Abstract
The prochlorophytes are oxygenic prokaryotes differing from other cyanobacteria by the presence of a light-harvesting system containing both chlorophylls (Chls) a and b and by the absence of phycobilins. We demonstrate here that the Chl a/b binding proteins from all three known prochlorophyte genera are closely related to IsiA, a cyanobacterial Chl a-binding protein induced by iron starvation, and to CP43, a constitutively expressed Chl a antenna protein of photosystem II. The prochlorophyte Chl a/b protein (pcb) genes do not belong to the extended gene family encoding eukaryotic Chl a/b and Chl a/c light-harvesting proteins. Although higher plants and prochlorophytes share common pigment complements, their light-harvesting systems have evolved independently.
The three known genera of prochlorophytes, Prochloron (1), Prochlorothrix (2), and Prochlorococcus (3), form a polyphyletic group within the cyanobacterial radiation on the basis of molecular phylogeny (4–6). None of the prochlorophytes appears to be more closely related to plant and green algal chloroplasts than are other cyanobacteria, despite the common presence of a chlorophyll (Chl) a/b antenna (5–7). The three prochlorophytes occupy different ecological niches and vary in pigment composition (8, 9). The light-harvesting antenna of Prochlorothrix hollandica, a filamentous fresh-water species, has a Chl a/b ratio of 2.5 to 4 (8, 9), whereas the antenna of Prochloron didemni, an obligate symbiont in ascidians, has a Chl a/b ratio of 2.4 and also contains a small amount of a Chl c-like pigment (10). Prochlorococcus, an open ocean genus, contains divinyl Chl a and b and a Chl c-like pigment, with the Chl a/b ratio of different strains ranging between 0.6 and 13 (11). The prochlorophyte Chl a/b antenna polypeptides are immunologically related to each other, but no relatedness to higher plant light-harvesting complex (LHC) polypeptides has been detected (8, 9).
The common presence of Chl b in chloroplasts and prochlorophytes is difficult to reconcile with the phylogenetic studies, leading to proposals that the ability to use Chl b as a light-harvesting pigment evolved independently more than once or was spread by lateral gene transfer from the organism where it originated (5, 6). To determine the origin of the prochlorophyte Chl a/b binding proteins, we isolated the genes encoding these proteins from members of all three prochlorophyte genera and discovered that they belong to a completely different family from genes encoding the Chl a/b-binding proteins of chloroplasts.
MATERIALS AND METHODS
Prochlorophytes and Chl a/b Proteins.
Prochlorococcus sp. (Med strain, CCMP1378), Prochlorococcus marinus (Sarg strain, CCMP1375), and P. hollandica were cultured in iron-replete media (11–13). P. didemni was harvested from its host Lissoclinum patella growing in shallow waters of the Great Barrier Reef (Australia). The Chl a/b complexes were purified by sucrose density gradient centrifugation (14) or by nondenaturing SDS/PAGE (13, 15), and the apoproteins were separated by denaturing gel electrophoresis for N-terminal or tryptic peptide sequencing (16).
Gene Isolation and Sequencing.
The genes from the Prochlorococcus spp. and P. didemni were amplified by polymerase chain reaction (PCR) using degenerate primers to the N-terminal sequence (1)MQTYGNP(7) obtained from Prochlorococcus sp. (Med strain) and a region conserved in both PsbC and IsiA proteins, (226)GGIWHI(231), with dITP substituted at positions of 4-fold degeneracy. A single 660-bp fragment, amplified from genomic DNA of the Med strain, was cloned, sequenced and used as a probe to isolate genomic DNA fragments (>3 kbp) containing the full-length pcb genes from both Med and Sarg strains. The P. didemni PCR products were cloned into pGEM-T (Promega), and size-selected clones were sequenced. An almost complete pcb sequence was subsequently obtained using the same first primer and a new primer based on the conserved GHLWHA near the C terminus. A 3.3-kb fragment was isolated from a P. hollandica genomic library using a psbC DNA probe and initially identified as containing an isiA-like gene. Complete sequencing of this fragment showed the presence of two complete and one partial pcb genes.
Sequence Analysis.
