Abstract
Mutations in GIGYF2 have recently been described as causative of Parkinson's disease in Europeans. In an attempt to replicate these results in independent populations, we sequenced the entire coding region of GIGYF2 in a large series of Portuguese and North American samples. We report the finding of two of the previously published mutations in neurologically normal Control individuals. This suggests that mutations in GIGYF2 are not strongly related to the development of the disease in either of these populations.
INTRODUCTION
Parkinson's disease [PD (MIM 168600)], the second most common form of neurodegeneration, is characterized by loss of dopaminergic neurons in the nigro-striatal pathway, leading to bradykinesia, resting tremor, muscular rigidity and postural instability (1).
The genetic factors underlying PD have been sought with considerable success in the past decade. Thus far, family-based studies, whole-genome linkage scans and candidate-gene analyses have yielded 14 chromosomal loci (PARK1–PARK14) in which mutations in seven genes are unequivocally linked to rare forms of PD (SNCA, PARK1 OMIM #168601 and PARK4, PRKN, PARK2 OMIM #602544; PINK1, PARK6 OMIM #605909; DJ-1, PARK7 OMIM #606324; LRRK2, ATP13A2, PARK9 OMIM #610513 and PLA2G6, PARK14 OMIM #603604) (2–8). Although the mechanism through which mutations in these genes exert their pathogenicity is not fully understood, SNCA and LRRK2 are known to cause autosomal-dominant disease, while the remaining cause autosomal-recessive PD. More controversial results have been obtained for loci such as PARK5 (9) and PARK13 (10,11).
PARK11 is located in chromosome 2q36–37 and was initially described by a whole-genome linkage analysis in a population of familial PD patients (12–14). Although conflicting results followed shortly (15), it was also detected in an earlier association analysis (16). The PARK11 locus spans 18cM encompassing 73 candidate genes (12–14) where the highest LOD score was obtained for the marker D2S206, located within intron 21 of GIGYF2, a gene encoding a 1320 amino acid protein (Grb10-Interacting GYF Protein 2, gigyf2).
Because of this and because gigyf2 has been shown to interact with grb10 and consequently have a potential role in insulin and insulin-like growth factor signaling (17), Lautier et al. (18) recently performed a screening of pathogenic mutations in GIGYF2 in a series of 249 familial PD Cases and 237 Controls from two different populations in Europe. The authors reported 10 different mutations spread in an even manner throughout the gene in PD patients but not in Controls, suggesting that these variants would be the cause of the disease in these patients.
These results prompted us to undertake a complete screening of GIGYF2 mutations in a large series of 724 Cases and 911 neurologically normal Controls from two different populations.
RESULTS
Portuguese series
The Portuguese cohort yielded 31 variants (Table 2), of these, seven were known polymorphisms present in dbSNP. Four non-synonymous variants were found only in Cases, while 16 non-synonymous variants were present only in Controls (Table 2). Of the remaining 24 variants not present in dbSNP, six were synonymous changes, suggesting no functional change for the protein. None of the variants present in both Cases and Controls showed association with disease. Although the variants rs12328151 and rs2289912 showed a P-value <0.05 after χ2, they failed to maintain the association after Bonferroni correction (P > 0.7).
Table 2.
