Abstract
Mutations in the gene CLN3 are responsible for the neurodegenerative disorder juvenile neuronal ceroid lipofuscinosis or Batten disease. CLN3 encodes a multi-spanning and hydrophobic transmembrane protein whose function is unclear. As a consequence, the cell biology that underlies the pathology of the disease is not well understood. We have developed a genetic gain-of-function system in Drosophila to identify functional pathways and interactions for CLN3. We have identified previously unknown interactions between CLN3 and the Notch and Jun N-terminal kinase signalling pathways and have uncovered a potential role for the RNA splicing and localization machinery in regulating CLN3 function.
INTRODUCTION
Juvenile neuronal ceroid lipofuscinosis (JNCL or Batten disease) is the most common of the NCLs, a collection of neurodegeneration disorders affecting children (1,2). In JNCL, retinal degeneration manifests at age 5–7 years usually followed by seizures and a progressive loss of motor skills and mental acuity. JNCL is invariably fatal, with death occurring usually by 25 years. Histopathologically, post-mortem brain material from JNCL patients shows widespread neuronal loss with accompanying gliosis (reviewed in 3). Common to all NCLs, both neuronal and peripheral tissues from JNCL patients show extensive accumulations of lysosomal storage material, indicative of some form of lysosomal dysfunction. Consequently, the NCLs are included in the larger family of metabolic disorders, the lysosomal storage disorders (4).
JNCL is an autosomal recessive disorder resulting from mutations in the gene CLN3 that encodes a multi-spanning transmembrane protein (5,6). CLN3 is expressed widely and many cell types from JNCL patients display lysosomal storage material. However, only neuronal pathology is seen: either the CLN3 protein has a function unique to neurons or neurons are particularly susceptible to loss of CLN3. Although major progress has been made using mouse models to examine the disease progression (7–10), our understanding is currently restricted by a lack of knowledge about the functions of, and even the precise location of, the CLN3 protein within cells.
Various approaches have been used to study the cell biology of CLN3. In vitro studies using CLN3 mutant cell lines generated from mouse models and human patients have suggested roles for CLN3 in many cellular processes including intracellular trafficking, endocytosis, apoptosis, autophagy and lipid cycling (reviewed in 3). Yeast strains mutant for the CLN3 homologue, BTN1, have also proved valuable: Saccharomyces cerevisiae and Schizosaccharomyces pombe mutant strains both show elevated vacuolar pH (11,12), whereas S. cerevisiae strains are defective in arginine transport into the vacuole, resulting in a disruption in nitric oxide metabolism (13–15). However, these varied approaches have failed to produce a consensus on the function of CLN3. Progress has also been hampered by the highly hydrophobic nature of the CLN3 protein that makes conventional biochemical approaches difficult and by the fact that it is seemingly expressed at low levels. There is a need for new model systems to study CLN3 biology and function to further our understanding of the disease. In particular, systems are required that will identify functional pathways without relying on biochemical techniques. Identifying such pathways may in turn reveal novel therapeutic targets for JNCL.
The fruit fly Drosophila is an attractive model system for studying neuronal function and neurodegeneration because of its relatively simple nervous system, the powerful genetic tools available and the ability to study neuronal and synapse biology at high resolution (reviewed in 16,17). Several human neurodegenerative disorders have been modelled successfully in Drosophila, including congenital and infantile forms of NCL (18,19). Here we describe the Drosophila homologue of CLN3 and introduce gain-of-function genetic approaches to identify pathways that require CLN3 function and novel interactions for CLN3. We demonstrate that ectopic CLN3 expression inhibits Notch signalling but activates the Jun N-terminal kinase (JNK) signalling pathway and identify novel and unexpected genetic interactions with a regulator of RNA translation, stability and localization.
RESULTS
Juvenile NCL is an autosomal recessive disorder caused by mutations in the gene CLN3 (5). The protein encoded is a multi-spanning transmembrane protein, the topology of which remains unclear. The most recent prediction based on a model constrained by experimental data suggests six transmembrane spans with both the N- and C-termini resident in the cytosol (20). The Drosophila genome encodes one likely orthologue—CG5582 at position 75A2. Sequence alignments show strong conservation with vertebrate CLN3 proteins in the predicted transmembrane regions, in the lumenal loops and in the C-terminal region thought to be intracellular (Supplementary Material, Fig. S1). Both lysosomal targeting sequences identified experimentally in the human protein are conserved in Drosophila CLN3.
