Abstract
Prader–Willi syndrome (PWS) is a complex genetic disorder characterized by hyperphagia, obesity and hypogonadotrophic hypogonadism, all highly suggestive of hypothalamic dysfunction. The NDN gene, encoding the MAGE family protein, necdin, maps to the PWS chromosome region and is highly expressed in mature hypothalamic neurons. Adult mice lacking necdin have reduced numbers of gonadotropin-releasing hormone (GnRH) neurons, but the mechanism for this reduction is unknown. Herein, we show that, although necdin is not expressed in an immature, migratory GnRH neuronal cell line (GN11), high levels are present in a mature GnRH neuronal cell line (GT1-7). Furthermore, overexpression of necdin activates GnRH transcription through cis elements bound by the homeodomain repressor Msx that are located in the enhancer and promoter of the GnRH gene, and knock-down of necdin expression reduces GnRH gene expression. In fact, overexpression of Necdin relieves Msx repression of GnRH transcription through these elements and necdin co-immunoprecipitates with Msx from GnRH neuronal cells, indicating that necdin may activate GnRH gene expression by preventing repression of GnRH gene expression by Msx. Finally, necdin is necessary for generation of the full complement of GnRH neurons during mouse development and extension of GnRH axons to the median eminence. Together, these results indicate that lack of necdin during development likely contributes to the hypogonadotrophic hypogonadal phenotype in individuals with PWS.
INTRODUCTION
Prader–Willi syndrome (PWS) is a multigenic, neurodevelopmental disorder affecting ∼1 in 15 000 live births (1). Diagnostic criteria include neonatal hypotonia, failure to thrive, hyperphagia, excessive weight gain, developmental delay and hypogonadotropic hypogonadism (2), all indicative of hypothalamic dysfunction. As a contiguous multigene disorder, PWS is caused by loss of expression of genes in the 15q11–13 chromosomal region. Several maternally silenced, imprinted genes lie within this region, including MKRN3, MAGEL2, NDN and SNURF/SNRPN/IC. Interestingly, single-gene mutations have not been found to cause PWS, suggesting that loss of multiple genes is required to produce this syndrome. However, it is likely that loss of expression of specific, individual genes may contribute to the various distinct phenotypes associated with this complex disorder. For this reason, it has been important to study the genes within the Prader-Willi interval individually. Both necdin and Magel2 are members of the Melanoma Antigen Gene (MAGE) family. Along with Dlxin (also termed Maged1 or NRAGE), they form a closely related subgroup of the type II subfamily of MAGE proteins, characterized by a conserved MAGE homology domain (3). The type II subfamily is expressed in normal tissues, as opposed to the type I MAGE genes that are mainly expressed in tumorigenic tissues (4).
Mouse chromosome region 7C is homologous to human 15q11–13, and therefore, mutant mice have been utilized as important models for the study of PWS. Several genetic alterations created at 7C have been proposed as PWS models with varying phenotypes (5–12). Specifically, four different necdin-null mutations have been studied (10,11,13–16). Gerard et al. (14), reported variable respiratory defects corresponding to the respiratory compromise seen in children with PWS. Muscatelli et al. (15), observed behavioral and hypothalamic abnormalities, including a reduction in hypothalamic neuron populations in the adult, including 25% fewer gonadotropin-releasing hormone (GnRH)-positive neurons in the medial preoptic area (mPOA). Kuwajima et al. (17) and Kuwako et al. (10) reported reduced differentiation of GABAergic neurons and sensory neuron defects in their strain of necdin-null mice, while we have documented defective axonal outgrowth in a variety of neuron populations including defective migration of sympathetic neurons in the superior cervical ganglia (18,19). Additionally, mice null for Nhlh1 and/or Nhlh2, transcriptional regulatory proteins known to directly activate necdin gene expression (20,21), have fertility defects and reduced numbers of GnRH neurons.
GnRH is the fundamental regulator of the hypothalamic/pituitary/gonadal axis, and thus, GnRH neurons are critical for reproductive function. GnRH mRNA is first detected in the olfactory placode at embryonic day 11.5 (e11.5) (22,23). At embryonic day 13.5 (e13.5), the neurons produce GnRH decapeptide and migrate across the nasal septum and cribriform plate, into the forebrain, where, by e16 (23,24), most migrating GnRH neurons reach their proper locations in the hypothalamus and extend their axons to the median eminence to secrete GnRH into the hypophyseal portal system. This migration and axon targeting are crucial for these neurons to execute their requisite role in reproductive function. In this way, GnRH neuron migration, differentiation and gene expression all occur coincidentally and, may, in fact, have interrelated molecular mechanisms (25).
We have previously shown that the Msx homeodomain transcriptional repressor proteins are expressed in the GnRH neurons in vitro and in vivo, regulate the GnRH gene, and inhibit the development and migration of GnRH neurons (26). Msx transcription factors bind the characterized rat GnRH enhancer and promoter at two ATTA sites in each region, to regulate transcriptional activity of the GnRH gene. Additionally, Msx1-null mouse embryos have increased numbers of GnRH-expressing neurons in the olfactory region at e12.5 and e13.5, an effect that resolves by e16.5. These actions are countered by Dlx homeodomain transcriptional activators (26), which are also expressed in GnRH neurons and bind to the same sequence recognition sites in the GnRH gene. Mouse embryos null for Dlx1 and Dlx2 have 30–35% fewer GnRH neurons. Thus, elimination of the repressor, Msx, increases the number of GnRH neurons, while elimination of the activator, Dlx, decreases the number of GnRH neurons, though the mechanism responsible for regulating their differential binding and activities is not known. Interestingly, Msx and Dlx proteins interact with two type II MAGE proteins, necdin and Dlxin-1 (4,27). Specifically, in muscle cells, necdin and Dlxin-1 were shown to relieve Msx repression of the Wnt1 promoter allowing the promotion of muscle cell differentiation (17,28). However, these studies were performed in vitro with synthetic proteins or in heterologous cells using highly overexpressed, exogenous proteins, leaving their interpretation of their physiological roles open (4).
The majority of individuals with PWS display defects in sexual development generally attributed to hypogonadotrophic hypogonadism (29), implicating the GnRH system. Adult necdin-null mice have reduced numbers of GnRH neurons (15), suggesting that loss of necdin could affect fertility in PWS patients through actions on GnRH neuron development and/or gene expression. Herein, we demonstrate that necdin activates GnRH gene expression by relieving Msx repression of GnRH transcription. We show that necdin is present in an endogenous protein complex with Msx1 and that Dlxin-1 is likely a component of this complex. Additionally, necdin is necessary for proper GnRH neuronal development in vivo. Therefore, we propose that the absence of necdin may result in a decrease in GnRH levels and/or the number of GnRH neurons, and thereby likely contributes to the etiology of hypogonadotrophic hypogonadism in PWS patients.
RESULTS
Necdin is expressed in GT1-7 but not GN11 cells
There are only ∼800–1000 GnRH neurons dispersed in the adult mouse septohypothalamic region (22), making analysis of their molecular mechanisms particularly difficult. Thus, we have utilized immortalized cell lines as in vitro models of GnRH neurons. GT1-7 cells were isolated from a tumor in the forebrain, which was created by targeted oncogenesis utilizing a transgene containing the rat GnRH regulatory region coupled to SV40 T antigen. The GT1-7 cells display characteristics of mature GnRH neurons (30–32), including several neuronal features, producing high levels of GnRH in the characteristic pulsatile pattern, and the ability to functionally substitute for GnRH neurons when implanted into the hypothalamus of an adult hypogonadal mouse. GN11 cells (33) were created in a similar manner using human GnRH regulatory region and derived from a tumor isolated from the olfactory region of the mouse. In contrast to GT1-7 cells, however, GN11 cells respond to migratory cues in culture and make low levels of GnRH, both qualities consistent with an immature GnRH neuron. The GN11 and GT1-7 cell lines have been used as models to identify and analyze transcription factors that regulate GnRH gene expression.
