Abstract
Spinal muscular atrophy (SMA), a common neuromuscular disorder, is caused by homozygous absence of the survival motor neuron gene 1 (SMN1), while the disease severity is mainly influenced by the number of SMN2 gene copies. This correlation is not absolute, suggesting the existence of yet unknown factors modulating disease progression. We demonstrate that the SMN2 gene is subject to gene silencing by DNA methylation. SMN2 contains four CpG islands which present highly conserved methylation patterns and little interindividual variations in SMN1-deleted SMA patients. The comprehensive analysis of SMN2 methylation in patients suffering from severe versus mild SMA carrying identical SMN2 copy numbers revealed a correlation of CpG methylation at the positions −290 and −296 with the disease severity and the activity of the first transcriptional start site of SMN2 at position −296. These results provide first evidence that SMN2 alleles are functionally not equivalent due to differences in DNA methylation. We demonstrate that the methyl-CpG-binding protein 2, a transcriptional repressor, binds to the critical SMN2 promoter region in a methylation-dependent manner. However, inhibition of SMN2 gene silencing conferred by DNA methylation might represent a promising strategy for pharmacologic SMA therapy. We identified histone deacetylase (HDAC) inhibitors including vorinostat and romidepsin which are able to bypass SMN2 gene silencing by DNA methylation, while others such as valproic acid and phenylbutyrate do not, due to HDAC isoenzyme specificities. These findings indicate that DNA methylation is functionally important regarding SMA disease progression and pharmacological SMN2 gene activation which might have implications for future SMA therapy regimens.
INTRODUCTION
Autosomal recessive proximal spinal muscular atrophy (SMA) is a severely progressing neuromuscular disorder and a major cause of inherited childhood lethality. SMA is characterized by the loss of lower motor neurons in the anterior horns of the spinal cord, causing symmetrical weakness and atrophy of voluntary muscles. Patients with SMA have been classified into four types depending on age of onset and progression of the disease: type I SMA is the most severe form with generalized muscle weakness and hypotonia and a disease onset within the first 6 months of life. The children are unable to sit or walk and usually die within the first 2 years of life. Type II SMA individuals are able to sit but unable to walk unaided. They usually present first symptoms after the first 6 months of life and survive beyond 2 years. Type III SMA patients are able to sit and walk, and the lifespan is not reduced. Disease onset before the age of 3 years is classified as type IIIa, whereas an age of onset beyond 3 years is classified as type IIIb SMA. Type IV SMA patients are mildly affected with an age of onset after the third decade of life (reviewed in 1).
The disease determining survival motor neuron gene 1 (SMN1) is homozygously absent in 96% of all SMA patients and intragenic SMN1 mutations are rare (2). Within the SMA region on chromosome 5q, the human survival motor neuron (SMN) gene exists in two copies, SMN1 and SMN2, which are ubiquitously expressed and encode identical proteins (3). Even though all SMA patients lacking SMN1 carry at least one SMN2 gene copy, the amount of functional SMN protein produced by SMN2 is not sufficient to prevent progressive α-motor neuron degeneration. This finding has been assigned to a single translationally silent ‘C’ to ‘T’ transition within exon 7, affecting the splicing of primary SMN transcripts (4). As a consequence, the disease determining SMN1 gene produces full-length transcripts only (FL-SMN), whereas the majority of SMN2 transcripts lack exon 7 due to alternative splicing (Δ7-SMN). Truncated Δ7-SMN proteins are reduced in their ability to self-oligomerize, which is essential for proper SMN function (5,6), and have been shown to ameliorate, but not to prevent, the SMA phenotype in vivo (7). However, several studies have revealed a strong inverse correlation between the number of SMN2 copies and SMA severity (8–11). Most type I SMA patients carry two SMN2 copies, whereas type II SMA patients carry three and type III SMA patients carry three or four SMN2 copies. Rarely, patients with two SMN2 copies show mild phenotypes (9), suggesting the influence of yet unidentified modifying factors modulating disease progression. Due to the disease modifying property of the SMN2 gene which has been verified in transgenic mouse models (12), SMN2 represents the major therapeutic target. Consequently, transcriptional SMN2 activation and/or modulation of the SMN2 splicing pattern to increase FL-SMN levels may be an effective strategy for SMA treatment.
Numerous small-molecule histone deacetylase (HDAC) inhibitors have been shown to increase SMN2-derived FL-SMN protein levels in vitro by transcriptional activation and/or by modulation of the SMN2 splicing pattern. These compounds include the fatty acids sodium butyrate (SB), phenylbutyrate (PB) and valproic acid (VPA) (13–17); the benzamide M344 (15,18) as well as the hydroxamic acids SAHA and trichostatin A (TSA) (15,19,20). The potential applicability of HDAC inhibitors for SMA therapy was confirmed in transgenic mice which mimic SMA-like features. By employing a knockout transgenic mouse model, Chang et al. (17) demonstrated that oral application of SB increased lifespan and SMN2-derived SMN protein levels in motor neurons of affected mice. Concordantly, Tsai et al. (21) demonstrated that oral application of VPA attenuates motor neuron death, increases spinal SMN protein levels and partially normalizes motor function in SMA-like mice. Clinical phase II trials have been initiated to evaluate the efficacy of the FDA-approved drugs PB and VPA for SMA therapy. Pilot trials with small numbers of patients treated with PB (22,23) or VPA (24,25) revealed elevated FL-SMN2 transcript levels in some patients and increased quantitative muscle strength and subjective muscle function, which might further emphasize the potential use of HDAC inhibitors for SMA treatment.
However, the efficacies of PB and VPA for SMA therapy await clinical confirmation and the mechanism(s) by which HDAC inhibitors elevate transcriptional SMN2 gene activity remain elusive. In general, HDAC inhibition promotes a more relaxed chromatin structure, allowing transcriptional activation. Given the fact that the fundamental mechanisms of epigenetic gene regulation, histone modification and DNA methylation have shown to be intimately interlinked (26,27), we hypothesized that DNA methylation plays a pivotal role in epigenetic SMN2 gene regulation and SMA pathogenesis. DNA methylation has been shown to be the most stable type of epigenetic modification modulating the transcriptional plasticity of mammalian genomes (28). However, the role of DNA methylation in SMN2 gene regulation has not been addressed so far. In this study, we demonstrate for the first time that the SMN2 gene is subject to gene silencing by DNA methylation, which might have major implications for SMA disease progression and upcoming pharmacological interventions for epigenetic SMA therapy.
RESULTS
SMN2 gene activity is reduced by DNA methylation
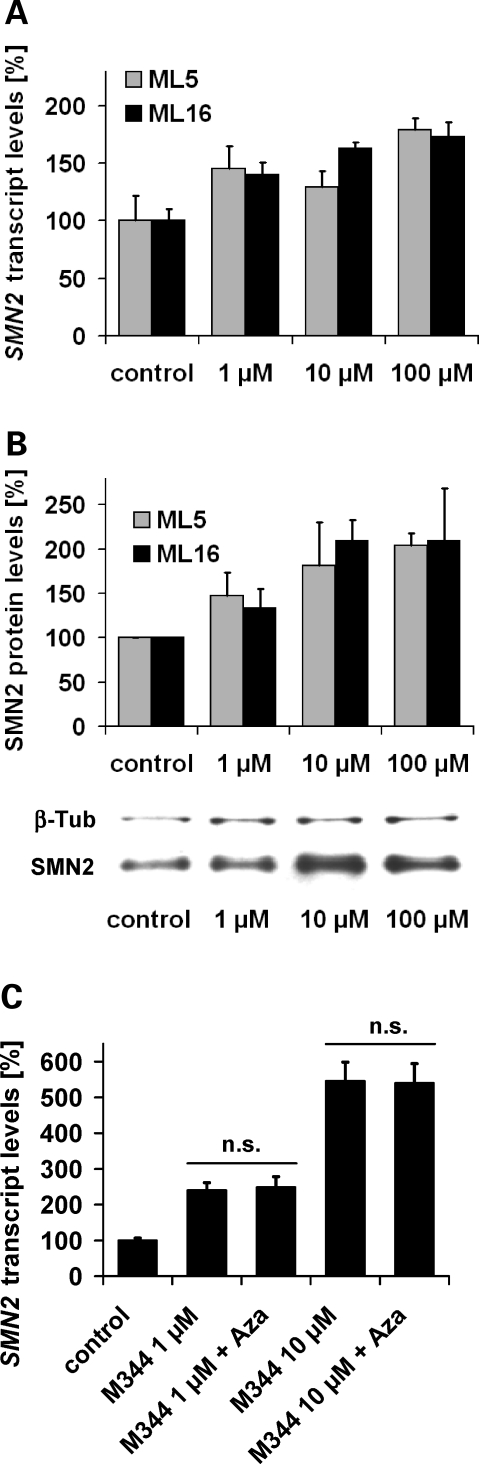
We raised the question whether the major target gene for SMA therapy, SMN2, is subject to gene silencing mediated by DNA methylation. To address this issue, SMN1-deleted fibroblast cell lines (ML5, ML16) derived from two independent SMA patients carrying three SMN2 copies were treated with various doses of 5-aza-2′-deoxycytidine (Aza), a well-established DNA-demethylating compound. Aza treatment increased SMN2-derived transcript and protein levels in a dose-dependent fashion in both SMA fibroblast cell lines (Fig. 1A and B), indicating that SMN2 gene activity is inversely correlated with DNA methylation. Using both cell lines, we previously have shown that HDAC inhibitors augment SMN2 protein levels by transcriptional SMN2 gene activation and increased SMN2 exon 7 inclusion (14,15,18). Concordantly, treatment of SMA fibroblasts (ML16) with effective doses of the experimental pan-HDAC inhibitor M344 significantly increased total SMN2 transcript levels (Fig. 1C). However, even though interference with either DNA methylation or HDAC activity was sufficient to increase SMN2 gene expression, there is no additive or synergistic effect when combined (Fig. 1C).