Protein sequences were aligned with the program macaw (version 2.03; ref. 17) using the Blosum 62 matrix with manual refinement. Gaps (hyphens) were included to optimize alignments. Hydrophobic domains and helix ends (arrows) were determined as in ref. 18. For phylogenetic analysis, sequences were aligned with clustal v (19), and distance and parsimony trees were generated using programs from the phylip package (20). All regions with gaps were excluded from the analysis, leaving 308 residues; however, the same configuration of branches resulted when gaps were included. To include the IsiA-like sequence of P. didemni in the analysis, trees were also calculated using only the 206 common amino acid positions.
Data bank accession numbers (brackets) of the sequences reported here are as follows. European Molecular Biology Laboratory: P. hollandica pcbA and pcbB (X97043), P. didemni pcb (Z72476) and isiA-like (Z72475). GenBank: Prochlorococcus sp. (Med strain) pcb (U57660), and P. marinus (Sarg strain) pcb (U57661). Additional GenBank sequences used in phylogenetic analysis were the isiAs of Synechocystis sp. PCC6803 (L26530), Synechococcus sp. PCC7942 (A30189), Synechococcus sp. PCC7002 (A47673), and Anabaena sp. PCC7120 (S42648), and psbC sequences of Synechococcus sp. PCC7942 (M20814), Synechocystis sp. PCC6803 (S06469), and P. hollandica (U40144).
RESULTS
To identify the genes for prochlorophyte Chl a/b proteins, N-terminal or internal peptide sequences were obtained from the major Chl a/b antenna polypeptides of Prochlorococcus sp. Med strain (ref. 12; F.P., J.L.R., K. Wyman, and P. G. Falkowski, unpublished work) and P. didemni (10), and from the 32- and 38-kDa antenna proteins of P. hollandica (13). One or more peptides from each prochlorophyte showed high similarity to the Chl a binding protein encoded by the cyanobacterial isiA gene, suggesting that the prochlorophyte Chl a/b proteins could be related to it. The IsiA Chl a protein is synthesized only during Fe starvation (21, 22) and is itself related to the constitutively expressed core Chl a protein (psbC gene product) present in photosystem II of all oxygenic phototrophs (21).
Gene cloning and sequencing confirmed that the prochlorophyte Chl a/b protein (pcb) genes were related to but distinct from isiA genes. P. hollandica was found to have three closely related pcb genes arranged in tandem and cotranscribed (data not shown), whereas only one gene was found in each of the two species of Prochlorococcus. Two sequences were obtained from P. didemni, one of which closely matched the tryptic peptide sequences. A comparison of amino acid sequences deduced from one complete pcb gene from P. hollandica and Prochlorococcus sp., the almost complete pcb gene sequence from P. didemni, and two cyanobacterial isiA sequences are shown in Fig. 1. The prochlorophyte Pcb sequences share an average of 54% identical residues with IsiA sequences (Table 1), and the degree of relatedness is even higher when conservative substitutions are taken into account (shaded areas in Fig. 1). The second P. didemni sequence, which was incomplete, had 78–83% identity with cyanobacterial IsiA sequences (Table 1) and did not match any of the peptide sequences. It was therefore considered an IsiA homologue. None of the Pcb sequences had any detectable relatedness to members of the eukaryotic Chl a/b antenna family.
Figure 1.

Alignment of protein sequences deduced from nucleotide sequences of the pcb genes of the prochlorophytes P. hollandica (pcbA), Prochlorococcus sp. CCMP 1378 (Med strain), and P. didemni, and the isiA genes of the cyanobacteria Synechocystis sp. PCC6803 and Synechococcus sp. PCC7942. Identical residues are in white type on a black background; gray squares are either conservative substitutions (within the groups FILMV, FYW, AG, HNQ, EQ, and DN) or columns where two different amino acids were found in both pcb and isiA proteins. Identity of cloned genes was confirmed by comparison with peptide sequences. Tryptic and N-terminal sequences determined from purified Chl a/b-binding protein P32 of P. hollandica were (2)ATTATPEYG(10), (112)GPEDLXQXDFEFA(124), (126)NFPFEWDDAAQA(137), (286)FSVAPYFVDTIDLPNGA(302), and (341)ALGFDFK(347). N-terminal sequence from the 32-kDa polypeptide of Prochlorococcus sp. (Med) was (1)MQTYGNPDVTYGXXAGN(17). Partial Lys-C digests of the 34-kDa Chl a/b protein of P. didemni gave the peptides (K)E(1)MQTYGNPDVEYGXXAGNSRLA and 118(K)EGPARAPKFDFDXGDGKXLGFI(140). The two additional amino acids consistently detected in the first peptide suggest that the P. didemni protein has an N-terminal extension not found in any other members of the family. The numbers flanking the peptide sequences correspond to residue numbers in the alignment. Double-headed arrows are predicted membrane-spanning helices.