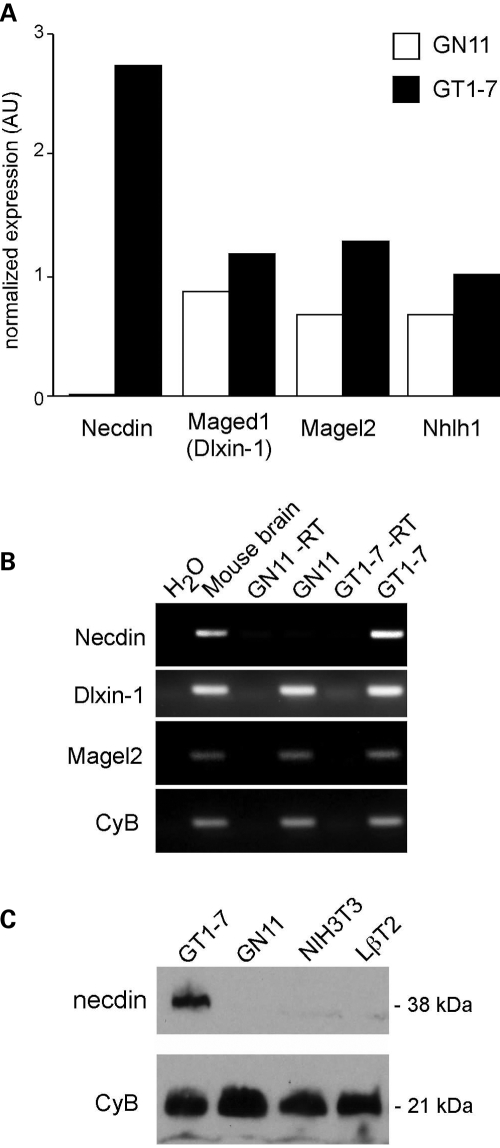
To identify candidate genes critical for GnRH neuron development, we performed an Affymetrix microarray screen comparing RNA from GN11 versus GT1-7 cells. Over 2000 transcripts showed significantly higher levels in GT1-7 versus GN11 cells. In particular, necdin mRNA levels were ∼1000-fold higher in GT1-7 than GN11 cells (P = 2 × 10−61) by VAMPIRE Bayesian variance modeling analysis (34). Interestingly, other MAGE family members, Maged1 (encoding Dlxin-1 protein) and Magel2, as well as a Necdin regulating gene, Nhlh1 (NSCL-1) (20), were not significantly different between the two cell lines (Fig. 1A). The expression of MAGE family members was confirmed using RT–PCR and primers specific for Necdin, Dlxin-1 and Magel2 (Fig. 1B). Necdin expression was not detected in the GN11 cells, but was readily detected in GT1-7 cells. Dlxin-1 and Magel2 transcripts were observed in both GN11 and GT1-7 cell lines. All three MAGE family members were expressed in the adult mouse brain positive control.
Figure 1.
Necdin is expressed in GT1-7 but not detected in GN11 cells. (A) RNA from GT1-7 versus GN11 cells was compared using Affymetrix microarray MOE430A. Fold difference between GT1-7 and Gn11 are shown for MAGE family members Necdin, Maged1 (encoding Dlxin-1) and Magel2, and for Nhlh1. Data were analyzed using GeneSpring software and the mean of two arrays is presented. (B) RT–PCR analysis to confirm expression of MAGE family members, necdin, Dlxin-1 and Magel2 in GN11 and GT1-7 cell lines. Adult mouse brain cDNA was used as a positive control for Mage mRNAs. Cyclophilin B (CyB) was used as a control for equal loading. Ethidium-stained 1% gel pictured. (C) Immunoblots of Necdin and cyclophilin B proteins from GT1-7, GN11, NIH3T3, and LβT2 cells.
The differential expression of necdin between these two cell lines is also evident at the protein level. Immunoblots of proteins from GT1-7, GN11, NIH3T3, and LβT2 [mouse pituitary gonadotrope (35)] cells show that GT1-7 cells express high levels of necdin, while it is undetectable in the other cells (Fig. 1C). Thus, necdin is differentially expressed between immature and mature GnRH neuronal model cell lines and absent from non-neuronal cell lines, suggesting that it could be important for the development of these cells and expression of GnRH.
Necdin activates GnRH gene expression and relieves Msx repression of GnRH gene expression
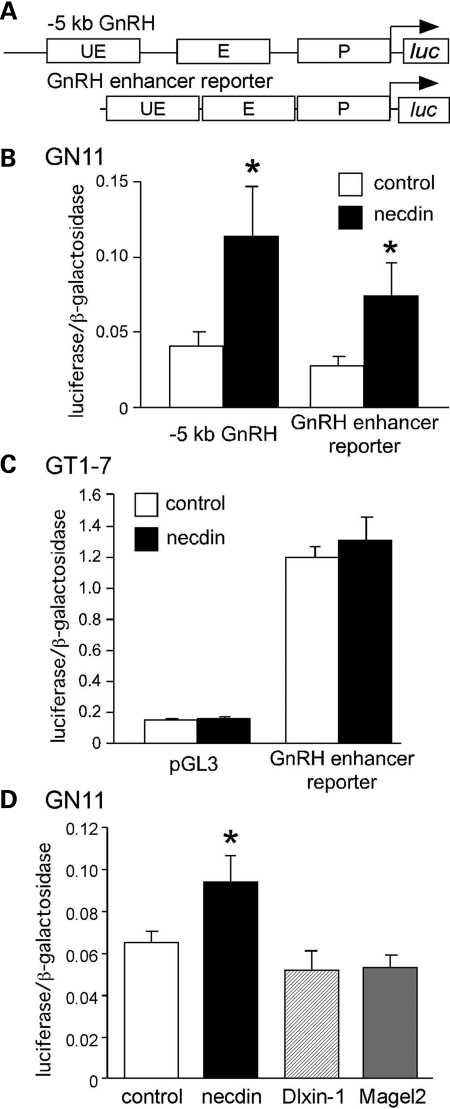
To determine whether necdin regulates GnRH expression, we transiently co-transfected GN11 cells with a Necdin expression vector and rat GnRH-luciferase reporter plasmids. We utilized GN11 cells due to their lack of endogenous necdin. GT1-7 cells have such high levels of necdin that a Necdin expression vector would not be expected to significantly increase levels. Both a luciferase reporter containing 5 kb of the upstream region of the rat GnRH gene and a reporter driven by a condensed combination of the three most well-characterized rat GnRH regulatory elements (36): upstream enhancer, enhancer and promoter (GnRH enhancer reporter, Fig. 2A) were significantly induced by necdin (Fig. 2B). As expected, when necdin was overexpressed in GT1-7 cells (which already have high levels of endogenous necdin, see Fig. 1), significant activation did not result (Fig. 2C). Additionally, overexpression of Magel2 (another Prader-Willi candidate gene and MAGE family member) did not affect GnRH gene expression, nor did the related MAGE protein, Dlxin-1 (Fig. 2D). Consequently, necdin is sufficient to activate GnRH gene expression in GN11 cells and this activation localizes to the known enhancer and promoter regions.
Figure 2.
Necdin activates GnRH gene expression. (A) Two luciferase reporters were studied: −5 kb GnRH and GnRH enhancer reporter, which is a condensed combination of the three well-characterized rat GnRH regulatory elements: upstream enhancer (UE), enhancer (E) and promoter (P) (36). (B) Each reporter was co-transfected into GN11 cells with a necdin expression vector (necdin) or empty vector (control). (C) GT1-7 cells were co-transfected with pGL3-luciferase or GnRH enhancer reporter and either necdin expression vector or empty vector control. (D) GN11 cells were co-transfected with GnRH enhancer reporter and either necdin, Magel2, Dlxin-1, or empty vector control. Experiments were performed in quadruplicate and repeated at least three times. Results shown are mean ± SEM. The means were compared by one-way ANOVA and Tukey–Kramer HSD. *indicates significant difference (P < 0.05) relative to empty vector control.
Since MAGE family members interact with, and regulate gene expression through, Msx and Dlx homeodomain proteins (27), and Msx and Dlx proteins regulate GnRH transcription during development (26), we investigated the effect of necdin on Msx and Dlx regulation of the GnRH gene. As we previously demonstrated (26), overexpression of Msx1 significantly decreases GnRH reporter gene transcription in GN11 cells. Interestingly, however, co-transfection of necdin prevents this repression by Msx1 (Fig. 3A). In contrast, confirming our previous studies (26), Dlx proteins significantly increase GnRH reporter activity. However, co-expression of necdin did not alter the effects of Dlx on GnRH activity (Fig. 3B), indicating that, in this case, necdin selectively acts through Msx homeodomain proteins.
Figure 3.
Necdin specifically disrupts Msx repression of GnRH gene expression. (A) The GnRH enhancer reporter was co-transfected into GN11 cells with either empty vector control or Msx1 expression vector ± necdin expression vector. *Significant difference (P < 0.05) from empty vector control, while #Significant difference (P < 0.05) relative to Msx1 alone, as indicated by brackets. (B) The GnRH enhancer reporter was co-transfected with either empty vector control, or with Dlx1, Dlx2 and Dlx5 expression vectors (Dlx) ± necdin expression vector. Experiments were performed in quadruplicate and repeated at least three times. Results shown are mean ± SEM. One-way ANOVA and Tukey–Kramer HSD compared the means.