Figure 1.
Transcriptional survival motor neuron gene 2 (SMN2) activation by the DNA-demethylating drug 5-aza-2′-deoxycytidine (Aza) in SMN1-deleted SMA (spinal muscular atrophy) fibroblasts. Bar graphs show SMN2 transcript (A) and SMN2 protein levels (B) in ML5 and ML16 fibroblast cell lines treated with various doses of Aza for 72 h (mean ± SEM). Expression levels in solvent- and time-matched controls were set to 100%. Increased SMN2 transcript levels (normalized to ß-actin) were observed following Aza treatment (1, 10, 100 µm) and a dose-dependent SMN2 gene activation was confirmed on protein level (normalized to ß-tubulin). A representative western blot analysis (ML16) is shown. (C) Quantification of SMN2 transcript levels (normalized to ß-actin) in ML16 cells after treatment with different doses of the SMN2-activating HDAC (histone deacetylase) inhibitor M344 either alone or in combination with 10 µm Aza for 48 h. Transcriptional SMN2 gene activation either by M344 alone or in combination with Aza (10 µm) reached levels of significance in all cases (P < 0.05, t-test). A >5-fold induction was observed following treatment with 10 µm M344 (546 ± 53%), while no additive effects were observed after co-treatment with 10 µm Aza (P > 0.05, t-test).
SMN2 is a methylated gene containing four CpG islands
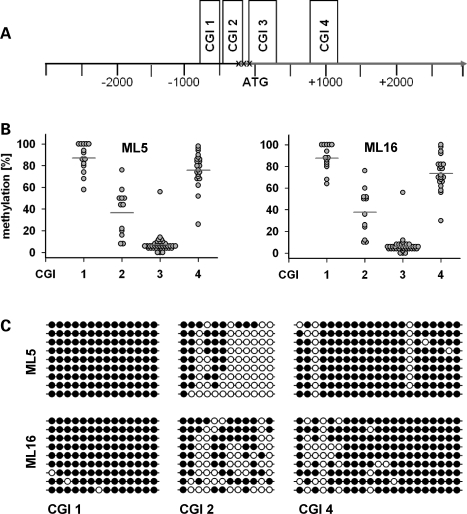
To identify putative CpG islands (CGIs) involved in epigenetic SMN2 gene regulation by DNA methylation, we analysed the genomic region 3000 nucleotides (nt) upstream and downstream of the translational SMN2 start site (defined as nt position +1). By employing the CGI finder algorithm (www.EBI.ac.uk/emboss) we identified four putative CGIs that localize within a ∼2 kb genomic region (nt −896 to +1146) surrounding the translational SMN2 start site (Fig. 2A). These four putative CGIs, subsequently referred to as SMN2CGIs, contain a total of 85 CpG dinucleotides, 14 of which are located in SMN2CGI 1 (nt −896 to −645), 12 in SMN2CGI 2 (nt −469 to −247), 38 in SMN2CGI 3 (nt −151 to +295) and 21 in SMN2CGI 4 (nt +844 to +1146). To quantify DNA methylation levels of each CpG dinucleotide located within the four SMN2CGIs, we performed bisulphite genomic sequencing using pyrosequencing technology (29). The method is based on the selective deamination of unmethylated cytosine to uracil by treatment with bisulphite, while 5-methylcytosine remains unchanged. Pyrosequencing of subsequently generated PCR (polymerase chain reaction) products allows for both the detection and the quantification of cytosine methylation. Using genomic DNA isolated from SMN1-deleted SMA fibroblasts (ML5, ML16), bisulphite genomic sequencing revealed almost identical SMN2 methylation patterns in both SMA fibroblast cell lines (Fig. 2B). SMN2CGI 1 and SMN2CGI 4 were hypermethylated in both cell lines with mean methylation levels >70%. SMN2CGI 3 was hypomethylated (mean methylation levels <10%) with a single outlier at nt position +89, while SMN2CGI 2 displayed intermediate mean methylation levels. Using bisulphite-treated DNA derived from ML5 and ML16 SMA fibroblasts as templates, cloning and sequencing of PCR products revealed similar results. Again, SMN2CGI 1 and SMN2CGI 4 were almost completely methylated, SMN2CGI 3 was nearly unmethylated (data not shown), while SMN2CGI 2 showed intermediate methylation levels (Fig. 2C).
Figure 2.
Survival motor neuron gene 2 (SMN2) is a methylated gene containing four CpG islands (CGIs). Putative CGIs within the genomic region 3000 nucleotides (nt) upstream and downstream the translational SMN2 start site (NCBI36: 5:70254287:70248287) were identified using the CGI finder and plotting tool (www.EBI.ac.uk/emboss). This algorithm identified four putative CGIs within the respective genomic region (SMN2CGI 1: nt −896 to −645, SMN2CGI 2: nt −469 to −247, SMN2CGI 3: nt −151 to +295, SMN2CGI 4: nt +844 to +1146). SMN2CGI 2 contains the first out of three transcriptional start sites (TSS) of SMN2 at nt −296, while the second TSS at nt position −242 is located close to the downstream border of SMN2CGI 2 (30,31). (A) Schematically illustrates the localizations of all four SMN2CGIs within the analysed genomic region. The localizations of the TSS at nt positions −296, −242 and −163 are indicated (30,31). (B) Frequency plots illustrating the methylation levels of each CpG dinucleotide within each SMN2CGI in the respective SMA(Spinal muscular atrophy)-fibroblasts. The mean methylation levels of each SMN2CGI, indicated by a horizontal line, are as follows: ML5: SMN2CGI 1: 87.1%, SMN2CGI 2: 36.6%, SMN2CGI 3: 6.9%, SMN2CGI 4: 75.9%; ML16: SMN2CGI 1: 87.7%, SMN2CGI 2: 37.7%, SMN2CGI 3: 6.2%, SMN2CGI 4: 73.4%. (C) Methylation analysis of SMN2CGI 1, SMN2CGI 2 and SMN2CGI 4 in ML5 and ML16 fibroblasts cell lines by bisulphite treatment, followed by PCR amplification of the respective SMN2CGI, cloning of PCR products, and sequencing. The methylation patterns of nine independent clones for each SMN2CGI and each cell line are shown. Empty circles represent unmethylated CpGs, whereas full circles correspond to their methylated status.
SMN2 gene methylation patterns are highly conserved and correlate with SMA severity
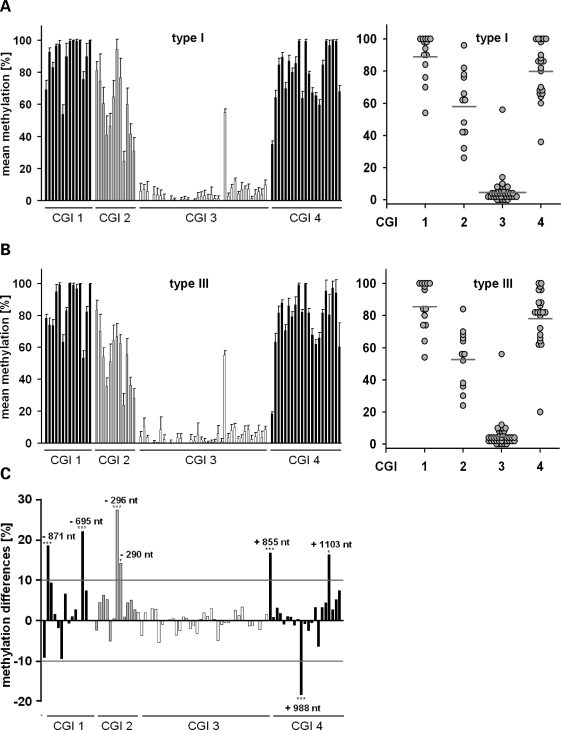
The vast majority of SMN1-deleted SMA patients carrying two SMN2 copies suffer from type I or II SMA, while only few patients (∼2%) develop a mild (type III SMA) phenotype (9), suggesting the existence of a yet unknown factor counteracting disease progression. To address whether SMN2 gene methylation correlates with the disease severity, we analysed methylation levels of each CpG dinucleotide within the SMN2CGIs using DNA isolated from blood samples taken from 10 unrelated, SMN1-deleted type I SMA patients carrying two SMN2 copies (six females, four males). Bisulphite genomic sequencing identified SMN2 methylation patterns similar to those observed in SMA fibroblasts (Fig. 3A), suggesting that SMN2 gene methylation patterns do not essentially differ between tissues. Again, SMN2CGI 1 and SMN2CGI 4 appeared to be hypermethylated, SMN2CGI 3 hypomethylated with a single outlier at position +89, while SMN2CGI 2 displayed intermediate mean methylation levels. An unexpected observation was that methylation levels of each single CpG dinucleotide showed only little interindividual variations between the 10 type I SMA patients (Fig. 3A). The subsequent analysis of DNA isolated from blood samples taken from seven available SMN1-deleted type III SMA patients carrying two SMN2 copies (four females, three males) again revealed little interindividual variations (Fig. 3B). However, the comparison of methylation levels of each CpG dinucleotide identified significant correlations between CpG methylation levels and the disease severity at seven CpGs located in SMN2CGI 1 (nt −871, −695), SMN2CGI 2 (nt −296, −290), and SMN2CGI 4 (nt +855, +988, +1103) (Fig. 3C). At six sites (nt −871, −695, −296, −290, +855, +1103), significantly lower methylation frequencies were observed in type III compared with type I SMA patients. A striking finding at this point was that the most pronounced differences in CpG methylation levels were observed at nt position −296 (SMN2CGI 2; type I SMA: 94.1 ± 6.3%; type III SMA: 66.7 ± 8.0%, P = 0.000084, t-test), which exactly represents the first out of three transcriptional start sites (TSS) of SMN2 (Fig. 2A) (30,31). The correlation of CpG methylation levels of all 17 patients with their gender (10 females, seven males) did not reveal any significant results (data not shown), which is in line with a previous comprehensive study showing that DNA methylation is not affected by gender (28).