Variants found in PD Cases and Controls from the Portuguese series
| Exon | Variant cDNA | Protein | Frequency affected (%) | Frequency unaffected (%) | CHISQ | P-value | OR | 95% CI |
|---|---|---|---|---|---|---|---|---|
| IVS3 | rs11555646 | – | 0.322 | 0.322 | 3.84E−05 | 0.9951 | 1.001 | 0.7928–1.263 |
| 5 | c.C129T | p.Y43Y | 0 | 0.001 | – | – | – | – |
| 10 | c.G566A | p.P189L | 0 | 0.001 | – | – | – | – |
| 12 | c.C924T | p.P308P | 0 | 0.001 | – | – | – | – |
| 13 | c.A1113T | p.D371E | 0.004 | 0 | – | – | – | – |
| 15 | c.C1397A | p.A466D | 0 | 0.001 | – | – | – | – |
| 15 | c.C1418T | p.P473L | 0 | 0.001 | – | – | – | – |
| 15 | c.T1433Ga | p.N478Ta | 0 | 0.001 | – | – | – | – |
| 15 | rs2289912 | p.P481T | 0.007 | 0.021 | 3.921 | 0.04768 | 0.3507 | 0.1187–1.037 |
| 15 | c.T1444A | p.T482S | 0.002 | 0 | – | – | – | – |
| 16 | rs2305138 | p.E539E | 0.051 | 0.068 | 1.698 | 0.1925 | 0.7342 | 0.4606–1.17 |
| 17 | c.G1779T | p.A593A | 0.009 | 0.009 | 0.009129 | 0.9239 | 1.056 | 0.3438–3.245 |
| 23 | c.C2470A | p.A824S | 0.002 | 0 | – | – | – | – |
| 25 | c.G2898T | p.S966S | 0.004 | 0 | – | – | – | – |
| 26 | rs3816334 | p.Q1001Q | 0.318 | 0.32 | 0.01092 | 0.9168 | 0.9877 | 0.7826–1.247 |
| 26 | c.3004insCAGCAG | p.Q1002_Q1003insQQ | 0 | 0.001 | – | – | – | – |
| 26 | c.3127insCAG | p.Q1042_Q1043insQ | 0 | 0.001 | – | – | – | – |
| 26 | c.C3059T | p.T1020M | 0 | 0.001 | – | – | – | – |
| 28 | c.A3410T | p.E1137V | 0 | 0.001 | – | – | – | – |
| IVS28 | rs2305137 | – | 0.33 | 0.333 | 0.0212 | 0.8842 | 0.9832 | 0.7832–1.234 |
| 29 | c.A3575G | p.H1192R | 0 | 0.002 | – | – | – | – |
| 29 | c.3693–3695del3 | p.Q1232del | 0.395 | 0.407 | 0.1855 | 0.6667 | 0.9531 | 0.766–1.186 |
| 29 | 3693–3694insCAGCAG | p.P1231_Q1232insQQ | 0.006 | 0.009 | 0.4711 | 0.4925 | 0.6299 | 0.1664–2.385 |
| 29 | c.3689insAGC | p.L1230_P1231insS | 0 | 0.001 | – | – | – | – |
| 29 | c.3689–3709del21 | p.L1230_Q1236del | 0.047 | 0.068 | 2.612 | 0.1061 | 0.6755 | 0.4187–1.09 |
| 29 | 3689–3708del24a | p.L1230_Q1237dela | 0.006 | 0.01 | 0.7755 | 0.3785 | 0.5593 | 0.1508–2.075 |
| 29 | rs12328151 | p.P1238P | 0.29 | 0.348 | 5.043 | 0.02473 | 0.767 | 0.6083–0.9671 |
| 30 | c.A3812G | p.N1271S | 0 | 0.002 | – | – | – | – |
| 30 | c.A3816G | p.Q1272Q | 0 | 0.001 | – | – | – | – |
| 30 | c.G3837A | p.V1279V | 0 | 0.001 | – | – | – | – |
| 31 | rs34424361 | p.S1306S | 0 | 0.002 | – | – | – | – |
χ2 values, P-values, odds ratios and 95% confidence interval (CI) were calculated for each variant.
aThese variants were described as pathogenic by Lautier et al.
Interestingly, one of the variants present only in the Control group (p.N478T) had previously been reported as a pathogenic mutation (18). In addition, the exon 29 p.L1230_Q1237del, which had also been suggested to be disease-causing, is present in this population with an increased frequency in the Control group.
We failed to find any of the remaining 8 variants previously associated with disease in our cohort.
US series
The US series harbored 40 different variants (Table 3) including seven described SNPs in exons 15, 16, 26, 29 and 31; and introns 3 and 28. None of these polymorphisms showed association with PD after χ2 test of association. Although rs2305138 showed a P-value of 0.04, this is not considered significant after multiple-test correction.
Table 3.
