaires. Population controls were adult blood bank donors with no history of TB [mean age 30 (16–66), 25.0% female]. Ethical clearance was obtained from local ethics committees in St Petersburg, Russia, and Cambridge, UK, and written informed consent was provided by all participating subjects. Patients with extra-pulmonary TB and HIV-positive subjects were excluded. Further details on the Russian case–control group are given in Nejentsev et al. (16).
Variants selected for genotyping; genetic analyses
The variants, rs-numbers and PCR amplification primers as well as sensor/anchor nucleotides for genotyping are summarized in Table 6. First, seven MCP-1 variants were selected for genotyping, among them five promoter, one exonic and one 3′ UTR variants. Three of the variants (rs1024611, rs4586, rs3917891) were tagging SNPs extracted from the HapMap database for the West African ethnicity of Yoruba (YRI), and four promoter variants (rs1024610, rs3917885, rs3091332, rs2857656) were chosen because of their high frequencies in Africans (http://innateimmunity.net//PGAs/InnateImmunity/CCL2/). As an additional exploratory analysis, genotypes were imputed for untyped HapMap (release 2) SNPs (YRI) of the MCP-1 region using the software PLINK 1.0.3 (http://pngu.mgh.harvard.edu/~purcell/plink/download.shtml/). After imputation, two additional variants (rs9889296, rs991804) were selected because of an increased association with the phenotype studied.
Table 6.
Variants selected for genotyping
| MCP-1 variant | rs number | Selection criterion | Primer oligonucleotides | Sensor/anchor oligonucleotides |
|---|---|---|---|---|
| −11822G/A | rs9889296 | Imputation | F-AGACTAGGCACTTAATTACTGTT | S-CTGGCATTTACTTATTCATTCATTTGCCTATTTAT |
| R-CTTTGCATTCCTTTATGACAGC | A-CTTCTTTGAATCTATCTTAGCACCTAGGACAGGG | |||
| −2581A/G | rs1024611 | HapMap | F-TTCTCTCACGCCAGCAC | S-AAAGTGACAGCTGTCTGCCTC |
| R-TGACTTGGCCTTTGCATATATC | A-CACTTCTGCTCTGTCAAGAAAAGATGCCCTCCCCC | |||
| −2138A/T | rs1024610a | Frequency | F-CAGCATCTTTCAGCTTGT | S-GGGAAACCTCTCTCTAATCAGTTAGTGC |
| R-GTGTAGGAATTTCTTCCTAGGC | A-TCCTTTACCATGAACTTGGTGGACCGCATTCAATT | |||
| −2134T/G | rs3917885a | Frequency | F-CAGCATCTTTCAGCTTGT | S-GGGAAACCTCTCTCTAATCAGTTAGTGC |
| R-GTGTAGGAATTTCTTCCTAGGC | A-TCCTTTACCATGAACTTGGTGGACCGCATTCAATT | |||
| −1549A/T | rs3091332 | Frequency | F-GCACATCTACTATTCTGTCTGAGTTA | S-AAAGTATGTCAGCACCATACCTGACTCCCTGAATG |
| R-AGAAAACACTTTTCACTACACTTG | A-ACTCAACAATGCCATTACTGACCAC | |||
| −362G/C | rs2857656 | Frequency | F-GAGCCTGACATGCTTTCATCTA | S-TTCGCTTCACAGAAAGCAGAATCCTTA |
| R-TTTCCATTCACTGCTGAGAC | A-AAATAACCCTCTTAGTTCACATCTGTGGTCAGTCT | |||
| +900C/T | rs4586 | HapMap | F-TAAGATCAGAATAATCCAGTTCATCC | S-TCACCTGCTGCTATAACTTCACCAA |
| R-GCTGGTGATTCTTCTATAGCTC | A-AGGAAGATCTCAGTGCAGAGGCTCGC | |||
| +3318C/T | rs3917891 | HapMap | F-CTGGCAAGAAGCACACT | S-ACTCGCTTTGTCAGTCAAGACAGGTCAGATATTCT |
| R-CCTCTGCAACTCCAGTTAG | A-AGCCTACATCGATCATACAGGTATGATAAT | |||
| +5356C/T | rs991804 | Imputation | F-GCTCCTCTTCCCATTGC | S-CAGCCAGTCCTGGTAACCTCTG |
| R-GTCAGAACAAAGGACACTATGAAA | A-TCCTCAGTTCTCTCATATTCAGGTCATTGGAGCCA |
Primer pairs and sensor/anchor oligonucleotides for LightTyper-based MCP-1 genotyping.