Necdin activates GnRH through Msx/Dlx-binding sites in the GnRH enhancer and promoter regions
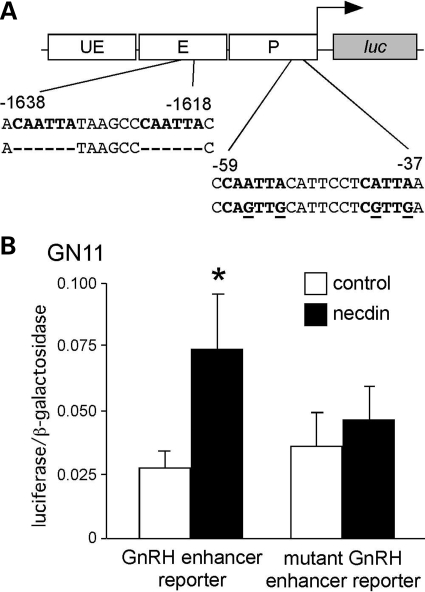
If necdin activates through prevention of Msx repression, the characterized Msx DNA-binding sites in the GnRH regulatory sequences should be necessary for activation by necdin. The GnRH enhancer and promoter each have two Q50 homeodomain-binding elements at −1637, −1624, −58, and −42 bp, respectively, all of which bind Msx and Dlx family members (26). We have previously shown that mutation of all four characterized Msx/Dlx-binding sites eliminates the repressive effect of Msx (26). When necdin was transiently co-transfected into GN11 cells with the GnRH enhancer reporter carrying these mutations (Fig. 4A), there was no significant activation by necdin (Fig. 4B), demonstrating that the regulation by necdin requires these Msx-binding sites in the GnRH enhancer and promoter for activation of the GnRH gene.
Figure 4.
Necdin activation requires the Msx-binding sites in the GnRH enhancer and promoter. (A) The rat GnRH enhancer reporter contains four Msx/Dlx-binding sites (bold) in the GnRH promoter and enhancer at −1637, −1624, −58, and −42 bp relative to the mRNA start site. Mutations are shown as deletions (-) or base changes (underline) in the below sequence. (B) Necdin expression vector was co-transfected into GN11 cells with either GnRH enhancer reporter or mutant GnRH enhancer reporter. Experiments were performed in quadruplicate and repeated at least three times. Results shown are mean ± SEM. Means were compared by Student’s t-test and *Significant values (P < 0.05) relative to empty vector control.
Necdin was reported to act as a transcriptional repressor by binding to guanosine-rich clusters in DNA (37). To distinguish whether GnRH is acting directly or indirectly though the Msx-binding sites, which are not guanosine-rich (CAATTA), we utilized electromobility shift assays with both in vitro-translated necdin and GT1-7 nuclear extracts and necdin antibody to detect any supershift. However, in neither instance was necdin detected as bound to the DNA (data not shown).
Necdin is necessary for GnRH gene expression in GT1-7 cells
To determine whether necdin is required for GnRH gene expression, we knocked down necdin protein levels in GT1-7 cells. siRNA duplex pools targeting necdin, cyclophilin B, or a non-targeting control were transfected into GT1-7 cells, which normally express high levels of endogenous necdin and GnRH. Immunoblots show that knockdown of necdin and cyclophilin B control proteins were specific to the appropriate siRNA duplex pool (Fig. 5A).
Figure 5.
Knockdown of necdin in GT1-7 cells decreases GnRH mRNA and gene expression. (A) GT1-7 cells were either mock-transfected or transfected with non-targeting (control), cyclophilin B (CyB) or necdin (Ndn) siRNA duplexes. Specific knockdown of necdin and cyclophilin B proteins was detected by immunoblot. (B) GT1-7 cells were transfected as in (A) followed by RNA isolation. GnRH was measured by quantitative RT–PCR and data are shown as pg GnRH mRNA when normalized to CyB mRNA. Results are the means of three experiments performed in triplicate and *indicates significantly different from control by Student’s t-test (P < 0.05). (C) GT1-7 cells were co-transfected with control or Ndn siRNA duplexes and either GnRH enhancer reporter or mutant GnRH enhancer reporter. Experiments were performed in quadruplicate and repeated at least three times. Results shown are mean ± SEM. One-way ANOVA and Tukey–Kramer HSD compared the means. *P < 0.05.
We next examined endogenous GnRH mRNA levels in GT1-7 cells in which necdin had been knocked down. RNA levels were analyzed from mock, non-targeting control and necdin siRNA duplex pool transfected GT1-7 cells by quantitative RT–PCR. GT1-7 cells subjected to siRNA knockdown of necdin expressed significantly less GnRH mRNA than did cells transfected with control siRNAs (Fig. 5B). Thus, necdin is necessary for normal GnRH gene expression in GT1-7 cells.
To further establish that necdin knockdown affected GnRH expression at the transcriptional level, we co-transfected GT1-7 cells with siRNA duplex pools and either wild-type or mutant GnRH enhancer reporters. Activity of the GnRH enhancer reporter was reduced 37% when necdin protein was knocked down (Fig. 5C). Importantly, however, the mutant GnRH enhancer reporter showed no decrease in activity, confirming that regulation of GnRH by necdin requires the Msx/Dlx-binding sites.
Necdin interacts with Msx
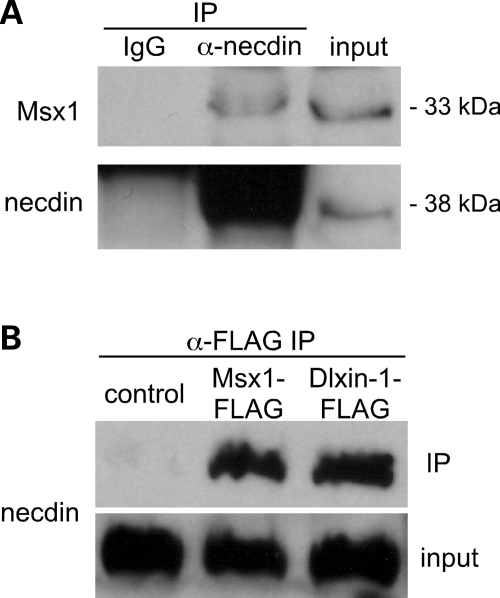
Since necdin increases GnRH gene expression through Msx-binding sites, we examined whether these proteins interact within the GT1-7 cells by co-immunoprecipitating endogenous necdin and Msx1 from GT1-7 cells. Msx1 was immunoprecipitated with anti-necdin antibody, but not with the IgG control, demonstrating an association between endogenous necdin and Msx1 proteins in GT1-7 cells (Fig. 6A).
Figure 6.
Necdin interacts with Msx in GT1-7 cells. (A) Proteins from GT1-7 cells were immunoprecipitated (IP) with either anti-necdin antibody or rabbit IgG control. Msx1 and necdin were detected by immunoblotting the immunoprecipitated proteins. Ten percent of the input was used for Msx1 and 2% for necdin. (B) GT1-7 cells were transiently transfected with Msx1-FLAG, Dlxin1-FLAG expression vector or empty vector control. Cell extract was immunoprecipitated with anti-FLAG affinity gel. Immunoprecipitates and 4% of the input were immunoblotted for necdin.
Though Grg family co-repressors have been shown to interact with Msx to augment their repressor function in GnRH transcription in GT1-7 cells (38), Grg proteins were not co-immunoprecipitated with necdin and Grg repression of GnRH gene expression was not affected by necdin, nor did mutation of the engrailed homology domain within Msx [the putative site of Grg interaction (38)] affect its interaction with necdin (data not shown). However, Grg proteins also act as co-repressors for Oct-1, which acts through other binding sites in the GnRH gene and might also act through additional GnRH gene regulatory factors and regions.
Another MAGE family member, Dlxin-1 (encoded by the Maged1 gene), has also been shown to complex with necdin and Msx/Dlx family members (17). Msx1-FLAG and Dlxin-1-FLAG expression vectors were transfected into GT1-7 cells, then proteins were immunoprecipitated with anti-FLAG antibody. Immunoblots for necdin demonstrated that necdin associates with both Msx1 and Dlxin-1 in GT1-7 cells (Fig. 6B). Consequently, Dlxin-1 may also be in the Msx/necdin complex that regulates GnRH gene expression.