Variants found in PD Cases and Controls from the US series
| Exon | Variant | Minor allele | Frequency affected (%) | Frequency unaffected (%) | CHISQ | P-value | OR | 95% CI | |
|---|---|---|---|---|---|---|---|---|---|
| cDNA | Protein | ||||||||
| IVS3 | rs11555646 | – | C | 33.62 | 30.41 | 1.829 | 0.1763 | 1.16 | 0.9356–1.437 |
| 7 | c.322G>C | p.G108R | C | 0.1202 | 0.2315 | 0.2973 | 0.5856 | 0.5187 | 0.04694–5.731 |
| 12 | c.894G>C | p.G298G | C | 0.1351 | 0 | – | – | – | – |
| 14 | c.1183T>A | p.S395T | A | 0 | 0.1471 | – | – | – | – |
| 15 | c.1358A>G | p.D453G | G | 0.1241 | 0 | – | – | – | – |
| 15 | c.1380G>C | p.Q460Q | C | 0.1241 | 0 | – | – | – | – |
| 15 | rs2289912 | p.P481T | A | 1.489 | 1.942 | 0.4951 | 0.4817 | 0.7632 | 0.3588–1.624 |
| 15 | c.1467A>G | p.P489P | G | 0.1241 | 0 | – | – | – | – |
| 16 | rs2305138 | p.E539E | A | 4.4 | 6.824 | 4.186 | 0.04075 | 0.6284 | 0.4013–0.984 |
| 16 | c.1620C>T | p.H540H | T | 0 | 0.1312 | – | – | – | – |
| 17 | c.1757T>C | p.M586T | C | 0.1285 | 0 | – | – | – | – |
| 17 | c.1779G>T | p.A593A | T | 0.3856 | 0.5714 | 0.2699 | 0.6034 | 0.6735 | 0.1502–3.02 |
| 17 | c.1793C>T | p.F598F | T | 0.1285 | 0 | – | – | – | – |
| 17 | c.1795C>T | p.Q599K | T | 0.1285 | 0 | – | – | – | – |
| 18 | c.1935C>T | p.H645H | T | 0 | 0.1126 | – | – | – | – |
| 21 | c.2186G>A | p.S729G | A | 0.1229 | 0 | – | – | – | – |
| 21 | c.2268A>G | p.A756A | G | 0 | 0.1232 | – | – | – | – |
| 22 | c.2332A>G | p.R778G | G | 0.2445 | 0 | – | – | – | – |
| 24 | c.2683A>G | p.R895G | G | 0.2551 | 0 | – | – | – | – |
| 25 | c.2898G>T | p.S966S | T | 0.2924 | 0.7576 | 1.473 | 0.2248 | 0.3842 | 0.07728–1.91 |
| 25 | c.2907A>G | p.E969E | G | 0.1462 | 0 | – | – | – | – |
| 25 | c.2945G>A | p.R982Q | A | 0 | 0.1263 | – | – | – | – |
| 26 | c.2985T>C | p.A995A | C | 0 | 0.2375 | – | – | – | – |
| 26 | rs3816334 | p.Q1001Q | A | 31.16 | 31.47 | 0.01905 | 0.8902 | 0.9855 | 0.8014–1.212 |
| 26 | c.3045A>G | p.K1015K | G | 0 | 0.2375 | – | – | – | – |
| 27 | c.3167C>G | p.S1056C | G | 0.2421 | 0 | – | – | – | – |
| 27 | c.3174G>A | p.L1058L | A | 0 | 0.1205 | – | – | – | – |
| IVS28 | rs2305137 | – | G | 35.99 | 33.46 | 1.113 | 0.2915 | 1.118 | 0.9084–1.377 |
| 29 | c.3575A>G | p.H1192R | G | 0.5195 | 0.1238 | 1.955 | 0.1621 | 4.214 | 0.47–37.79 |
| 29 | c.3693–3695del3 | p.Q1232del | W | 47.53 | 50 | 0.9608 | 0.327 | 0.9059 | 0.7435–1.104 |
| 29 | rs12328151 | p.P1238P | A | 29.15 | 30.74 | 0.4796 | 0.4886 | 0.9267 | 0.7472–1.149 |
| 29 | c.3698–3709del21 | p.L1230_Q1236del | M | 5.829 | 5.693 | 0.01344 | 0.9077 | 1.025 | 0.6715–1.566 |
| 29 | c.3693_3694insCAGCAG | p.P1231_Q1232insQQ | M | 0.3886 | 0.1238 | 1.097 | 0.295 | 3.148 | 0.3268–30.33 |
| 29 | c.3689–3712del24a | p.L1230_Q1237dela | M | 1.036 | 0.6188 | 0.8431 | 0.3585 | 1.682 | 0.5477–5.163 |
| 29 | c.3711_3712insCAG | p.Q1237_P1238insQ | M | 0 | 0.1188 | – | – | – | – |
| 30 | c.3806A>G | p.S1269G | G | 0 | 0.1171 | – | – | – | – |
| 30 | c.3808A>G | p.N1270D | G | 0.2564 | 0 | – | – | – | – |
| 30 | c.3814C>G | p.Q1272E | G | 0 | 0.1404 | – | – | – | – |
| 31 | rs34424361 | p.S1306S | A | 1.285 | 1.991 | 1.321 | 0.2503 | 0.6409 | 0.2984–1.377 |
| 31 | c.3940A>G | p.I1314V | G | 0.1168 | 0 | – | – | – | – |
Minor allele, frequency of the minor allele in Cases and Controls, χ2 values, P-values, odds ratios and 95% confidence interval (CI) were calculated for each variant.