Table 1.
Identity matrix for IsiA, Pcb, and PsbC proteins of prochlorophytes and cyanobacteria
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. PcbA P. hollandica | 65 | 45 | 50 | 60 | 58 | 56 | 58 | 59 | 54 | 51 | 50 | 51 | |
| 2. PcbB P. hollandica | 70 | 45 | 52 | 59 | 57 | 54 | 54 | 59 | 55 | 47 | 50 | 54 | |
| 3. Pcb P. marinus (Sarg) | 48 | 46 | 75 | 48 | 47 | 47 | 45 | 46 | 45 | 40 | 39 | 39 | |
| 4. Pcb Prochlorococcus sp. (Med) | 51 | 52 | 76 | 55 | 55 | 53 | 52 | 52 | 54 | 45 | 45 | 47 | |
| 5. Pcb P. didemni | 62 | 60 | 48 | 51 | 60 | 56 | 61 | 59 | 60 | 56 | 56 | 56 | |
| 6. IsiA-like P. didemni | 56 | 55 | 46 | 53 | 59 | 78 | 83 | 78 | 76 | 57 | 56 | 59 | |
| 7. IsiA Synechocystis sp. PCC6803 | 58 | 54 | 49 | 51 | 57 | 76 | 76 | 74 | 73 | 51 | 49 | 53 | |
| 8. IsiA Synechococcus sp. PCC7942 | 58 | 54 | 48 | 51 | 61 | 81 | 76 | 76 | 74 | 52 | 52 | 54 | |
| 9. IsiA Anabaena sp. PCC7120 | 60 | 58 | 47 | 51 | 60 | 77 | 73 | 75 | 74 | 54 | 56 | 56 | |
| 10. IsiA Synechococcus sp. PCC7002 | 55 | 53 | 48 | 53 | 59 | 76 | 75 | 75 | 72 | 55 | 56 | 58 | |
| 11. PsbC P. hollandica | 46 | 43 | 36 | 40 | 52 | 56 | 49 | 49 | 50 | 52 | 78 | 80 | |
| 12. PsbC Synechococcus sp. PCC6803 | 46 | 46 | 37 | 43 | 54 | 55 | 49 | 51 | 53 | 53 | 81 | 88 | |
| 13. PsbC Synechococcus sp. PCC7942 | 45 | 47 | 37 | 43 | 53 | 58 | 50 | 50 | 52 | 53 | 79 | 87 |
Pairwise amino acid identity (%) for protein sequences derived from pcb genes of P. hollandica, P. didemni, Prochlorococcus sp. Med and Sarg strains, and isiA and psbC from cyanobacteria and P. hollandica. Sequences were aligned using clustal v (19). (Lower) % identity calculated on basis of the number of residues in the shorter sequence; (Upper) % identity calculated on basis of the 206 common residues from all sequences.
We constructed phylogenetic trees (20, 23) using the protein sequences deduced from all the prochlorophyte pcb genes, the available isiA sequences, and several psbC sequences. The neighbor-joining distance matrix tree (Fig. 2A) shows that IsiA and PsbC sequences form two separate clusters, well-supported by bootstrap resampling. The prochlorophyte Pcb sequences also cluster, but not as strongly. Similar results were obtained with a maximum parsimony algorithm (20) except that the Pcb grouping had lower bootstrap support (Fig. 2, values in parenthesis). Sequences from the two Prochlorococcus sp. are more closely related to each other than they are to the P. didemni or P. hollandica sequences. The similarity between the PcbA and PcbB sequences of P. hollandica suggests that they arose as the result of a gene duplication, after the divergence of the prochlorophyte genera. Overall, the prochlorophyte Pcb sequences are much more divergent than the IsiA or PsbC clades. The partial IsiA-like sequence from P. didemni clustered strongly with the other IsiAs when it was included in the analysis (Fig. 2B).
Figure 2.
