Abstract
Autosomal Emery–Dreifuss muscular dystrophy and related disorders with dilated cardiomyopathy and variable skeletal muscle involvement are caused by mutations in LMNA, which encodes A-type nuclear lamins. How alterations in A-type lamins, intermediate filament proteins of the nuclear envelope expressed in most differentiated somatic cells, cause cardiomyopathy is only poorly understood. We demonstrated previously abnormal activation of the extracellular signal-regulated kinase (ERK) branch of the mitogen-activated protein kinase (MAPK) signaling cascade in hearts of Lmna H222P ‘knock in’ mice, a model of autosomal Emery–Dreifuss muscular dystrophy. We therefore treated LmnaH222P/H222P mice that develop cardiomyopathy with PD98059, an inhibitor of ERK activation. Systemic treatment of LmnaH222P/H222P mice with PD98059 inhibited ERK phosphorylation and blocked the activation of downstream genes in heart. It also blocked increased expression of RNAs encoding natriuretic peptide precursors and proteins involved in sarcomere organization that occurred in placebo-treated mice. Histological analysis and echocardiography demonstrated that treatment with PD98059 delayed the development of left ventricular dilatation. PD98059-treated LmnaH222P/H222P mice had normal cardiac ejection fractions assessed by echocardiography when placebo-treated mice had a 30% decrease. These results emphasize the role of ERK activation in the development of cardiomyopathy caused by LMNA mutations. They further provide proof of principle for ERK inhibition as a therapeutic option to prevent or delay heart failure in humans with Emery–Dreifuss muscular dystrophy and related disorders caused by mutations in LMNA.
INTRODUCTION
Mutations in LMNA encoding A-type nuclear lamins cause several diverse diseases often referred to as laminopathies (1). Autosomal-dominant and recessive Emery–Dreifuss muscular dystrophy, dilated cardiomyopathy type 1A and limb-girdle muscular dystrophy type 1B are a subset of the laminopathies that affect striated muscle (2–5). A common feature of these disorders is cardiomyopathy, and 8% of familial and sporadic cardiomyopathies may be caused by mutations in LMNA (6). Although implantable pacemakers and defibrillators can prevent complications of cardiac dysrhythmias that occur early in these disorders, affected individuals eventually develop heart failure for which there is no curative treatment and cardiac transplantation is ultimately necessary (6–8). How alterations in A-type lamins cause cardiomyopathy is only poorly understood and the few hypotheses that have been raised have not been tested in physiologically relevant vertebrate animal models.
We have demonstrated previously abnormal activation of the extracellular signal-regulated kinase (ERK) branch of the mitogen-activated protein kinase (MAPK) signaling cascade in hearts of Lmna H222P ‘knock in’ mice, a model of autosomal Emery–Dreifuss muscular dystrophy (9). Male LmnaH222P/H222P mice develop left ventricular (LV) dilatation and depressed contractile function starting at ∼8–10 weeks of age and invariably develop LV dilatation and decreased cardiac contractility at 16 weeks of age, typically dying between 16 and 36 weeks (10). On the basis of our observations that ERK is activated in these mice prior to the onset of clinically detectable cardiomyopathy as well as our demonstration that lamin A variants that cause striated muscle disease activate ERK when expressed in cultured cells, we hypothesized that the activation of ERK plays a primary pathogenic role in the development of cardiomyopathy (9). We further hypothesized that pharmacological inhibition of ERK would prevent or delay the development of dilated cardiomyopathy in LmnaH222P/H222P mice. To test this hypothesis, we treated LmnaH222P/H222P with PD98059, a compound that inhibits MAPK/ERK kinase (MEK), thereby preventing phosphorylation (activation) of ERK (11).
RESULTS
Systemic treatment of LmnaH222P/H222P mice with PD98050 inhibits ERK activity in the heart
We administered PD98059, at a dose of 3 mg/kg/day, or placebo (dimethylsulfoxide—DMSO) by intraperitoneal injection 5 days a week to male LmnaH222P/H222P mice. A comparable dose of PD98059 administered systematically has been shown to inhibit ERK activity in rat hearts (12). Treatment was initiated at 8 weeks of age and continued until the mice were 16 weeks of age. At 16 weeks of age, the mice were analyzed by echocardiography and then sacrificed for histological and biochemical studies. Untreated male Lmna+/+ and LmnaH222P/H222P mice were similarly analyzed for comparisons.