F, forward primer; R, reverse primer; S, sensor; A, anchor; Imputation, SNPs selected owing to an increased association with the phenotype after imputation of genotypes of so far untyped SNPs; HapMap, tagging SNPs from the HapMap database of the Yoruba ethnicity; Frequency, SNPs chosen according to high differences in frequencies in Caucasian and Asian ethnicities.
aDetected by the same PCR and genotyping assay.
DNA was extracted from whole blood according to standard methods. Genotypes of MCP-1 variants were determined by dynamic allele-specific hybridization with fluorescence resonance energy transfer (LightTyper®; Roche Diagnostics, Mannheim, Germany). The sequences of sensor and anchor nucleotides of LightTyper-based genotyping are given in Table 6.
Databases and statistical analyses
Demographic data, self-reported signs and symptoms and primary laboratory results were double-entered into a 4th Dimension database (San José, CA, USA). Microbiological data were provided as datasheets. All data were locked before using them in a pseudonymized form for analyses.
Power calculation was performed with the public CATS software (http://www.sph.umich.edu/csg/abecasis/CaTS/). The STATA 9 software (Stata Corporation, College Station, TX, USA) was used to calculate HWE and ORs of MCP-1 allele and genotype frequencies. Corrections of P-values were performed according to the Bonferroni–Holm procedure (17). Notably, MCP-1 −1549 (rs3091332) and +3318 (rs3917891) were in almost perfect linkage (r2 = 0.98; Fig. 1).
Logistic regression analyses (STATA 9) were applied to adjust for gender, age and ethnicity and to calculate SNP–SNP effects. Haplotype frequencies and OR with global and adjusted P-values (10 000 permutations) were estimated and compared with the public UNPHASED software (version 3.0.12; http://www.mrc-bsu.cam.ac.uk/personal/frank/software/unphased/). LDs were calculated and graphically displayed with the Haploview 4.1 software (www.broad.mit.edu/mpg/haploview/). A logistic regression model was applied in an SNP–SNP effect and interaction analysis to test whether the effect of MCP-1 −362 was independent of that of MCP-1 −2581. Different models of single variants and the combination of two variants were compared with an LR test. Modified TDT with the implementation of maximum likelihood analyses were calculated to avoid biases caused by ethnic stratification (UNPHASED software). Genotypes of 207 full trios (affected child plus both unaffected parents) and 125 parent–child pairs (affected child plus a single unaffected parent, mostly the mother) were included in the TDT.
A meta-analysis of results available for the MCP-1 −2581 genotype, including the present study of Ghanaians and Russians as well as the case–control studies of Mexican and Korean individuals (4) and of Hong Kong Chinese TB patients and controls (5), was conducted (STATA 9). The test of heterogeneity between the studies was assessed by a χ2 statistic. The DerSimonian and Laird random effect model was applied estimating the pooled OR and CI.
FUNDING
This work was supported by the German Federal Ministry of Education and Research, German National Genome Research Network (NGFN1, Project 01GS0162; NGFN2, NIE-S17T20). The Diabetes and Inflammation Laboratory is funded by the Juvenile Diabetes Research Foundation and the Wellcome Trust. S.N. holds the Royal Society University Research Fellowship.
ACKNOWLEDGEMENTS
The authors are indebted to all participating individuals in Ghana and Russia. We thank all field workers, nurses and physicians involved in the recruitment of patients and controls and gratefully acknowledge the excellent technical assistance of Lincoln Gankpala, Emmanuel Abbeyquaye and Gerd Ruge and appreciate the logistics provided by the staff of the Kumasi Centre for Collaborative Research in Tropical Medicine (KCCR), Kumasi, Ghana. We thank Professor John A. Todd for his help and advice in establishing the Russian TB sample collection and Jeffrey Szeszko for DNA extraction.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Takiff H.E. Host genetics and susceptibility. In: Palomino J.C., Leão S.C., Ritacco V., editors. Tuberculosis 2007. From Basic Science to Patient Care. 2007. pp. 207–262. Available at www.tuberculosistextbook.com/ [Google Scholar]
