Abstract
Cleidocranial dysplasia (CCD) in humans is an autosomal-dominant skeletal disease that results from mutations in the bone-specific transcription factor RUNX2 (CBFA1/AML3). However, distinct RUNX2 mutations in CCD do not correlate with the severity of the disease. Here we generated a new mouse model with a hypomorphic Runx2 mutant allele (Runx2neo7), in which only part of the transcript is processed to full-length (wild-type) Runx2 mRNA. Homozygous Runx2neo7/neo7 mice express a reduced level of wild-type Runx2 mRNA (55–70%) and protein. This mouse model allowed us to establish the minimal requirement of functional Runx2 for normal bone development. Runx2neo7/neo7 mice have grossly normal skeletons with no abnormalities observed in the growth plate, but do exhibit developmental defects in calvaria and clavicles that persist through post-natal growth. Clavicle defects are caused by disrupted endochondral bone formation during embryogenesis. These hypomorphic mice have altered calvarial bone volume, as observed by histology and microCT imaging, and decreased expression of osteoblast marker genes. The bone phenotype of the heterozygous mice, which have 79–84% of wild-type Runx2 mRNA, is normal. These results show there is a critical gene dosage requirement of functional Runx2 for the formation of intramembranous bone tissues during embryogenesis. A decrease to 70% of wild-type Runx2 levels results in the CCD syndrome, whereas levels >79% produce a normal skeleton. Our findings suggest that the range of bone phenotypes in CCD patients is attributable to quantitative reduction in the functional activity of RUNX2.
INTRODUCTION
Skeletogenesis occurs by two distinct osteogenic processes: intramembranous and endochondral bone formation. Both processes require mesenchymal cell condensation leading to the formation of cartilage and bone tissues, respectively. Craniofacial and clavicle development are highly dependent on normal intramembranous bone formation, whereas the axial skeleton develops by endochondral bone formation. The RUNX2 transcription factor is well established to be required for both types of bone formation and is implicated in mesenchymal–epithelial interactions in tooth development (1–4). Null mutation of Runx2 in mice results in neonatal lethality with a complete absence of bone tissue, defective hypertrophic chondrocytes and lack of differentiated osteoblasts (5–8). RUNX2 mutations in humans are responsible for cleidocranial dysplasia (CCD), an autosomal-dominant heritable skeletal disease characterized by open or delayed closure of calvarial sutures, hypoplastic or aplastic clavicles and supernumerary teeth (9,10). Heterozygous disruption of the Runx2 locus in mice produces a CCD phenotype except for the teeth (6,8). These and other studies of human CCD have identified multiple phenotypes owing to mutations in the RUNX2 gene (10,11).
Runx2 is characterized by a highly conserved runt homology DNA-binding domain (RHD) in the N-terminus, and the C-terminus contains a nuclear matrix-associated regulatory domain (nuclear matrix-targeting signal—NMTS) that targets Runx2 to subnuclear foci (12). Runx2 association with the nuclear scaffold facilitates interaction with many co-regulatory proteins and chromatin-modifying complexes for the regulation of gene transcription. Mice homozygous for the deletion of the NMTS domain in exon 8 (Runx2ΔC) do not form bone owing to the maturational arrest of osteoblasts (7). Heterozygotes exhibit the absence of clavicles (7,13) and a delay in calvarial bone formation. These phenotypes indicate that the C-terminal domain is as critical for the function of Runx2 in skeletogenesis as the DNA-binding domain.
In CCD patients, many monoallelic mutations of RUNX2 have been identified, including deletion, missense, nonsense and frameshift mutations (9–11). The majority of these mutations are clustered in the N-terminal RHD, and several positions emerge as mutational hotspots (11). Most of these RHD mutations result in premature termination and presumably defective DNA binding. Other mutants can express full-length RUNX2 (FL-Runx2) but are predicted to exhibit impaired binding of the Cbfβ partner protein to the runt domain structure. It has been suggested that the CCD mutations create nonfunctional, dominant-negative or partially defective RUNX2 proteins (11,14). Recent studies indicate that some skeletal and dental defects, as represented by short stature and supernumerary teeth, show significant differences in individual CCD patients that are related to the residual transactivation potential that remains in mutant RUNX2 proteins (11). Because all CCD patients exhibit craniofacial defects, it has been proposed that intramembranous bone formation may require a high level of functional RUNX2. However, what range or extent of functional loss in RUNX2 causes the CCD defects has remained obscure.
We have performed gene-targeting experiments in mouse embryonic stem (ES) cells and have generated a novel, hypomorphic Runx2 mutant allele (Runx2neo7) through the insertion of a neomycin cassette (neo) into intron 7. In the resulting mice, the normal splicing of Runx2 is partly disrupted and a Runx2-neo chimeric mRNA encoding a truncated protein is produced. Homozygous mutant mice (Runx2neo7/neo7) express only 55–70% of the normal amount of correctly spliced mRNA and thus a decreased amount of Runx2 full-length protein, resulting in a CCD phenotype. Skeletal defects do not occur in heterozygous mice (Runx2+/neo7) with 79–84% of wild-type Runx2 levels. Studies reveal that a critical threshold is crossed between 79% (normal phenotype) and 70% (CCD phenotype) of the wild-type Runx2 activity, which results in CCD phenotypes. Our studies have established the Runx2 gene-dosage requirement for the normal bone development.
RESULTS
Generation of a hypomorphic Runx2 allele in mice
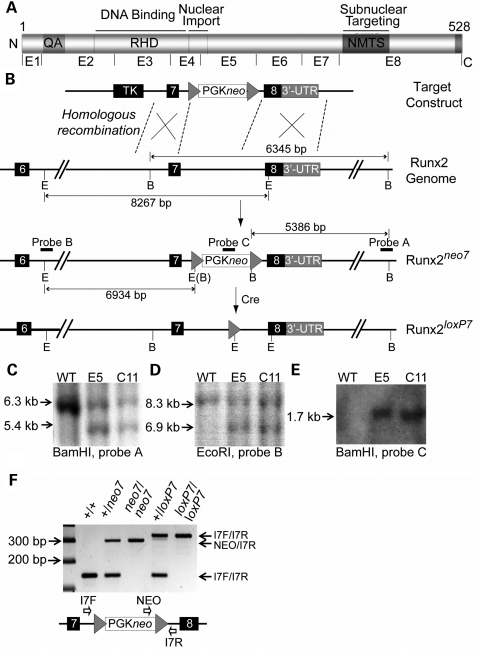
Previous gene-targeting experiments in mice have shown that the insertion of a PGK-driven neomycin resistance cassette (PGKneo) into an intron can interfere with normal splicing (15–17), thereby reducing the wild-type mRNA expression level of a target gene. In generating mouse models to study Runx2 functional domains, we identified a hypomorphic allele resulting from the insertion of the PGKneo cassette in intron 7 (Runx2neo7) (Fig. 1). PGKneo was subsequently removed and the Runx2loxp7 allele was generated. Figure 1B shows a schematic diagram of the Runx2 genomic locus and the targeting strategy. The targeted mouse ES cells were identified by a series of Southern blots using different probes (Fig. 1C, D and E). Genotypes of animals were determined routinely by PCR (Fig. 1F) and confirmed by Southern Blot analysis using the probes described (data not shown). The heterozygous and homozygous mutant mice were viable and fertile.
Figure 1.
Generation of the Runx2neo7 and Runx2loxp7 alleles. (A) Organization of the functional domains of Runx2 protein. (B) Schematic diagram of the C-terminus of the Runx2 gene and strategy to create Runx2neo7 and Runx2loxp7 alleles; loxP sites are represented by arrowheads. The relative positions of PGKneo and TK cassettes are shown. E, EcoRI restriction site; B, BamHI restriction site. The sizes of digested fragments are illustrated. The locations of probes for Southern blot are indicated as black horizontal bars. (C–E) Probes A, B and C [indicated in (B)] were used for Southern blot analysis, respectively, and the expected fragment sizes were detected for the wild-type and mutant alleles in two correctly targeted ES cell clones (E5 and C11). (F) Genotyping by PCR analysis of mouse tail genomic DNA from WT (+/+), neo7 heterozygous (+/neo7), neo7 homozygous (neo7/neo7), LoxP7 heterozygous (+/loxP7) and LoxP7 homozygous (loxP7/loxP7) animals. Bands corresponding to amplification products for the I7F/I7R and NEO/I7R primers are indicated on the right. The diagram shows the primer locations, and the sequences are listed in Table 2.