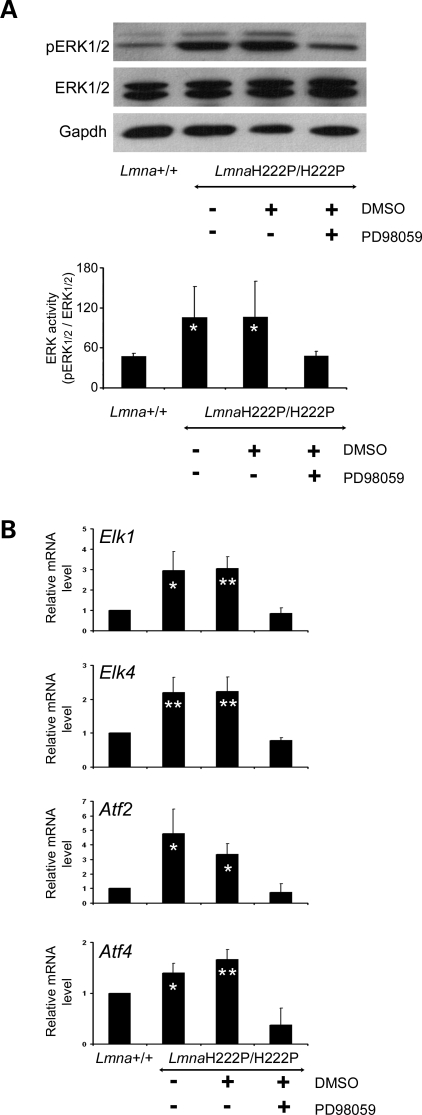
Systemic administration of PD98059 to mice inhibited phosphorylation of ERK1 and ERK2 in hearts, as shown by immunoblotting of proteins in tissue homogenates with antibodies against phosphorylated ERK1/2 and total ERK1/2 (Fig. 1A). The inhibition was specific of ERK1/2 relative to Jun N-terminal kinase (JNK), as at a dose of 3 mg/kg/day we did not observe the inhibition of JNK signaling in the heart (data not shown). To confirm the inhibition of ERK1/2 signaling, we monitored the expression of selected downstream genes activated by the kinases using real-time RT–PCR. As expected, the inhibition of phosphorylation of ERK1/2 led to decreased expression of Elk1, Elk4, Atf2 and Atf4 (Fig. 1B).
Figure 1.
Treatment of LmnaH222P/H222P mice with MEK inhibitor PD98059 inhibits phosphorylation of ERK1/2 and activation of downstream target genes. (A) Representative immunoblots using antibodies against phosphorylated ERK1/2 (pERK1/2) and antibodies against total ERK1/2 using proteins extracted from hearts from LmnaH222P/H222P mice treated with PD98059 or placebo (DMSO). Results of hearts from Lmna+/+ mice and untreated LmnaH222P/H222P mice are shown for comparison. Data in bar graphs are the quantification of phosphorylated ERK1/2 compared with total ERK1/2 measured by scanning the immunoblots and using Scion Image software (Scion Corporation). Values are means ± standard deviations for n = 3 samples from different animals per group. Results were compared using a two-tailed t-test (*P < 0.05). (B) Quantitative real-time RT–PCR showing the expression of RNAs of selected downstream target genes (Elk1, Elk4, Atf2, Atf4) of ERK-signaling cascade in hearts from LmnaH222P/H222P mice treated with PD98059 or placebo (DMSO). Results from hearts from Lmna+/+ mice and untreated LmnaH222P/H222P mice are shown for comparison. Bars indicate the fold overexpression of the indicated mRNA in hearts. Values are means ± standard deviations for n = 4 samples from different animals per group. Reactions were performed in triplicate for each different RNA sample. Results were compared using a two-tailed t-test (*P < 0.05, **P < 0.005).
PD98050 treatment prevents the development of cardiomyopathy in LmnaH222P/H222P mice
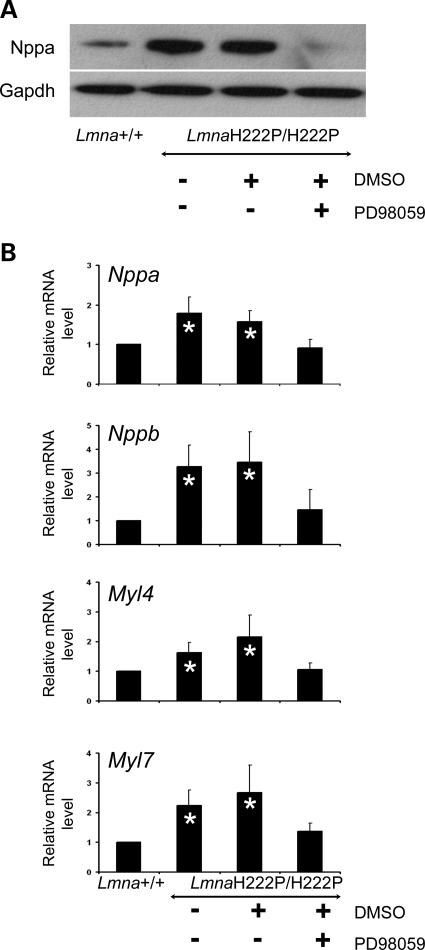
A feature of dilated cardiomyopathy is the upregulation of cardiac hormones such as natriuretic peptides (13–15). The upregulation of genes involved in sarcomere organization also occurs in dilated cardiomyopathies (15–17). In hearts from untreated LmnaH222P/H222P mice and those treated with placebo, the expression of natriuretic peptide precursor A was significantly increased (Fig. 2A). In contrast, PD98059-treated LmnaH222P/H222P mice had a cardiac expression of this peptide similar to Lmna+/+ mice (Fig. 2A). In hearts from untreated LmnaH222P/H222P mice and LmnaH222P/H222P mice treated with placebo, the expression of Nppa and Nppb mRNAs encoding natriuretic peptide precursors as well as Myl4 and Myl7 mRNAs encoding myosin light chains was significantly increased (Fig. 2B). In contrast, PD98059-treated LmnaH222P/H222P mice had a cardiac expression of Nppa, Nppb, Myl4 and Myl7 similar to Lmna+/+ mice (Fig. 2B).
Figure 2.