aThis variant was described as pathogenic by Lautier et al.
Interestingly, two of the variants Lautier et al. described as mutations in their series were found in this cohort in both Cases and Controls. These variants (c.3689-3712del24 and p.H1192R) were not associated with PD in our cohort after χ2 test for association (P = 0.385 and 0.1621, respectively). In addition, we have found 26 novel variants of which 16 were non-synonymous. Of these, three (p.G108R, p.L1230_Q1236del and p.P1231_Q1232insQQ) were found in both Cases and Controls but were not associated with disease (P = 0.58, 0.90 and 0.29, respectively). The remaining variants were found in either a single Control sample (p.S395T, p.R982Q, p.S1269G and p.Q1272E) or a single PD Case (p.D453G, p.M586T, p.Q599K, p.S729G, p.R778G, p.R895G, p.S1056C, p.N1270D and p.I1314V) (Table 3).
Except for p.A995A and p.K1015K which were found in homozygous state in a single Control sample, the rest of silent changes were found in heterozygous state in only one sample.
DISCUSSION
We present herein a detailed analysis of the genetic variability in the GIGYF2 gene and its association with PD in two large sets of Cases and age-matched Controls, from two geographically distinct populations. The sample originating from Portugal comprised a total of 267 PD samples and 451 healthy age-matched Controls, while the US series comprised 460 Cases and 460 Controls. A significant difference for the previously published study on GIGYF2 variants and PD is the fact that we used a different transcript—isoform ‘a' (NM_001103147.1)—whereas the transcript studied before was isoform ‘b' (NM_015575.3), which lacks one exon when compared with the former. Due to this difference, the present mutation numbering differs from the work previously published.
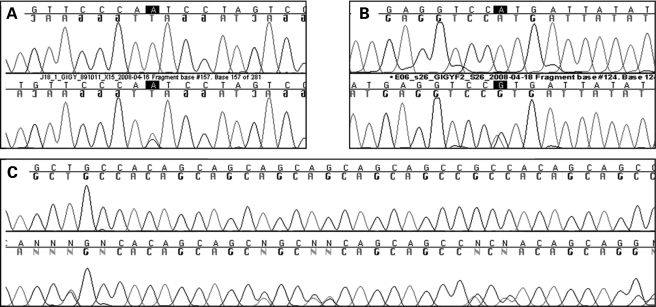
The combined analysis of both cohorts yielded 46 variants of which seven are SNPs already present in dbSNP. Most of the remaining variants were present in both Case and Control groups and, additionally, none was shown to be statistically associated with PD. Lautier et al. presented a list of 10 mutations that their results suggested to be pathogenic, and an additional mutation present in one Control individual. Although we failed to find all these mutations in our combined series, we did find three of them: p.L1230_Q1237del (described as Del LPQQQQQQ 1209–1216 by Lautier et al.), p.N478T (first described as Asn457Thr) and p.H1992R (described as His1171Arg). Although p.L1230_Q1237del was found in a similar number of Cases and Controls in both populations (P = 0.37 and 0.84 in the Portuguese and US series respectively), p.N487T was only found in one Portuguese Control sample, and p.H1192R, identified by Lautier et al. in one Control individual only; it was present in both Cases and Controls in our US series, suggesting it to be a benign variant (Fig. 1).
Figure 1.