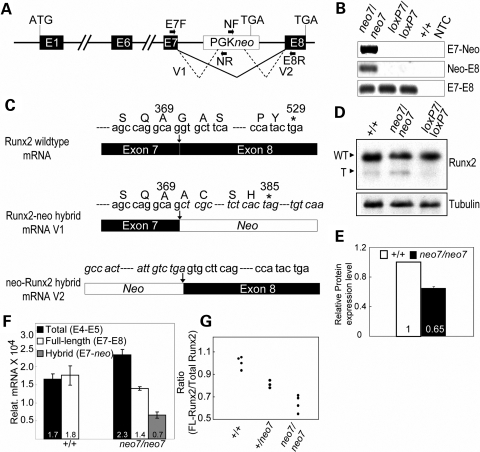
To examine the extent of alternative splicing and the products generated by the Runx2neo7 allele, we used reverse transcription–polymerase chain reaction (RT–PCR) analysis (Fig. 2A) of calvarial mRNAs. We identified alternatively spliced RNA between exon 7 and neo (V1), as well as between neo and exon 8 (V2) (Fig. 2A) in Runx2neo7/neo7 homozygotes (Fig. 2B). These hybrid transcripts were not present either in Runx2loxp7/loxp7 homozygotes or in wild-type animals (Fig. 2B). Normal splice products between exon7 and exon8 were detected in all the mice (Fig. 2B bottom panel). Nucleotide sequences of the V1 and V2 hybrid mRNAs were analyzed by directly sequencing RT–PCR products. The V1 hybrid mRNA presumably encodes a truncated Runx2 protein according to the reading frame of the sequence (Fig. 2C, middle panel). The truncated protein was identified by western blot in Runx2neo7/neo7 homozygous mice (Fig. 2D). The FL-Runx2 protein was visible but no truncated Runx2 was found in Runx2loxp7/loxp7 homozygous or wild-type mice (Fig. 2D). The quantitative results (measured by density of bands) show that the expression of FL-Runx2 in Runx2neo7/neo7 homozygous mice was decreased to ∼65% of that in wild-type mice (Fig. 2E). Taken together, these data indicate that the introduction of PGKneo into intron7 results in alternative splicing between neo and exons 7 and/or 8 of Runx2, leading to the expression of a mutant Runx2 protein with a deleted C-terminus.
Figure 2.
The neo cassette causes decreased expression of wild-type Runx2. (A) Genomic structure of the Runx2neo7 locus. The black line and dotted line represent normal and alternative splicing, respectively. The locations of primers used for RT–PCR analysis of alternative splicing are indicated as black horizontal arrows (E7F, NF, NR and E8R; the sequences are listed in Table 2). (B) RT–PCR analysis of RNA isolated from calvaria of 6-week-old wild-type (+/+), Runx2neo7 homozygous (neo7/neo7) and Runx2loxP7 homozygous (loxP7/loxP7) mice. NTC, No-DNA template control. Top: RT–PCR analysis is used to identify the alternative splicing between Runx2 exon 7 and neo (E7-neo); middle: RT–PCR analysis identifies alternative splicing between neo and Runx2 exon 8 (neo-E8); bottom: RT–PCR analysis shows the normal splicing between Runx2 exon 7 and exon 8 (E7–E8). (C) Schematic diagram of nucleotide sequence analysis for wild-type and Runx2-neo hybrid mRNA. Asterisk indicates the stop codon in the reading frame. The italicized letters represent the nucleotide sequence of PGKneo. (D) Immunoblot analysis of Runx2 protein from calvaria of 12-week-old wild-type, Runx2neo7/neo7 and Runx2loxp7/loxp7 mice. WT, wild-type Runx2; T, truncated Runx2. The band located near the “T” position in the +/+ lane is non-specific. Tubulin (shown at the bottom) was used as a loading control. (E) The relative protein expression level was quantified by ImageJ software. (F) Real-time quantitative RT–PCR analysis for the Runx2 isoforms using isolated osteoblasts (at confluency) from calvaria of newborn mice. All the measured data were normalized to 28S RNA. Values are the mean (indicated in each column) ± the SD of independent samples (n = 4 mice/group). E4–E5: primers span exons 4 and 5; E7–E8: primers span exons 7 and 8; E7-neo: primers span exon 7 and neo cassette. (G) The ratio of full-length (wild-type) Runx2/total Runx2 mRNA was calculated for individual wild-type (+/+) (n = 4), heterozygous (+/neo7) (n = 3) and homozygous (neo7/neo7) (n = 4) mice. Duplicate analysis of each sample was performed by real-time quantitative RT–PCR (see Materials and Methods). The SD for each sample was in the range of 0.002–0.02.
In order to quantify the relative abundance of hybrid and full-length mRNAs resulting from mutant or WT alleles, we performed real-time quantitative PCR using cDNA prepared from calvarial osteoblast cells at confluency (Fig. 2F). In wild-type mice, the level of total (exon 4–exon 5) and full-length (exon 7–exon 8) Runx2 mRNA is similar, 1.7 × 104 ± 0.2 × 104 and 1.8 × 104 ± 0.3 × 104, respectively, and no hybrid mRNA was detected. However in homozygous Runx2neo7/neo7 mice, the FL-Runx2 mRNA (1.4 × 104 ± 0.1 × 104) was decreased to ∼61% of the total (exon 4–exon 5) Runx2 mRNA (2.3 × 104 ± 0.2 × 104), and hybrid Runx2-neo mRNA was present. We then measured the percent of wild-type Runx2 mRNA in individual Runx2neo7/neo7, Runx2+/neo7 and wild-type mice. The result shows that wild-type Runx2 mRNA was reduced to 55–70% in homozygous and 79–84% in heterozygous animals (Fig. 2G). Thus the PGKneo insertion into the Runx2 gene reduces the level of FL-Runx2 mRNA and causes dosage insufficiency of Runx2 in Runx2neo7/neo7 homozygous mice.
Runx2 dosage insufficiency affects calvaria and clavicle development
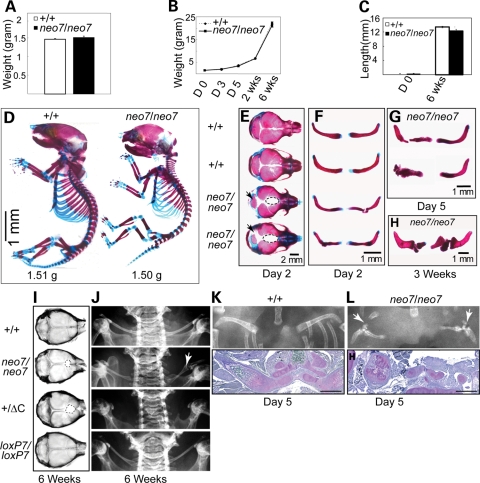
Although both Runx2 null and Runx2ΔC homozygous mice have perinatal lethal phenotypes owing to complete absence of osteoblasts and bone (5–8), Runx2neo7/neo7 transgenic mice survive after birth to adulthood. The weight differences are not significant between newborns of Runx2neo7 homozygous and wild-type mice (Fig. 3A), and no remarkable differences in weight gain were observed through 6 weeks of post-natal growth (Fig. 3B). Also, no statistically significant differences of long bone length between Runx2neo7/neo7 and wild-type mice were observed for newborns or at 6 weeks of age (Fig. 3C). These initial findings indicate that the 55–70% of wild-type Runx2 level remaining in the homozygotes supports mouse viability. In order to determine whether the loss of Runx2 activity produces a skeletal phenotype in these transgenic mice, we investigated bone structures beginning from the post-natal period.
Figure 3.
Runx2neo7 homozygous mice display defects of calvaria and clavicles during post-natal growth. (A) Weight of 2-day-old Runx2neo7/neo7 and wild-type mice (n = 5, each group). Values are the mean ± the SEM of independent samples. (B) Growth curves of Runx2neo7/neo7 and wild-type animals (n = 5, each group) were established by monitoring the weight from newborn (day 0) to 6 weeks olds at regular intervals. Values are the mean ± SEM of independent samples. (C) Femur length was measured in newborn (day 0) and 6-week-old Runx2neo7/neo7 (n = 4) and wild-type animals (n = 5). Values are the mean ± the SEM of independent samples. (D) Staining of whole skeletons from newborn mice with AB to detect cartilage, and AR to detect bone by standard procedures. (E and F) Calvaria and clavicles of 2-day-old mice were isolated and stained with AR and AB. Runx2neo7/neo7 calvaria show cranial defects including wide suture, decreased basisphenoid bone (arrow) and non-osseous tissue present in the junction of posterior frontal suture and coronal suture (dotted line). Examples of the hypoplastic clavicles found in Runx2neo7/neo7 mice are shown. (G and H) Clavicle defects persisted during post-natal development through day 5 and 3 weeks of age. (I) Radiographic images of calvaria from 6-week-old mice. Non-osseous tissue in the junction of the posterior frontal suture and coronal suture is indicated as a dotted line. (J) Radiographic images of clavicles from 6-week-old mice. Broken clavicle (arrow) is visualized in the Runx2neo7/neo7 mouse (neo7/neo7). The clavicle is completely absent in Runx2+/ΔC heterozygous mice (+/ΔC). (K) Radiographic image of clavicle from 5-day-old wild-type (+/+) (top panel) and demineralized paraffin-embedded clavicle section from the same mouse (lower panel) stained by toluidine blue show normal clavicle bone formation. (L) Runx2neo7/neo7 (neo7/neo7) radiograph and toluidine blue-stained tissue section of clavicle (as described in K) show disrupted bone formation of the clavicle with several segments. H: humerus. Bar in (K) and (L) represents 100 µm.