Effect of MEK inhibitor PD98059 on cardiac expression of natriuretic peptides and myosins in LmnaH222P/H222P mice. (A) Immunoblot showing the expression of natriuretic peptide precursor A (Nppa) in hearts from LmnaH222P/H222P mice treated with PD98059 or placebo (DMSO). Results using hearts from Lmna+/+ mice and untreated LmnaH222P/H222P mice are shown for comparison. Labeling with antibody against Gapdh is shown as a loading control. (B) Quantitative real-time RT–PCR showing the expression of RNAs from NppA and NppB genes, respectively, encoding natriuretic peptide precursors A and B, and Myl4 and Myl7 genes, encoding myosin light chains, in hearts from LmnaH222P/H222P mice treated with PD98059 or placebo (DMSO). Results from hearts from Lmna+/+ mice and untreated LmnaH222P/H222P mice are shown for comparison. Bars indicate the fold overexpression of the indicated mRNA in hearts as calculated by the CT method. Values are means ± standard deviations for n = 4 samples from different animals per group. Reactions were performed in triplicate for each different RNA sample. Results were compared using a two-tailed t-test (*P < 0.05).
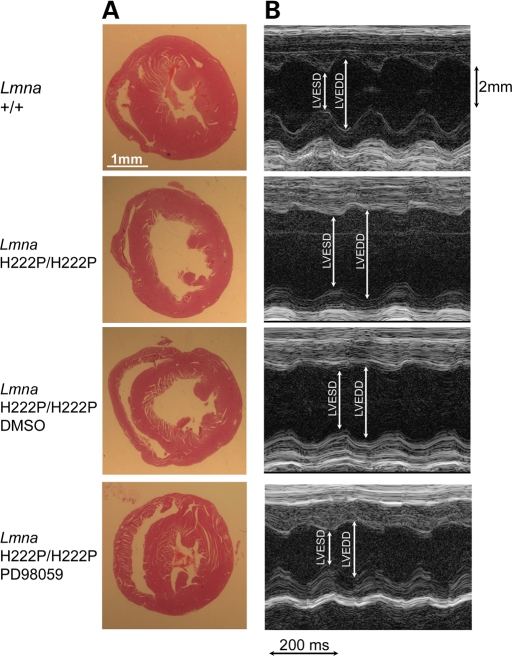
LV dilatation in male LmnaH222P/H222P mice at 16 weeks of age was demonstrated by histopathological analysis (Fig. 3A). At this age, there was no significant cardiomyocyte disarray or cardiac fibrosis on light microscopic examination (data not shown). M-mode transthoracic echocardiography showed increased LV end-systolic and end-diastolic diameters in LmnaH222P/H222P mice compared with Lmna+/+ mice (Fig. 3B). Treatment with PD98059 prevented the development of LV dilatation as measured by histopathology and echocardiography (Fig. 3).
Figure 3.
Treatment with the MEK inhibitor PD98059 prevents dilation and deterioration of dynamics of the left ventricle in LmnaH222P/H222P mice. (A) Histological analysis of heart sections stained with hematoxylin and eosin from LmnaH222P/H222P mice treated with PD98059 or placebo (DMSO). Hearts from Lmna+/+ mice and untreated LmnaH222P/H222P mice are shown for comparison. The left ventricle is dilated in LmnaH222P/H222P mice that were untreated or that received DMSO placebo, whereas hearts from LmnaH222P/H222P mice treated with PD98059 had an LV chamber diameter similar to Lmna+/+ mice. Scale bar: 1 mm. (B) Transthoracic M-mode echocardiographic tracings in LmnaH222P/H222P mice treated with PD98059 or placebo (DMSO). Tracings from Lmna+/+ mice and untreated LmnaH222P/H222P mice are shown for comparison. LV end-systolic diameter (LVESD) and LV end-diastolic diameter (LVEDD) are indicated. Note LVESD and LVEDD are similar in LmnaH222P/H222P mice treated with PD98059 and decreased in LmnaH222P/H222P mice that were untreated or that received DMSO placebo.
Cardiac structure and function were further assessed by echocardiography at 16 weeks of age in a total of 30 living mice in the four different groups studied (Table 1). Compared with Lmna+/+ mice, LmnaH222P/H222P mice had significantly increased LV end-diastolic and end-systolic diameters. They also had decreased cardiac contractility indicated by reduced ejection fraction and LV fractional shortening. Ejection fraction in LmnaH222P/H222P mice was decreased by ∼30% compared with Lmna+/+ mice at 16 weeks of age (73.12 ± 6.69 versus 50.78 ± 9.12%; P < 0.005). LmnaH222P/H222P mice treated with DMSO had ventricular chamber diameters, ejection fraction and LV fractional shortening similar to untreated LmnaH222P/H222P mice. LmnaH222P/H222P mice treated with PD98059 had normal cardiac contractility with ejection fraction and LV fractional shortening virtually identical to Lmna+/+ mice. With 100% accuracy in real-time, a ‘blinded’ echocardiographer unaware of the genotype or treatment received classified all Lmna+/+ mice and LmnaH222P/H222P mice receiving PD98059 as having normal cardiac function and all LmnaH222P/H222P mice that were untreated or treated with placebo as having abnormal cardiac function. Hence, treatment with PD98059 for 8 weeks prevented the development of LV dilatation and cardiac contractile dysfunction in LmnaH222P/H222P mice.
Table 1.