Drosophila CLN3 localizes to lysosomes, the plasma membrane and recycling vesicles
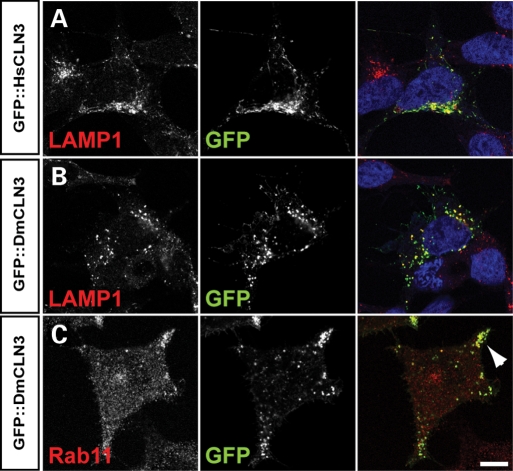
The exact localization of CLN3 remains undefined despite examination of its distribution in a number of studies (reviewed in 21). Owing to the absence of appropriate specific antisera, most studies have employed overexpression and epitope-tagging to get an indication of localization in cultured cells and neurons. A similar approach was used to examine the distribution of Drosophila CLN3 in cells. An N-terminal GFP-CLN3 construct was expressed in HEK293 cells and its distribution compared with various markers of cellular compartments and with that of the human CLN3. Like the human protein, the distribution of Drosophila CLN3 partially overlaps with that of LAMP1, a marker for late endosomes and lysosomes [compare Fig. 1A and B; (22,23)]. At higher expression levels, CLN3 is also seen on the plasma membrane (data not shown). Strong co-localization was also seen in a subset of transfected cells with anti-Rab11, particularly in large vesicles in the periphery of the cells (Fig. 1C), supporting previous data (24). Rab11 regulates the recycling of vesicles from the perinuclear recycling compartment to the plasma membrane, suggesting that CLN3 may be a component of, or trafficked by, these vesicles. No significant co-localization was seen with markers of other compartments or endosomes (data not shown).
Figure 1.
Localization of Drosophila CLN3 in HEK293 cells. N-terminal fusions of human (Hs) or Drosophila (Dm) CLN3 to GFP were expressed in cells and their distributions detected with an antibody to GFP (middle panels, green in merge). Lysosomes and late endosomes were detected with anti-LAMP, recycling vesicles with anti-Rab11 (left panels, red in merge). (A and B) Both human and Drosophila CLN3 partially co-localizes with LAMP1. (C) In a subset of cells, Drosophila CLN3 is seen to co-localize with Rab11-positive vesicles at the periphery of cells (arrowhead). Bar: 5 µm.
CLN3 overexpression in the eye leads to degeneration
Our understanding of CLN3 cell biology would be aided significantly if more of the signalling pathways and molecules that interact with CLN3 were known. The highly hydrophobic nature of the CLN3 protein means that some of the commonly used biochemical approaches for identifying interactions can be difficult. To overcome this problem, we decided to develop a strategy that would allow us to identify interacting molecules and functional signalling pathways using genetic approaches. We generated transgenic flies to express CLN3 under the control of the bipartite GAL4-UAS system that allows for highly regulated tissue-specific expression (25). We were then able to examine the phenotypic consequences of increasing levels of CLN3 function during organ and tissue development in the fly. These phenotypes were caused by a moderate increase in the expression of CLN3; for example, in our experiments, in the wing, we estimate that the levels of CLN3 are increased 2.86-fold (±0.42 SD) over basal levels of CLN3 expression (see Materials and Methods). Finally, we were able to use genetics to identify pathways and molecules that affect these phenotypes.