Skeletal phenotypes of newborn mice from Runx2neo7/neo7 homozygotes and wild-type animals were initially determined from alizarin red/alcian blue (AR/AB)-stained skeletons. Wild-type and homozygous animals displayed similar proportions of cartilage and mineralized bone tissues (i.e. ribs, vertebra and limbs) (Fig. 3D). These results suggest that endochondral bone formation was not severely impaired. In contrast, the calvarial bones show reduced ossification in Runx2neo7/neo7 mice (Fig. 3D). In order to clarify the phenotypes in calvaria and clavicles, these bones were excised from the AR/AB-stained mice (Fig. 3E–H). Wide sutures, reduced basisphenoid bone (Fig. 3E, arrow) and increased non-osseous tissue between the parietal bones (Fig. 3E, dotted line), were evident in all newborn Runx2neo7/neo7 littermates. The same mice (as in Fig. 3E) also displayed clavicle defects (Fig. 3F), which were retained during the post-natal growth period of 5 days (Fig. 3G) and 3 weeks (Fig. 3H). Broken and misshapen clavicles were observed in all of the homozygotes (n = 25). No remarkable defects of calvaria or clavicles were observed either in newborn Runx2+/neo7 heterozygotes or in Runx2loxP7/loxP7 mice, where the neo insertion was removed (data not shown). Taken together, these results indicate that Runx2 dosage insufficiency causes hypoplastic clavicles and defects in calvarial bone formation.
At 6 weeks of age, all of the Runx2neo7/neo7 mice (n = 13) displayed similar developmental defects as were observed at birth (Fig. 3I and J). Radiography of the calvaria shows non-osseous tissue at the junction of the posterior frontal suture and coronal suture (Fig. 3I, dotted line), with either short clavicles (as shown in Fig. 3H) and/or thin clavicles that had not broken in 6-week-old Runx2neo7/neo7 mice as shown in Fig. 3J (arrow). These same calvarial defects were noted in Runx2+/ΔC heterozygotes (Fig. 3I), but in contrast, no clavicles are observed in the Runx2+/ΔC animals (Fig. 3J). To better understand the partial formation of clavicles in the Runx2neo7/neo7 homozygotes, histological studies were performed on 5-day-old mice. Radiographs and toluidine blue-stained clavicle sections representative of n = 3 wild-type and Runx2neo7/neo7 mice are shown in Figure 3K and L. For wild-type mouse, normal endochondral and intramembranous bone development occurs at two ossification centers to form a single ossification which is observed in the clavicle at day 5 (Fig. 3K). However in the Runx2neo7/neo7 mice, truncations of clavicle development are observed. In the radiograph (Fig. 3L), a pseudoarthrosis joint is formed, as also seen in the 3 week clavicle (Fig. 3H), because either fusion of the two growth centers (lateral and medial) did not occur or the clavicle fractured. Histology revealed that segments of cartilaginous clavicle tissue initiated from the medial side, but there is very little intramembranous bone that forms on the lateral side of the clavicle throughout all the sections (Fig. 3L). The cartilage tissue contains a disorganized growth plate, but with hypertrophic chondrocytes. These results emphasize the differences in phenotypes of wild-type, Runx2neo7/neo7 (55–70% of wild-type Runx2 mRNA level) and Runx2+/ΔC mice (50% of wild-type Runx2 mRNA level). The latter is completely missing their clavicles (Fig. 3J). The histological appearance of the clavicle from Runx2neo7/neo7 mice suggests that intramembranous bone formation was disrupted at several times during clavicle development. The calvarial and clavicle defects that occur during embryonic development of the skeleton are not corrected during post-natal growth.
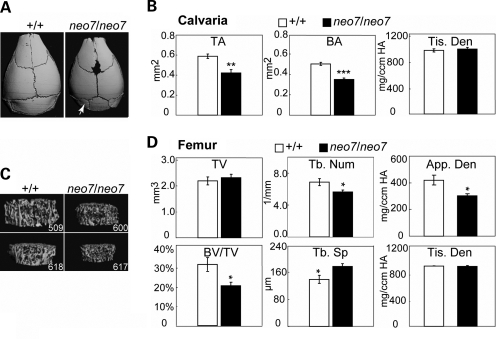
The phenotypes of Runx2neo7/neo7 mice were also evaluated by µCT analysis to determine changes in bone structure at 6 weeks of age (Fig. 4). The images show that the basisphenoid bone is smaller in Runx2neo7/neo7 animals than in wild-types, resulting in a prominent lambdoid suture at the base of the calvaria (Fig. 4A, arrow). A decrease in both total area (TA) and bone area (BA) of the calvaria (Fig. 4B) was observed in Runx2neo7/neo7 mice compared with wild-type, whereas mineralization of the remaining calvarial tissue was not affected, which is reflected by tissue density (Fig. 4B, Tis. Den). In order to determine whether the Runx2 dosage insufficiency caused a change in limb bones during post-natal development, femurs from 6-week-old adult Runx2neo7/neo7 and wild-type mice were selected for analysis. No femur abnormalities were apparent in Runx2neo7/neo7 mice by radiographic examination (data not shown). However, reductions in spongy bone were revealed by µCT imaging analyses (Fig. 4C) showing decreased bone volume/tissue volume (BV/TV) and trabecular number (Tb. Num), as well as increased trabecular spacing (Tb. Sp) (Fig. 4D). In femurs of these neomycin knock-in mice, the apparent density (Fig. 4D, App. Den) is moderately decreased but tissue density is normal (Fig. 4D, Tis. Den), which indicates that mineral content of mature bone is not impaired in Runx2neo7/neo7 mice. Moreover, in cortical bone of Runx2neo7/neo7 mice, we find no change from WT in cortical thickness, tissue density and bone porosity, which reflects bone resorption (data not shown). In the adult mouse, the decrease in bone tissue volume and area in calvaria, as well as decreased trabecular number in long bone, indicates that the 30–45% decrease in wild-type Runx2 in the homozygous mouse has an effect on bone architecture, whereas the ∼20% reduction in the heterozygous mouse does not (data not shown). Taken together, these findings suggest that during embryonic development, intramembranous bones (i.e. calvarium, clavicle) are more sensitive to requirements for sufficient Runx2 expression, and during post-natal growth, the reduced Runx2 levels continue to affect bone formation.
Figure 4.
Bone loss in calvaria of Runx2neo7 homozygous mice: (A) Three-dimensional microCT images of calvaria from 6-week-old wild-type (+/+) and Runx2neo7/neo7 (neo7/neo7) mice. The defects in posterior frontal suture and lambdoid suture (arrow) are visible. (B) The three-dimensional microCT parameters were measured in calvarial of wild-type and Runx2neo7/neo7 mice (n = 3 mice/group). TA: total area; BA: bone area; Tis. Den: tissue density. (C) Three-dimensional microCT images of trabecular bone above the femoral growth plate in 6-week-old wild-type and Runx2neo7/neo7 mice. 509 and 600 are from the same litter. 618 and 617 are littermates from another litter. (D) Three-dimensional microCT parameters were measured in femurs and selected parameters are shown. TV: total volume; BVF: BV/TV; Tb. Num: trabecular number; Tb. Sp: trabecular spacing; App. Den: apparent density; Tis. Den: tissue density. n = 3 mice/group. Values are the mean ± the SEM of independent samples. Student's t-test was applied for statistical analysis. *P < 0.1; **P < 0.05; ***P < 0.01. We also observed similar trends in a third litter from a different generation (data not shown).
Reduced Runx2 expression causes early defects in suture tissue that compromise calvaria
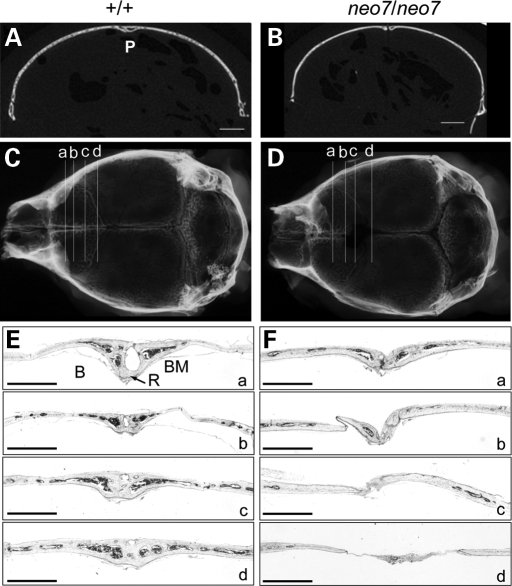
We further examined the defects in bone formation arising from the calvarial sutures in the Runx2neo7/neo7 mice. The two-dimensional coronal µCT images of the sagittal sutures (Fig. 5A) reveal that the right and left parietal bones of wild-type animals are interposed in the patent suture. However, in the Runx2neo7/neo7 mice, there is a wide gap between the parietal bones, and the typical patent sagittal suture is not formed (Fig. 5B). The histological appearance of the sutures was analyzed from serial coronal sections of calvaria taken from 6-week-old adult mice (Fig. 5C and D). The posterior frontal suture of wild-type animals has fused completely across its endocranial and ectocranial surfaces in all sections (Fig. 5Ea–d), as has been described previously (18). With deeper cut sections, the posterior frontal suture displayed normal bone and marrow tissue. In Runx2neo7/neo7 mice, the posterior frontal suture appears normal in radiographic images (Fig. 5D); however, histological analysis revealed irregular tissue between interparietal bones instead of the normal bony bridge structure (Fig. 5Ea versus 5Fa). In areas without osseous tissue, a thin layer of fibrous tissue connected the parietal bones, and this area was narrower in width than wild-type (Fig. 5E and F, compare b–d; see also Fig. 5A and B). In conclusion, the histological appearance of the suture at 6 weeks of age in Runx2neo7/neo7 mice provides evidence that 55–70% of wild-type Runx2 levels are not sufficient to support normal differentiation of sutural mesenchymal cells to osteoblasts in the developing embryo.