Echocardiographic data at 16 weeks of age for Lmna+/+ mice and LmnaH222P/H222P mice untreated, treated with DMSO placebo or treated with PD98059
| Genotype | n | LVEDD (mm) | LVESD (mm) | IVSD (mm) | EF (%) | FS (%) |
|---|---|---|---|---|---|---|
| Lmna+/+ | 12 | 3.45 ± 0.42 | 2.00 ± 0.36 | 0.70 ± 0.13 | 73.12 ± 6.69 | 41.72 ± 5.76 |
| LmnaH222P/H222P | 6 | 4.14 ± 0.27** | 3.25 ± 0.45** | 0.70 ± 0.05 | 50.78 ± 9.12** | 25.82 ± 5.70** |
| LmnaH222P/H222P (DMSO) | 5 | 3.89 ± 0.14* | 3.04 ± 0.32** | 0.69 ± 0.02 | 52.70 ± 9.03** | 26.96 ± 5.68** |
| LmnaH222P/H222P (PD98059) | 7 | 3.12 ± 0.20 | 1.82 ± 0.16 | 0.69 ± 0.06 | 73.52 ± 4.68 | 41.55 ± 4.29 |
LVEDD, left ventricular end-diastolic diameter; LVESD, left ventricular end-systolic diameter; IVSD, interventricular septum diameter; EF, ejection fraction; FS, fractional shortening. Values are means ± standard deviations. Comparison between groups was performed using one-way ANOVA and Tukey adjustment for post hoc multiple comparison (5% type I error). Conditions of homogeneity of variances were validated and non-parametric tests were performed to validate results.
*P < 0.05 versus Lmna+/+.
**P < 0.005 versus Lmna+/+.
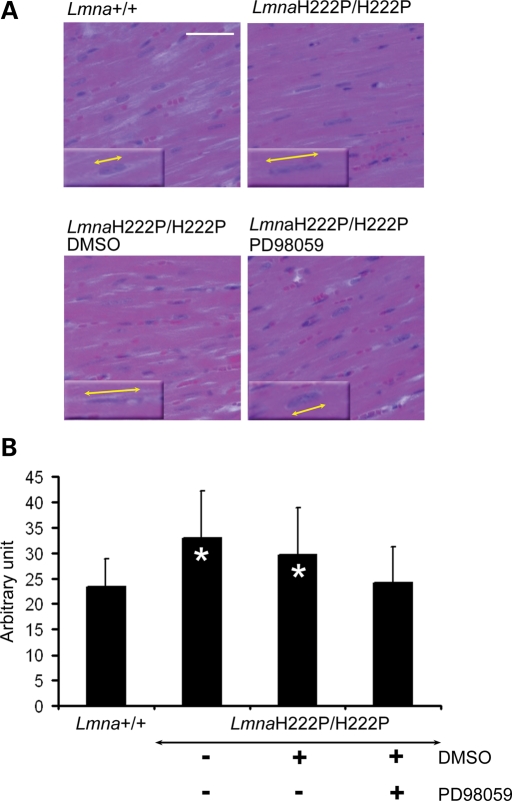
Alterations in nuclear morphology, including abnormal elongation of nuclei, have been described in hearts of mice deficient in A-type lamins that develop dilated cardiomyopathy (18). We observed similar elongation of nuclei in cardiomyocytes of 16-week-old LmnaH222P/H222P mice. Treatment with PD98059 prevented this alteration. Nuclei in cardiomyocytes of Lmna+/+ mice had a well-rounded oval shape, whereas nuclei in cardiomyocytes of LmnaH222P/H222P mice had an abnormally elongated shape (Fig. 4A). Cardiomyocyte nuclei in LmnaH222P/H222P mice treated with PD98059 but not placebo had an overall shape that was similar to those in Lmna+/+ mice (Fig. 4A). Mean lengths of cardiomyocyte nuclei in untreated and placebo-treated LmnaH222P/H222P mice were significantly longer than in Lmna+/+ mice and LmnaH222P/H222P mice treated with PD98059 (Fig. 4B).
Figure 4.
Treatment with PD98059 prevents abnormal elongation of cardiomyocyte nuclei in LmnaH222P/H222P mice. (A) Histological analysis of cross-sections of hearts from LmnaH222P/H222P mice treated with PD98059 or placebo (DMSO). Hearts from Lmna+/+ mice and untreated LmnaH222P/H222P mice were used for comparisons. Sections are stained with hematoxylin and eosin. Inserts with yellow lines with arrowheads demonstrate the measurement of nuclear length. Scale bar: 50 µm. (B) Quantification of nuclear elongation in cardiomyocytes from mice. Cardiomyocyte nuclei are measured along the yellow lines with arrowheads as shown as examples in (A). Bars indicate the length of cardiomyocyte nuclei in the indicated hearts. Values are means ± standard deviations for n = 400 cardiomyocytes (*P < 0.0005).
Overall, LV diameters, cardiomyocyte nuclear morphology and cardiac ejection fraction were normal in LmnaH222P/H222P mice treated with PD98059 at an age when untreated and placebo-treated mice had significant abnormalities in these parameters. Enhanced synthesis of natriuretic peptides and sarcomeric proteins was also prevented. Treatment with an inhibitor of ERK activation therefore delayed the development of significant cardiomyopathy in mice with an Lmna mutation that causes Emery–Dreifuss muscular dystrophy in humans.