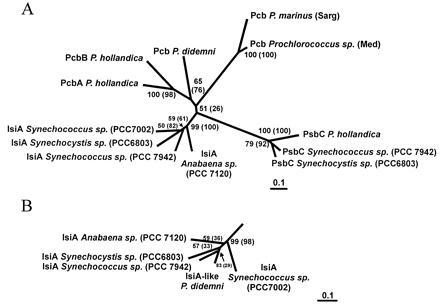
Phylogenetic distance tree (neighbor-joining method; ref. 23) of Pcb, IsiA, and PsbC proteins from prochlorophytes and cyanobacteria. Numbers at the branch points represent the bootstrap values for 100 replicate trees; numbers in parentheses are the bootstrap values for the parsimony analysis that generated a similar tree (not shown). The protein sequences were aligned using clustal v (19); trees were generated using protdist and neighbor (pam matrix) or protpars programs of the phylip package (20). (A) Tree using 308 common residues. All regions with gaps were excluded from the analysis, but the same configuration of branches resulted when gaps were included. (B) IsiA branch from trees that included the IsiA-like sequence from P. didemni, using the 206 common residues.
DISCUSSION
Sequence comparisons show that the newly identified pcb genes of prochlorophytes and the isiA genes are part of a family of related genes encoding proteins that can bind either Chl a+b or Chl a alone. The family also includes the psbC gene and the more distantly related psbB gene (24), which encode the proteins of the essential photosystem II Chl a–protein complexes CP43 and CP47 that are present in all oxygenic photosynthesizing organisms. The pcb genes of prochlorophytes are constitutively expressed like psbB and psbC, while the isiA genes are highly expressed only during iron deprivation (21, 22, 25). The expression of the isiA gene leads to the accumulation of high levels of a novel Chl a–protein complex (22), which does not appear to play a role in light harvesting but may act as a Chl storage protein to allow the rapid rebuilding of the photosynthetic apparatus when iron becomes available (22, 25).
At the primary structure level, the prochlorophyte Chl a/b proteins share no relatedness with the extended family (18) of eukaryotic light-harvesting Chl a, Chl a/b, and Chl a/c binding proteins (LHCs). However, it is noteworthy that within both the prochlorophytes and the green chloroplasts, the Chl a/b binding proteins belong to larger families of structurally related proteins whose members vary in pigment complement, size, and function (18, 26). Comparisons of the Chl a/b LHCs of green plants have shown that related protein sequences can bind different ratios of Chl a to Chl b (18). This suggests that only a relatively small number of changes in the primary sequence of the common ancestor of the Pcb and IsiA proteins would have been necessary to permit the binding of both Chl b and Chl a (or divinyl Chls a and b) as well as the Chl c-like pigment. The ability of members of two different protein families to bind a range of pigments might have facilitated the evolution of oxygen-evolving prokaryotes containing other types of chlorophylls, with or without phycobilins, such as the recently discovered Chl d-containing prokaryote Acaryochloris marina (27).
A further common factor in both gene families is the presence of members synthesized in response to environmental stresses. The LHC family includes higher plant and cyanobacterial relatives induced by high light stress and nutrient deprivation (18, 28, 29). Of particular interest is the cyanobacterial relative, the one-helix high light-inducible protein (HLIP) (28), which has been proposed as the ancestor of the eukaryotic LHCs (28, 29). The HLIPs have not been shown to bind Chl and are believed to function in photoprotection rather than light-harvesting (28), suggesting that the LHC family may have originated as high light stress response proteins rather than as light-harvesting antennas. Since HLIP genes have been found in all cyanobacteria examined to date (see ref. 29), it is likely that the prochlorophytes also have HLIPs, while their Chl a/b proteins are encoded by a totally unrelated gene family.
Two evolutionary scenarios could have led to the acquisition of Chl b synthesis by the three distantly related prochlorophytes. The progenitor of oxygenic photosynthetic bacteria may have been able to synthesize and use a wide variety of chlorophylls as well as phycobilins for light-harvesting. This array of pigments could have been lost to different degrees in various descendants (18, 30), with the prochlorophytes losing phycobilins but not the ability to make Chl b, and the other cyanobacteria losing Chl b while retaining phycobilins. It has recently been discovered that P. marinus (CCMP1375) possesses both the divinyl Chl a/b antenna and a type of phycoerythrin (31). We cannot tell if this prochlorophyte has lost other phycobilins as a result of a recent acquisition of Chl b, but in any case, this shows that it is possible for a Chl a/b antenna and phycobilins to coexist and function in the same cell.