Necdin does not change the cellular localization of Msx
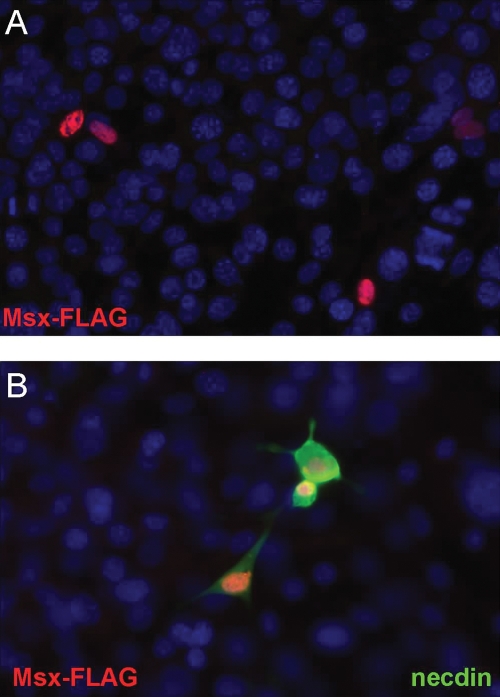
MAGE proteins have been observed to alter the cellular localization of other proteins to the nucleus or cytoplasm, depending on their presence or activation state (39). Thus, another mechanism by which necdin could relieve Msx repression of the GnRH gene, would be to change the nuclear localization of Msx, by sequestering it out of the nucleus, into the cytoplasm, such that it could no longer bind to DNA. We utilized GN11 cells, as their lack of endogenous necdin allowed control of its expression and detection of FLAG-tagged Msx1 (Msx1-FLAG) in cells with or without necdin co-expression. Without necdin expression, Msx1-FLAG was detected only within the nucleus of the GN11 cells (Fig. 7A). When co-transfected into the GN11 cells, necdin was localized to both cytoplasm and nucleus; however, Msx1-FLAG localization remained in the nucleus (Fig. 7B), suggesting that the presence of necdin does not cause Msx1 to migrate out of the nucleus. These results indicate that the mechanism for necdin interference with Msx repression of GnRH gene expression is unlikely to be due to changes in the cellular localization of Msx.
Figure 7.
Necdin expression does not change the cellular localization of Msx. (A) Msx1-FLAG overexpression in GN11 was detected with an anti-FLAG-Cy3 antibody conjugate (red). (B) GN11 cells were transiently transfected with both necdin and Msx1-FLAG expression vectors and then subjected to immunofluorescence using the anti-FLAG-Cy3 antibody (red) or anti-necdin (green). Blue staining is DAPI nuclear labeling. Image shown is at 40× magnification.
Necdin-null mice have fewer GnRH neurons during developmental migration
Adult necdin-null mice exhibit reduced numbers of GnRH neurons in the mPOA (15). However, the role of necdin during GnRH neuronal development has not been analyzed. GnRH immunohistochemistry was performed on sagittal sections of necdin-null embryos and their wild-type littermates and GnRH-positive cells were counted (Fig. 8). By e13.5, the full complement of GnRH neurons should be established predominantly in the nasal and cribriform plate regions (23) (Fig. 8A–E). Wild-type e13.5 embryos had 1053 ± 83 GnRH-positive cells in total (Fig. 8D), consistent with previous findings (26). In contrast, necdin-null littermates had significantly reduced numbers (563 ± 102) of GnRH neurons (Fig. 8D). The pathway for GnRH neuron migration was divided into three areas (depicted in Fig. 8B): nasal, cribriform plate and brain, to analyze progression of migration. Wild-type and necdin-null e13.5 mice had similar numbers of GnRH neurons in the nasal region, 351 ± 52 versus 361 ± 24, respectively. However, both the cribriform plate and brain regions had significantly reduced numbers of GnRH-positive cells in necdin-null animals (cribriform plate: WT 379 ± 61 versus null 161 ± 33; brain: 241 ± 31 WT versus 43 ± 29 null; Fig. 8E). Thus, e13.5 necdin-null mice have significantly fewer GnRH neurons than wild-type. Specifically, fewer GnRH neurons appeared to cross the cribriform plate into the brain, indicating that necdin plays a vital role in the developmental progression of GnRH neurons.
Figure 8.
Embryonic day 13.5 Necdin-null mouse embryos have significantly fewer GnRH neurons and lower density of GnRH fibers in the median eminence. (A) Diagram of e13 mouse head anatomy in sagittal showing location of migrating GnRH neurons and the orientation of all sections. Immunohistochemistry for GnRH in e13.5 wild-type embryos (B) or necdin-null embryos (C). Images are shown at 4×. (D) Average numbers of total GnRH neurons at e13.5. (E) Average numbers of GnRH neurons counted in the nasal, cribriform plate or brain regions at e13.5 as depicted in (B). e17.5 Necdin-null mice have fewer GnRH neurons in the brain. (F) Diagram of e17 mouse head anatomy in sagittal showing location of migrating GnRH neurons. The red box represents the field shown in images in (G) and (H), while the green box indicates the field shown in images in (K) and (L). All images are in the orientation shown in the illustration. Immunohistochemistry for GnRH in e17.5 wild-type embryos (G) or necdin-null embryos (H). Images are shown at 10×. (I) Average numbers of total GnRH neurons at e17.5. (J) Average numbers of GnRH neurons counted in the nasal, cribriform plate or brain regions at e17.5 as depicted in (B). Means were compared with wild-type littermates by Student’s t-test and *P < 0.05. Error bars are standard deviation. Sagittal sections of littermate e17.5 wild-type (K) and necdin-null (L) embryos were stained for GnRH immunoreactivity in the median eminence (ME). Sections also reveal the structure of the developing pituitary (P). Images are representative sections from those used for GnRH neuron quantitation.
We then examined the number and location of GnRH-positive neurons in necdin-null and wild-type e17.5 embryos (Fig. 8F–J). Interestingly, the total number of GnRH neurons was comparable in the necdin-null mice and their wild-type littermates (wild-type 1175 ± 309 and necdin-null 1045 ± 77; Fig. 8I). However, the number of GnRH-positive neurons located in the brain was 31% significantly lower in necdin-null (520 ± 27) compared with wild-type embryos (758 ± 120; Fig. 8J). While the numbers of GnRH neurons in the cribriform plate region were the same at e17.5, regardless of necdin expression, the number of GnRH expressing cells in the nasal region trended higher in the necdin-null mice (P = 0.08). Since sections were taken and analyzed through the majority of the heads of these animals, we can ensure that the neurons are not mis-migrating to an area not being observed for staining and represent the total number of GnRH expressing neurons in the animal. These data indicate that the loss of necdin causes a more dramatic effect on neuron numbers in early GnRH neuronal migration (46% decrease at e13.5) than at the later stage (e17.5) and may also indicate a delay in the birth and migration of this specialized population. However, even at e17.5, the 31% reduction in GnRH neurons in the brain is consistent with the ∼25% reduction previously noted in the adult mPOA (15).
We then assessed the effect of loss of necdin on axonal outgrowth to the median eminence at e17.5, a stage of development at which GnRH can easily be detected there due to the targeting of GnRH neuronal axons. Figure 8K and L shows the median eminence in e17.5 wild-type versus necdin-null embryos. Strong GnRH immunostaining was evident in all wild-type embryos examined, while the necdin-null embryos exhibited only weak GnRH immunostaining indicative of few fibers reaching the median eminence. This indicates that the axonal outgrowth from the GnRH neurons that have reached the hypothalamic region is defective or severely delayed in necdin-null mice. Together these studies show that necdin regulates both the expression of the GnRH gene and GnRH neurons during their development.
DISCUSSION
Hypogonadism is one of the major diagnostic criteria for PWS (2). It is usually hypogonadotrophic and at least partially attributed to hypothalamic dysfunction (40). Several genes residing within the human 15q11–13 region are inactivated in PWS, including two MAGE family genes, NDN and MAGEL2. Hypothalamic insufficiency has been suggested in necdin-null mice (15), making necdin an intriguing candidate for contribution to the hypogonadal phenotype. Our investigation of this hypothesis demonstrates that necdin regulates GnRH gene expression and GnRH neurons during development, strongly implicating lack of necdin in the etiology of PWS-associated infertility.