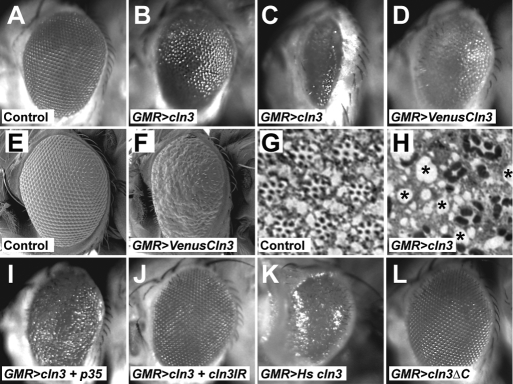
Initially, we decided to focus on overexpression in the developing fly eye. The development of the eye is heavily dependent on endocytic and vesicle trafficking events (26–28), both suggested functions for CLN3. Expression of CLN3 in the eye produced a significant glazing of the compound eye indicative of degeneration (Fig. 2A versus B and C). The severity of the phenotype varied between lines, presumably reflecting differing levels of expression of the CLN3 transgene from different chromosomal locations. Expression of a YFP-CLN3 fusion produced a similar phenotype (Fig. 2D), as did the expression of human CLN3 (Fig. 2K). Scanning electron microscopy (SEM) shows loss of the regular crystalline array of ommatidia, fusions of ommatidia and alterations in bristle patterns (Fig. 2E versus F). Sections through the eyes show an extensive degeneration (Fig. 2G versus H). To confirm that this phenotype is specific to CLN3 overexpression, we also overexpressed the lysosomal proteins, LAMP and spinster, and the late endosomal protein, Hook, in the eye. These controls displayed no phenotype, suggesting the CLN3 phenotypes are specific and not a consequence of overloading the endosomal-lysosomal membrane system (data not shown).
Figure 2.
Overexpression of CLN3 in the developing Drosophila eye results in degeneration. (A–D) Light micrographs of eyes from adult flies. (A) Control eye (GMRgal4/+); (B and C) Overexpression of CLN3 at lower (B) and higher (C) levels under the control of the eye-specific GMRgal4 causes glazing and loss of tissue (both denoted GMR>cln3). (D) Overexpression of a VenusYFP-CLN3 fusion results in a similar phenotype. (E and F) SEM of the surface of adult eyes. (E) Control eyes show a regular array of ommatidia. (F) Overexpression of a VenusYFP-CLN3 fusion results in misshapen and fused ommatidia (denoted GMR>VenusCln3). (G and H) Sections of adult retinas. (G) In control retinas, ommatidia-containing groups of dark staining photoreceptors are arrayed evenly. (H) Overexpression of CLN3 (denoted GMR>CLN3) results in disruption to ommatidial organization and widespread vacuolation indicative of degeneration (asterisks). (I) The principal eye phenotype is not due to apoptosis, as co-expression of the baculovirus apoptosis inhibitor, p35, with CLN3 does not rescue the phenotype. (J) The eye phenotype is suppressed by co-expression of dsRNA construct targeting CLN3 (denoted cln3IR), indicating that it is due to CLN3 overexpression. (K) Overexpression of human CLN3 results in a similar phenotype (GMR>Hs cln3). (L) Overexpression of a C-terminally truncated form of CLN3 shows no phenotype (GMR>cln3ΔC).
The external glazing of the Drosophila eye can be caused by widespread apoptosis during eye development. However, in this case, blocking apoptosis by co-expressing the baculovirus inhibitor p35 (29) failed to rescue the phenotype (Fig. 2I), suggesting apoptosis is not the primary cause. Co-expression of an RNAi construct designed for CLN3 knockdown did revert the phenotype back towards wild-type, indicating that the initial phenotype is specifically because of the overexpression of CLN3 (Fig. 2J).
Notch-like phenotype resulting from overexpression of CLN3
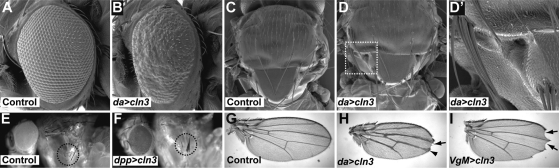
Having observed strong phenotypes in the eye, CLN3 was expressed ubiquitously throughout Drosophila development or selectively in imaginal tissues which give rise to particular structures of the adult fly during metamorphosis. In both cases, phenotypes reminiscent of Notch loss-of-function resulted (28). Ubiquitous overexpression using the daughterless Gal4 driver was semi-lethal. Escaping adults were noticeably smaller than controls (Supplementary Material, Fig. S2) and displayed roughened eyes (compare Fig. 3A versus B; overexpression denoted by da>cln3). Overexpression also led to duplication of macrochaetae (the larger sensory bristles) on the thorax of the adult fly (Fig. 3C versus D, boxed region enlarged in Fig. 3D′). Wings displayed thickened veins (compare veins in Fig. 3G versus H), vein deltas and notches in the wing margin (Fig. 3H).