Figure 3.
Survival motor neuron gene 2 (SMN2) methylation patterns are highly conserved in blood samples derived from spinal muscular atrophy (SMA) patients. Bar charts show the mean methylation levels (±SEM) of each CpG dinucleotide within the respective SMN2CGIs in DNA isolated from blood samples drawn from SMN1-deleted SMA patients carrying two SMN2 copies (A, B). Type I (n = 10) and type III SMA patients (n = 7) show highly conserved SMN2 methylation patterns as can be deduced from the comparatively low SEM for the methylation levels of each CpG dinucleotide. The mean methylation levels of each SMN2CGI (see frequency plot) in both groups are as follows: type I SMA – SMN2CGI 1: 89.1%, SMN2CGI 2: 58.0%, SMN2CGI 3: 4.6%, SMN2CGI 4: 79.9%; type III SMA – SMN2CGI 1: 85.5%, SMN2CGI 2: 52.6%, SMN2CGI 3: 4.8%, SMN2CGI 4: 78.1%. SMN2 methylation correlates with the disease severity. Bar chart shows differences in DNA methylation at each CpG dinucleotide in type I versus type III SMA patients (C). Seven CpG dinucleotides (nt −871, −695, −296, −290, +855, +988, +1103) were identified whose methylation frequencies differ by at least 10% in type I versus type III SMA patients. Correlations between CpG methylation frequencies and disease severity reached levels of significance in all seven cases. Three levels of statistical significance were discriminated: *P < 0.05, **P < 0.01, ***P < 0.001 (t-test).
Reduced methylation at positions −296 and −290 of the SMN2 gene is associated with mild SMA in SMA fibroblasts
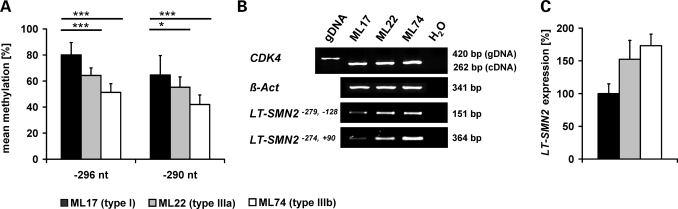
To validate a correlation between SMN2 gene methylation and the disease severity we analysed fibroblasts cell lines derived from independent SMN1-deleted SMA patients, also carrying two SMN2 copies. Repeated quantification of CpG methylation levels at nt positions −296 and −290 (both SMN2CGI 2) corroborated a significant correlation between CpG methylation and disease severity in type I (ML17), type IIIa (ML22) and type IIIb SMA fibroblasts (ML74) (Fig. 4A). No significant differences were observed at the remaining five positions (−871, −695, +855, +988, +1103, data not shown) demonstrated to be differentially methylated in DNA isolated from blood (Fig. 3C). Given the fact that the nearby CpG dinucleotides at nt positions −296 and −290 co-localize with the first TSS of SMN2 located at nt position −296, it appears evident that methylation levels at these sites modulate the expression of SMN2 transcripts derived from this TSS (subsequently designated as the long transcript SMN2, LT-SMN2). Using different primer combinations which specifically amplify LT-SMN2, an inverse correlation between −296 and −290 methylation levels and LT-SMN2 transcript levels was observed in type I, type IIIa and type IIIb SMA fibroblast cell lines (Fig. 4B and C). However, no significant differences in total SMN2 protein levels were detected in ML17, ML22 and ML74 fibroblasts due to the finding that LT-SMN2 transcripts account for <5% of total SMN2 transcripts in these cells (data not shown).
Figure 4.
Low survival motor neuron gene 2 (SMN2) methylation levels at the positions −296 and −290 are associated with mild spinal muscular atrophy (SMA) in SMA fibroblasts. (A) Bar chart showing the mean methylation levels (±SEM) at positions −296 and −290 in fibroblasts derived from SMN1-deleted SMA patients carrying two SMN2 copies. At both sites, type I SMA fibroblasts (ML17) show significantly higher methylation levels than fibroblasts derived from type IIIa (ML22) and type IIIb (ML74) patients. Three levels of statistical significance were discriminated: *P < 0.05, **P < 0.01, ***P < 0.001 (t-test). Methylation levels at positions −296 and −290 correlate with the activity of the transcriptional start site (TSS) at position −296 in SMA fibroblasts. Relative to type I SMA fibroblasts (ML17), the low methylation levels at positions −296 and −290 observed in type IIIa and type IIIb SMA fibroblasts are associated with high SMN2 transcripts levels originated from the TSS at position −296 (LT-SMN2) as shown by semi-quantitative RT–PCR analyses using two different primer combinations. (B) ß-Actin primers were used as internal control to verify equal loading of cDNA. CDK4 primers were used to detect potential contaminations with genomic DNA (amplicon size cDNA: 262 bp, amplicon size gDNA: 420 bp). (C) Differences in LT-SMN2 expression levels were confirmed by real-time PCR (normalized to ß-actin) using the LT-SMN2−279,−128 primer pair. Data are given as mean percentage (±SEM) relative to LT-SMN2 transcript levels in type I SMA fibroblasts (ML17) which were set to 100%.
HDAC inhibitor treatment bypasses LT-SMN2 gene silencing mediated by DNA methylation
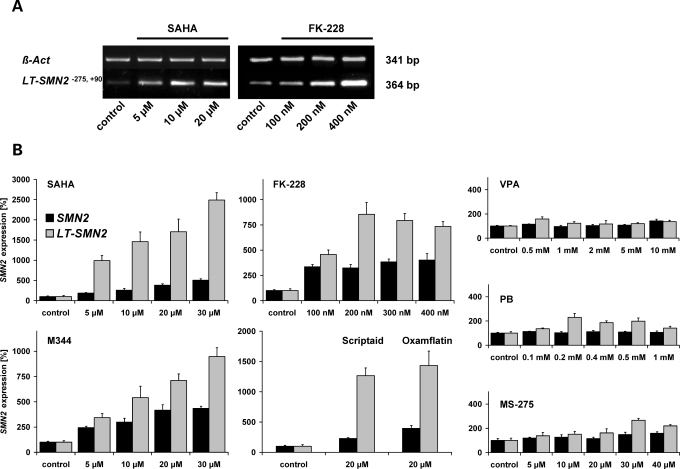
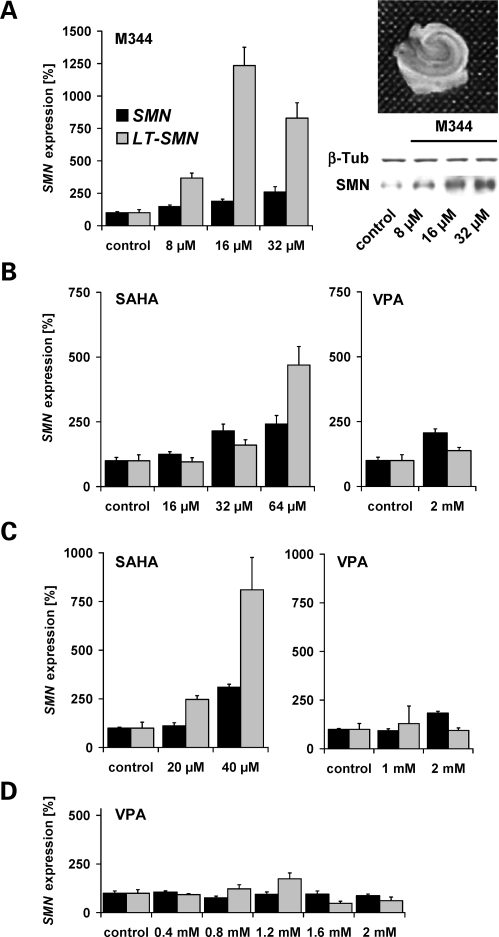
Based on the observation that DNA methylation restricts SMN2 gene activity (Fig. 1A and B), we hypothesized that the abolition of SMN2 gene silencing conferred by DNA methylation might represent a promising strategy for pharmacologic SMA therapy. In 1998, Jones et al. (26) have shown that the experimental pan-HDAC inhibitor TSA is able to bypass gene silencing mediated by DNA methylation. To address this issue, type I SMA fibroblasts (ML17) showing high methylation levels at positions −296 and −290 associated with low LT-SMN2 mRNA expression (Fig. 4) were treated with effective doses of highly potent HDAC inhibitors of different chemical classes. Specifically, cells were exposed to SAHA, M344, scriptaid, oxamflatin (hydroxamic acids), or romidepsin (FK-228, cyclic tetrapeptide) for 48 h. All five test compounds dramatically increased LT-SMN2 transcript levels dose dependently, indicating that HDAC inhibitors are able to relieve LT-SMN2 gene silencing mediated by DNA methylation (Fig. 5A and B). In all analyses, the dose-dependent inductions of LT-SMN2 expression were associated with robustly increased total SMN2 transcript levels. For example, treatment with 30 µm doses of SAHA results in a ∼25-fold induction of LT-SMN2 and a 5-fold induction of total SMN2 transcript levels (Fig. 5B). However, the hitherto analysed HDAC inhibitors possess negligible HDAC isoenzyme selectivities and concordantly we previously have shown that these compounds are able to fully antagonize HDAC activity in vitro (32). Thus, we next assessed whether isoenzyme selective HDAC inhibitors such as MS-275 as well as VPA and PB, which are both under clinical evaluation for SMA therapy, show similar effects. Interestingly, neither the benzamide MS-275 nor the fatty acids VPA and PB were able to substantially elevate LT-SMN2 transcript levels even at excessive doses (Fig. 5B), indicating that MS-275, VPA and PB were not able to overcome LT-SMN2 gene silencing conferred by DNA methylation in this experimental setting. To confirm these findings in a neuroectodermal tissue we subsequently employed human hippocampal brain slice cultures (OHSCs) derived from epilepsy surgery (four individuals, Fig. 6A–D). Even though LT-SMN transcripts derived from the SMN1 or SMN2 genes were not distinguishable in this experimental paradigm due to identical promoter sequences (33), a pronounced induction of LT-SMN transcription was observed following treatment with M344 (Fig. 6A) or SAHA (Fig. 6B and C), while VPA again had no or only moderate effects (Fig. 6B–D). A dose-dependent induction of SMN expression following treatment with M344 was confirmed on protein level (Fig. 6A).