Figure 5.
Histological appearance of calvarial morphogenesis in Runx2neo7 homozygous mice. (A and B) Two-dimensional microCT images in coronal section of sagittal suture from 6-week-old wild-type (+/+) and Runx2neo7/neo7 (neo7/neo7) mice. Bar represents 1 mm. P: patent suture. (C and D) Radiographic images of calvaria from 6-week-old wild-type and Runx2neo7/neo7 mice. The horizontal lines a, b, c and d represent respective sections that are displayed in (E) and (F). (E and F) Sections from the posterior frontal suture of the same wild-type and Runx2neo7/neo7 mice shown in (A–D) were stained with toluidine blue. Bar represents 1 mm. R: endocranial ridging; B: bony bridge; BM: bone marrow.
Reduction of functional Runx2 dosage alters the expression of osteoblast marker genes and delays cellular differentiation
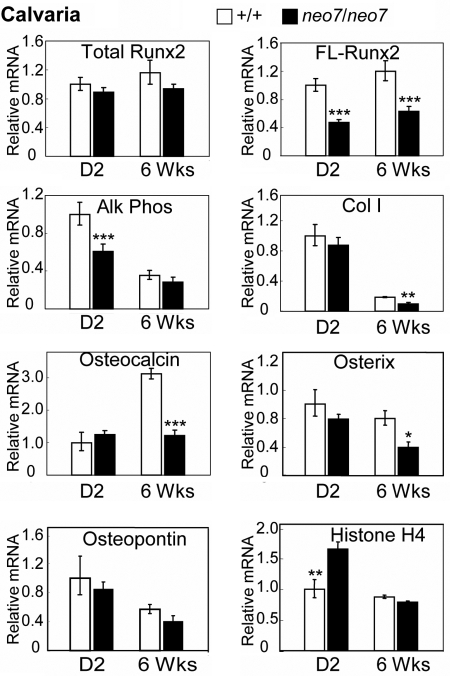
The levels of total and FL-Runx2 mRNAs were compared using whole calvarial tissue from wild-type and Runx2neo7/neo7 mice at two different stages of growth (day 2 and 6 weeks). No significant changes were observed in total Runx2 mRNA between wild-type and Runx2neo7/neo7 mice, whereas the FL-Runx2 mRNA level in Runx2neo7/neo7 mice is reduced to 40–50% of that found in whole calvaria of wild-type mice at both ages (Fig. 6). The results suggest that the Runx2 dosage insufficiency persists during post-natal growth.
Figure 6.
Dosage insufficiency of Runx2 alters the expression of osteoblast marker genes in post-natal and adult calvarial tissue. Real-time quantitative RT–PCR analyses were carried out for the mRNA levels of total Runx2 (Fig. 2E), FL-Runx2, Alk Phos, Col I, OC, Osx, osteopontin and histone H4 in the whole calvaria (cleaned from soft tissue) of wild-type (+/+) and Runx2neo7/neo7 (neo7/neo7) mice at 2 days (D2) (n = 5/group) and 6 weeks (6 Wks) (n = 4/group) of age, respectively. All measured data were normalized to mHPRT. Values are the mean ± the SEM of independent samples. Student's t-test was applied for statistical analysis. *P < 0.1; **P < 0.05; ***P < 0.01.
To investigate the molecular mechanism(s) causing the calvarial developmental defects of Runx2neo7/neo7 mice, we selected several osteogenic markers and quantified their levels in the same calvarial tissue in which Runx2 expression was assayed. Age-dependent changes of these mRNAs in wild-type mice were similar to previous studies reflecting osteoblast activity (Fig. 6) (4,19). In 2-day-old Runx2neo7/neo7 mice, alkaline phosphatase (Alk Phos), an early marker of the post-proliferative osteoblast phenotype, decreased to ∼60% of normal, but no significant differences in Alk Phos levels were observed between Runx2neo7/neo7 and WT mice at 6 weeks when Alk Phos expression is reduced to low levels (Fig. 6). Compared with wild-type mice, the expression of type I collagen (Col I), a marker of matrix maturation, and osteocalcin (OC), a marker of mature mineralized tissue, was normal in calvaria of 2-day-old Runx2neo7/neo7 mice, but significantly decreased in 6-week-old Runx2neo7/neo7 mice (Fig. 6). Osterix (Osx), a transcription factor that is also essential for osteoblast differentiation and tissue mineralization (20) and regulated by Runx2 (21,22), was also reduced at 6 weeks (Fig. 6). There was no significant change in osteopontin levels between wild-type and mutant mice (Fig. 6), consistent with other findings that Runx2 is not a strong regulator of osteopontin (reviewed in 23). Because differentiation markers are often inversely related to cell proliferation, we examined histone H4 expression which is coupled to DNA synthesis. We find a higher expression level of histone H4 in 2-day-old Runx2neo7/neo7 mice, whereas histone H4 expression is equivalent in adult mice (Fig. 6). Overall, these changes in the gene expression of the Runx2neo7/neo7 mice indicate the impaired differentiation of osteoblasts in the calvarial tissue.
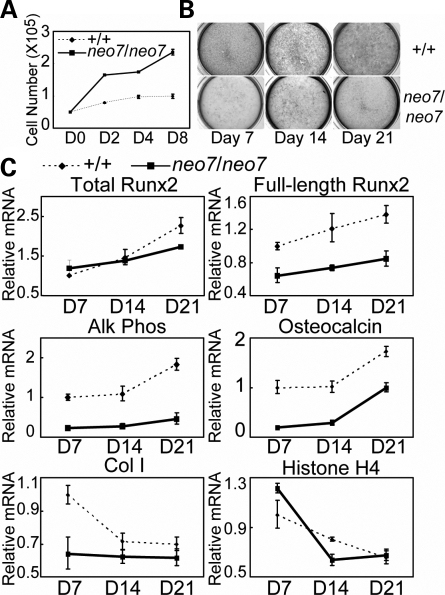
Next, we determined cell autonomous abnormalities of osteoblast differentiation and proliferation ex vivo using calvaria-derived osteoblasts. Cells from Runx2neo7/neo7 mice demonstrated a significantly enhanced proliferation compared with wild-type cells (Fig. 7A), analogous to the phenotype of Runx2 null mouse models (24). The differentiation of cultured osteoblasts from Runx2neo7/neo7 mice was reduced relative to those from wild type, as measured by Alk Phos staining (Fig. 7B) as well as by the expression of Alk Phos, OC and Col I mRNAs at days 7, 14 and 21 in culture (Fig. 7C). During the differentiation time course, the expression of FL-Runx2 in Runx2neo7/neo7 osteoblast cells was maintained at ∼60% of that in wild-type osteoblasts (Fig. 7C). We also find the expression of histone H4 mRNA in Runx2neo7/neo7 osteoblasts was enhanced at the early stage of differentiation (day 7), analogous to in vivo studies (Fig. 6), but dropped to normal levels in the mineralization stage (Fig. 7C).
Figure 7.
Cell autonomous defects in osteoblast growth and differentiation in Runx2neo7/neo7mice. (A) Growth profiles of calvarial osteoblast cultures from newborn wild-type (+/+) and Runx2neo7/neo7 (neo7/neo7) mice (n = 6 for each) were established by counting cell number at regular intervals. (B) Cultured osteoblast cells of newborn wild-type and Runx2neo7/neo7 mice were fixed at D7, D14 and D21 after initiation of differentiation. Immunocytochemistry for the early marker Alk Phos was carried out as described in Materials and Methods. (C) Real-time quantitative RT–PCR analyses were carried out for the mRNA levels of total Runx2, FL-Runx2, Alk Phos, Col I, OC and histone H4 in calvarial osteoblasts from newborn wild-type and Runx2neo7/neo7 mice at the indicated differentiation time points. All measured data were normalized to mGAPDH. Values are the mean ± the SD of n = 3 independent samples (wells).
Taken together, our results suggest: (i) Runx2 dosage insufficiency causes enhanced proliferation of osteoblasts, which is consistent with its role in contributing to exit from the cell cycle (24,25); (ii) a decrease of Runx2-dependent osteoblast differentiation at the neonatal age is directly reflected by decreased Alk Phos and OC expression in both calvarial tissue and isolated osteoblast cells and may represent decreased numbers of committed osteoprogenitors as suggested from µCT parameters; and (iii) the decreased expression of Col I, OC and Osx reflects the histology of calvaria of 6-week-old Runx2neo7/neo7 mice, which have less bone tissue present compared with wild type.