Figure 3.
Notch-like phenotypes in Drosophila overexpressing CLN3. (A–D) SEM of Drosophila eyes and thoraces. (A and C) Control flies (daGal4/+) display a regular crystalline array of ommatidia in the eye and individual macrochaetae on the thorax. (B and D) Overexpression of CLN3 (denoted da>cln3) leads to a roughening of the eye and multiple duplications of the macrochaetae (boxed region enlarged in D′). The microchaetae on the thorax are unaffected. (E and F) A similar duplication of the sternopleural bristles is seen when CLN3 is overexpressed with the dppGal4 driver (dpp>cln3, bristles circled). Overexpression of CLN3 also causes Notch-like phenotypes in the adult wing. (G) Control wing. (H) Overexpression throughout the wing (da>cln3) causes thickening of veins, vein deltas (arrowhead) and notching in the margin (arrow). (I) Overexpression in the wing margin (VgM>cln3) leads to significant notching (arrows).
Using different Gal4 driver lines to express CLN3 in subsets of cells in the developing fly resulted in similar phenotypes. Expression driven by dppGal4 driver resulted in duplications in the sternopleural bristles on the side of the adult fly (Fig. 3E versus F, bristles circled, denoted dpp>cln3). Finally, expression driven along the wing margin under the control of the VgM gal4 line caused large notches to appear in the wing blade (Fig. 3I, arrowheads, denoted VgM>cln3). Taken together, these phenotypes strongly suggest that overexpression of CLN3 leads to loss of Notch function. Notch acts as a receptor for the Delta/Serrate/LAG-2 ligand family which functions in a variety of contexts to allow communication between cells whereby transcription of target genes are activated in the Notch expressing cell. This signalling pathway requires proteolytic cleavage of the Notch receptor and its endocytosis into the cell (reviewed in 30).
If Notch signalling was indeed affected by CLN3 overexpression, we reasoned that introducing a mutant allele of the Notch ligand, Delta, should enhance the effect. We focused on duplication of the macrochaetae on the scutellum of the adult fly. In control flies, there are invariably 4.0 macrochaetae (n = 26, see Materials and Methods for genotypes). When CLN3 was overexpressed in the posterior of the imaginal wing disc using the engrailed GAL4 driver, duplications of one or more of the bristles were common and the mean number of macrochaetae per scutellum increased to 4.65 (n = 17). When CLN3 was overexpressed in a genetic background heterozygote for a strong allele of Delta, the frequency of duplication increased further to a mean of 6.29 macrochaetae per scutellum (n = 24, P < 0.0001). These data indicate that the Notch signalling pathway does interact genetically with the gain-of-function phenotypes seen with CLN3 overexpression.