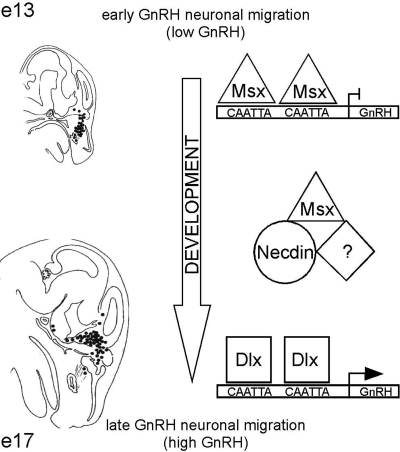
The microarray comparison of mRNAs in immature GN11, versus mature GT1-7, GnRH cell lines revealed that necdin was the most differentially expressed transcript, suggesting a critical role for necdin in the maturation of GnRH neurons. Necdin is a marker of mature CNS neurons and is thought to play a role in neuronal differentiation (41,42). During development, the level of necdin increases in neurons as they differentiate and exit the cell cycle to become post-mitotic (42,43), consistent with its high expression in GT1-7 and absence from GN11 cells. In fact, we find that transfection of necdin into immature GN11 cells results in significant activation of GnRH transcription. Since necdin is known to interact with Msx repressor and Dlx activator homeodomain proteins in the regulation of the Wnt1 promoter in muscle cells (4,27) and we had previously shown that Msx repression and Dlx activation contribute to differential regulation of GnRH gene expression during developmental migration (26), we addressed the interaction of necdin with Msx and Dlx in GnRH gene expression. Necdin activation of GnRH transcription was dependent upon the four known Msx/Dlx-binding sites, indicating that necdin may not directly bind to the GnRH regulatory sequence, but instead function as a co-factor. siRNA knockdown of necdin in GT1-7 cells reduces both endogenous GnRH mRNA and GnRH reporter gene transcription, and this effect is also dependent upon the four Msx/Dlx-binding elements. In fact, necdin relieves Msx repression of GnRH gene transcription, while Dlx activation of GnRH is not affected, indicating that necdin is specifically preventing Msx repression through the GnRH enhancer and promoter binding sites. Furthermore, Msx1 co-immunoprecipitates from GT1-7 cells with necdin, demonstrating that a complex between the endogenous Msx1 and necdin proteins forms in these GnRH neurons. Another MAGE family member, Dlxin-1, also has been shown to interact with Msx and Dlx, in the presence of necdin (4,27). Dlxin-1-FLAG transfected into GT1-7 cells did produce a complex with necdin. However, Dlxin-1 did not affect GnRH gene expression, nor did the related protein, Magel2. These studies indicate that necdin forms a complex with Msx1 that interferes with Msx repression of GnRH transcription, while Magel2 and/or Dlxin are either not necessary or their endogenous levels are sufficient for the formation and action of the Msx/necdin complex. Inactivation of Msx by necdin may then permit Dlx activators to play their role in the induction of GnRH gene expression as the GnRH neuron migrates, matures and increases synthesis of GnRH (Fig. 9).
Figure 9.
A model for necdin action on GnRH gene expression. Early in their development, the majority of GnRH neurons is located in the nasal septum and is migrating toward the brain. During this spatiotemporal period, Msx homeodomain repressors are expressed in vivo and may be bound to specific elements within the GnRH promoter and enhancer to repress GnRH, resulting in low GnRH expression (26). Late in their development, most GnRH neurons have crossed into the forebrain and hypothalamus (areas of Dlx activator expression) and are expressing high levels of GnRH. It is thought that Dlx proteins are then binding to the same homeodomain elements within the GnRH regulatory region, to increase GnRH expression (26). The ‘switch’ from Msx to Dlx factors on the GnRH promoter may be the result of the interaction of Msx repressors with the MAGE protein, necdin, which is being expressed along the GnRH migratory route at this time, resulting in an inactivation of Msx repression and allows Dlx activators to exert their function. Necdin may require co-factors such as MAGE family member Dlxin-1 for this functional complex.
Necdin interference with Msx repression of GnRH gene expression requires the Msx-binding elements in the GnRH gene and appears to occur through formation of an Msx/necdin protein complex. This interaction with necdin might prevent Msx repression through one or more mechanisms such as: necdin might prevent Msx from recruiting a specific co-repressor, necdin association might alter Msx protein stability or localization to the nucleus, or perhaps necdin interferes with Msx binding to DNA. However, the only known co-repressors for Msx in GT1-7 cells, the Grg proteins, were not detected in complexes immunoprecipitated with necdin antibody. In addition, Msx proteins were not reduced in level or detected outside of the nucleus in GN11 cells transfected with necdin, indicating that necdin does not act by altering the localization or stability of Msx proteins. The binding of Msx to DNA is critical for its repression of the GnRH gene and yet Dlx binds the same DNA sites and is important for activation of the GnRH gene. Therefore, one can hypothesize that necdin interaction with Msx either blocks or decreases its affinity for DNA or otherwise isolates it away from the DNA, thus, allowing Dlx homeodomain activators greater access to their shared DNA-binding elements. The more plausible molecular interactions underlying necdin activation of GnRH gene transcription are illustrated in Figure 9.
GnRH transcriptional activity is thought to be dependent on the developmental stage of the embryo (25), with mRNA levels increasing as the neurons mature. Thus, the action of necdin in the GnRH neurons as they develop and migrate into the brain is consistent with its activity in counteracting the repression by Msx, allowing the GnRH gene to increase transcription as the neurons differentiate. Additionally, recent evidence points to a role for necdin in the migration of a population of sympathetic neurons during late embryogenesis (19). In support of our hypothesis, necdin appears important for GnRH neuron maturation in vivo as the neurons migrate into the brain. At e13.5, though the numbers of GnRH neurons remained the same in the nasal area of the necdin null embryos, ∼58% fewer GnRH-positive neurons were observed in the cribriform plate region and 72% fewer entering the brain. By e17.5, there were still ∼31% fewer GnRH neurons in the brain. While it is unclear whether the reduction in GnRH neuron numbers is a result of cell death or loss of GnRH expression, it is likely that there are fewer functional GnRH neurons in these animals. These results are consistent with the report that adult necdin-null mice have reduced numbers of GnRH neurons in the mPOA (15). The lack of necdin in the null animals may affect GnRH neurons only after they have reached the cribriform plate and primarily affect those having reached the brain. GnRH neurons are thought to begin expressing necdin mRNA at e12 (41), as they are migrating toward the cribriform plate. Thus, our findings suggest that the lack of necdin results in Msx maintaining repression of GnRH gene expression later in development, resulting in lower levels of GnRH transcription in the GnRH neuron and perhaps a delay in GnRH neuronal migration or maturation resulting in fewer functional GnRH neurons ultimately reaching the hypothalamus. In this way, necdin could be involved in ensuring that GnRH gene expression is activated within a specific temporal and spatial window, allowing the developmental migration of the GnRH neuron.
Necdin has also been detected in the developing pituitary, in Rathke’s pouch (44), yet we do not detect it in the gonadotrope cell line, LβT2. Though four different necdin-null mouse lines have been characterized, none of these have noted any pituitary defects. In addition, the structure and size of the developing pituitary in the 17.5 embryos (Fig. 8) was normal. Thus, it is possible that necdin has a role in pituitary development, however, the effects on GnRH neurons in the necdin-null mice are not due to pituitary gonadotropin defects since GnRH neurons have been shown to develop, migrate and target the median eminence normally in gonadotrope-ablated animals (45,46) and, indeed, in mice lacking either GnRH receptor or GnRH itself (47).
Though the fertility of necdin-null mice has not been investigated in detail, a reduction by as much as 66% of GnRH neurons due to other mutations still results in fertile female mice (48). Thus, lack of necdin in PWS patients could result in infertility if: (i) axon targeting to the median eminence is defective even in the adult, (ii) the reduction in GnRH neurons due to the absence of necdin was more dramatic in humans than mice, (iii) a 30% reduction in GnRH neurons was sufficient for infertility in humans, or (iv) additional gene deficiencies in the PWS interval augment the reduction of GnRH neurons. Additionally, since a genetic defect has been identified in only 30% of idiopathic hypothalamic hypogonadism patients (49), it will be important to determine whether mutations in the necdin gene might be found in cases of idiopathic hypothalamic hypogonadism of unknown genetic etiology.
In summary, we have identified a MAGE protein, necdin, as a key regulator of GnRH gene expression both in vitro and in vivo. Necdin gene expression is inactivated in PWS in which patients are typically infertile. Lack of necdin reduces GnRH gene expression, results in decreased numbers of GnRH neurons, and decreased targeting of GnRH axons to the median eminence during development, actions that likely contribute to hypogonadotrophic hypogonadism and infertility in PWS.
MATERIALS AND METHODS
Cell culture and transfections
GT1-7, GN11, NIH3T3, and LβT-2 cells were cultured in Dulbecco’s modified Eagle’s medium with 4.5% glucose, 10% fetal bovine serum and 1× penicillin–streptomycin in 5% CO2 at 37°C. Cells in 24-well plates were transfected at 90 000 and 50 000 cells per well, respectively, with 400 ng/well of rat GnRH enhancer luciferase reporter or mutant GnRH enhancer reporter and 100 ng/well of pHismaxC-Necdin, pCB6+Msx1, pCAGG-Dlx1, pCB6+Dlx2, or pcDNA3Dlx5. All transfections included thymidine kinase-β-galactosidase as an internal control, pGL3 empty reporter and appropriate empty vectors as parallel controls. Transfections used FuGENE 6 (Roche) according to manufacturer’s protocols. The mutant GnRH enhancer reporter contains mutations as previously described (26).
Transfections were harvested at 48 h in lysis buffer (100 mm potassium phosphate and 0.2% TritonX-100, pH 7.8) unless otherwise noted. Luciferase assays were performed as previously described (26) and β-galactosidase assays were performed as directed by the manufacturer (Tropix). Luciferase values were normalized to internal control β-galactosidase values and were always compared with empty vector control. Experiments were performed in quadruplicate, at least 3×. Data represent mean ± SEM of at least three independent experiments.