GIGIF2 variants described originally as mutations by Lautier et al. and found both in Control and PD samples in our series. The upper sequence in each of the panels (A–C) shows the wild-type sequence; while the lower sequence shows the mutation in heterozygous state. (A) p.N478T variant found in a Control sample from the Portuguese series. (B) p.H1192R variant found in both Cases and Controls from the US series. (C) p.L1230_Q1237del variant found in both Cases and Controls from the US and Portuguese series.
Interestingly we have found 37 new single nucleotide variants in our cohorts of which 26 are non-synonymous, and five deletion or insertion mutations in both PD and Control groups (Tables 2 and 3); this high number is probably reflective of the repeat rich nature of exons 26 and 29.
In order to test if there was an enrichment of rare non-synonymous mutations in Cases when compared with Controls, we compared the collective frequency of non-synonymous alterations that were identified only in Cases, with the collective frequency of non-synonymous changes found only in Controls; a 2 × 2 Fischer exact test of association showed no statistically significant difference (Table 4).
Table 4.
Comparison of the collective frequency of non-synonymous alterations that were identified only in Cases, with the collective frequency of non-synonymous changes found only in Controls (a Fischer exact test of association was performed)
| Presenting non-synonymous variants | Without non-synonymous variants | ||
|---|---|---|---|
| Portuguese cohort | |||
| PD | 4 | 263 | |
| Control | 16 | 435 | P = 0.15 |
| US cohort | |||
| PD | 11 | 451 | |
| Control | 4 | 456 | P = 0.12 |
In comparison with the report of Lautier et al., some differences should be noted. The first is the ethnicity of the cohorts studied. Although Lautier et al. screened samples originating in Italy and France, our study included samples from Portugal and the United States. It is possible, although we believe unlikely, that differences in the genetic background between these cohorts, result in disparities in the pathogenicity of the variants. A second difference to the previous study is the sample selection criteria. Although Lautier et al. selected samples with positive family history, we included samples with and without family history representative of PD of their respective populations. Although the number of samples with family history may be smaller in our combined cohort, the effect of pathogenic mutations in GIGYF2 in these samples would be evident.
Another possibility is that the healthy individuals harboring mutations may in future convert to disease; the fact that we used age-matched Controls and that there is no enrichment of these mutations in the Case group support our supposition that this is also unlikely.
The literature is clearly scarce in results relating genetic variability of GIGYF2 with PD. We present the first follow-up study to the results published by Lautier et al. Although we cannot rule out a small genetic contribution of GIGYF2 to PD, our data seem to point in the direction that the pathogenic variants previously published are rare polymorphisms. We support this statement based on the fact that two of such mutations were found in our Control groups and that SNPs across GIGYF2 did not show any association with PD; we cannot of course unequivocally rule out the other previously identified mutations from having a role in disease.
The previous study used a rather small Control group to verify the presence of the variants (n = 227), and thus rare variants may have been missed. We believe this is a critical and important finding; this gene has already been assigned a PARK designation (http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=607688) and we feel, given the evidence that we present here, that such a designation may be premature or misleading.
MATERIALS AND METHODS
Portuguese series
The series originating from Portugal comprised 267 PD patients and 451 healthy Controls, their characteristics are presented in Table 1. Patients were selected in a consecutive manner, in the Movement Disorder clinic of the University of Coimbra Hospital. Diagnosis followed the UK Brain Bank criteria (19). Control samples were collected from healthy unrelated individuals from the same population and geographical regions. All Controls were subjected to a neurological examination and found free of any symptoms suggestive of parkinsonism.
Table 1.
Characteristics of the Portuguese cohort
| Characteristics | PD (n = 267) | Control (n = 451) |
|---|---|---|
| Age at collection (mean ± SD) | 65 ± 11.6 | 66 ± 12.2 |
| Age at onset (mean ± SD) | 55 ± 12.7 | N/A |
| Range of age at onset | 24–89 | N/A |
| Family history | ||
| Positive | 78 | N/A |
| Negative | 189 | N/A |
US series
The US series were taken directly from pre-compiled panels from the National Institute of Neurological Disorders and Stroke (NINDS) funded Neurogenetics repository hosted by the Coriell Institute for research (NJ, USA). All participants provided written informed consent.