Runx2 dosage insufficiency does not disrupt growth plate maturation
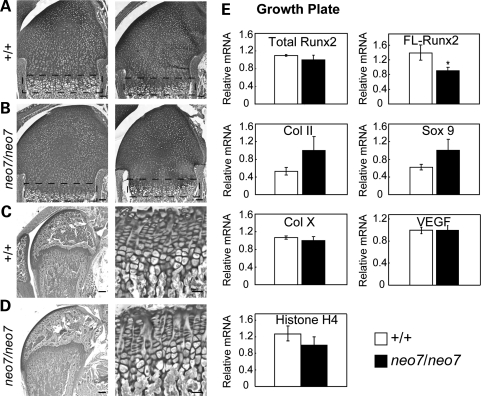
Because Runx2 is important for hypertrophic chondrocyte maturation, growth plate histology was examined in neonatal pups (2-day-old) and adult (6-week-old) mice to determine whether loss of Runx2 functional activity in Runx2neo7/neo7 mice will affect normal chondrocyte differentiation and maturation. The epiphyses of long bones from both wild-type and Runx2neo7/neo7 mice display normal cartilage tissue and organization of growth plate chondrocytes at both ages (Fig. 8A–D) and in both sexes (data not shown). To be certain that no molecular differences occurred during chondrocyte maturation, we isolated the growth plate zone from long bones of day 5 mice (Fig. 8A and B) for the analysis of the levels of cartilage maturation markers as well as FL-Runx2 (exon 7–exon 8). The FL-Runx2 in homozygous Runx2neo7/neo7mice was ∼60% of that in wild-type mice (Fig. 8E). We found no significant differences in the Runx2 target genes type X collagen and VEGF (Fig. 8E), which are important for endochondral bone formation (26,27). We conclude that during endochondral ossification, reduction of functional Runx2 levels to 55–70% of normal does not affect the sequence of chondrocyte maturation, matrix synthesis by chondrocytes or mineralization of cartilage matrix.
Figure 8.
Histological appearance of growth plate organization and expression of chondrocyte marker genes in Runx2neo7 homozygous mice. (A and B) Sections from the long bone of the 2-day-old wild-type (+/+) (A) and Runx2neo7/neo7 mice (neo7/neo7) (B) (n = 2 mice/group) are stained by toluidine blue. The growth plate zone is indicated by the dotted rectangle. Bar represents 0.2 mm. (C and D) Toluidine blue-stained sections from the long bones of 6-week-old wild-type (C) and Runx2neo7/neo7 mice (D) are shown at low (×2.5) and at higher (×40) magnifications to display normal growth plate organization in both groups. Bar represents 1 mm (left) and 0.1 mm (right). (E) Real-time quantitative RT–PCR analyses were carried out for the mRNA levels of total Runx2, FL-Runx2, type II collagen (Col II), sox9, type X collagen (Col X), VEGF and histone H4 in the growth plate zones (A and B) from long bones of 5-day-old wild-type (n = 2) and Runx2neo7/neo7 (n = 3) mice. All measured data were normalized to mHPRT. Values are the mean ± the SEM of independent samples.
Further evidence for a minimum requirement for Runx2 at early stages of osteoblastogenesis
In the transition of a pluripotent mesenchymal cell to a committed osteoprogenitor cell, Runx2 levels must increase. From studies of the hypomorphic Runx2neo7/neo7 mouse, we have determined that heterozygotes with 79–84% of wild-type Runx2 have a normal skeleton, whereas the 30–45% reduction in the Runx2neo7/neo7 mouse results in a CCD phenotype. Thus we have defined a critical threshold level which is >70% of wild-type Runx2, which is required for normal skeletal development. We have summarized the CCD-like phenotypic features of all Runx2-deficient mouse models, together with the percentage of wild-type Runx2 mRNA levels (Table 1).
Table 1.
Wild-type Runx2 dosage and skeletal phenotypes
| Genotype | Percentage of wild-type Runx2 mRNA | Phenotype |
|---|---|---|
| +/+ | 100a | Normal |
| −/− | 0a | No bone formation |
| +/− | 50a | CCD syndromeb |
| ΔC/ΔC | 0a | No bone formation |
| +/ΔC | 50a | No clavicle, calvarial defects |
| neo7/neo7 | 55–70c | CCD syndromeb |
| +/neo7 | 79–84c | Normal |
aTheoretical value based on published work.
bFor the Runx2 null heterozygous mouse (+/−), the CCD syndrome was described as hypo-plastic clavicle and abnormal calvarial development as well as a nasal bone defect. The Runx2neo7/neo7 homozygous mouse (neo7/neo7) has clavicle and calvarial defects of CCD syndrome, but no nasal bone abnormality was observed.
cMeasured value based on quantitative PCR analysis using an isolated osteoblast population; the percentage of wild-type Runx2 mRNA was calculated on the basis of FL-Runx2 mRNA (measured by RT–PCR of exon 7–exon 8) relative to total Runx2 mRNA (measured by RT–PCR of exon 4–exon 5).
DISCUSSION
Our study describes an informative hypomorphic mouse model that reveals the relationship between RUNX2 gene dosage and distinguishable CCD phenotypic features found in patients with CCD (reviewed in 10). Genetic and molecular data demonstrate that the insertion of a neomycin cassette into intron 7 (neo7) has generated a hypomorphic Runx2 allele that reduces the expression of wild-type Runx2. The Runx2+/neo7 heterozygote mouse exhibits ∼20% reduction in Runx2 but no phenotype is observed. However, the Runx2neo7/neo7 homozygote with ≥30% reduction of Runx2 levels has CCD-like defects in calvaria and clavicle tissues and subtle defects in the amount of trabecular bone tissue in the femur of 6-week-old mice. Thus, a decrease in wild-type Runx2 expression over a narrow interval from ∼20–30% defines the difference between normal and defective bone formation and establishes a critical level of Runx2 functional activity that must be maintained during development to prevent bone abnormalities and during post-natal growth to maintain adequate bone formation. Significantly, cell autonomous defects in osteoblast proliferation (enhanced) and differentiation (reduced) were confirmed in ex vivo calvarial isolated cells. Our studies suggest that the spectrum of bone abnormalities in CCD patients is related to the level of functional activity of RUNX2, which is determined by both the amount of the mutant protein and the type of mutation (10,14,28).
The main clinical features of human CCD include persistently open skull sutures, hypoplasia or aplasia of the clavicles, dental anomalies and a delay in vertebral formation (10,29). Previous studies have demonstrated that the human CCD phenotype can be caused by various RUNX2 gene mutations (10,11,29–34). Whereas 61% of the mutations identified reside in the Runt domain, 23% occur in the C-terminus including the NMTS-activating domain of RUNX2 (30–32). All of the mutations in the Runt homology domain result in a CCD phenotype, but the various RHD mutations could not be correlated with the severity of these multiple phenotypes as reported in numerous studies from different countries (11,30,33–35). However, from phenotypic information of patients with RUNX2 C-terminal mutations, a hypothetical genotype–phenotype correlation was suggested (28). Considering that the RUNX2 C-terminus contains the subnuclear matrix-targeting signal and activation domain which is necessary to direct RUNX2 to subnuclear locations for maximal transactivation and organization of RUNX2 with co-regulatory proteins (e.g. Smads) essential for normal bone formation (4,7,36), different mutations or deletions in the RUNX2 C-terminus will cause different levels of reduction in RUNX2 biological activities which may be related to the phenotypic variations in CCD.
Our comparison of the existing Runx2 null and heterozygous mouse models with a newly generated hypomorphic mouse (Table 1) suggests that the level of RUNX2 function may be the critical determinant for the correlation to the severity of the human CCD phenotype. From a clinical perspective, it has been reported that in some CCD patients, a RUNX2 mutation could not be detected (11). This finding implies that in those patients, the level of RUNX2 activity may be decreased as a result of a defect in promoter activity or in the interaction between RUNX2 and one of its co-activator proteins such as the Cbfβ DNA-binding partner (34–36).
A small percent of patients have complete absence of clavicles (10,29), analogous to the Runx2+/− and Runx2+/ΔC mouse models (5,7), but the majority of CCD patients display hypoplastic or aplastic clavicles. The clavicle has two ossification centers; one medial which condenses first into cartilage, and the lateral center which undergoes intramembranous bone formation (37–39). Our hypomorphic Runx2neo7/neo7 mouse initiates the development of the cartilaginous center of clavicles, but the clavicles are partially disrupted in formation during development and are abnormally thin compared with wild type. Intramembranous bone formation of the clavicle did not progress in the mutant embryo. Thus different patterns of abnormally shaped clavicles are found in the Runx2neo7/neo7 mice, in contrast to Runx2+/− mice (50% loss of Runx2) with completely missing clavicles (5,40). The clavicle is the first bone to begin the process of ossification during embryogenesis, but also takes the longest time to complete maturation (39). Our study suggests that a low level of functional Runx2 protein can continually interrupt clavicle ossification during normal embryogenesis and early post-natal growth, and these defects are retained in adult mice. Thus the Runx2neo7/neo7 mouse provides a better understanding of the biochemical and molecular basis of the human CCD disease related to gene dosage effects that more severely affect intramembranous bone formation during embryonic development.