Figure 5.
Histone deacetylase (HDAC) inhibitor treatment bypasses LT-SMN2 gene silencing mediated by DNA methylation. Treatment of type I SMA (spinal muscular atrophy) fibroblasts (ML17) with increasing doses of SAHA (hydroxamic acid) or FK-228 (cyclic tetrapeptide) elevates LT-SMN2 transcript levels in a dose-dependent manner as shown by semi-quantitative RT–PCR analyses. (A) ß-Actin primers were used as internal control to verify equal loading of cDNA. LT-SMN2 and total SMN2 transcript levels in type I SMA fibroblasts (ML17) following treatment with HDAC inhibitors for 48 h were further analysed by real-time PCR. (B) LT-SMN2 and total SMN2 transcript levels (both normalized to ß-actin) following HDAC inhibitor treatment are given as mean percentages (±SEM) relative to LT-SMN2 and total SMN2 transcript levels in solvent- and time-matched controls which were set to 100%.
Figure 6.
HDAC (histone deacetylase) inhibitor treatment elevates LT-SMN expression in human organotypic hippocampal brain slice cultures (OHSCs). (A–D) Bar charts showing LT-SMN and total survival motor neuron (SMN) transcript levels in human OHSCs derived from four different individuals. OHSCs were treated with the indicated doses of M344, SAHA or VPA (valproic acid) for 48 h. LT-SMN and total SMN transcript levels (both normalized to ß-actin) are given as mean percentages (±SEM) relative to LT-SMN and total SMN transcript levels in solvent- and time-matched controls which were set to 100%. Using OHSCs derived from the first patient (A), a dose-dependent SMN induction was confirmed on protein level (relative to ß-tubulin; 8 µm: 138 ± 2%; 16 µm: 188 ± 30%; 32 µm: 219 ± 41%). A representative human OHSC and a representative western blot analysis are shown. Induction of SMN protein expression by M344 reached levels of significance at all indicated doses (P < 0.05, t-test).
Transcriptional LT-SMN2 induction by HDAC inhibitors is not mediated by DNA-demethylation of the SMN2 gene
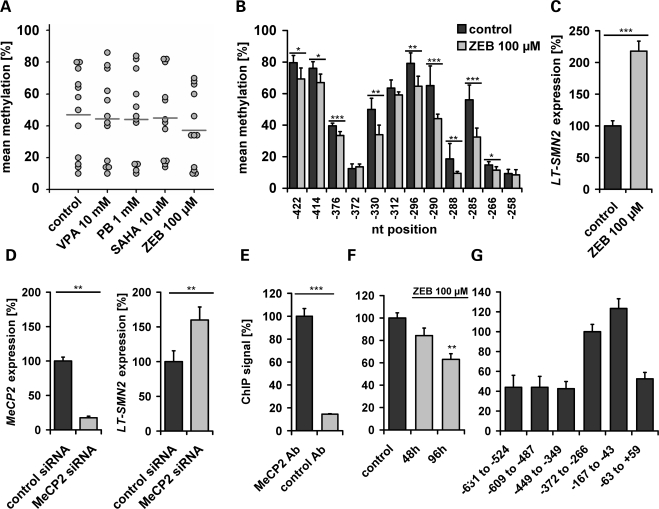
The mechanism by which pan-HDAC inhibitors such as SAHA and romidepsin (FK-228) bypass LT-SMN2 silencing by DNA methylation remains elusive. In single cell cultures, TSA and VPA have shown to trigger DNA demethylation in a replication-independent manner (34–36) and gene-specific DNA demethylating activities of VPA, MS-275 and TSA have subsequently been confirmed in vivo (37,38). To assess whether HDAC inhibitors elevate LT-SMN2 transcript levels due to their propensity to promote DNA demethylation, we next analysed the methylation levels of SMN2CGI 2 in type I SMA fibroblasts (ML17) following HDAC inhibitor treatment for 48 h. Using 10 µm doses of SAHA, this experimental setting has shown to substantially elevate LT-SMN2 and total SMN2 transcript levels (Fig. 5A and B). However, SAHA did not promote SMN2CGI 2 demethylation and neither did VPA or PB as shown by bisulphite genomic sequencing (Fig. 7A). In contrast, treatment of ML17 cells with zebularine, a DNA-demethylating drug structurally related to Aza, induced SMN2CGI 2 demethylation (Fig. 7A and B), and consistent with previous findings, reduced SMN2CGI 2 methylation levels including the nt positions −296 and −290 associated with significantly increased LT-SMN2 transcript levels (Fig. 7B and C). These results suggest that transcriptional LT-SMN2 induction by HDAC inhibitors is not mediated by DNA-demethylation of the SMN2 gene promoter.
Figure 7.
LT-SMN2 induction by HDAC (histone deacetylase) inhibitors is not mediated by survival motor neuron gene 2 (SMN2) promoter demethylation. (A) Treatment of type I spinal muscular atrophy (SMA) fibroblasts (ML17) with valproic acid (VPA) (10 mm), phenylbutyrate (PB) (1 mm) or SAHA (10 µm) for 48 h did not affect SMN2CGI 2 methylation levels. Frequency plot illustrating methylation levels of each CpG dinucleotide within the SMN2CGI 2 in treated ML17 cells compared with time- and solvent-matched ML17 control fibroblasts. The DNA-demethylating drug zebularine (ZEB, 100 µm) was used as positive control. The mean methylation levels of SMN2CGI 2, indicated by horizontal lines, are as follows – control: 47.0%, VPA: 44.5%, PB 44.1%, SAHA: 45.0%, ZEB: 37.3%. Demethylation of CpG dinucleotides at positions −296 and −290 is associated with increased LT-SMN2 expression. Treatment of type I SMA fibroblasts (ML17) with ZEB (100 µm, 48 h) results in a significant demethylation of nine out of 12 CpG dinucleotides located in SMN2CGI 2, including the nt positions −296 and −290. (B) ZEB-induced SMN2CGI 2 demethylation is associated with increased LT-SMN2 levels as shown by quantitative real-time PCR. LT-SMN2 transcript levels (normalized to ß-actin) are given as mean percentages (±SEM) relative to LT-SMN2 expression levels in solvent- and time-matched controls set to 100%. (C) siRNA-mediated knockdown of MeCP2 elevates LT-SMN2 transcript levels in ML17 SMA fibroblasts. (D) MeCP2 and LT-SMN2 transcript levels (normalized to ß-actin) are given as mean percentages (±SEM) relative to MeCP2 or LT-SMN2 expression levels in time-matched controls transfected with AllStars negative control siRNA. The transcriptional co-repressor methyl-CpG-binding-protein 2 (MeCP2) is associated with the SMN2 promoter region and binds in a methylation-dependent fashion. (E, F) Using SMN1-deleted SMA fibroblasts cells (ML17), a significant binding of MeCP2 to the SMN2 promoter region was observed by chromatin immunoprecipitation (ChIP) analysis using an anti-MeCP2 antibody and primers amplifying the genomic SMN2 promoter region from nt −372 to −266. An unrelated antibody (negative Ctrl IgG, rabbit, Diagenode) was used as negative control. Treatment of ML17 SMA fibroblast cells with 100 µm ZEB resulted in a significant reduction of MeCP2 binding to the SMN2 promoter region. MeCP2 is enriched at a 286 bp SMN2 promoter region. (G) ChIP analyses using ML17 fibroblasts and six different primer pairs covering the genomic SMN2 promoter region from nt −631 to +59. Enrichment after MeCP2 ChIP was detectable for all primer pairs while two primer pairs showed considerably higher ChIP-PCR signal intensities. Three levels of statistical significance were discriminated: *P < 0.05, **P < 0.01, ***P < 0.001 (t-test).