DISCUSSION
Our results support the hypothesis that ERK activation induced by abnormalities in A-type lamins is a pathogenic mechanism in the generation of cardiomyopathy. We demonstrated the inhibition of ERK phosphorylation and attenuated activation of downstream genes when PD98059 was administered systemically to LmnaH222P/H222P mice. Concurrent with this inactivation of ERK signaling in the heart, we documented normal LV diameters, normal cardiomyocyte nuclear morphology and normal cardiac ejection fraction in LmnaH222P/H222P mice treated with PD98059 at an age when untreated and placebo-treated mice had significant abnormalities in these parameters. These results are consistent with our previous findings that ERK is abnormally activated in cardiomyocytes of LmnaH222P/H222P mice and cells expressing lamin A variants found in human subjects with cardiomyopathy (9). Results from Favreau et al. (19) using cultured myoblasts also suggest that the nuclear lamina may serve as scaffold for substrates of the MEK-ERK pathway and that this may be impeded by A-type lamin alterations resulting from LMNA mutations that cause Emery–Dreifuss muscular dystrophy.
PD98059 shows high specificity for MEK over other serine/threonine kinases (11,20). However, it also has activity against cyclooxygenase-1 and cyclooxygenase-2 (21). It is therefore possible that the beneficial effects of PD98059 in LmnaH222P/H222P mice could in part be due to cyclooxygenase inhibition. We do not however consider cyclooxygenase inhibition to be a major mechanism of action given the widespread use of non-steroidal anti-inflammatory drugs in clinical practice and the absence of data showing any utility in preventing heart failure. In fact, retrospective populations cohort studies suggest that the use of both cyclooxygenase-2 inhibitors and non-selective cyclooxygenase inhibitors is associated with the exacerbation of heart failure in humans (22,23). Nonetheless, future controlled experimental testing of cyclooxygenase inhibition in LmnaH222P/H222P mice would be useful in determining whether it has any beneficial effect in delaying or preventing cardiomyopathy.
Similar to LmnaH222P/H222P mice, we have shown previously abnormal activation of ERK signaling in hearts of Emd−/y mice lacking the integral inner nuclear membrane protein emerin that binds to A-type lamins (24). In humans, EMD mutations resulting in the lack of or reduced emerin in the nuclear envelope cause X-linked Emery–Dreifuss muscular dystrophy (25–27). Like the autosomally inherited form of the disease caused by LMNA mutations, dilated cardiomyopathy is a major feature of X-linked Emery–Dreifuss muscular dystrophy. Therefore, the present results in LmnaH222P/H222P mice may also be relevant to cardiomyopathy caused by emerin deficiency. However, because the clinical phenotype of first-degree heart block in Emd−/y mice >40 weeks of age is very subtle and not readily measurable without intensive electrophysiologically monitoring (28,29), we have deferred a trail of an ERK inhibitor in this animal model.
Our results provide initial proof of principle for ERK inhibition as a therapeutic option to prevent or delay the onset of heart failure in cardiomyopathy caused by LMNA mutation. The only other way of improving an abnormal phenotype caused by mutations in the gene encoding A-type lamins in mammals is the use of a protein farnesyltransferase inhibitor to block the prenylation of truncated prelamin A in mice carrying a mutation that causes Hutchinson–Gilford progeria syndrome (30,31). In the present study, treatment with an MEK inhibitor at an age when LmnaH222P/H222P mice first begin to develop cardiac abnormalities maintained LV function at normal levels, whereas untreated mice had an ∼30% reduction in ejection fraction over a time period of 8 weeks. In humans, the progression of cardiomyopathy caused by LMNA mutations is often rapid compared with other primary cardiomyopathies (6). Therefore, pharmacological interventions to slow progression could be clinically beneficial. Although further preclinical investigation, including, for example, an analysis of effects on different tissues, skeletal myopathy and overall activity, is necessary to determine the safety and efficacy of ERK inhibition as a therapeutic intervention for dilated cardiomyopathy caused by LMNA mutations, it is worth noting that oral MEK inhibitors have already been safely administered to humans (32,33).
MATERIALS AND METHODS
Mice
LmnaH222P mice were generated and genotyped as described (10). Genotyping was performed by PCR using oligonucleotides 5′-CAGCCATCACCTCTCCTTTG-3′ and 5′-AGCACCAGGGAGAGGACAGG-3′. Mice were fed a chow diet and housed in a barrier facility. The Institutional Animal Care and Use Committee at the Columbia University Medical Center approved the use of animals and the study protocol.
MEK inhibitor
PD98059 (Calbiochem) was dissolved in DMSO (Sigma) at a concentration of 0.5 mg/ml and was delivered to a dose of 3 mg/kg/day for 5 days a week. The inhibitory activity of PD98059 is specific for MEK and does not inhibit JNK (11; data not shown). The placebo control consisted of DMSO alone and was delivered in the same volume. Placebo and PD98059 were administered by intraperitoneal injection using a 275/8-gauge syringe. Treatment was started when mice were 8 weeks of age and continued until 16 weeks of age.
Protein extraction and immunoblotting
Hearts were excised from mice at 16 weeks of age and were homogenized in RIPA extraction buffer (Cell Signaling) containing protease inhibitors (25 mg/ml aprotinin and 10 mg/ml leupeptin). Protein samples were subjected to SDS–PAGE, transferred to nitrocellulose membranes and blotted with primary antibodies against ERK1/2 (Santa Cruz), phosphorylated ERK1/2 (Cell Signaling), natriuretic peptide precursor A (Santa Cruz) and Gapdh (Ambion). Secondary antibodies were horseradish peroxidate-conjugated (Amersham). Recognized proteins were visualized by enhanced chemiluminescence (ECL, Amersham). The signal generated using antibody against Gapdh was used as internal control to normalize the amounts of protein between immunoblots.