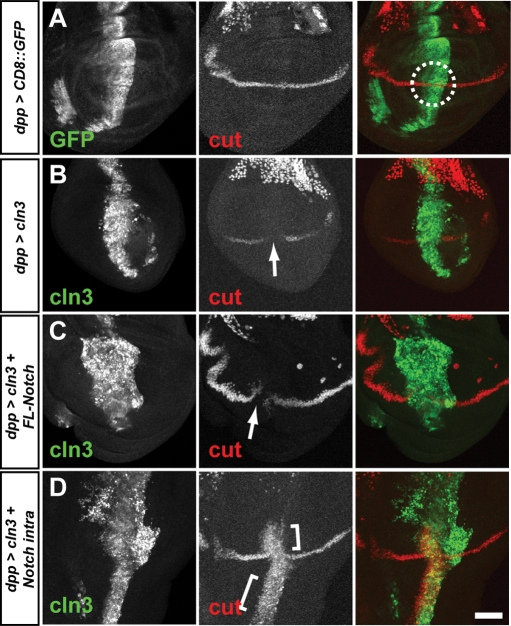
Several of the phenotype described earlier can also result from alterations in EGF, BMP and other signalling pathways (31). To demonstrate further that that CLN3 overexpression does indeed inhibit Notch signalling, transcription of the cut gene was used as readout for the Notch pathway. cut encodes a homeodomain protein structurally related to human CDP and mouse cux proteins (32). cut is transcribed in a narrow strip of cells at the dorsal–ventral margin of the developing Drosophila imaginal wing disc, and this transcription requires Notch activity (33). We made use of the fact that dppGal4 drives expression along the anterior–posterior compartment boundary of the wing disc which intersects at only one region with the dorsal–ventral stripe of cut expression. This is demonstrated in Figure 4A, where a membrane-tethered GFP was expressed under the control dppGal4 (dpp>CD8::GFP). Discs were processed to visualize GFP (green) and Cut protein (red). The overlapping regions of expression are circled in the merged panel. When CLN3 was overexpressed under the control of dppGal4 (dpp>CLN3), a clear loss of Cut protein was seen at the intersection of the expression domains, indicating a loss of Notch activity (Fig. 4B, arrow).
Figure 4.
Loss of Notch activity in the developing wing. (A) dppGal4 drives expression of membrane-tethered CD8::GFP in a stripe along the A-P compartment boundary (left panel and green in merge). This stripe intersects with the restricted expression of the Notch transcriptional target, cut (middle panel and red in merge, intersecting region circled). (B) When CLN3 expression is driven by dppGal4, cut expression is lost at the intersection, indicating loss of Notch transcriptional activity (arrow). (C) Co-expression of a full-length Notch construct fails to rescue cut expression completely (arrow). (D) Co-expression of the dominant active Notch intracellular domain (Notchintra) rescues cut expression and also induces ectopic cut expression along the A-P boundary (bracketed). Posterior is right. Bar: 10 µm.
To try to understand the mechanism of inhibition of Notch, a full-length Notch construct was co-expressed with CLN3. This construct failed to reconstitute the Cut stripe fully (Fig. 4C, arrow), indicating that the inhibition is likely to be at the level of the Notch protein function rather than its expression. This was confirmed by immunostaining for Notch protein which indicated no changes in Notch protein levels in the wing disc where CLN3 was expressed (data not shown). We also asked whether CLN3 overexpression was actually inhibiting downstream targets of Notch transcription, rather than Notch itself. Notch is cleaved upon ligand binding, releasing the intracellular part of the protein which translocates to the nucleus to act on transcription. The Notchintra construct does not require ligand binding or cleavage and so acts as a dominant active molecule (34). Co-expression of this construct alongside CLN3 restored cut transcription and also induced ectopic cut elsewhere along the dppGal4 domain of expression (Fig. 4D). Clearly, the cells expressing CLN3 remain competent for signalling downstream of Notch.
Activation of JNK signalling by CLN3
Mutants that affect Notch glycosylation or cleavage cause a global inhibition of Notch function, for examples, see references (35,36). These mutants display neurogenic phenotypes in embryos and duplications of the smaller sensory bristles (the microchaetae) on the thorax in addition to the types of phenotypes described earlier. CLN3 overexpression does not affect microchaetae (Fig. 3D and D′) and there is no neurogenic phenotype (data not shown). Hence, the mechanism of inhibition is unlikely to be via these processes but we considered that elucidating the mechanism might shed light on normal CLN3 function. We focused on two observations that suggested CLN3 may inhibit Notch via activation of JNK signalling.