In vivo, we find that the size of the skull bone is not affected in the Runx2neo7/neo7 mouse, but mice develop a clearly defined non-mineralized area between the coronal and posterior frontal suture of the calvarium. This area represents a small percent of the total bone (Fig. 5). Histology shows that bone tissue on either side of the suture is not well developed, which is consistent with a reduction in the early osteoblast markers Osx and Alk Phos, and in OC, a late mineralization marker in calvarium. These changes are likely reflecting the increased amount of non-osseous tissue throughout the calvarial posterior frontal suture of the Runx2neo7/neo7 mouse compared with wild-type mouse, and possibly impaired osteoblast differentiation from decreased Runx2 expression. The decreased Runx2 levels appear to have the greatest effect during the early development of intramembranous bones (calvaria and clavicle) when mesenchymal cells in the suture require a Runx2-generated signal to commit to the osteoblast lineage. In other studies, high expression of Runx2 was found in sutural mesenchyme, osteogenic fronts, and in the critical area of closure of cranial sutures of the parietal bones (41,42). Notably, other skeletal inherited diseases, such as Apert, Beare–Stevenson, Crouzon and Pfeiffer syndromes, are characterized by craniosynostosis, which results in premature suture closure, skull deformity and symmetric bony syndactyly of the hands and feet (43). In those patients, the level of functional RUNX2 is abnormally induced because of activating mutations in the FGF-receptor signal pathway. Thus, combined with our findings, the phenotypes of loss or gain of functional RUNX2 in human disorders suggest that the normal development of the skeleton, especially of calvaria, is highly dependent on the level of functional Runx2 protein. Together, these observations indicate that a sufficient dosage of Runx2 is essential for the normal mesenchymal cell differentiation and osteoblastic maturation at the osteogenic fronts interfacing with mesenchyme.
A cell autonomous defect in osteoblasts of the Runx2neo7/neo7 hypomorph is suggested by the decreased trabecular number found by µCT studies. As reported previously for Runx2 null or Runx2ΔC calvarial osteoblasts (24,44), osteoblasts from Runx2neo7/neo7 homozygous mice also showed an enhanced proliferation rate, indicating that the role of Runx2 in supporting exit from the cell cycle is critical. Another important role for Runx2 in early development is an epigenetic function to support phenotype stability in osteoprogenitor cells through Runx2 association with mitotic chromosomes (45). Impaired osteogenesis is revealed by decreased expression of several bone markers in calvarial tissue and cultured osteoblasts. The 55–70% remaining functional wild-type Runx2 in homozygous Runx2neo7/neo7 mice is apparently not sufficient to commit an adequate number of osteoprogenitors to mature osteoblasts and to drive osteoblasts to the final differentiation stage ex vivo. However an adequate number of osteoblasts are produced in the late embryo and post-natal mouse to form skeletal structures. Interestingly, we did not find abnormalities in the morphology of the growth plate for the progression of endochondral bone formation. However, some CCD patients have shorter limbs, and a severe growth plate abnormality was characterized in a fetus with CCD (46).
In conclusion, we have described a Runx2 dosage-insufficiency mouse model with typical CCD phenotype. Our genetic analyses provide new insight into the connection between modest changes in Runx2 levels and the display of the CCD phenotype. Our mouse model together with previous clinical studies suggests that the defects of skeletal structures in CCD patients are consistent with the extent to which mutants reduce the level of RUNX2 functional activity.
MATERIALS AND METHODS
Mice
A genomic fragment containing the 3′-end of the mouse Runx2 locus was obtained from the genomic DNA of AB2.2 mouse ES cells. The whole fragment was confirmed by sequencing and was then subcloned into pBluescript SK+ vector (Invitrogen). Subsequently, the PGKneo-cassette flanked by loxP sites (47,48) was inserted into intron 7 with NotI site, and a thymidine kinase (TK) gene was inserted into intron 6 with NheI site. The final target vector was linearized by SalI and electroporated into AB2.2 ES cells. We grew them under double selection. We digested DNA of individual double-resistant clones with BamHI or EcoRI and then probed them with different external and internal probes as described in Fig. 1B. Two independently identified homologous recombinant clones were used for blastocyst injection. Chimeric mice were bred to C57BL/6 mice to yield heterozygous mice which were then interbred to yield homozygous mutant animals.
Female heterozygous mice were crossed with male protamine-Cre recombinase transgenic mice (49) to remove the PGKneo-cassette from the Runx2 genome. All of the generated offspring were identified by Southern blot as described earlier and PCR genotyping with the primers described in Fig. 1F. The sequence of primers is listed in Table 2. All animals in the study were housed in pathogen-free facilities and monitored carefully.
Table 2.
Nucleotide sequence of primers used in this study
| Name | Sequence (5′–3′) |
|---|---|
| I7Fa | TTCGGGAGTTAGACAGCAGAAG |
| I7Ra | ACAGTCAGAGCCTTGATGAG |
| NEOa | GAAGACAATAGCAGGCATGCTG |
| E7Fb | TCAGTAAGAAGAGCCAGGCAGG |
| E8Rb | GTACCATTGGGAACTGATAGGATG |
| NRb | TTGACAAAAAGAACCGGGCGCCCCTGCGCT |
| NFb | GCCACTCCCACTGTCCTTT |
| Rx2E7-Fc | GATGACACTGCCACCTCTGA |
| Rx2E8-Rc | ATGAAATGCTTGGGAACTGC |
| Rx2-Fc | CGGCCCTCCCTGAACTCT |
| Rx2-Rc | TGCCTGCCTGGGATCTGTA |
| OC-Fc | CTGACA AAGCCTTCATGTCC |
| OC-Rc | GCGCCGGAGTCTGTTCAC |
| AP-Fc | TTGTGCGAGAGAAAGAGAGAGA |
| AP-Rc | GTTTCAGGGCATTTTTCAAGGT |
| Col1-Fc | CCCAAGGAAAAGAAGCACGTC |
| Col1-Rc | AGGTCAGCTGGATAGCGACATC |
| HPRT-Fc | CAGGCCAGACTTTGTTGGAT |
| HPRT-Rc | TTGCGCTCATCTTAGGCTTT |
| Osterix-Fc | TATGCTCCGACCTCCTCAACT |
| Osterix-Rc | TCCTATTTGCCGTTTTCCCGA |
aPrimer sequence for PCR genotyping.
bPrimer sequence for Runx2 splicing isoform analysis.
cPrimers sequence for real-time quantitative PCR analysis.
RT–PCR and western analysis
Total RNA was isolated from calvaria tissue using TRIzol reagents (Invitrogen). One microgram of RNA was treated with RNase-free DNaseI (Zymo Research) and reverse transcribed into cDNA using SuperScipt II Reverse Transcriptase (Invitrogen). The following PCR reactions were performed with the following primer pairs: E7 and E8, E7 and NR, NF and E8 (sequence as in Table 2). The PCR parameters are 95°C (3 min), followed by 35 cycles of 95°C (1 min), 56°C (1 min) and 72°C (1 min), then ended by 72°C (10 min). PCR products were purified and sequenced to identify the Runx2-neo chimeric transcript. Total protein was isolated directly from calvaria of 12-week-old mice. The protein lysates were resolved by 8% SDS/PAGE, and western blot analysis was performed by using a mouse monoclonal Runx2 antibody (31), followed by incubation with a goat peroxidase-tagged anti-mouse IgG secondary antibody. Bands were visualized by ECL reagents (Perkin Elmer).
Skeletal preparation and radiograph examination
Mice at different ages were eviscerated and fixed in 100% ethanol. Skeletal morphology was analyzed by AR and AB staining, followed by tissue clarification with KOH by standard procedures (50). To compare the morphology, the selected bones were isolated from the whole skeleton and photographed using a dissection microscope (Leica, CLS 150) with a digital camera (Zeiss, Axiocam HRC). For radiographic analysis, the adult animals were scarified, de-skinned and organs were removed. The calvaria were isolated. All of the specimens were fixed in 100% ethanol. After 4 days, the specimens were placed on X-ray film (Kozak) and X-rays were taken by Faxitron Specimen Radiography System Model Mx-20. The parameters were 20 s at 20 kV for whole body and 5 s at 20 kV for calvaria.
MicroCT
MicroCT was performed on selected bone specimens using a group of three wild-type (+/+) and three homozygous (neo7/neo7) 6-week-old male mice. All the data and images of microCT were collected by MicroCT Facility, University of Connecticut Health Center. Data were analyzed for statistical significance using Student’s t-test.
Histological examination
Calvaria, clavicle and long bone isolated from mice of different ages were fixed in phosphate-buffered saline (pH 7.4) containing 4% paraformaldehyde and embedded in paraffin. The tissues from adult mice were decalcified by incubation in 18% EDTA (pH 7.4) for 3–4 weeks prior to embedding. All of the embedded tissues were cut to 6 µm thick sections and dried on Superfrost Plus slides. Sections were stained with Toluidine Blue by the standard procedure and viewed with a microscope (Zeiss, Axionskop 40). All images were captured with a digital camera (Zeiss, Axiocam HRC).
Quantitative real-time RT–PCR analysis
Total RNA was isolated from growth plate, calvaria tissue or isolated calvarial cells grown to confluency using TRIzol Reagent (Invitrogen) and reverse-transcribed by the method mentioned earlier. Quantitative real-time RT–PCR analysis was performed in an ABI PRISM 7000 Sequence Detector (Applied Biosystem) with the following parameters: 50°C (2 min), 95°C (10 min), followed by 40 cycle of 95°C (15 s) and 60°C (1 min). The gene expression levels of mouse calvaria tissue and growth plate were normalized to mHPRT and compared by the ddCT method. The molecular level of wild-type Runx2 and neo-Runx2 chimeric transcripts in cultured osteoblast cells was measured by the absolute standard curve method (51). Student's t-test was performed for significant difference comparison. Primers sequences for total Runx2, FL-Runx2 and other osteogenic marker genes can be found in Table 2.