Transcriptional repressor MeCP2 binds to the SMN2 promoter and mediates transcriptional LT-SMN2 silencing
Two basic models evolved regarding the mechanism by which DNA methylation affects transcription (39). DNA methylation can directly repress transcription by blocking transcriptional activators from binding to cognate DNA sequences, alternatively methyl-CpG DNA-binding proteins (MBPs) recognize methylated DNA and recruit co-repressors to silence gene expression directly. Since HDAC inhibitors do not promote SMN2 gene demethylation in our experimental setting (Fig. 7A), and are able to bypass LT-SMN2 gene silencing by DNA methylation (Figs 5 and 6), we hypothesized that DNA methylation at the SMN2 promoter region is recognized by an MBP which recruits HDAC activity to mediate epigenetic gene silencing. Previous studies revealed that two global mechanisms of epigenetic gene regulation, DNA methylation and histone deacetylation, can be linked by the methyl-CpG-binding-protein 2 (MeCP2) (27). MeCP2 is an abundant chromosomal protein that specifically binds to methylated DNA and requires HDAC activity to repress transcription from methylated promoters (26,27). Thus, we hypothesized that the methylated SMN2 promoter is recognized by MeCP2. In line with this hypothesis, siRNA-mediated knockdown of MeCP2 resulted in significantly increased LT-SMN2 transcript levels in ML17 type I SMA fibroblast cells (Fig. 7D). To validate the role of MeCP2 in LT-SMN2 expression, we next analysed the potential binding of MeCP2 to the SMN2 promoter region by chromatin immunoprecipitation (ChIP). Initial analyses using SMN2CGI 2-specific primers covering the genomic region from nt −372 to −266 revealed a clear association of MeCP2 with the SMN2 promoter region in ML17 SMA fibroblasts (Fig. 7E). Elevated LT-SMN2 transcript levels following treatment of ML17 fibroblasts with zebularine (Fig. 7C) are associated with a decrease in the MeCP2 binding to the SMN2 promoter (Fig. 7F), indicating that MeCP2 recognizes the SMN2 promoter in a methylation-dependent fashion. Moreover, increased LT-SMN2 transcript levels following siRNA-mediated knockdown of MeCP2 (Fig. 7D) are associated with decreased MeCP2 binding to the SMN2 promoter (77.8 ± 4.6%, P < 0.05, t-test). These data indicate a methylation-dependent binding of the transcriptional repressor MeCP2 to the SMN2 promoter region resulting in LT-SMN2 silencing. To determine the binding pattern of MeCP2 on the SMN2 promoter we performed MeCP2 ChIP analyses using ML17 SMA fibroblasts and six different primer pairs covering the genomic SMN2 promoter region from nt −631 to +59, which comprises SMN2CGI 2 (nt −469 to −247) and adjacent sequences. Although MeCP2 binding was detectable in all cases (Fig. 7G), ChIP-PCR signal intensities were substantially higher at the genomic region from nt −349 and −63, suggesting enrichment at this site. This particular 286 bp region contains all three transcriptional SMN2 start sites at positions −296, −242 and −163 and the differentially methylated CpG dinucleotides at positions −296 and −290, suggesting a probable role of MeCP2 in SMN2 gene silencing.
DISCUSSION
Here, we show for the first time that SMN2, the major disease modifier and target gene for pharmacological SMA treatment, is epigenetically regulated by DNA methylation given by the fact that DNA-demethylating compounds increase SMN2 gene activity (Figs 1A and B, and 7C). We demonstrate that SMN2 contains four putative CGIs (SMN2CGIs) located within the genomic region 896 nt upstream and 1146 nt downstream of its translational start site (Fig. 2A). Analysis of DNA isolated from fibroblast cell lines and blood samples derived from SMN1-deleted SMA patients identified SMN2CGI 1 and SMN2CGI 4 to be hypermethylated, SMN2CGI 3 appeared to be almost methylation-free, while SMN2CGI 2 showed intermediate methylation levels (Figs 2B and C, and 3A–C). Since CGIs are usually defined as short unmethylated DNA patches (29), solely SMN2CGI 3 (446 bp, GC content 62.1%) can be regarded as a ‘classical’ CGI. However, recent data categorize SMN2CGI 1 (252 bp, GC content 54.8%), SMN2CGI 2 (223 bp, GC content 58.3%) and SMN2CGI 4 (303 bp, GC content 59.4%) as ‘weak’ CGIs due to their small size and comparatively low GC content (40). Differences between the ‘classical’ SMN2CGI 3 and the ‘weak’ SMN2CGIs 1, 2 and 4 become more apparent when calculating the CpG density, which is defined as the ratio between observed versus expected CpG dinucleotides (40). Although SMN2CGI 3 possesses a high CpG density (0.94) usually found in unmethylated CGIs, the remaining SMN2CGIs show intermediate CpG densities (SMN2CGI 1: 0.74, SMN2CGI 2: 0.79, SMN2CGI 4: 0.79) characteristic for weak CGIs (29). In line with our data (Figs 2B and C, and 3A and B), weak CGIs usually show high frequencies of DNA methylation (40). Moreover, SMN2 appears to be a typical gene regarding its CGI distribution. Approximately 70% of human genes are linked to promoter CGIs (41) and about half of all CGIs were found at the TSS of an annotated gene (such as SMN2CGI 2) (29). Among the four SMN2CGIs analysed, we provide first evidence that particularly the genomic region comprising the weak SMN2CGI 2 is functionally important regarding SMA disease progression and pharmacological SMA therapy.
Even though SMN2 has been identified as the major SMA disease modifier, differences in disease progression have frequently been observed in the presence of identical SMN1 mutations and SMN2 copy numbers (9,42). In the presence of two SMN2 copies, homozygous absence of SMN1 causes type I SMA in approximately two-thirds of all patients, while milder phenotypes are consequently also quite common (9,42). These findings led to the hypothesis that SMN2 genes are not functionally equivalent. Using DNA derived from comparatively easily accessible sources such as blood and skin we identified significantly lower methylation levels at two adjacent CpG dinucleotides in type III SMA patients compared with patients suffering from severe type I SMA (nt positions −296 and −290, both SMN2CGI 2, Figs 3C and 4A), suggesting that the capability of the SMN2 gene copies to counteract SMA disease progression is modulated by DNA methylation. The functional relevance of DNA methylation at nt positions −296 and −290 is further highlighted by the finding that these CpG dinucleotides co-localize with the first TSS of SMN2 at nt position −296 (Fig. 2A). We demonstrate that methylation levels at nt positions −296 and −290 inversely correlate with the activity of the first TSS of SMN2 (Figs 4A–C, and 7B and C) indicating that DNA methylation at these nt positions is linked with transcriptional LT-SMN2 silencing. In summary, our data provide first evidence that SMN2 gene copies are not functionally equivalent due to differences in DNA methylation affecting LT-SMN2 expression. However, LT-SMN2 contributes only moderately to total SMN2 transcript levels in SMA fibroblasts (described earlier). To address the question whether LT-SMN2 expression is regulated in a tissue-dependent fashion, we subsequently quantified LT-SMN2 and total SMN2 transcript levels in various tissues derived from adult transgenic mice carrying the human SMN2 gene (Smn−/− SMN2+/+) (43). Consistent with previous findings, total SMN2 transcript levels are higher in CNS tissues such as cerebrum and spinal cord compared with mesodermal and endodermal tissues such as skeletal muscle, heart and liver (19). Similarly, LT-SMN2 transcripts display considerably higher expression levels in CNS tissues with a >3-fold higher expression in spinal cord compared with muscle, heart and liver, suggesting that the amounts of SMN protein derived from LT-SMN2 are sufficient to modulate motor neuron degradation (Supplementary Material, Fig. S1). However, SMA disease severities appear to be modified by multiple mechanisms. In rare cases, haploidentical SMN1-deleted siblings show highly discordant SMA phenotypes ranging from affected to unaffected (44). In these families it has been shown that the SMA phenotype can be influenced by elevated expression of an independent modifying gene, Plastin 3, which protects against SMA in females (45). Exemplary analysis of nt positions −296 and −290 in two of these families (#34, #482) (44) revealed no differences in DNA methylation between haploidentical affected and unaffected individuals (data not shown).
Despite the apparent correlation between the CpG methylation levels at nt positions −296 and −290 and the disease severity, the finding that LT-SMN2 expression is silenced by DNA methylation is especially important regarding pharmacological SMA therapy using HDAC inhibitors. The fundamental mechanisms of epigenetic gene regulation, DNA methylation and histone acetylation, have shown to be interlinked (26,27). Consistent with these findings we demonstrate that a panel of five pan-HDAC inhibitors including the hydoxamic acid vorinostat (SAHA) as well as the cyclic tetrapeptide romidepsin (FK-228) are able to bypass LT-SMN2 gene silencing in SMA fibroblasts (Fig. 5) and human OHSCs (Fig. 6), resulting in up to 5-fold inductions of total SMN2 transcript levels. In contrast, the HDAC isoenzyme selective inhibitors MS-275, VPA and PB display only moderate effects (Figs 5 and 6). These findings highlight functional differences between HDAC inhibitors regarding pharmacological SMA therapy and suggest that pan-HDAC inhibitors possess superior functional capacities to activate SMN2. Given the fact that the pan-HDAC inhibitors vorinostat (SAHA) and romidepsin (FK-228) are either FDA approved or under clinical phase II evaluation for cancer treatment, our data further highlight that these readily available compounds might be considered as candidate drugs for SMA therapy in case the ongoing clinical trials using VPA or PB do not show the desired efficacies. Especially vorinostat appears to be promising due to the fact that this molecule has shown to cross the blood-brain barrier and ongoing clinical evaluation in cancer patients exemplifies a good oral bioavailability and biological activity (46).