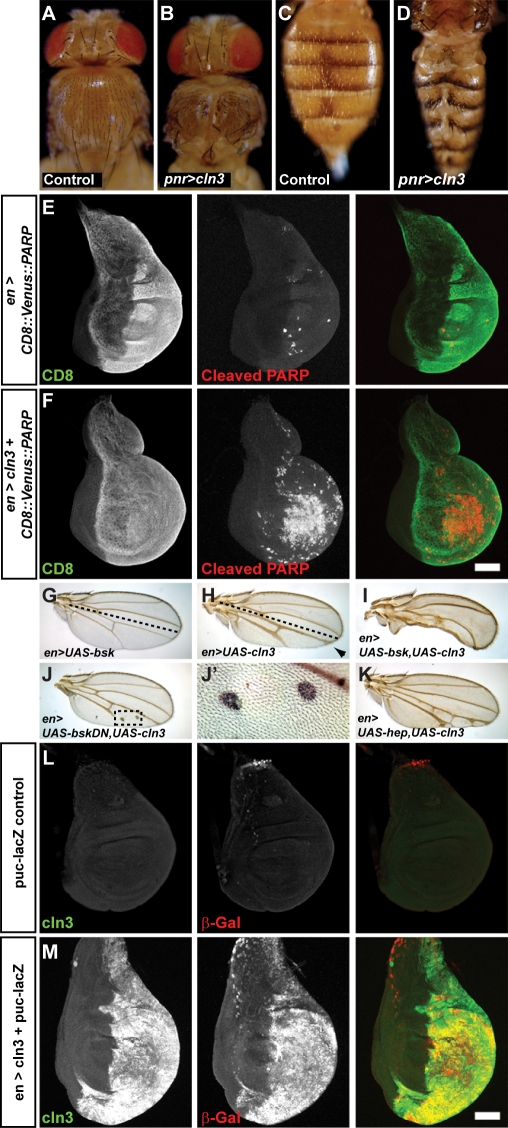
First, we observed that CLN3 overexpression specifically in the dorsal midline led to dorsal closure phenotypes in both the thorax (Fig. 5A versus B) and the abdomen (Fig. 5C versus D) of adult flies. In both the embryo and during metamorphosis, the dorsal surface is closed by migration of epithelia towards the midline and subsequent fusion. Dorsal closure phenotypes are characteristic of mutations in components of the JNK signalling pathway (37–39). Importantly, a previous study has reported genetic interactions in dorsal closure events between Notch and bsk (the Drosophila homologue of JNK). In dorsal closure events, increased levels of phospho-JNK are seen in Notch mutants, suggesting crosstalk between the two signalling pathways (40). The mechanism is not known, although the interaction appears to be independent of ligand binding to Notch.
Figure 5.
CLN3 interacts genetically with and ectopically activates JNK signalling. Thoraces and abdomens of adult Drosophila. (A and C) Control thorax and abdomen. (B and D) Overexpression of CLN3 along the dorsal midline by pnr-Gal4 leads to dorsal closure defects in the thorax (B) and abdomen (D), indicating a possible affect on JNK signalling. (E and F) CLN3 expression increases apoptosis in the wing disc. To visualize apoptosis, a CD8::Venus::PARP membrane-tethered probe for caspase activity was expressed in the posterior half of third instar wing discs (CD8 immunostaining, left panels and green in merge). The PARP probe is cleaved by active caspases, so apoptotic cells can be detected with an antibody specific to the cleaved form of PARP (cleaved PARP, middle panels and red in merge). (E) Few apoptotic cells are seen when the probe is expressed alone. (F) Co-expression of the probe with CLN3 leads to a large increase in apoptosis, particularly in the central wing pouch region (middle panels). Posterior is right. Bar: 5 µm. (G–J) Expression of bsk, a Drosophila JNK, enhances the effect of CLN3. (G) Expression of bsk in the posterior half of the wing (below dashed line, denoted en>UAS-bsk) does not alter the morphology. (H) Expression of CLN3 in the posterior half (en>UAS-cln3) leads to thickened veins, vein deltas (arrowhead) and loss of tissue. (I) Co-expression of CLN3 with bsk leads to almost complete loss of tissue in the posterior half, indicating a strong genetic interaction (en>UAS-cln3+UAS-bsk). (J) Co-expression of a dominant negative bsk construct (UAS-bskDN) with CLN3 leads to vein truncations and clones of cells with multiple bristles in the posterior half of the wing (boxed area enlarged in J′). (K) Co-expression of CLN3 with hep (a Drosophila JNKK) also enhances the CLN3 phenotype (en>UAS-hep,UAS-cln3). (L and M) Overexpression of CLN3 leads to ectopic activation of JNK signalling. puc-lacZ was used as reporter for activated JNK signalling (β-Gal, middle panels and red in merge). (L) Control wing discs show little JNK activity. (M) Overexpression of CLN3 in the posterior half (CLN3, left panels and green in merge) results in a dramatic ectopic activation of JNK signalling (M). Posterior is right. Bar: 5 µm.