Alternatively, the ability to synthesize Chl b may have arisen several times during the evolution of oxygenic photosynthetic bacteria and the ancestral chloroplast (5, 6). Although the enzyme responsible for the conversion of the 3-methyl group of Chl a (or one of its precursors) to the formyl group of Chl b has not been isolated, the reaction has been shown to require molecular oxygen, typical of a monooxygenase-catalyzed reaction (32). Since there are many monooxygenases in the cell, Chl b synthases could have been recruited on several different occasions (over two billion years) from other metabolic pathways by gene duplication and divergence. The phycobilins could have been lost subsequent to the acquisition of Chl b.
Regardless of which scenario prevailed for the evolution of Chl b synthesis, the similarity of Pcb and IsiA proteins strongly suggests that they originated by gene duplication in a common ancestor. Assuming that PsbB and PsbC were integral components (24) of an ancestral photosystem II in the early evolution of oxygenic photosynthesis, one can speculate that both Pcb and IsiA may have originated from a deletion of the large lumenal loop between helices 5 and 6 in the PsbC protein (21, 24). Whether a single ancestral gene gave rise to separate clades of pcb and isiA genes or whether there were separate duplications in each of the prochlorophyte lineages cannot be determined at present. Although the isiA-like and pcb genes in P. didemni form separate clades, lending support to the former more parsimonious hypothesis, we have yet to establish that other prochlorophytes also contain both genes. Furthermore, the clustering of the Pcb branches in Fig. 2 is not strongly supported by bootstrap analysis, so the apparent Pcb clade might be an artifact of long branches attracting.
Our demonstration that the Chl a/b binding proteins of prochlorophytes are structurally related to cyanobacterial Chl a binding proteins rather than to the Chl a/b binding proteins of green chloroplasts reconciles the apparent discrepancy between pigment composition and previous phylogenetic studies (4–7). Furthermore, it provides a model for the origin of the prochlorophyte proteins from an ancestral protein already binding at least one type of Chl. These findings strongly support the suggestion that the presence of Chl a/b light-harvesting proteins in both prochlorophytes and green chloroplasts must be the result of convergent evolution (33).
Acknowledgments
We thank Dr. D. G. Durnford for helpful discussions and Dr. T. Cavalier-Smith for reviewing the manuscript. Financial support was provided by the Natural Sciences and Engineering Research Council of Canada (B.R.G.), by the National Science Foundation (S.S.G. and R.A.), by the Department of Energy (J.L.R.), by the Australian Research Council (A.W.D.L. and R.G.H.), and by the Centre National de la Recherche Scientifique-Sciences de l’Univers (F.P.).
Footnotes
Abbreviations: Chl, chlorophyll; LHC, light-harvesting complex; HLIP, high light-inducible protein.
References
- 1.Lewin R A. Nature (London) 1976;261:697–698. doi: 10.1038/261697b0. [DOI] [PubMed] [Google Scholar]
- 2.Burger-Wiersma T, Veenhuis M, Korthals H J, Van de Wiel C C M, Mur L R. Nature (London) 1986;320:262–264. [Google Scholar]
- 3.Chisholm S W, Olson R J, Zettler E R, Goericke R, Waterbury J B, Welschmeyer N A. Nature (London) 1988;334:340–343. [Google Scholar]
- 4.Wilmotte A. In: The Molecular Biology of Cyanobacteria. Bryant D A, editor. Dordrecht, The Netherlands: Kluwer; 1995. pp. 1–25. [Google Scholar]
- 5.Palenik B, Haselkorn R. Nature (London) 1992;355:265–267. doi: 10.1038/355265a0. [DOI] [PubMed] [Google Scholar]
- 6.Urbach E, Robertson D L, Chisholm S W. Nature (London) 1992;355:267–270. doi: 10.1038/355267a0. [DOI] [PubMed] [Google Scholar]