Moreover, we provide evidence of how LT-SMN2 gene silencing by DNA methylation is regulated. ChIP experiments using ML17 SMA fibroblasts revealed that the methyl-CpG-binding protein 2 (MeCP2) is localized at a 286 bp SMN2 promoter region (nt −349 to −63) containing all the three transcriptional SMN2 start sites at nt positions −296, −242 and −163 as well as the differentially methylated CpG dinucleotides at positions −296 and −290 (Fig. 7G). MeCP2 binds to the SMN2 promoter region in a methylation-dependent fashion (Fig. 7F) and MeCP2 knockdown induced LT-SMN2 mRNA expression (Fig. 7D). Following binding to methylated DNA, MeCP2 is known to recruit HDACs to chromatin (26,47), suggesting that the regulatory component MeCP2 might be involved in mediating SMN2 gene silencing by DNA methylation. Since the isoenzyme selective HDAC inhibitors MS-275 and VPA have shown to inhibit HDAC1, HDAC2, HDAC3 (both MS-275 and VPA) and HDAC9 (MS-275) at doses used in our study (32), the finding that these drugs do not counteract LT-SMN2 silencing suggests that the LT-SMN2 induction observed following pan-HDAC inhibitor treatment was due to inhibition of the remaining classical HDACs (HDACs 4–8, 10, 11). Thus, further analysis of the specific roles of MeCP2 and HDAC isoenzymes in LT-SMN2 expression represent important steps to develop optimized HDAC inhibitors targeted to overcome LT-SMN2 silencing.
Regarding the apparent correlation between SMN2CGI 2 methylation levels and disease severity, continuative studies are mandatory to assess whether the accurate quantification of DNA methylation levels at nt positions −296 and −290 represents a valuable tool to predict SMA disease progression in the presence of identical SMN1 mutations and SMN2 copy numbers. Furthermore, initial SMN1 promoter methylation analyses of DNA samples derived from SMN2-deleted and unaffected individuals revealed that SMN1 is subject to promoter methylation very similar to SMN2 (Supplementary Material, Fig. S2). Based on these findings one might speculate whether aberrant SMN1 promoter methylation may be causative for SMA in patients showing at least one non-mutated SMN1 gene copy.
MATERIALS AND METHODS
Chemicals
The following chromatin-remodelling compounds have been used: M344 (382149, Calbiochem, San Diego, CA, USA), MS-275 (382147, Calbiochem), PB (567616, Calbiochem), zebularine (691400, Calbiochem), SAHA (270–288, Axxora, Lörrach, Germany), VPA (P-4543, Sigma-Aldrich, St Louis, MO, USA), scriptaid (S7817, Sigma-Aldrich), oxamflatin (O3139, Sigma-Aldrich) and Aza (A-3656, Sigma-Aldrich). Romidepsin (FK-228) was kindly provided by Gloucester Pharmaceuticals (Cambridge, MA, USA). All test compounds were dissolved in 100% DMSO or H2O (VPA).
SMA patient samples and DNA isolation
Informed consent was obtained from all SMA patients or their parents. All patients fulfilled the diagnostic criteria for SMA (48); SMA disease severities (type I SMA, MIM #253300; type II SMA, MIM #253550; type IIIa/IIIb, MIM #253400) were classified as described (49). Genomic DNA was isolated from venous blood samples drawn for diagnostic purposes using the salting-out method (50) and SMN1/SMN2 gene copy numbers were determined as described previously (9). The fibroblast cell lines used in this study have been established from skin biopsies and were cultured as described (14). Fibroblasts were cultured for the indicated time spans and harvested by trypsinization. DNA was isolated from fibroblast cell cultures using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol.
Human OHSCs
Human hippocampi were obtained from patients with chronic intractable temporal lobe epilepsy who underwent surgical treatment in the Epilepsy Surgery Program at the Department of Neurosurgery (University of Erlangen). For scientific use of tissue specimens, informed and written consent was obtained from all patients and studies were approved by the local Ethics Committee of the University of Erlangen. Surgical specimens were prepared and processed according to the interface technique (51). In brief, hippocampal brain samples were cut into 350 µm thick horizontal slices on a vibratome (Leica Microsystems, Wetzlar, Germany). Each brain slice was transferred into culture plate insert membranes (BD Biosciences, San Jose, CA, USA) and thereafter into 6-well culture dishes (BD Biosciences) containing 1.2 ml culture medium as described in detail (52). A day after preparation, culture medium was changed, human OHSCs were exposed to the test compounds or solvent only for 48 h and snap-frozen in liquid nitrogen.
Protein sample preparation and western blot analysis
Human OHSCs and fibroblast cell culture cell pellets were resuspended in RIPA buffer (150 mm NaCl, 1% NP40, 0.5% DOC, 0.1% SDS, 50 mm Tris, pH 8.0) to obtain whole cell protein extracts. Denatured protein (7.5 µg) of each sample were resolved by SDS–PAGE on 4–12% Bis–Tris Gels (Invitrogen, Carlsbad, CA, USA) and transferred to nitrocellulose membrane by overnight wet blotting. Immunostaining of the membrane was performed according to standard protocols using the following antibodies: mouse monoclonal anti-ß-tubulin (T 4026, Sigma-Aldrich, dilution 1:20 000), mouse monoclonal anti-SMN (610647, BD Biosciences, dilution 1:1,000) and horseradish peroxidase conjugated goat anti-mouse IgG (115-035-003, Dianova, dilution 1:10 000) as secondary antibody. Detection of signals was carried out using the Super Signal West Pico chemiluminescence reagent (Pierce, Rockford, IL, USA). Densitometric analyses were performed using the Quantity One 1-D Analysis Software (Bio-Rad, Hercules, CA, USA).
RNA isolation and quantitative RT–PCR analysis
Isolation of total RNA was performed using the RNeasy Mini Kit and QIAshredder columns according to the manufacture’s protocol (Qiagen). RNA concentrations were determined spectrophotometrically. For semiquantitative PCR analyses, reverse transcription (RT) was performed using oligo dT primers and 1 µg of total RNA by applying the SuperScript First-Strand Synthesis System (Invitrogen). Two microlitres of each RT reaction was used for each RT–PCR. cDNAs were amplified using primers of ß-actin (ß-actin-fwd: 5′-AAC GGC TCC GGC ATG TGC AA-3′, ß-actin-rev: 5′-CTC AAA CAT GAT CTG GGT CAT CTT-3′), CDK-4 (CDK-4-fwd: 5′-CTA TGG GAC AGT GTA CAA GG-3′, CDK-4-rev: 5′-GAT ATG TCC TTA GGT CCT GG-3′), LT-SMN2 (LT-SMN2−279-fwd: 5′-ACT CCA GCC TGA GCG ACA-3′, LT-SMN2−274-fwd: 5′-AGC CTG AGC GAC AGG GCGA-3′, LT-SMN2−128-rev: 5′-TCT ACG AGT GGT TAT CGC-3′, LT-SMN2+90-rev: 5′-ATC ATC GCT CTG GCC TGT GCC-3′). The following PCR conditions were used: 5 min initial denaturation (95°C), followed by 25–37 cycles (95°C for 15 s, 68°C for 30 s and 72°C for 35 s and final extension step (72°C for 5 min). PCR products were separated on 2% agarose gels and visualized by ethidium bromide staining. For real-time quantification of target gene expression, one step RT–PCR was preformed using the QuantiTect SYBR Green RT–PCR Kit (Qiagen) on an Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems, Darmstadt, Germany). Each 20 µl RT–PCR mix contained 10 ng total RNA (4 ng/µl), 2 µl of the primer dilution, 10 µl ×2 QuantiTect SYBR Green RT-Master Mix and 0.2 µl QuantiTect RT Mix. One-step RT–PCR reactions were carried out in 96-well optical reaction plates, covered with Optical Adhensive Covers (Applied Biosystems). Cycling conditions were as follows: 50°C for 30 min (RT step), 95°C for 15 min and 40 cycles of 94°C for 15 s, 55°C for 30 s and 72°C for 35 s. Real-time RT–PCR was conducted four times for each gene and each RNA sample and results is given as mean ± SEM. The comparative method of relative quantification (2−ΔΔCt) was used to calculate the relative expression levels of each target gene (normalized to ß-actin) compared with the non-treated control samples. RT–PCR specificity of each reaction was verified by melting curve analysis and confirmed by 2% agarose gel electrophoresis. Whenever possible, amplicons have been designed to span exon borders to exclude false positive detection of genomic contaminations. The following primers have been used for quantitative real-time RT–PCR: LT-SMN2-fwd 5′-ACT CCA GCC TGA GCG ACA-3′, LT-SMN2-rev 5′-TCT ACG AGT GGT TAT CGC-3′. SMN2: Quantitect primer assay QT01673679 (Qiagen), ß-actin: Quantitect primer assay QT00095431 (Qiagen), MeCP2: Quantitect primer assay QT00039361 (Qiagen). On cDNA level, full-length SMN transcripts derived from SMN1 and SMN2 are identical except for two silent nt differences in exons 7 and 8. SMN2 Quantitect primer assay QT01673679 amplifies exons 5 and 6. Please note that the SMN2 primers used also detect SMN1 when present.