RNA isolation and quantitative real-time RT–PCR analysis
Total RNA was extracted using the RNeasy isolation kit (Qiagen) as described previously (9). cDNA was synthesized as described previously (9) using Omniscript Reverse Transcriptase (Qiagen) on total cellular RNA. For each replicate in each experiment, RNA from tissue samples of different animals was used. Primers were designed that correspond to mouse RNA sequences using Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). The real-time RT–PCR reaction contained iQ SYBR green super mix (Bio-Rad), 200 nm of each primer and 0.2 µl of template in a 25 µl reaction volume. Amplification was carried out using appropriate primers and the MyiQ Single-Color Real-Time PCR Detection System (Bio-Rad) with an initial denaturation at 95°C for 2 min, followed by 50 cycles at 95°C for 30 s and 62°C for 30 s. Relative levels of mRNA expression were calculated using the CT method (34). Individual expression values were normalized by comparison with Gapdh mRNA.
Pathological analysis of hearts
Mice were sacrificed at 16 weeks of age and freshly removed hearts were fixed in 4% formaldehyde for 48 h, embedded in paraffin, sectioned at 5 µm and stained with hematoxylin and eosin and Masson’s trichrome. Representative stained sections were photographed using a Microphot SA (Nikon) light microscope attached to a Spot RT Slide camera (Diagnostic Instruments). Images were processed using Adobe Photoshop 6.0 (Adobe Systems). The length of cardiomyocytes was measured using Scion Image software (Scion Corporation). Data were reported as means ± standard deviations and are compared with respective controls using a two-tailed t-test.
Transthoracic echocardiography
At 16 weeks of age, mice were anesthetized with 1.5% isoflurane in O2 and placed on a heating pad (37°C). Cardiac function was assessed by echocardiography with a Visualsonics Vevo 770 ultrasound with a 30 MHz transducer applied to the chest wall. Cardiac ventricular dimensions and ejection fraction were measured in 2D mode and M-mode three times for the number of animals indicated. A ‘blinded’ echocardiographer (J.S.), unaware of the genotype or treatment, performed the examinations.
Statistical analysis
To determine significant differences between groups of animals analyzed by echocardiography, we used one-way analysis of variance (ANOVA). For each parameter, there was a global effect between different groups (P < 0.001). This indicated that at least one group had significantly different results than another. We then used a Tukey adjustment for post hoc multiple comparisons (5% type I error) to determine which groups were significantly different. Homogeneity of variances between groups was validated using Levene test (alpha = 0.05). The normality of residuals was validated using Shapiro–Wilk test. To validate all results, non-parametric tests (Kruskal–Wallis and Mann–Whitney) were performed and concordance checked. Other statistical methods used are described in the figure legends.
FUNDING
This work was supported by grants from the Muscular Dystrophy Association (MDA 4287) and National Institutes of Health (AR048997). A.M. was supported in part by fellowships from Association Française contre les Myopathies and la Fondation pour la Recherche Médicale.
ACKNOWLEDGEMENTS
We thank Dr Andrew R. Marks for critically reviewing the manuscript and advice, Dr Wei Wu for assistance with the figures and Marie Thomas for help with statistical analysis. This work is dedicated to the memory of Dr Ketty Schwartz.
Conflict of Interest statement. H.J.W. and A.M. are inventors on a provisional US patent application filed by The Trustees of Columbia University in the City of New York based in part on the results presented in this paper.
REFERENCES
- 1.Worman H.J., Bonne G. ‘Laminopathies’: a wide spectrum of human diseases. Exp. Cell Res. 2007;313:2121–2133. doi: 10.1016/j.yexcr.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonne G., Di Barletta M.R., Varnous S., Bécane H.M., Hammouda E.H., Merlini L., Muntoni F., Greenberg C.R., Gary F., Urtizberea J.A., et al. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery–Dreifuss muscular dystrophy. Nat. Genet. 1999;21:285–288. doi: 10.1038/6799. [DOI] [PubMed] [Google Scholar]
- 3.Fatkin D., MacRae C., Sasaki T., Wolff M.R., Porcu M., Frenneaux M., Atherton J., Vidaillet H.J., Jr, Spudich S., De Girolami U., et al. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N. Engl. J. Med. 1999;341:1715–1724. doi: 10.1056/NEJM199912023412302. [DOI] [PubMed] [Google Scholar]
- 4.Muchir A., Bonne G., van der Kooi A.J., van Meegen M., Baas F., Bolhuis P.A., de Visser M., Schwartz K. Identification of mutations in the gene encoding lamins A/C in autosomal dominant limb girdle muscular dystrophy with atrioventricular conduction disturbances (LGMD1B) Hum. Mol. Genet. 2000;9:1453–1459. doi: 10.1093/hmg/9.9.1453. [DOI] [PubMed] [Google Scholar]