Second, we observed that CLN3 overexpression caused an induction of apoptosis in the developing wing disc (Fig. 5E and F). Activation of JNK signalling can commonly lead to apoptosis (reviewed in 41). Apoptotic cells were identified through expression of a reporter for caspase activity—a membrane-tethered CD8::PARP fusion protein (42). In apoptotic cells, PARP is cleaved by caspases and this event can be detected with an antibody specific to the cleaved form. We expressed the PARP reporter with or without CLN3 in the posterior half of the developing wing disc using the engrailed GAL4 driver. Few apoptotic cells were visible in wing discs expressing the reporter alone (Fig. 5E). In contrast, when CLN3 was co-expressed, a large increase in apoptosis was seen (Fig. 5F), especially in the wing pouch.
Taken together, these data suggested that CLN3 overexpression may affect JNK signalling, but to examine this more directly, we looked for genetic interactions between the two. Overexpressing CLN3 in the posterior half of the wing disc during development leads to thickened veins and notches in the posterior half of adult wings (Fig. 5G versus H, posterior half shown below the dashed line in H). In contrast, overexpressing either the Drosophila JNK homologue encoded by bsk (en>UAS-bsk) or a dominant negative JNK (bskDN) construct has no effect on the wing (Fig. 5G and data not shown). However, expressing both CLN3 and bsk leads to a dramatic increase in phenotype, with almost complete loss of tissue in the posterior half suggesting extensive apoptosis and demonstrating a strong genetic interaction (en>UAS-bsk,UAS-cln3, Fig. 5I). Co-expression of CLN3 and bskDN produces clones of cells in the wings with multiple bristles (en>UAS-bskDN,UAS-CLN3, Fig. 5J and boxed region enlarged in 5J′). Presumably, these are cells with complete inhibition of Notch function that would usually die but are rescued from apoptosis by dominant negative JNK. Co-expression of hep, the Drosophila JNKK, also enhanced the CLN3 overexpression phenotype (en>UAS-hep,UAS-cln3, Fig. 5K).
We next examined direct effects of CLN3 overexpression on JNK signalling itself in the wing disc. Using a puc-lacZ reporter as a readout for JNK-signalling activity (39), ectopic activation of JNK signalling could clearly be seen in the posterior half of wing discs when CLN3 was overexpressed in that compartment [Fig. 5L versus M, compare anterior (left) and posterior (right) compartments]. This activation of JNK signalling does not appear to be due to an increase in JNK levels but rather an increase in active JNK, as there is no observable increase in JNK protein levels in the CLN3 overexpressing cells (data not shown). In contrast, in a similar experiment, CLN3 overexpression had no effect on EGF receptor signalling in the posterior half of wing discs, visualized using an argos-lacZ reporter (data not shown). It therefore seems unlikely that the activation of JNK signalling is due to a generalized effect on ligand–receptor complex trafficking or degradation.
Dominant modification of the CLN3 overexpression eye phenotype
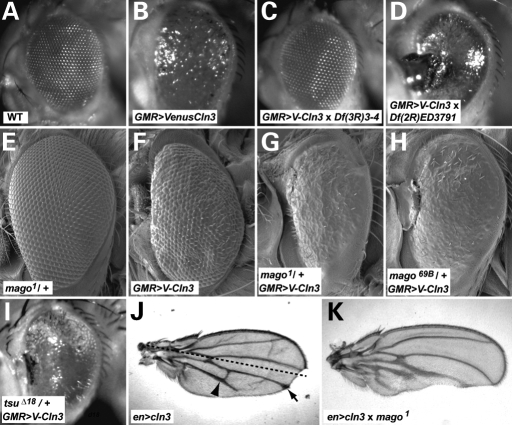
Alterations of the appearance of the Drosophila eye and wing are commonly used for genetic screens. Neither organ is essential for viability, and modifications of the phenotype are easily scoreable. We tested the feasibility of using the degenerative eye and phenotype caused by overexpression of CLN3 as the basis of a screen to identify dominant genetic interactions. Briefly, a collection of defined chromosomal deficiencies, each with a group of genes deleted, were mated individually to flies overexpressing CLN3 in the developing eye. In each case, the progeny resulting from the cross carry only half the gene dosage for each gene lying within the deficiency. The progeny were then scored for modification of the starting phenotype: either a reversion back towards wild-type or a more severe phenotype. Modification indicates that one of the genes whose dose has been reduced interacts genetically with CLN3. A combination of the DrosDel collection of defined deficiencies and lines from the Bloomington Stock Centre deficiency kits were used to screen the large majority of the two main Drosophila autosomes. As expected, the deficiency that deletes the CLN3 locus suppressed the eye phenotype by reducing the gene dosage. In addition, five other deficiencies caused significant suppression of the phenotype and four deficiencies caused significant enhancement (Fig. 6A–D and Supplementary Material, Table S1).