Neurologically normal Controls were derived from five different panels of DNA: NDPT002, NDPT006, NDPT009, NDPT022 and NDPT024, containing DNA from total of 460 unrelated individuals from North America, including 225 males and 235 females. All individuals were Caucasian and lacked history of Alzheimer's disease, amyotrophic lateral sclerosis, ataxia, autism, bipolar disorder, brain aneurism, dementia, dystonia, or PD. None had any first-degree relative with a known primary neurological disorder and the mean age of participants was 68.57 (range 55–95).
PD Cases were taken from five panels of DNA: NDPT001, NDPT005, NDPT007, NDPT017 and NDPT018. These panels contain DNA from 460 unrelated Caucasian individuals from North America with PD, including 258 males and 202 females. The mean age at onset is 66.36 years (range 50–87) and they all showed at least one of the main clinical signs of PD such as resting tremor, rigidity, bradykinesia, gait disorder and postural instability at the disease onset. All subjects were questioned regarding family history of parkinsonism, dementia, tremor, gait disorders and other neurological dysfunction. Subjects both with and without a reported family history of PD were included. None were included who had three or more relatives with parkinsonism, nor with clear Mendelian inheritance of PD.
A more detailed description of both Case and Control samples, can be found at http://ccr.coriell.org/Sections/Collections/NINDS/DNAPanels.aspx?PgId=195&coll=ND
Sequencing analysis
Screening of GIGYF2 was carried out using genomic DNA of a total of 724 Cases and 911 neurologically normal Controls from two different populations. Polymerase chain reaction (PCR) amplification was performed in a final volume of 16 µl containing 10 ng of genomic DNA, 10 pmol of forward and reverse primers (sequence available upon request) and 12 µl of FastStart PCR Master mix (Roche). Primers for all coding exons and intron/exon boundaries were designed using ExonPrimer (http://ihg2.helmholtz-muenchen.de/ihg/ExonPrimer.html) for isoform a (NM_001103147.1) of GIGYF2, which is the longest transcript of the gene, encoding 31 exons. Note that this isoform is different than that sequenced by Lautier et al. in their series (NM_015575.3). Therefore amino acid and cDNA positions are different.
Each purified product was sequenced using Applied Biosystems BigDye terminator v3.1 sequencing chemistry as per manufacturer's instructions (Applied Biosystems, Foster City, CA, USA). The resulting reactions were purified and resolved on an ABI3730XL genetic analyzer (Applied Biosystems) and analyzed with Sequencher software v4.1.4 (Gene Codes Corporation, VA, USA).
Statistical analysis
Statistical analyses included Hardy–Weinberg equilibrium, χ2 and Bonferroni correction tests and were performed using PLINK v1.03 (20).
FUNDING
This work was supported by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, Department of Health and Human Services, Annual Report number Z01-AG000957-05 and Fundação para a Ciência e Tecnologia grant (SFRH/BD/29647/2006).
ACKNOWLEDGEMENTS
We would like to thank the subjects for participating in this research and the submitters for depositing samples at the National Institute on Neurological Disorders and Stroke (NINDS) Neurogenetics repository. The samples for this study are derived from the NINDS Neurogenetics repository at Coriell Cell Repositories, supported by an extramural NINDS contract.
Conflict of Interest statement. The authors declare that they have no competing financial interests.