- 7.Nelissen B, Van de Peer Y, Wilmotte A, De Wachter R. Mol Biol Evol. 1995;12:1166–1173. doi: 10.1093/oxfordjournals.molbev.a040289. [DOI] [PubMed] [Google Scholar]
- 8.Matthijs H C P, van der Staay G W M, Mur L R. In: The Molecular Biology of Cyanobacteria. Bryant D A, editor. Dordrecht, The Netherlands: Kluwer; 1995. pp. 49–64. [Google Scholar]
- 9.Bullerjahn G S, Post A F. Crit Rev Microbiol. 1993;19:43–59. doi: 10.3109/10408419309113522. [DOI] [PubMed] [Google Scholar]
- 10.Larkum A W D, Scaramuzzi C, Cox G C, Hiller R G, Turner A G. Proc Natl Acad Sci USA. 1994;91:679–683. doi: 10.1073/pnas.91.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Partensky F, Hoepffner N, Li W K W, Ulloa O, Vaulot D. Plant Physiol. 1993;101:285–296. doi: 10.1104/pp.101.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lichtlé C, Thomas J C, Spilar A, Partensky F. J Phycol. 1995;31:934–941. [Google Scholar]
- 13.van der Staay G W M, Staehelin L A. J Biol Chem. 1994;269:24834–24844. [PubMed] [Google Scholar]
- 14.La Roche J, Partensky F, Falkowski P. In: Photosynthesis: From Light to Biosphere. Mathis P, editor. Vol. 1. Dordrecht, The Netherlands: Kluwer; 1995. pp. 171–174. [Google Scholar]
- 15.van der Staay G W M, Ducret A, Aebersold R, Li R, Golden S S, Hiller R G, Wrench P M, Larkum A W D, Green B R. In: Photosynthesis: From Light to Biosphere. Mathis P, editor. Vol. 1. Dordrecht, The Netherlands: Kluwer; 1995. pp. 175–178. [Google Scholar]
- 16.Aebersold R, Leavitt J, Hood L E, Kent S B H. Proc Natl Acad Sci USA. 1987;84:6970–6974. doi: 10.1073/pnas.84.20.6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuler G D, Altschul S F, Lipman D J. Proteins Struct Funct Genet. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- 18.Green B R, Pichersky E. Photosynth Res. 1994;39:149–162. doi: 10.1007/BF00029382. [DOI] [PubMed] [Google Scholar]
- 19.Higgins D G, Sharp P M. Comput Appl Biol Sci. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- 20.Felsenstein, J. (1995) phylip, Phylogeny Inference Package (Univ. of Washington, Seattle).
- 21.Laudenbach D E, Straus N A. J Bacteriol. 1988;170:5018–5026. doi: 10.1128/jb.170.11.5018-5026.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burnap R L, Troyan T, Sherman L A. Plant Physiol. 1993;103:893–902. doi: 10.1104/pp.103.3.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 24.Bricker T M. Photosynth Res. 1990;24:1–13. doi: 10.1007/BF00032639. [DOI] [PubMed] [Google Scholar]
- 25.Falk S, Samson G, Bruce D, Huner N P A, Laudenbach D E. Photosynth Res. 1995;45:51–60. doi: 10.1007/BF00032235. [DOI] [PubMed] [Google Scholar]
- 26.Green B R, Durnford D G. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:685–714. doi: 10.1146/annurev.arplant.47.1.685. [DOI] [PubMed] [Google Scholar]
- 27.Miyashita H, Ikemoto H, Kurano N, Chihara M, Miyachi S. Nature (London) 1996;383:402. [Google Scholar]
- 28.Dolganov N A M, Bhaya D, Grossman A R. Proc Natl Acad Sci USA. 1995;92:636–640. doi: 10.1073/pnas.92.2.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Green B R, Kühlbrandt W. Photosynth Res. 1995;44:139–148. doi: 10.1007/BF00018304. [DOI] [PubMed] [Google Scholar]
- 30.Bryant D A. Curr Biol. 1993;2:240–242. doi: 10.1016/0960-9822(92)90361-d. [DOI] [PubMed] [Google Scholar]
- 31.Hess W R, Partensky F, van der Staay G W M, Garcia-Fernandez J M, Borner T, Vaulot D. Proc Natl Acad Sci USA. 1996;93:11126–11130. doi: 10.1073/pnas.93.20.11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Wettstein D, Gough S, Kannangara C G. Plant Cell. 1995;7:1039–1057. doi: 10.1105/tpc.7.7.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cavalier-Smith T. Biol J Linnean Soc. 1982;17:289–306. [Google Scholar]