- 5.Di Barletta M.R., Ricci E., Galluzzi G., Tonali P., Mora M., Morandi L., Romorini A., Voit T., Orstavik K.H., Merlini L., et al. Different mutations in the LMNA gene cause autosomal dominant and autosomal recessive Emery–Dreifuss muscular dystrophy. Am. J. Hum. Genet. 2000;66:1407–1412. doi: 10.1086/302869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor M.R., Fain P.R., Sinagra G., Robinson M.L., Robertson A.D., Carniel E., Di Lenarda A., Bohlmeyer T.J., Ferguson D.A., Brodsky G.L., et al. Natural history of dilated cardiomyopathy due to lamin A/C gene mutations. J. Am. Coll. Cardiol. 2003;41:771–780. doi: 10.1016/s0735-1097(02)02954-6. [DOI] [PubMed] [Google Scholar]
- 7.Bécane H.M., Bonne G., Varnous S., Muchir A., Ortega V., Hammouda E.H., Urtizberea J.A., Lavergne T., Fardeau M., Eymard B., et al. High incidence of sudden death with conduction system and myocardial disease due to lamins A and C gene mutation. Pacing Clin. Electrophysiol. 2000;23:1661–1666. doi: 10.1046/j.1460-9592.2000.01661.x. [DOI] [PubMed] [Google Scholar]
- 8.Meune C., Van Berlo J.H., Anselme F., Bonne G., Pinto Y.M., Duboc D. Primary prevention of sudden death in patients with lamin A/C gene mutations. N. Engl. J. Med. 2006;354:209–210. doi: 10.1056/NEJMc052632. [DOI] [PubMed] [Google Scholar]
- 9.Muchir A., Pavlidis P., Decostre V., Herron A.J., Arimura T., Bonne G., Worman H.J. Activation of MAPK pathways links LMNA mutations to cardiomyopathy in Emery–Dreifuss muscular dystrophy. J. Clin. Invest. 2007;117:1282–1293. doi: 10.1172/JCI29042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arimura T., Helbling-Leclerc A., Massart C., Varnous S., Niel F., Lacène E., Fromes Y., Toussaint M., Mura A.M., Keller D.I., et al. Mouse model carrying H222P-Lmna mutation develops muscular dystrophy and dilated cardiomyopathy similar to human striated muscle laminopathies. Hum. Mol. Genet. 2005;14:155–169. doi: 10.1093/hmg/ddi017. [DOI] [PubMed] [Google Scholar]
- 11.Dudley D.T., Pang L., Decker S.J., Bridges A.J., Saltiel A.R. A synthetic inhibitor of the mitogen-activated protein kinase cascade. Proc. Natl Acad. Sci. USA. 1995;92:7686–7689. doi: 10.1073/pnas.92.17.7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanada S., Node K., Minamino T., Takashima S., Ogai A., Asanuma H., Ogita H., Liao Y., Asakura M., Kim J., et al. Long-acting Ca2+ blockers prevent myocardial remodeling induced by chronic NO inhibition in rats. Hypertension. 2003;41:963–967. doi: 10.1161/01.HYP.0000062881.36813.7A. [DOI] [PubMed] [Google Scholar]
- 13.Barrans J.D., Allen P.D., Stamatiou D., Dzau V.J., Liew C.C. Global gene expression profiling of end-stage dilated cardiomyopathy using a human cardiovascular-based cDNA microarray. Am. J. Pathol. 2002;160:2035–2043. doi: 10.1016/S0002-9440(10)61153-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mizuno Y., Yoshimura M., Harada E., Nakayama M., Sakamoto T., Shimasaki Y., Ogawa H., Kugiyama K., Saito Y., Nakao K., et al. Plasma levels of A- and B-type natriuretic peptides in patients with hypertrophic cardiomyopathy or idiopathic dilated cardiomyopathy. Am. J. Cardiol. 2000;86:1036–1040. doi: 10.1016/s0002-9149(00)01147-4. [DOI] [PubMed] [Google Scholar]
- 15.Wang J., Xu N., Feng X., Hou N., Zhang J., Cheng X., Chen Y., Zhang Y., Yang X. Targeted disruption of Smad4 in cardiomyocytes results in cardiac hypertrophy and heart failure. Circ. Res. 2005;97:821–828. doi: 10.1161/01.RES.0000185833.42544.06. [DOI] [PubMed] [Google Scholar]
- 16.Abraham W.T., Gilbert E.M., Lowes B.D., Minobe W.A., Larrabee P., Roden R.L., Dutcher D., Sederberg J., Lindenfeld J.A., Wolfel E.E., et al. Coordinate changes in myosin heavy chain isoform gene expression are selectively associated with alterations in dilated cardiomyopathy phenotype. Mol. Med. 2002;8:750–760. [PMC free article] [PubMed] [Google Scholar]
- 17.Grzeskowiak R., Witt H., Drungowski M., Thermann R., Hennig S., Perrot A., Osterziel K.J., Klingbiel D., Scheid S., Spang R., et al. Expression profiling of human idiopathic dilated cardiomyopathy. Cardiovasc. Res. 2003;59:400–411. doi: 10.1016/s0008-6363(03)00426-7. [DOI] [PubMed] [Google Scholar]
- 18.Nikolova V., Leimena C., McMahon A.C., Tan J.C., Chandar S., Jogia D., Kesteven S.H., Michalicek J., Otway R., Verheyen F., et al. Defects in nuclear structure and function promote dilated cardiomyopathy in lamin A/C-deficient mice. J. Clin. Invest. 2004;113:357–369. doi: 10.1172/JCI19448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Favreau C., Delbarre E., Courvalin J.C., Buendia B. Differentiation of C2C12 myoblasts expressing lamin A mutated at a site responsible for Emery–Dreifuss muscular dystrophy is improved by inhibition of the MEK-ERK pathway and stimulation of the PI3-kinase pathway. Exp. Cell Res. 2008;314:1392–1405. doi: 10.1016/j.yexcr.2008.01.018. [DOI] [PubMed] [Google Scholar]