RNA isolation, microarray and RT–PCR
RNA was extracted using Ultraspec™ (Biotecx Laboratories Inc.), according to manufacturer’s instructions. RNA was re-precipitated from 3 m NaOAC, pH 5.2, and then ethanol before submission to the UCSD-VA GeneChip Core for analysis using Affymetrix MOE430A microarrays. Two arrays were performed using RNA from independent batches of cells. Data were analyzed using GeneSpring (Silicon Genetics) for all genes on chip and VAMPIRE Bayesian variance modeling (34) for those with statistically different expression between. The statistical approach in VAMPIRE finds many genes that are missed by the other methods (50,51) and can be more robust than ANOVA-based procedures at low sample number.
RNA for RT–PCR analysis was isolated using the RNeasy mini kit (QIAGEN). cDNA was generated using 2 µg RNA and the SuperScript III First-Strand Synthesis System (Invitrogen), with the exception of the adult mouse brain cDNA (Zyagen) used as a positive control. Primers sequences are as follows:
Necdin forward: 5′-AGCAGGACTTAACAGCAACGCA-3′.
Necdin reverse: 5′-TGCCTACACTGAGAACAGTCCA-3′.
Dlxin-1 forward: 5′-AGACGAGAGCTATGGCTCAGAAAC-3′.
Dlxin-1 reverse: 5′-TCCATCAAGGTCTGCACAAGCAAG-3′.
Magel2 forward: 5′-GACCAAGCCAAGGTGCCTGTCCAG-3′.
Magel2 reverse: 5′-CAGACAGTATTTTACCGATGAGTC-3′.
cyclophilin B forward: 5′-CGTGGCCAACGATAAGAAGA-3′.
cyclophilin B reverse: 5′-GAAGTCTCCACCCTGGATCA-3′.
Immunoblot and co-immunoprecipitation
Whole cell extract was prepared by lysis [50 mm Tris–HCl, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, protease-inhibitor cocktail (Sigma) and 1 mm PMSF, pH 7.4] directly on tissue culture dishes. Lysate was rocked at 4°C for 15 min then centrifuged for 10 min. For immunoblotting, 20 µg of protein was electrophoresed on 12% polyacrylamide mini-gels, transferred to PVDF and blocked overnight in 5% non-fat dry milk in Tris-buffered saline with 0.1% Tween-20. Anti-necdin (Abcam ab18554) and anti-cyclophilin B antibodies (Abcam ab16045) were diluted at 1:1000, and anti-Msx1 was diluted at 1:1600 (Aviva ARP37094) in blocking buffer. Anti-rabbit HRP secondary (1:5000) (Amersham NA934V) was diluted in Tris-buffered saline with 0.1% Tween-20. Chemiluminescence was detected (Pierce SuperSignal West Pico) by exposure to film.
For co-immunoprecipitation experiments, 200 µg of pre-cleared GT1-7 lysate was incubated with 4 µg of either anti-necdin or anti-rabbit IgG (Santa Cruz sc-2027) at 4°C 1 h. Twenty-five microliters of Protein A Magnetic Beads (NEB) were added and rocked overnight at 4°C. Bead/protein complexes were washed 5× then eluted in 2× SDS sample buffer at 70°C for 5 min. Two immunoblots were run with each sample for detection of Msx or necdin. For GT1-7 cells transfected with FLAG-Msx1 or FLAG-Dlxin1 (Fig. 6B), 250 µg of extract was immunoprecipitated using EZ-View Red FLAG Affinity Gel (Sigma) per manufacturer’s instructions. Proteins were dissociated from beads by boiling for 5 min in 2× SDS sample buffer then immunoblotted for necdin.
siRNA knockdown and quantitative RT–PCR
Necdin (ON-TARGETplus SMARTpool for mouse, Dharmacon), Non-Targeting control (ON-TARGETplus siCONTROL pool, Dharmacon) and Cyclophilin B (ON-TARGETplus, siCONTROL pool for mouse, Dharmacon) siRNA duplex pools were transfected into GT1-7 cells using DharmaFect3 reagent (Dharmacon). Proteins were extracted at 72 h and immunoblotted for cyclophilin B and necdin.
RNA was harvested at 48 h post-transfection using the RNeasy mini kit (QIAGEN). cDNA was generated using 2 µg RNA and the SuperScript III First-Strand Synthesis System (Invitrogen), was diluted 1:10 to yield an equivalent of 50 ng starting RNA per reaction. All quantitative RT–PCR reactions were performed in a 25 µl volume, using either iQ SYBR Green Supermix (BioRad) or Absolute Blue SYBR Green Fluorescein Mix (Thermo Scientific). Primers: GnRH forward: TGCTGACTGTGTGTTTGGAAGGCT, GnRH reverse: TTTGATCCACCTCCTTGCGACTCA (cyclophilin B primers are as described earlier); cycling conditions: 15 min at 95°C, 40 cycles of 95° for 15 s, 55°C for 30 s and 72°C for 30 s. Additionally, an 81-cycle step at 55°C for 30 s was performed for melting temperature analysis to confirm purity. Serial dilutions of Necdin or cyclophilin B cDNA plasmids were used in parallel for standard curves. PCR was performed in triplicate and repeated 4× on a BioRad iQ5. Values were determined using the standard curves based on threshold cycle (Ct) values. Replicates were averaged and divided by the mean of cyclophilin B within the same sample.
Co-transfections were performed as described earlier, except siRNA duplex pools for a non-targeting negative control or Necdin were co-transfected with reporter plasmids using DharmaFect3 transfection reagent. Co-transfections also included thymidine kinase-β-galactosidase as an internal control.
Immunocytochemistry
GN11 cells were cultured as described earlier in 2-well Lab-Tek II Chamber Slides (Fisher). Cells were fixed in 3.7% formaldehyde for 10 min, permeabilized for 20 min in 0.2% NP40, 1% BSA, 10% goat serum in PBS, avidin blocked for 30 min in 20% goat serum, 5% BSA, avidin solution from Avidin/Biotin blocking kit per manufacturer’s instructions (Vector Labs) in PBS, biotin blocked for 30 min according to manufacturer’s instructions (Vector Labs), and incubated overnight at 4°C in anti-necdin (Abcam ab18554, 1:1000) and anti-FLAG Cy3 (Sigma, 1:100) antibody diluted in blocking solution (20% goat serum, 5% BSA). Cells were incubated for 30 min with biotinylated goat-anti-rabbit secondary antibody (Molecular Probes) used at 1:500 in blocking solution. Strepavidin-Alexa488 fluorescent conjugate (1:200 in PBS) in was incubated with cells for 1 h in the dark. Slides were mounted with coverslips using VectaShield Hard Set Mounting Media with DAPI (Vector Labs). All incubations were done at room temperature unless otherwise noted. Each antibody individually and no primary antibody controls were also performed in parallel. Fluorescence was visualized with a Nikon Eclipse TE2000-U microscope.
Immunohistochemistry
Necdin-null mice (14) were generated by heterozygous crosses, then pregnant females were euthanized and embryos were harvested at e13.5 and 17.5 (e17.5). Genotypes were confirmed as previously described (13). Whole embryos (e13.5) or embryo heads (e17.5) were fixed in 10% acetic acid, 30% formaldehyde, 60% ethanol, overnight at 4°C and dehydrated in ethanol/water washes prior to embedding in paraffin. Ten micrometer sagittal sections were floated onto SuperFrost Plus slides (Fisher) and dried overnight at 37°C. Approximately 120–200 sections were processed and stained for GnRH per head depending on the developmental stage. Prior to staining, slides were incubated at 60°C for 30 min. Slides were deparaffinized in xylene washes, then rehydrated in ethanol/water washes. Antigens were retrieved by boiling for 10 min in 10 mm sodium citrate. After cooling and washing 2× in water, endogenous peroxidase was quenched with 0.3% hydrogen peroxide for 10 min. Slides were blocked in PBS with 5% goat serum and 0.3% Triton X-100 for 45 min. Slides were incubated overnight at 4°C in anti-GnRH antibody (Affinity BioReagents PA1-121; diluted 1:1000 in blocking buffer), then in biotinylated goat-anti-rabbit IgG (Vector Labs) at 1:300 for 30 min. GnRH peptide was visualized using the Vectastain ABC elite kit and VIP peroxidase kit (Vector Labs). Sections were counterstained using methyl green (Vector Labs). GnRH neurons were counted (double-blind) in all sections of three or more embryos per time point and genotype. Cells were divided into nasal, cribriform plate and brain regions, and the mean calculated.
FUNDING
This work was supported by National Institutes of Health [R01 DK044838, U54 HD012303 to P.L.M., T32 GM08666 to N.L.G.M.]; and Canadian Institutes of Health Research [MOP-81290 to R.W.].