Cell culture and Alk Phos cytochemistry
Calvarial osteoblasts were isolated from wild-type (+/+) and homozygous (neo7/neo7) 1-day-old mice. Osteoblast cells were obtained and maintained as described previously (24,52). Cells were plated at a density of 1 × 106 cells/six-well plate. When the cells reached confluence, regular growth medium (αMEM supplemented with 10% FBS) was replaced with osteogenic media (BGJb supplemented with 10% FBS, 10 mm of β-glycerol phosphate and 25 µg/ml of ascorbic acid, first feeding, or 50 µg/ml of ascorbic acid, subsequent feeding) for differentiation assay. Cultured cells were stained for Alk Phos as described previously (53) at day 7, day 14 and day 21. The stained cells in individual well were visualized using a dissection microscope (Leica, CLS 150).
FUNDING
National Institutes of Health (AR048818, AR039588). The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
ACKNOWLEDGEMENTS
We thank Marilyn Keeler in the UMass Transgenic Animal Core, Douglas J. Adams at the University of Connecticut Health Center for microCT image and data collection, Judy Rask for manuscript preparation and other members of the Stein/Lian Laboratory for many helpful discussions.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Ryoo H.M., Wang X.P. Control of tooth morphogenesis by Runx2. Crit. Rev. Eukaryot. Gene Expr. 2006;16:143–154. doi: 10.1615/critreveukargeneexpr.v16.i2.30. [DOI] [PubMed] [Google Scholar]
- 2.Aberg T., Wang X.P., Kim J.H., Yamashiro T., Bei M., Rice R., Ryoo H.M., Thesleff I. Runx2 mediates FGF signaling from epithelium to mesenchyme during tooth morphogenesis. Dev. Biol. 2004;270:76–93. doi: 10.1016/j.ydbio.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 3.James M.J., Jarvinen E., Wang X.P., Thesleff I. Different roles of Runx2 during early neural crest-derived bone and tooth development. J. Bone Miner. Res. 2006;21:1034–1044. doi: 10.1359/jbmr.060413. [DOI] [PubMed] [Google Scholar]
- 4.Lian J.B., Javed A., Zaidi S.K., Lengner C., Montecino M., van Wijnen A.J., Stein J.L., Stein G.S. Regulatory controls for osteoblast growth and differentiation: role of Runx/Cbfa/AML factors. Crit. Rev. Eukaryot. Gene Expr. 2004;14:1–41. [PubMed] [Google Scholar]
- 5.Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R.T., Gao Y.-H., Inada M., et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 6.Mundlos S., Otto F., Mundlos C., Mulliken J.B., Aylsworth A.S., Albright S., Lindhout D., Cole W.G., Henn W., Knoll J.H.M., et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- 7.Choi J.-Y., Pratap J., Javed A., Zaidi S.K., Xing L., Balint E., Dalamangas S., Boyce B., van Wijnen A.J., Lian J.B., et al. Subnuclear targeting of Runx/Cbfa/AML factors is essential for tissue-specific differentiation during embryonic development. Proc. Natl Acad. Sci., USA. 2001;98:8650–8655. doi: 10.1073/pnas.151236498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Otto F., Thornell A.P., Crompton T., Denzel A., Gilmour K.C., Rosewell I.R., Stamp G.W.H., Beddington R.S.P., Mundlos S., Olsen B.R., et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 9.Lee B., Thirunavukkarasu K., Zhou L., Pastore L., Baldini A., Hecht J., Geoffroy V., Ducy P., Karsenty G. Missense mutations abolishing DNA binding of the osteoblast-specific transcription factor OSF2/CBFA1 in cleidocranial dysplasia. Nat. Genet. 1997;16:307–310. doi: 10.1038/ng0797-307. [DOI] [PubMed] [Google Scholar]
- 10.Otto F., Kanegane H., Mundlos S. Mutations in the RUNX2 gene in patients with cleidocranial dysplasia. Hum. Mutat. 2002;19:209–216. doi: 10.1002/humu.10043. [DOI] [PubMed] [Google Scholar]
- 11.Yoshida T., Kanegane H., Osato M., Yanagida M., Miyawaki T., Ito Y., Shigesada K. Functional analysis of RUNX2 mutations in Japanese patients with cleidocranial dysplasia demonstrates novel genotype–phenotype correlations. Am. J. Hum. Genet. 2002;71:724–738. doi: 10.1086/342717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stein G.S., Lian J.B., van Wijnen A.J., Stein J.L., Montecino M., Javed A., Zaidi S.K., Young D.W., Choi J.Y., Pockwinse S.M. Runx2 control of organization, assembly and activity of the regulatory machinery for skeletal gene expression. Oncogene. 2004;23:4315–4329. doi: 10.1038/sj.onc.1207676. [DOI] [PubMed] [Google Scholar]
- 13.Smith N., Dong Y., Lian J.B., Pratap J., Kingsley P.D., van Wijnen A.J., Stein J.L., Schwarz E.M., O’Keefe R.J., Stein G.S., Drissi M.H. Overlapping expression of Runx1(Cbfa2) and Runx2(Cbfa1) transcription factors supports cooperative induction of skeletal development. J. Cell Pysiol. 2005;203:133–143. doi: 10.1002/jcp.20210. [DOI] [PubMed] [Google Scholar]
- 14.Matheny C.J., Speck M.E., Cushing P.R., Zhou Y., Corpora T., Regan M., Newman M., Roudaia L., Speck C.L., Gu T.L., et al. Disease mutations in RUNX1 and RUNX2 create nonfunctional, dominant-negative, or hypomorphic alleles. EMBO J. 2007;26:1163–1175. doi: 10.1038/sj.emboj.7601568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carmeliet P., Ferreira V., Breier G., Pollefeyt S., Kieckens L., Gertsenstein M., Fahrig M., Vandenhoeck A., Harpal K., Eberhardt C., et al. Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature. 1996;380:435–439. doi: 10.1038/380435a0. [DOI] [PubMed] [Google Scholar]
- 16.Meyers E.N., Lewandoski M., Martin G.R. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat. Genet. 1998;18:136–141. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- 17.Nagy A., Moens C., Ivanyi E., Pawling J., Gertsenstein M., Hadjantonakis A.K., Pirity M., Rossant J. Dissecting the role of N-myc in development using a single targeting vector to generate a series of alleles. Curr. Biol. 1998;8:661–664. doi: 10.1016/s0960-9822(98)70254-4. [DOI] [PubMed] [Google Scholar]
- 18.Recinos R.F., Hanger C.C., Schaefer R.B., Dawson C.A., Gosain A.K. Microfocal CT: a method for evaluating murine cranial sutures in situ. J. Surg. Res. 2004;116:322–329. doi: 10.1016/j.jss.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 19.Shalhoub V., Jackson M.E., Paradise C., Stein G.S., Lian J.B., Marks S.C., Jr Heterogeneity of colony stimulating factor-1 gene expression in the skeleton of four osteopetrotic mutations in rats and mice. J. Cell. Physiol. 1996;166:340–350. doi: 10.1002/(SICI)1097-4652(199602)166:2<340::AID-JCP12>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 20.Nakashima K., Zhou X., Kunkel G., Zhang Z., Deng J.M., Behringer R.R., de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 21.Nishio Y., Dong Y., Paris M., O’Keefe R.J., Schwarz E.M., Drissi H. Runx2-mediated regulation of the zinc finger Osterix/Sp7 gene. Gene. 2006;372:62–70. doi: 10.1016/j.gene.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 22.Kim Y.J., Kim H.N., Park E.K., Lee B.H., Ryoo H.M., Kim S.Y., Kim I.S., Stein J.L., Lian J.B., Stein G.S., et al. The bone-related Zn finger transcription factor Osterix promotes proliferation of mesenchymal cells. Gene. 2006;366:145–151. doi: 10.1016/j.gene.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 23.Denhardt D.T., Mistretta D., Chambers A.F., Krishna S., Porter J.F., Raghuram S., Rittling S.R. Transcriptional regulation of osteopontin and the metastatic phenotype: evidence for a Ras-activated enhancer in the human OPN promoter. Clin. Exp. Metastasis. 2003;20:77–84. doi: 10.1023/a:1022550721404. [DOI] [PubMed] [Google Scholar]
- 24.Pratap J., Galindo M., Zaidi S.K., Vradii D., Bhat B.M., Robinson J.A., Choi J.-Y., Komori T., Stein J.L., Lian J.B., et al. Cell growth regulatory role of Runx2 during proliferative expansion of pre-osteoblasts. Cancer Res. 2003;63:5357–5362. [PubMed] [Google Scholar]