- 2.Blackwell J.M., Black G.F., Peacock C.S., Miller E.N., Sibthorpe D., Gnananandha D., Shaw J.J., Silveira F., Lins-Lainson Z., Ramos F., et al. Immunogenetics of leishmanial and mycobacterial infections: the Belem Family Study. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 1997;352:1331–1345. doi: 10.1098/rstb.1997.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jamieson S.E., Miller E.N., Black G.F., Peacock C.S., Cordell H.J., Howson J.M., Shaw M.A., Burgner D., Xu W., Lins-Lainson Z., et al. Evidence for a cluster of genes on chromosome 17q11–q21 controlling susceptibility to tuberculosis and leprosy in Brazilians. Genes Immun. 2004;5:46–57. doi: 10.1038/sj.gene.6364029. [DOI] [PubMed] [Google Scholar]
- 4.Flores-Villanueva P.O., Ruiz-Morales J.A., Song C.H., Flores L.M., Jo E.K., Montaño M., Barnes P.F., Selman M., Granados J. A functional promoter polymorphism in monocyte chemoattractant protein-1 is associated with increased susceptibility to pulmonary tuberculosis. J. Exp. Med. 2005;202:1649–1658. doi: 10.1084/jem.20050126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu S.F., Tam C.M., Wong H.S., Kam K.M., Lau Y.L., Chiang A.K. Association between RANTES functional polymorphisms and tuberculosis in Hong Kong Chinese. Genes Immun. 2007;8:475–479. doi: 10.1038/sj.gene.6364412. [DOI] [PubMed] [Google Scholar]
- 6.Serbina N.V., Jia T., Hohl T.M., Pamer E.G. Monocyte-mediated defense against microbial pathogens. Annu. Rev. Immunol. 2008;26:421–452. doi: 10.1146/annurev.immunol.26.021607.090326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin Y., Gong J., Zhang M., Xue W., Barnes P.F. Production of monocyte chemoattractant protein1 in tuberculosis patients. Infect. Immun. 1998;66:2319–2322. doi: 10.1128/iai.66.5.2319-2322.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu L., Tseng S., Horner R.M., Tam C., Loda M., Rollins B.J. Control of TH2 polarization by the chemokine monocyte chemoattractant protein-1. Nature. 2000;404:407–411. doi: 10.1038/35006097. [DOI] [PubMed] [Google Scholar]
- 9.Kamerbeek J., Schouls L., Kolk A., van Agterveld M., van Soolingen D., Kuijper S., Bunschoten A., Molhuizen H., Shaw R., Goyal M., van Embden J. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Embden J.D., Cave M.D., Crawford J.T., Dale J.W., Eisenach K.D., Gicquel B., Hermans P., Martin C., McAdam R., Shinnick T.M., et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niemann S., Kubica T., Bange F.C., Adjei O., Browne E.N., Chinbuah M.A., Diel R., Gyapong J., Horstmann R.D., Joloba M.L., et al. The species Mycobacterium africanum in the light of new molecular markers. J. Clin. Microbiol. 2004;42:3958–3962. doi: 10.1128/JCM.42.9.3958-3962.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Owusu-Dabo E., Adjei O., Meyer C.G., Horstmann R.D., Enimil A., Kruppa T.F., Bonsu F., Browne E.N., Chinbuah M.A., Osei I., et al. Mycobacterium tuberculosis drug resistance, Ghana. Emerg. Infect. Dis. 2006;12:1171–1172. doi: 10.3201/eid1207.051028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thye T., Browne E.N., Chinbuah M.A., Gyapong J., Osei I., Owusu-Dabo E., Niemann S., Rüsch-Gerdes S., Horstmann R.D., Meyer C.G. No associations of human pulmonary tuberculosis with Sp110 variants. J. Med. Genet. 2006;43:e32. doi: 10.1136/jmg.2005.037960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herb F., Thye T., Niemann S., Browne E.N., Chinbuah M.A., Gyapong J., Osei I., Owusu-Dabo E., Werz O., Rüsch-Gerdes S., et al. ALOX5 variants associated with susceptibility to human pulmonary tuberculosis. Hum. Mol. Genet. 2008;17:1052–1060. doi: 10.1093/hmg/ddm378. [DOI] [PubMed] [Google Scholar]
- 15.Meyer C.G., Scarisbrick G., Niemann S., Browne E.N.L., Chinbuah M.A., Gyapong J., Osei I., Owusu-Dabo E., Kubica T., Rüsch-Gerdes S., et al. Pulmonary tuberculosis: virulence of Mycobacterium africanum and relevance in HIV co-infection. Tuberculosis. 2008;88:482–489. doi: 10.1016/j.tube.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Nejentsev S., Thye T., Szeszko J.S., Stevens H., Balabanova Y., Chinbuah A.M., Hibberd M., van de Vosse E., Alisjahbana B., van Crevel R., et al. Analysis of association of the TIRAP (MAL) S180L variant and tuberculosis in three populations. Nat. Genet. 2008;40:261–262. doi: 10.1038/ng0308-261. [DOI] [PubMed] [Google Scholar]
- 17.Holm S. A simple sequentially rejective Bonferroni test procedure. Scand. J. Stat. 1979;6:65–70. [Google Scholar]