ACKNOWLEDGEMENTS
We thank J. Bischof for tissue preparation, C. Abate-Shen for Msx1 and Dlx2 plasmids, J.L.R. Rubenstein for Dlx1 and Dlx5 plasmids, K. Watanabe for the Dlxin1 plasmid, and S. Radovick for the GN11 cells. We appreciate assistance from N.J.G. Webster with the microarray analysis and M.A. Lawson for statistical analysis. We are indebted to M.L. Givens for early advice, R. Larder for critical reading and members of the Mellon Lab for scientific discussions and support throughout this work.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Goldstone A.P. Prader–Willi syndrome: advances in genetics, pathophysiology and treatment. Trends Endocrinol. Metab. 2004;15:12–20. doi: 10.1016/j.tem.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Gunay-Aygun M., Schwartz S., Heeger S., O’Riordan M.A., Cassidy S.B. The changing purpose of Prader–Willi syndrome clinical diagnostic criteria and proposed revised criteria. Pediatrics. 2001;108:E92. doi: 10.1542/peds.108.5.e92. [DOI] [PubMed] [Google Scholar]
- 3.Taniura H., Kobayashi M., Yoshikawa K. Functional domains of necdin for protein–protein interaction, nuclear matrix targeting, and cell growth suppression. J. Cell. Biochem. 2005;94:804–815. doi: 10.1002/jcb.20345. [DOI] [PubMed] [Google Scholar]
- 4.Barker P.A., Salehi A. The MAGE proteins: emerging roles in cell cycle progression, apoptosis, and neurogenetic disease. J. Neurosci. Res. 2002;67:705–712. doi: 10.1002/jnr.10160. [DOI] [PubMed] [Google Scholar]
- 5.Ding F., Prints Y., Dhar M.S., Johnson D.K., Garnacho-Montero C., Nicholls R.D., Francke U. Lack of Pwcr1/MBII-85 snoRNA is critical for neonatal lethality in Prader–Willi syndrome mouse models. Mamm. Genome. 2005;16:424–431. doi: 10.1007/s00335-005-2460-2. [DOI] [PubMed] [Google Scholar]
- 6.Stefan M., Ji H., Simmons R.A., Cummings D.E., Ahima R.S., Friedman M.I., Nicholls R.D. Hormonal and metabolic defects in a Prader–Willi syndrome mouse model with neonatal failure to thrive. Endocrinology. 2005;146:4377–4385. doi: 10.1210/en.2005-0371. [DOI] [PubMed] [Google Scholar]
- 7.Gabriel J.M., Merchant M., Ohta T., Ji Y., Caldwell R.G., Ramsey M.J., Tucker J.D., Longnecker R., Nicholls R.D. A transgene insertion creating a heritable chromosome deletion mouse model of Prader-Willi and angelman syndromes. Proc. Natl Acad. Sci. USA. 1999;96:9258–9263. doi: 10.1073/pnas.96.16.9258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chamberlain S.J., Johnstone K.A., DuBose A.J., Simon T.A., Bartolomei M.S., Resnick J.L., Brannan C.I. Evidence for genetic modifiers of postnatal lethality in PWS-IC deletion mice. Hum. Mol. Genet. 2004;13:2971–2977. doi: 10.1093/hmg/ddh314. [DOI] [PubMed] [Google Scholar]
- 9.Yang T., Adamson T.E., Resnick J.L., Leff S., Wevrick R., Francke U., Jenkins N.A., Copeland N.G., Brannan C.I. A mouse model for Prader–Willi syndrome imprinting-centre mutations. Nat. Genet. 1998;19:25–31. doi: 10.1038/ng0598-25. [DOI] [PubMed] [Google Scholar]
- 10.Kuwako K., Hosokawa A., Nishimura I., Uetsuki T., Yamada M., Nada S., Okada M., Yoshikawa K. Disruption of the paternal necdin gene diminishes TrkA signaling for sensory neuron survival. J. Neurosci. 2005;25:7090–7099. doi: 10.1523/JNEUROSCI.2083-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takazaki R., Nishimura I., Yoshikawa K. Necdin is required for terminal differentiation and survival of primary dorsal root ganglion neurons. Exp. Cell Res. 2002;277:220–322. doi: 10.1006/excr.2002.5558. [DOI] [PubMed] [Google Scholar]
- 12.Skryabin B.V., Gubar L.V., Seeger B., Pfeiffer J., Handel S., Robeck T., Karpova E., Rozhdestvensky T.S., Brosius J. Deletion of the MBII-85 snoRNA gene cluster in mice results in postnatal growth retardation. PLoS Genet. 2007;3:e235. doi: 10.1371/journal.pgen.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren J., Lee S., Pagliardini S., Gerard M., Stewart C.L., Greer J.J., Wevrick R. Absence of Ndn, encoding the Prader–Willi syndrome-deleted gene necdin, results in congenital deficiency of central respiratory drive in neonatal mice. J. Neurosci. 2003;23:1569–1573. doi: 10.1523/JNEUROSCI.23-05-01569.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerard M., Hernandez L., Wevrick R., Stewart C.L. Disruption of the mouse necdin gene results in early post-natal lethality. Nat. Genet. 1999;23:199–202. doi: 10.1038/13828. [DOI] [PubMed] [Google Scholar]
- 15.Muscatelli F., Abrous D.N., Massacrier A., Boccaccio I., Le Moal M., Cau P., Cremer H. Disruption of the mouse Necdin gene results in hypothalamic and behavioral alterations reminiscent of the human Prader–Willi syndrome. Hum. Mol. Genet. 2000;9:3101–3110. doi: 10.1093/hmg/9.20.3101. [DOI] [PubMed] [Google Scholar]
- 16.Tsai T.F., Armstrong D., Beaudet A.L. Necdin-deficient mice do not show lethality or the obesity and infertility of Prader–Willi syndrome. Nat. Genet. 1999;22:15–16. doi: 10.1038/8722. [DOI] [PubMed] [Google Scholar]
- 17.Kuwajima T., Taniura H., Nishimura I., Yoshikawa K. Necdin interacts with the Msx2 homeodomain protein via MAGE-D1 to promote myogenic differentiation of C2C12 cells. J. Biol. Chem. 2004;279:40484–40493. doi: 10.1074/jbc.M404143200. [DOI] [PubMed] [Google Scholar]
- 18.Lee S., Walker C.L., Karten B., Kuny S.L., Tennese A.A., O’Neill M.A., Wevrick R. Essential role for the Prader–Willi syndrome protein necdin in axonal outgrowth. Hum. Mol. Genet. 2005;14:627–637. doi: 10.1093/hmg/ddi059. [DOI] [PubMed] [Google Scholar]
- 19.Tennese A.A., Gee C.B., Wevrick R. Loss of the Prader–Willi syndrome protein necdin causes defective migration, axonal outgrowth, and survival of embryonic sympathetic neurons. Dev. Dyn. 2008;237:1935–1943. doi: 10.1002/dvdy.21615. [DOI] [PubMed] [Google Scholar]
- 20.Kruger M., Ruschke K., Braun T. NSCL-1 and NSCL-2 synergistically determine the fate of GnRH-1 neurons and control necdin gene expression. EMBO J. 2004;23:4353–6443. doi: 10.1038/sj.emboj.7600431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Good D.J., Porter F.D., Mahon K.A., Parlow A.F., Westphal H., Kirsch I.R. Hypogonadism and obesity in mice with a targeted deletion of the Nhlh2 gene. Nat. Genet. 1997;15:397–401. doi: 10.1038/ng0497-397. [DOI] [PubMed] [Google Scholar]
- 22.Wray S., Grant P., Gainer H. Evidence that cells expressing luteinizing hormone-releasing hormone mRNA in the mouse are derived from progenitor cells in the olfactory placode. Proc. Natl Acad. Sci. USA. 1989;86:8132–8136. doi: 10.1073/pnas.86.20.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwanzel-Fukuda M., Pfaff D.W. Origin of luteinizing hormone-releasing hormone neurons. Nature. 1989;338:161–164. doi: 10.1038/338161a0. [DOI] [PubMed] [Google Scholar]
- 24.Schwanzel-Fukuda M., Jorgenson K.L., Bergen H.T., Weesner G.D., Pfaff D.W. Biology of normal luteinizing hormone-releasing hormone neurons during and after their migration from olfactory placode. Endocrinol. Rev. 1992;13:623–634. doi: 10.1210/edrv-13-4-623. [DOI] [PubMed] [Google Scholar]
- 25.Simonian S.X., Herbison A.E. Regulation of gonadotropin-releasing hormone (GnRH) gene expression during GnRH neuron migration in the mouse. Neuroendocrinology. 2001;73:149–156. doi: 10.1159/000054631. [DOI] [PubMed] [Google Scholar]