Bisulphite genomic sequencing
DNA methylation analysis was carried out by bisulphite genomic sequencing using the pyrosequencing technology as described in detail (53). Bisulphite conversion of DNA (1 µg per sample) was performed using the EpiTect Bisulfite Kit (Qiagen) according to the manufacturer's protocol. Regions of interest were amplified by nested PCR using AmpliTaq Gold (Applied Biosystems) and biotinylated fwd/rev primer for the inner PCRs. The PCR conditions were as follows: 95°C for 5 min, followed by 40–50 cycles (95°C for 15 s, 52–55°C for 45 s, 72°C for 1 min) and a final extension step of 5 min at 72°C. The biotinylated PCR products (25 µl) were cleaned up using Streptavidin Sepharose™ High Performance (GE Healthcare, Uppsala, Sweden) and a PyroMark Vacuum Prep Workstation (Biotage, Uppsala, Sweden). The purified PCR products were used for the pyrosequencing reaction according to the manufacturer’s protocol using Pyro Gold Reagents and the PSQ96 MA System (Biotage). Sequence analysis and quantification of CpG-dinucleotide methylation was performed using Pyro Q-CpG software (Biotage). Cloning of PCR products was performed using the TOPO TA Cloning Kit with One Shot TOP10 chemically competent Escherichia coli and the pcDNA3.1/V5-His Vector (Invitrogen). Plasmids of at least nine positive clones were isolated using the QIAprep Spin Miniprep Kit (Qiagen) according to the manufacturer’s protocol. Inserts were sequenced using the BigDye Terminator Cycle Sequencing Kit V1.1 (Applied Biosystems) and the T7 forward primer. The primers used for bisulphite genomic sequencing (amplification, cloning, sequencing) are given in the Supplementary Material, Table S1. Please note that all primers given in the Supplementary Material, Table S1 detect SMN1 and SMN2 due to identical promoter sequences.
siRNA-mediated knockdown of MeCP2
siRNA-mediated knockdown was performed as described previously (54). Briefly, 1 day before transfection 1 × 105 SMA fibroblast cells were plated out in each well of a 6-well culture dish. Cells were transfected with MeCP2 siRNA (Qiagen, Hs_MeCP2_7, SI02664893, target sequence: ACG GAG CGG ATT GCA AAG CAA) or AllStars negative Control siRNA (Qiagen, 1027281) at a final concentration of 50 nM using DharmaFECT 1 siRNA Transfection Reagent (Dharmacon, Bonn, Germany). Transfection efficacy was controlled by transfection with siControl TOX transfection control (Dharmacon, D-001500-01-20). Cells were harvested after 48 h. RNA isolation and real-time PCR analyses were performed as described earlier.
Chromatin immunoprecipitation
ChIP experiments were carried out using the LowCell# ChIP Kit (Diagenode, Liège, Belgium) according to the manufacturer's instructions. Briefly, 3 × 105 fibroblast cells were used for each analysis. Cells were harvested by trypsinization. Proteins were cross-linked to DNA by adding formaldehyde to a final concentration of 1% for 8 min at room temperature to the trypsinized cells. The fixation reaction was stopped by adding glycine to a final concentration of 0.125 m and subsequent incubation for 10 min at room temperature. Chromatin was sheared using a Bioruptor (Diagenode), utilizing maximum power for 15 min with 30 s on/off cycles at 4°C. Five microgram of the MeCP2 antibody (Diagenode) per reaction was bound to the Protein A-coated paramagnetic beads which were included in the Kit. One hundered microlitre of sheared chromatin were added to the antibody-coated paramagnetic beads and incubated overnight at 4°C with rotation. Chromatin was immunoprecipitated and washed by using a Magnetic Rack (Diagenode). Thereafter DNA was purified from the antibody coated beads by using DNA purifying slurry and incubation of the samples in boiling water for 10 min. This step was followed by addition of proteinase K (Diagenode) and incubation at 55°C for 30 min in a thermomixer (1000 rpm). Subsequently the samples were incubated again in boiling water for 10 min to elute the DNA. DNA was analysed by real-time PCR on a 7500 Real-Time PCR System (Applied Biosystems), using Power SYBR Green PCR Master Mix (Applied Biosystems) and the primers listed in the Supplementary Material, Table S1. All PCRs were performed at least in triplicates.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG Online.
Conflict of Interest statement. None declared.
FUNDING
This work was kindly supported by the ‘Initiative Forschung und Therapie für SMA’ (to E.H. and to B.W.), the ‘Köln Fortune Program/Faculty of Medicine, University of Cologne’ (to E.H.), the ‘Deutsche Forschungsgemeinschaft (DFG)’ (to E.H., to I.B. and to B.W.), the ‘Wilhelm Sander Foundation’ (to E.H., I.Y.E. and I.B.), the ‘Families of Spinal Muscular Atrophy’ (to B.W. and E.H.) and the ‘Center for Molecular Medicine Cologne’ (to B.W. and E.H.). The ‘Monash International Post graduate Research Scholarship (MIPRS)’ and the ‘Monash Graduate Scholarship (MGS)’ (to S.L.). Funding to pay the Open Access charge was provided by the ‘Initiative Forschung und Therapie für SMA’.
Supplementary Material
REFERENCES
- 1.Wirth B., Brichta L., Hahnen E. Spinal muscular atrophy: from gene to therapy. Semin. Pediatr. Neurol. 2006;13:121–131. doi: 10.1016/j.spen.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Wirth B. An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA) Hum. Mutat. 2000;15:228–237. doi: 10.1002/(SICI)1098-1004(200003)15:3<228::AID-HUMU3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 3.Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 4.Lorson C.L., Hahnen E., Androphy E.J., Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl Acad. Sci. USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lorson C.L., Strasswimmer J., Yao J.M., Baleja J.D., Hahnen E., Wirth B., Le T., Burghes A.H., Androphy E.J. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat. Genet. 1998;19:63–66. doi: 10.1038/ng0598-63. [DOI] [PubMed] [Google Scholar]
- 6.Wolstencroft E.C., Mattis V., Bajer A.A., Young P.J., Lorson C.L. A non-sequence-specific requirement for SMN protein activity: the role of aminoglycosides in inducing elevated SMN protein levels. Hum. Mol. Genet. 2005;14:1199–1210. doi: 10.1093/hmg/ddi131. [DOI] [PubMed] [Google Scholar]
- 7.Le T.T., Pham L.T., Butchbach M.E., Zhang H.L., Monani U.R., Coovert D.D., Gavrilina T.O., Xing L., Bassell G.J., Burghes A.H. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum. Mol. Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- 8.McAndrew P.E., Parsons D.W., Simard L.R., Rochette C., Ray P.N., Mendell J.R., Prior T.W., Burghes A.H. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am. J. Hum. Genet. 1997;60:1411–1422. doi: 10.1086/515465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feldkotter M., Schwarzer V., Wirth R., Wienker T.F., Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am. J. Hum. Genet. 2002;70:358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wirth B., Brichta L., Schrank B., Lochmuller H., Blick S., Baasner A., Heller R. Mildly affected patients with spinal muscular atrophy are partially protected by an increased SMN2 copy number. Hum. Genet. 2006;119:422–428. doi: 10.1007/s00439-006-0156-7. [DOI] [PubMed] [Google Scholar]
- 11.Brahe C. Copies of the survival motor neuron gene in spinal muscular atrophy: the more, the better. Neuromuscul. Disord. 2000;10:274–275. doi: 10.1016/s0960-8966(99)00137-6. [DOI] [PubMed] [Google Scholar]
- 12.Monani U.R., Coovert D.D., Burghes A.H. Animal models of spinal muscular atrophy. Hum. Mol. Genet. 2000;9:2451–2457. doi: 10.1093/hmg/9.16.2451. [DOI] [PubMed] [Google Scholar]
- 13.Sumner C.J., Huynh T.N., Markowitz J.A., Perhac J.S., Hill B., Coovert D.D., Schussler K., Chen X., Jarecki J., Burghes A.H., et al. Valproic acid increases SMN levels in spinal muscular atrophy patient cells. Ann. Neurol. 2003;54:647–654. doi: 10.1002/ana.10743. [DOI] [PubMed] [Google Scholar]
- 14.Brichta L., Hofmann Y., Hahnen E., Siebzehnrubl F.A., Raschke H., Blumcke I., Eyupoglu I.Y., Wirth B. Valproic acid increases the SMN2 protein level: a well-known drug as a potential therapy for spinal muscular atrophy. Hum. Mol. Genet. 2003;12:2481–2489. doi: 10.1093/hmg/ddg256. [DOI] [PubMed] [Google Scholar]
- 15.Hahnen E., Eyupoglu I.Y., Brichta L., Haastert K., Trankle C., Siebzehnrubl F.A., Riessland M., Holker I., Claus P., Romstock J., et al. In vitro and ex vivo evaluation of second-generation histone deacetylase inhibitors for the treatment of spinal muscular atrophy. J. Neurochem. 2006;98:193–202. doi: 10.1111/j.1471-4159.2006.03868.x. [DOI] [PubMed] [Google Scholar]
- 16.Andreassi C., Angelozzi C., Tiziano F.D., Vitali T., De Vincenzi E., Boninsegna A., Villanova M., Bertini E., Pini A., Neri G., et al. Phenylbutyrate increases SMN expression in vitro: relevance for treatment of spinal muscular atrophy. Eur. J. Hum. Genet. 2004;12:59–65. doi: 10.1038/sj.ejhg.5201102. [DOI] [PubMed] [Google Scholar]