REFERENCES
- 1.Lang A.E., Lozano A.M. Parkinson's disease. First of two parts. N. Engl. J. Med. 1998;339:1044–1053. doi: 10.1056/NEJM199810083391506. [DOI] [PubMed] [Google Scholar]
- 2.Polymeropoulos M.H., Lavedan C., Leroy E., Ide S.E., Dehejia A., Dutra A., Pike B., Root H., Rubenstein J., Boyer R., et al. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 3.Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N. Mutations in the parkin gene cause autossomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 4.Valente E.M., Abou-Sleiman P.M., Caputo V., Muqit M.M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A.R., Healy D.G., et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 5.Bonifati V., Rizzu P., van Baren M.J., Schaap O., Breedveld G.J., Krieger E., Dekker M.C., Squitieri F., Ibanez P., Joosse M., et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 6.Paisan-Ruiz C., Jain S., Evans E.W., Gilks W.P., Simon J., van der Brug M., Lopez de Munain A., Aparicio S., Gil A.M., Khan N., et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 7.Ramirez A., Heimbach A., Grundemann J., Stiller B., Hampshire D., Cid L.P., Goebel I., Mubaidin A.F., Wriekat A.L., Roeper J., et al. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat. Genet. 2006;38:1184–1191. doi: 10.1038/ng1884. [DOI] [PubMed] [Google Scholar]
- 8.Paisan-Ruiz C., Bhatia K.P., Li A., Hernandez D., Davis M., Wood N.W., Hardy J., Houlden H., Singleton A., Schneider S.A. Characterization of PLA2G6 as a locus for dystonia-parkinsonism. Ann. Neurol. 2008 doi: 10.1002/ana.21415. in press; DOI: 10.1002/ana.21415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leroy E., Boyer R., Auburger G., Leube B., Ulm G., Mezey E., Harta G., Brownstein M.J., Jonnalagada S., Chernova T., et al. The ubiquitin pathway in Parkinson's disease. Nature. 1998;395:451–452. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- 10.Simon-Sanchez J., Singleton A.B. Sequencing analysis of OMI/HTRA2 shows previously reported pathogenic mutations in neurologically normal controls. Hum. Mol. Genet. 2008;17:1988–1993. doi: 10.1093/hmg/ddn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strauss K.M., Martins L.M., Plun-Favreau H., Marx F.P., Kautzmann S., Berg D., Gasser T., Wszolek Z., Muller T., Bornemann A., et al. Loss of function mutations in the gene encoding Omi/HtrA2 in Parkinson's disease. Hum. Mol. Genet. 2005;14:2099–2111. doi: 10.1093/hmg/ddi215. [DOI] [PubMed] [Google Scholar]
- 12.Pankratz N., Nichols W.C., Uniacke S.K., Halter C., Rudolph A., Shults C., Conneally P.M., Foroud T. Significant linkage of Parkinson disease to chromosome 2q36–37. Am. J. Hum. Genet. 2003;72:1053–1057. doi: 10.1086/374383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pankratz N., Nichols W.C., Uniacke S.K., Halter C., Murrell J., Rudolph A., Shults C.W., Conneally P.M., Foroud T. Genome-wide linkage analysis and evidence of gene-by-gene interactions in a sample of 362 multiplex Parkinson disease families. Hum. Mol. Genet. 2003;12:2599–2608. doi: 10.1093/hmg/ddg270. [DOI] [PubMed] [Google Scholar]
- 14.Pankratz N., Nichols W.C., Uniacke S.K., Halter C., Rudolph A., Shults C., Conneally P.M., Foroud T. Genome screen to identify susceptibility genes for Parkinson disease in a sample without parkin mutations. Am. J. Hum. Genet. 2002;71:124–135. doi: 10.1086/341282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prestel J., Sharma M., Leitner P., Zimprich A., Vaughan J.R., Durr A., Bonifati V., De Michele G., Hanagasi H.A., Farrer M., et al. PARK11 is not linked with Parkinson's disease in European families. Eur. J. Hum. Genet. 2005;13:193–197. doi: 10.1038/sj.ejhg.5201317. [DOI] [PubMed] [Google Scholar]
- 16.Maraganore D.M., de Andrade M., Lesnick T.G., Strain K.J., Farrer M.J., Rocca W.A., Pant P.V., Frazer K.A., Cox D.R., Ballinger D.G. High-resolution whole-genome association study of Parkinson disease. Am. J. Hum. Genet. 2005;77:685–693. doi: 10.1086/496902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giovannone B., Lee E., Laviola L., Giorgino F., Cleveland K.A., Smith R.J. Two novel proteins that are linked to insulin-like growth factor (IGF-I) receptors by the Grb10 adapter and modulate IGF-I signaling. J. Biol. Chem. 2003;278:31564–31573. doi: 10.1074/jbc.M211572200. [DOI] [PubMed] [Google Scholar]
- 18.Lautier C., Goldwurm S., Durr A., Giovannone B., Tsiaras W.G., Pezzoli G., Brice A., Smith R.J. Mutations in the GIGYF2 (TNRC15) gene at the PARK11 locus in familial Parkinson disease. Am. J. Hum. Genet. 2008;82:822–833. doi: 10.1016/j.ajhg.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes A.J., Daniel S.E., Lees A.J. Improved accuracy of clinical diagnosis of Lewy body Parkinson's disease. Neurology. 2001;57:1497–1499. doi: 10.1212/wnl.57.8.1497. [DOI] [PubMed] [Google Scholar]
- 20.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]