- 20.Alessi D.R., Cuenda A., Cohen P., Dudley D.T., Saltiel A.R. PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J. Biol. Chem. 1995;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- 21.Börsch-Haubold A.G., Pasquet S., Watson S.P. Direct inhibition of cyclooxygenase-1 and -2 by the kinase inhibitors SB 203580 and PD 98059. SB 203580 also inhibits thromboxane synthase. J. Biol. Chem. 1998;273:28766–28772. doi: 10.1074/jbc.273.44.28766. [DOI] [PubMed] [Google Scholar]
- 22.Mamdani M., Juurlink D.N., Lee D.S., Rochon P.A., Kopp A., Naglie G., Austin P.C., Laupacis A., Stukel T.A. Cyclo-oxygenase-2 inhibitors versus non-selective non-steroidal anti-inflammatory drugs and congestive heart failure outcomes in elderly patients: a population-based cohort study. Lancet. 2004;363:1751–1756. doi: 10.1016/S0140-6736(04)16299-5. [DOI] [PubMed] [Google Scholar]
- 23.Hudson M., Richard H., Pilote L. Differences in outcomes of patients with congestive heart failure prescribed celecoxib, rofecoxib, or non-steroidal anti-inflammatory drugs: population based study. BMJ. 2005;330:1370. doi: 10.1136/bmj.330.7504.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muchir A., Pavlidis P., Bonne G., Hayashi Y.K., Worman H.J. Activation of MAPK in hearts of EMD null mice: similarities between mouse models of X-linked and autosomal dominant Emery–Dreifuss muscular dystrophy. Hum. Mol. Genet. 2007;16:1884–1895. doi: 10.1093/hmg/ddm137. [DOI] [PubMed] [Google Scholar]
- 25.Bione S., Maestrini E., Rivella S., Mancini M., Regis S., Romeo G., Toniolo D. Identification of a novel X-linked gene responsible for Emery–Dreifuss muscular dystrophy. Nat. Genet. 1994;8:323–327. doi: 10.1038/ng1294-323. [DOI] [PubMed] [Google Scholar]
- 26.Nagano A., Koga R., Ogawa M., Kurano Y., Kawada J., Okada R., Hayashi Y.K., Tsukahara T., Arahata K. Emerin deficiency at the nuclear membrane in patients with Emery–Dreifuss muscular dystrophy. Nat. Genet. 1996;12:254–259. doi: 10.1038/ng0396-254. [DOI] [PubMed] [Google Scholar]
- 27.Manilal S., Nguyen T.M., Sewry C.A., Morris G.E. The Emery–Dreifuss muscular dystrophy protein, emerin, is a nuclear membrane protein. Hum. Mol. Genet. 1996;5:801–808. doi: 10.1093/hmg/5.6.801. [DOI] [PubMed] [Google Scholar]
- 28.Melcon G., Kozlov S., Cutler D.A., Sullivan T., Hernandez L., Zhao P., Mitchell S., Nader G., Bakay M., Rottman J.N., et al. Loss of emerin at the nuclear envelope disrupts the Rb1/E2F and MyoD pathways during muscle regeneration. Hum. Mol. Genet. 2006;15:637–651. doi: 10.1093/hmg/ddi479. [DOI] [PubMed] [Google Scholar]
- 29.Ozawa R., Hayashi Y.K., Ogawa M., Kurokawa R., Matsumoto H., Noguchi S., Nonaka I., Nishino I. Emerin-lacking mice show minimal motor and cardiac dysfunctions with nuclear-associated vacuoles. Am. J. Pathol. 2006;168:907–917. doi: 10.2353/ajpath.2006.050564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang S.H., Meta M., Qiao X., Frost D., Bauch J., Coffinier C., Majumdar S., Bergo M.O., Young S.G., Fong L.G. A farnesyltransferase inhibitor improves disease phenotypes in mice with a Hutchinson–Gilford progeria syndrome mutation. J. Clin. Invest. 2006;116:2115–2121. doi: 10.1172/JCI28968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang S.H., Qiao X., Fong L.G., Young S.G. Treatment with a farnesyltransferase inhibitor improves survival in mice with a Hutchinson–Gilford progeria syndrome mutation. Biochim. Biophys. Acta. 2008;1781:36–39. doi: 10.1016/j.bbalip.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lo Russo P.M., Adjei A.A., Varterasian M., Gadgeel S., Reid J., Mitchell D.Y., Hanson L., DeLuca P., Bruzek L., Piens J., et al. Phase I and pharmacodynamic study of the oral MEK inhibitor CI-1040 in patients with advanced malignancies. J. Clin. Oncol. 2005;23:5281–5293. doi: 10.1200/JCO.2005.14.415. [DOI] [PubMed] [Google Scholar]
- 33.Adjei A.A., Cohen R.B., Franklin W., Morris C., Wilson D., Molina J.R., Hanson L.J., Gore L., Chow L., Leong S., et al. Phase I pharmacokinetic and pharmacodynamic study of the oral, small-molecule mitogen-activated protein kinase kinase 1/2 inhibitor AZD6244 (ARRY-142886) in patients with advanced cancers. J. Clin. Oncol. 2008;26:2139–2146. doi: 10.1200/JCO.2007.14.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ponchel F., Toomes C., Bransfield K., Leong F.T., Douglas S.H., Field S.L., Bell S.M., Combaret V., Puisieux A., Mighell A.J., et al. Real-time PCR based on SYBR-Green I fluorescence: an alternative to the TaqMan assay for a relative quantification of gene rearrangements, gene amplifications and micro gene deletions. BMC Biotechnol. 2003;3:18. doi: 10.1186/1472-6750-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]