- 17.Chang J.G., Hsieh-Li H.M., Jong Y.J., Wang N.M., Tsai C.H., Li H. Treatment of spinal muscular atrophy by sodium butyrate. Proc. Natl Acad. Sci. USA. 2001;98:9808–9813. doi: 10.1073/pnas.171105098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riessland M., Brichta L., Hahnen E., Wirth B. The benzamide M344, a novel histone deacetylase inhibitor, significantly increases SMN2 RNA/protein levels in spinal muscular atrophy cells. Hum. Genet. 2006;120:101–110. doi: 10.1007/s00439-006-0186-1. [DOI] [PubMed] [Google Scholar]
- 19.Kernochan L.E., Russo M.L., Woodling N.S., Huynh T.N., Avila A.M., Fischbeck K.H., Sumner C.J. The role of histone acetylation in SMN gene expression. Hum. Mol. Genet. 2005;14:1171–1182. doi: 10.1093/hmg/ddi130. [DOI] [PubMed] [Google Scholar]
- 20.Avila A.M., Burnett B.G., Taye A.A., Gabanella F., Knight M.A., Hartenstein P., Cizman Z., Di Prospero N.A., Pellizzoni L., Fischbeck K.H., et al. Trichostatin A increases SMN expression and survival in a mouse model of spinal muscular atrophy. J. Clin. Invest. 2007;117:659–671. doi: 10.1172/JCI29562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai L.K., Tsai M.S., Lin T.B., Hwu W.L., Li H. Establishing a standardized therapeutic testing protocol for spinal muscular atrophy. Neurobiol. Dis. 2006;24:286–295. doi: 10.1016/j.nbd.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Mercuri E., Bertini E., Messina S., Pelliccioni M., D’Amico A., Colitto F., Mirabella M., Tiziano F.D., Vitali T., Angelozzi C., et al. Pilot trial of phenylbutyrate in spinal muscular atrophy. Neuromuscul. Disord. 2004;14:130–135. doi: 10.1016/j.nmd.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 23.Brahe C., Vitali T., Tiziano F.D., Angelozzi C., Pinto A.M., Borgo F., Moscato U., Bertini E., Mercuri E., Neri G. Phenylbutyrate increases SMN gene expression in spinal muscular atrophy patients. Eur. J. Hum. Genet. 2005;13:256–259. doi: 10.1038/sj.ejhg.5201320. [DOI] [PubMed] [Google Scholar]
- 24.Weihl C.C., Connolly A.M., Pestronk A. Valproate may improve strength and function in patients with type III/IV spinal muscle atrophy. Neurology. 2006;67:500–501. doi: 10.1212/01.wnl.0000231139.26253.d0. [DOI] [PubMed] [Google Scholar]
- 25.Brichta L., Holker I., Haug K., Klockgether T., Wirth B. In vivo activation of SMN in spinal muscular atrophy carriers and patients treated with valproate. Ann. Neurol. 2006;59:970–975. doi: 10.1002/ana.20836. [DOI] [PubMed] [Google Scholar]
- 26.Jones P.L., Veenstra G.J., Wade P.A., Vermaak D., Kass S.U., Landsberger N., Strouboulis J., Wolffe A.P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 27.Nan X., Ng H.H., Johnson C.A., Laherty C.D., Turner B.M., Eisenman R.N., Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 28.Eckhardt F., Lewin J., Cortese R., Rakyan V.K., Attwood J., Burger M., Burton J., Cox T.V., Davies R., Down T.A., et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat. Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki M.M., Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 30.Monani U.R., McPherson J.D., Burghes A.H. Promoter analysis of the human centromeric and telomeric survival motor neuron genes (SMNC and SMNT) Biochim. Biophys. Acta. 1999;1445:330–336. doi: 10.1016/s0167-4781(99)00060-3. [DOI] [PubMed] [Google Scholar]
- 31.Rouget R., Vigneault F., Codio C., Rochette C., Paradis I., Drouin R., Simard L.R. Characterization of the survival motor neuron (SMN) promoter provides evidence for complex combinatorial regulation in undifferentiated and differentiated P19 cells. Biochem. J. 2005;385:433–443. doi: 10.1042/BJ20041024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hahnen E., Hauke J., Trankle C., Eyupoglu I.Y., Wirth B., Blumcke I. Histone deacetylase inhibitors: possible implications for neurodegenerative disorders. Expert. Opin. Investig. Drugs. 2008;17:169–184. doi: 10.1517/13543784.17.2.169. [DOI] [PubMed] [Google Scholar]
- 33.Boda B., Mas C., Giudicelli C., Nepote V., Guimiot F., Levacher B., Zvara A., Santha M., LeGall I., Simonneau M. Survival motor neuron SMN1 and SMN2 gene promoters: identical sequences and differential expression in neurons and non-neuronal cells. Eur. J. Hum. Genet. 2004;12:729–737. doi: 10.1038/sj.ejhg.5201217. [DOI] [PubMed] [Google Scholar]
- 34.Cervoni N., Szyf M. Demethylase activity is directed by histone acetylation. J. Biol. Chem. 2001;276:40778–40787. doi: 10.1074/jbc.M103921200. [DOI] [PubMed] [Google Scholar]
- 35.Detich N., Bovenzi V., Szyf M. Valproate induces replication-independent active DNA demethylation. J. Biol. Chem. 2003;278:27586–27592. doi: 10.1074/jbc.M303740200. [DOI] [PubMed] [Google Scholar]
- 36.Milutinovic S., D’Alessio A.C., Detich N., Szyf M. Valproate induces widespread epigenetic reprogramming which involves demethylation of specific genes. Carcinogenesis. 2007;28:560–571. doi: 10.1093/carcin/bgl167. [DOI] [PubMed] [Google Scholar]
- 37.Dong E., Guidotti A., Grayson D.R., Costa E. Histone hyperacetylation induces demethylation of reelin and 67-kDa glutamic acid decarboxylase promoters. Proc. Natl Acad. Sci. USA. 2007;104:4676–4681. doi: 10.1073/pnas.0700529104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weaver I.C., Cervoni N., Champagne F.A., D’Alessio A.C., Sharma S., Seckl J.R., Dymov S., Szyf M., Meaney M.J. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 39.Klose R.J., Bird A.P. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 40.Weber M., Hellmann I., Stadler M.B., Ramos L., Paabo S., Rebhan M., Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 41.Saxonov S., Berg P., Brutlag D.L. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl Acad. Sci. USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cusco I., Barcelo M.J., Rojas-Garcia R., Illa I., Gamez J., Cervera C., Pou A., Izquierdo G., Baiget M., Tizzano E.F. SMN2 copy number predicts acute or chronic spinal muscular atrophy but does not account for intrafamilial variability in siblings. J. Neurol. 2006;253:21–25. doi: 10.1007/s00415-005-0912-y. [DOI] [PubMed] [Google Scholar]
- 43.Hsieh-Li H.M., Chang J.G., Jong Y.J., Wu M.H., Wang N.M., Tsai C.H., Li H. A mouse model for spinal muscular atrophy. Nat. Genet. 2000;24:66–70. doi: 10.1038/71709. [DOI] [PubMed] [Google Scholar]
- 44.Hahnen E., Forkert R., Marke C., Rudnik-Schoneborn S., Schonling J., Zerres K., Wirth B. Molecular analysis of candidate genes on chromosome 5q13 in autosomal recessive spinal muscular atrophy: evidence of homozygous deletions of the SMN gene in unaffected individuals. Hum. Mol. Genet. 1995;4:1927–1933. doi: 10.1093/hmg/4.10.1927. [DOI] [PubMed] [Google Scholar]
- 45.Oprea G.E., Krober S., McWhorter M.L., Rossoll W., Muller S., Krawczak M., Bassell G.J., Beattie C.E., Wirth B. Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science. 2008;320:524–527. doi: 10.1126/science.1155085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marks P.A., Richon V.M., Miller T., Kelly W.K. Histone deacetylase inhibitors. Adv. Cancer. Res. 2004;91:137–168. doi: 10.1016/S0065-230X(04)91004-4. [DOI] [PubMed] [Google Scholar]
- 47.Harikrishnan K.N., Chow M.Z., Baker E.K., Pal S., Bassal S., Brasacchio D., Wang L., Craig J.M., Jones P.L., Sif S., et al. Brahma links the SWI/SNF chromatin-remodeling complex with MeCP2-dependent transcriptional silencing. Nat. Genet. 2005;37:254–264. doi: 10.1038/ng1516. [DOI] [PubMed] [Google Scholar]
- 48.Munsat T.L., Davies K.E. International SMA consortium meeting. (26–28 June 1992, Bonn, Germany) Neuromuscul. Disord. 1992;2:423–428. doi: 10.1016/s0960-8966(06)80015-5. [DOI] [PubMed] [Google Scholar]
- 49.Zerres K., Rudnik-Schoneborn S. Natural history in proximal spinal muscular atrophy. Clinical analysis of 445 patients and suggestions for a modification of existing classifications. Arch. Neurol. 1995;52:518–523. doi: 10.1001/archneur.1995.00540290108025. [DOI] [PubMed] [Google Scholar]
- 50.Miller S.A., Dykes D.D., Polesky H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stoppini L., Buchs P.A., Muller D. A simple method for organotypic cultures of nervous tissue. J. Neurosci. Methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- 52.Eyupoglu I.Y., Hahnen E., Buslei R., Siebzehnrubl F.A., Savaskan N.E., Luders M., Trankle C., Wick W., Weller M., Fahlbusch R., et al. Suberoylanilide hydroxamic acid (SAHA) has potent anti-glioma properties in vitro, ex vivo and in vivo. J. Neurochem. 2005;93:992–999. doi: 10.1111/j.1471-4159.2005.03098.x. [DOI] [PubMed] [Google Scholar]
- 53.Dupont J.M., Tost J., Jammes H., Gut I.G. De novo quantitative bisulfite sequencing using the pyrosequencing technology. Anal. Biochem. 2004;333:119–127. doi: 10.1016/j.ab.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 54.Brichta L., Garbes L., Jedrzejowska M., Grellscheid S.N., Holker I., Zimmermann K., Wirth B. Nonsense-mediated messenger RNA decay of survival motor neuron 1 causes spinal muscular atrophy. Hum. Genet. 2008;123:141–153. doi: 10.1007/s00439-007-0455-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.