- 26.Givens M.L., Rave-Harel N., Goonewardena V.D., Kurotani R., Berdy S.E., Swan C.H., Rubenstein J.L., Robert B., Mellon P.L. Developmental regulation of gonadotropin-releasing hormone gene expression by the MSX and DLX homeodomain protein families. J. Biol. Chem. 2005;280:19156–19165. doi: 10.1074/jbc.M502004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masuda Y., Sasaki A., Shibuya H., Ueno N., Ikeda K., Watanabe K. Dlxin-1, a novel protein that binds Dlx5 and regulates its transcriptional function. J. Biol. Chem. 2001;276:5331–5338. doi: 10.1074/jbc.M008590200. [DOI] [PubMed] [Google Scholar]
- 28.Brunelli S., Tagliafico E., De Angelis F.G., Tonlorenzi R., Baesso S., Ferrari S., Niinobe M., Yoshikawa K., Schwartz R.J., Bozzoni I., et al. Msx2 and necdin combined activities are required for smooth muscle differentiation in mesoangioblast stem cells. Circ. Res. 2004;94:1571–1578. doi: 10.1161/01.RES.0000132747.12860.10. [DOI] [PubMed] [Google Scholar]
- 29.Burman P., Ritzen E.M., Lindgren A.C. Endocrine dysfunction in Prader–Willi syndrome: a review with special reference to GH. Endocr. Rev. 2001;22:787–799. doi: 10.1210/edrv.22.6.0447. [DOI] [PubMed] [Google Scholar]
- 30.Mellon P.L., Windle J.J., Goldsmith P., Pedula C., Roberts J., Weiner R.I. Immortalization of hypothalamic GnRH neurons by genetically targeted tumorigenesis. Neuron. 1990;5:1–10. doi: 10.1016/0896-6273(90)90028-e. [DOI] [PubMed] [Google Scholar]
- 31.Wetsel W.C., Valença M.M., Merchenthaler I., Liposits Z., López F.J., Weiner R.I., Mellon P.L., Negro-Vilar A. Intrinsic pulsatile secretory activity of immortalized LHRH secreting neurons. Proc. Natl Acad. Sci. USA. 1992;89:4149–4153. doi: 10.1073/pnas.89.9.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silverman A.J., Roberts J.L., Dong K.W., Miller G.M., Gibson M.J. Intrahypothalamic injection of a cell line secreting gonadotropin-releasing hormone results in cellular differentiation and reversal of hypogonadism in mutant mice. Proc. Natl Acad. Sci. USA. 1992;89:10668–10672. doi: 10.1073/pnas.89.22.10668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Radovick S., Wray S., Lee E., Nicols D.K., Nakayama Y., Weintraub B.D., Westphal H., Cutler GB, Jr, Wondisford F.E. Migratory arrest of gonadotropin-releasing hormone neurons in transgenic mice. Proc. Natl Acad. Sci. USA. 1991;88:3402–3406. doi: 10.1073/pnas.88.8.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsiao A., Worrall D.S., Olefsky J.M., Subramaniam S. Variance-modeled posterior inference of microarray data: gene-expression changes in 3T3-L1 adipocytes. Bioinformatics. 2004;20:3108–3127. doi: 10.1093/bioinformatics/bth371. [DOI] [PubMed] [Google Scholar]
- 35.Alarid E.T., Windle J.J., Whyte D.B., Mellon P.L. Immortalization of pituitary cells at discrete stages of development by directed oncogenesis in transgenic mice. Development. 1996;122:3319–3329. doi: 10.1242/dev.122.10.3319. [DOI] [PubMed] [Google Scholar]
- 36.Givens M.L., Kurotani R., Rave Harel N., Miller N.L.G., Mellon P.L. Phylogenetic footprinting reveals functional upstream regions of the gonadotropin-releasing hormone gene that enhance cell-specific expression. Mol. Endocrinol. 2004;18:2950–2966. doi: 10.1210/me.2003-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsumoto K., Taniura H., Uetsuki T., Yoshikawa K. Necdin acts as a transcriptional repressor that interacts with multiple guanosine clusters. Gene. 2001;272:173–179. doi: 10.1016/s0378-1119(01)00544-3. [DOI] [PubMed] [Google Scholar]
- 38.Rave-Harel N., Miller N.L.G., Givens M.L., Mellon P.L. The Groucho-related gene family regulates the gonadotropin-releasing hormone gene through interaction with the homeodomain proteins MSX1 and OCT1. J. Biol. Chem. 2005;280:30975–30983. doi: 10.1074/jbc.M502315200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuda T., Suzuki H., Oishi I., Kani S., Kuroda Y., Komori T., Sasaki A., Watanabe K., Minami Y. The receptor tyrosine kinase Ror2 associates with the melanoma-associated antigen (MAGE) family protein Dlxin-1 and regulates its intracellular distribution. J. Biol. Chem. 2003;278:29057–29064. doi: 10.1074/jbc.M302199200. [DOI] [PubMed] [Google Scholar]
- 40.Eiholzer U., l’Allemand D., Rousson V., Schlumpf M., Gasser T., Girard J., Gruters A., Simoni M. Hypothalamic and gonadal components of hypogonadism in boys with Prader–Labhart–Willi syndrome. J. Clin. Endocrinol. Metab. 2006;91:892–898. doi: 10.1210/jc.2005-0902. [DOI] [PubMed] [Google Scholar]
- 41.Andrieu D., Watrin F., Niinobe M., Yoshikawa K., Muscatelli F., Fernandez P.A. Expression of the Prader-Willi gene Necdin during mouse nervous system development correlates with neuronal differentiation and p75NTR expression. Gene Expr. Patterns. 2003;3:761–765. doi: 10.1016/s1567-133x(03)00138-8. [DOI] [PubMed] [Google Scholar]
- 42.Yoshikawa K. Cell cycle regulators in neural stem cells and postmitotic neurons. Neurosci. Res. 2000;37:1–14. doi: 10.1016/s0168-0102(00)00101-2. [DOI] [PubMed] [Google Scholar]
- 43.Aizawa T., Maruyama K., Kondo H., Yoshikawa K. Expression of necdin, an embryonal carcinoma-derived nuclear protein, in developing mouse brain. Brain Res. Dev. Brain Res. 1992;68:265–274. doi: 10.1016/0165-3806(92)90069-9. [DOI] [PubMed] [Google Scholar]
- 44.Lee S., Walker C.L., Wevrick R. Prader–Willi syndrome transcripts are expressed in phenotypically significant regions of the developing mouse brain. Gene Expr. Patterns. 2003;3:599–609. doi: 10.1016/s1567-133x(03)00113-3. [DOI] [PubMed] [Google Scholar]
- 45.Kendall S.K., Samuelson L.C., Saunders T.L., Wood R.I., Camper S.A. Targeted disruption of the pituitary glycoprotein hormone α-subunit produces hypogonadal and hypothyroid mice. Genes Dev. 1995;9:2007–2018. doi: 10.1101/gad.9.16.2007. [DOI] [PubMed] [Google Scholar]
- 46.Luo X., Ikeda Y., Parker K.L. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell. 1994;77:481–490. doi: 10.1016/0092-8674(94)90211-9. [DOI] [PubMed] [Google Scholar]
- 47.Gill J.C., Wadas B., Chen P., Portillo W., Reyna A., Jorgensen E., Mani S., Schwarting G.A., Moenter S.M., Tobet S., et al. The gonadotropin-releasing hormone (GnRH) neuronal population is normal in size and distribution in GnRH-deficient and GnRH receptor-mutant hypogonadal mice. Endocrinology. 2008;149:4596–4604. doi: 10.1210/en.2008-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herbison A.E., Porteous R., Pape J.R., Mora J.M., Hurst P.R. Gonadotropin-releasing hormone (GnRH) neuron requirements for puberty, ovulation and fertility. Endocrinology. 2008;149:597–604. doi: 10.1210/en.2007-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pitteloud N., Quinton R., Pearce S., Raivio T., Acierno J., Dwyer A., Plummer L., Hughes V., Seminara S., Cheng Y.Z., et al. Digenic mutations account for variable phenotypes in idiopathic hypogonadotropic hypogonadism. J. Clin. Invest. 2007;117:457–463. doi: 10.1172/JCI29884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang H., Bailey J.S., Coss D., Lin B., Tsutsumi R., Lawson M.A., Mellon P.L., Webster N.J. Activin modulates the transcriptional response of LβT2 cells to GnRH and alters cellular proliferation. Mol. Endocrinol. 2006;20:2909–2930. doi: 10.1210/me.2006-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lawson M.A., Tsutsumi R., Zhang H., Talukdar I., Butler B.K., Santos S.J., Mellon P.L., Webster N.J. Pulse sensitivity of the luteinizing hormone beta promoter is determined by a negative feedback loop Involving early growth response-1 and Ngfi-A binding protein 1 and 2. Mol. Endocrinol. 2007;21:1175–1191. doi: 10.1210/me.2006-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]