- 25.Galindo M., Pratap J., Young D.W., Hovhannisyan H., Im H.J., Choi J.Y., Lian J.B., Stein J.L., Stein G.S., van Wijnen A.J. The bone-specific expression of RUNX2 oscillates during the cell cycle to support a G1 related anti-proliferative function in osteoblasts. J. Biol. Chem. 2005;280:20274–20285. doi: 10.1074/jbc.M413665200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leboy P., Grasso-Knight G., D’Angelo M., Volk S.W., Lian J.V., Drissi H., Stein G.S., Adams S.L. Smad–Runx interactions during chondrocyte maturation. J. Bone Joint Surg. Am. 2001;83-A(Suppl. 1):S15–S22. [PubMed] [Google Scholar]
- 27.Drissi M.H., Li X., Sheu T.J., Zuscik M.J., Schwarz E.M., Puzas J.E., Rosier R.N., O’Keefe R.J. Runx2/Cbfa1 stimulation by retinoic acid is potentiated by BMP2 signaling through interaction with Smad1 on the collagen X promoter in chondrocytes. J. Cell Biochem. 2003;90:1287–1298. doi: 10.1002/jcb.10677. [DOI] [PubMed] [Google Scholar]
- 28.Quack I., Vonderstrass B., Stock M., Aylsworth A.S., Becker A., Brueton L., Lee P.J., Majewski F., Mulliken J.B., Suri M., et al. Mutation analysis of core binding factor A1 in patients with cleidocranial dysplasia. Am. J. Hum. Genet. 1999;65:1268–1278. doi: 10.1086/302622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooper S.C., Flaitz C.M., Johnston D.A., Lee B., Hecht J.T. A natural history of cleidocranial dysplasia. Am. J. Med. Genet. 2001;104:1–6. doi: 10.1002/ajmg.10024. [DOI] [PubMed] [Google Scholar]
- 30.Cunningham M.L., Seto M.L., Hing A.V., Bull M.J., Hopkin R.J., Leppig K.A. Cleidocranial dysplasia with severe parietal bone dysplasia: C-terminal RUNX2 mutations. Birth Defects Res. A Clin. Mol. Teratol. 2006;76:78–85. doi: 10.1002/bdra.20231. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y.W., Yasui N., Ito K., Huang G., Fujii M., Hanai J., Nogami H., Ochi T., Miyazono K., Ito Y. A RUNX2/PEBP2aA/CBFA1 mutation displaying impaired transactivation and Smad interaction in cleidocranial dysplasia. Proc. Natl Acad. Sci. USA. 2000;97:10549–10554. doi: 10.1073/pnas.180309597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim H.J., Nam S.H., Kim H.J., Park H.S., Ryoo H.M., Kim S.Y., Cho T.J., Kim S.G., Bae S.C., Kim I.S., et al. Four novel RUNX2 mutations including a splice donor site result in the cleidocranial dysplasia phenotype. J. Cell Physiol. 2006;207:114–122. doi: 10.1002/jcp.20552. [DOI] [PubMed] [Google Scholar]
- 33.Tessa A., Salvi S., Casali C., Garavelli L., Digilio M.C., Dotti M.T., Di G.S., Valoppi M., Grieco G.S., Comanducci G., et al. Six novel mutations of the RUNX2 gene in Italian patients with cleidocranial dysplasia. Hum. Mutat. 2003;22:104. doi: 10.1002/humu.9155. [DOI] [PubMed] [Google Scholar]
- 34.Machuca-Tzili L., Monroy-Jaramillo N., Gonzalez-del Angel A., Kofman-Alfaro S. New mutations in the CBFA1 gene in two Mexican patients with cleidocranial dysplasia. Clin. Genet. 2002;61:349–353. doi: 10.1034/j.1399-0004.2002.610505.x. [DOI] [PubMed] [Google Scholar]
- 35.Zhou G., Chen Y., Zhou L., Thirunavukkarasu K., Hecht J., Chitayat D., Gelb B.D., Pirinen S., Berry S.A., Greenberg C.R., et al. CBFA1 mutation analysis and functional correlation with phenotypic variability in cleidocranial dysplasia. Hum. Mol. Genet. 1999;8:2311–2316. doi: 10.1093/hmg/8.12.2311. [DOI] [PubMed] [Google Scholar]
- 36.Javed A., Bae J.S., Afzal F., Gutierrez S., Pratap J., Zaidi S.K., Lou Y., van Wijnen A.J., Stein J.L., Stein G.S., Lian J.B. Structural coupling of Smad and Runx2 for execution of the BMP2 osteogenic signal. J. Biol. Chem. 2008;283:8412–8422. doi: 10.1074/jbc.M705578200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogata S., Uhthoff H.K. The early development and ossification of the human clavicle—an embryologic study. Acta Orthop. Scand. 1990;61:330–334. doi: 10.3109/17453679008993529. [DOI] [PubMed] [Google Scholar]
- 38.Black S., Scheuer L. Age changes in the clavicle: from the early neonatal period to skeletal maturity. Int. J. Osteoarchaeol. 1996;6:425–434. [Google Scholar]
- 39.Hall B.K. Development of the clavicles in birds and mammals. J. Exp. Zool. 2001;289:153–161. doi: 10.1002/1097-010x(20010215)289:3<153::aid-jez1>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 40.Huang L.F., Fukai N., Selby P.B., Olsen B.R., Mundlos S. Mouse clavicular development: analysis of wild-type and cleidocranial dysplasia mutant mice. Dev. Dyn. 1997;210:33–40. doi: 10.1002/(SICI)1097-0177(199709)210:1<33::AID-AJA4>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 41.Park M.H., Shin H.I., Choi J.Y., Nam S.H., Kim Y.J., Kim H.J., Ryoo H.M. Differential expression patterns of Runx2 isoforms in cranial suture morphogenesis. J. Bone Miner. Res. 2001;16:885–892. doi: 10.1359/jbmr.2001.16.5.885. [DOI] [PubMed] [Google Scholar]
- 42.Rice D.P., Rice R., Thesleff I. Molecular mechanisms in calvarial bone and suture development, and their relation to craniosynostosis. Eur. J. Orthod. 2003;25:139–148. doi: 10.1093/ejo/25.2.139. [DOI] [PubMed] [Google Scholar]
- 43.Bonaventure J., El G.V. Molecular and cellular bases of syndromic craniosynostoses. Expert Rev. Mol. Med. 2003;5:1–17. doi: 10.1017/S1462399403005751. [DOI] [PubMed] [Google Scholar]
- 44.Zaidi S.K., Pande S., Pratap J., Gaur T., Grigoriu S., Ali S.A., Stein J.L., Lian J.B., van Wijnen A.J., Stein G.S. Runx2 deficiency and defective subnuclear targeting bypass senescence to promote immortalization and tumorigenic potential. Proc. Natl Acad. Sci. USA. 2007;104:19861–19866. doi: 10.1073/pnas.0709650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Young D.W., Hassan M.Q., Yang X.-Q., Galindo M., Javed A., Zaidi S.K., Furcinitti P., Lapointe D., Montecino M., Lian J.B., et al. Mitotic retention of gene expression patterns by the cell fate determining transcription factor Runx2. Proc. Natl Acad. Sci. USA. 2007;104:3189–3194. doi: 10.1073/pnas.0611419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng Q., Sebald E., Zhou G., Chen Y., Wilcox W., Lee B., Krakow D. Dysregulation of chondrogenesis in human cleidocranial dysplasia. Am. J. Hum. Genet. 2005;77:305–312. doi: 10.1086/432261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones S.N., Roe A.E., Donehower L.A., Bradley A. Rescue of embryonic lethality in Mdm2 deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 48.Ramirez-Solis R., Liu P., Bradley A. Chromosome engineering in mice. Nature. 1995;378:720–724. doi: 10.1038/378720a0. [DOI] [PubMed] [Google Scholar]
- 49.O’Gorman S., Dagenais N.A., Qian M., Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc. Natl Acad. Sci. USA. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lufkin T., Mark M., Hart C.P., Dolle P., LeMeur M., Chambon P. Homeotic transformation of the occipital bones of the skull by ectopic expression of a homeobox gene. Nature. 1992;359:835–841. doi: 10.1038/359835a0. [DOI] [PubMed] [Google Scholar]
- 51.Vandenbroucke I.I., Vandesompele J., Paepe A.D., Messiaen L. Quantification of splice variants using real-time PCR. Nucleic Acids Res. 2001;29:E68. doi: 10.1093/nar/29.13.e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Owen T.A., Aronow M., Shalhoub V., Barone L.M., Wilming L., Tassinari M.S., Kennedy M.B., Pockwinse S., Lian J.B., Stein G.S. Progressive development of the rat osteoblast phenotype in vitro: reciprocal relationships in expression of genes associated with osteoblast proliferation and differentiation during formation of the bone extracellular matrix. J. Cell. Physiol. 1990;143:420–430. doi: 10.1002/jcp.1041430304. [DOI] [PubMed] [Google Scholar]
- 53.Bae J.S., Gutierrez S., Narla R., Pratap J., Devados R., van Wijnen A.J., Stein J.L., Stein G.S., Lian J.B., Javed A. Reconstitution of Runx2/Cbfa1-null cells identifies a requirement for BMP2 signaling through a Runx2 functional domain during osteoblast differentiation. J. Cell Biochem. 2007;100:434–449. doi: 10.1002/jcb.21039. [DOI] [PubMed] [Google Scholar]