Figure 6.
Components of the EJC interact genetically with CLN3. (A–D) Light micrographs of adult Drosophila eyes. (A) Wild-type eye. (B) Degeneration in the eye due to overexpression of a VenusYFP fusion with CLN3 (denoted VenusCln3 or V-Cln3). Examples of eyes isolated from a screen of chromosomal deficiencies to identify dominant modifiers of the CLN3 overexpression phenotype. (C) Deficiency Df(3R)3-4 suppresses and (D) Df(2R)ED3791 enhances the phenotype. The EJC component mago nashi was shown to be the gene in ED3791 responsible for the enhancement. (E–H) SEM of adult eyes. (E) mago nashi heterozygote flies show no eye phenotype (mago1/+). (F) Degeneration caused by overexpression of a Venus::CLN3 fusion (GMR>V-Cln3). (G and H) Venus::CLN3 overexpression in flies that are heterozygote for either of two mago nashi alleles enhances the phenotype and results in the same type of necrosis at the anterior margin of the eye as in (D). (I) Overexpression of CLN3 in a background heterozygote for tsunagi/Y14, the dimerization partner of Mago in the EJC results in a similar enhancement of the phenotype (tsuΔ18/+). (J and K) mago nashi also interacts genetically with CLN3 in the wing. (J) Expression of CLN3 in the posterior half of the wing (below the dashed line) results in thickened veins, deltas (arrowhead), occasionally vein truncations (arrow) and loss of tissue. (K) The phenotype is enhanced in a mago nashi heterozygote background. Anterior is left in (A–I) and top in (J and K).
Identification of a novel genetic interactions between CLN3 and an RNA-binding protein
The deficiency Df(2R)ED3791 that dominantly enhanced the eye phenotype (Fig. 6D) was selected for further study to identify the gene responsible. The ED3791 deficiency deletes 79 genes. The same mating strategy was repeated using smaller overlapping deficiencies within the same region. One Df(2R)F36 that deletes only nine genes similarly enhanced the eye degeneration (data not shown). Subsequently, alleles of one of the nine genes, mago nashi, displayed a similar dominant enhancement when the gene dosage was halved (Fig. 6E–H), indicating that it was the gene responsible. mago nashi alleles also showed dominant enhancement of the wing phenotypes caused by CLN3 overexpression (Fig. 6J versus K). mago nashi alleles did not show a dominant phenotype with the GMRgal4 driver alone in the absence of CLN3 overexpression (data not shown).
This discovery highlights the power of this CLN3 gain-of-function system for identifying novel and unexpected interactions: mago nashi encodes a highly conserved component of the exon–exon junction complex (EJC) that regulates RNA stability and has roles in RNA and organelle transport and cell polarity in many systems (43). Mago nashi functions as a dimer with Tsunagi/Y14 (44,45). An allele of tsunagi also enhances the CLN3 overexpression eye phenotype (Fig. 6I), demonstrating the veracity of the interaction with CLN3.
CLN3-induced phenotypes require a fully functional protein
The majority of human JNCL patients are homozygous for a 1 kb deletion within the CLN3 gene that is predicted to encode a protein truncated after the third transmembrane domain (6). We made use of this truncation to verify that the phenotypes described earlier are specific to full-length CLN3. We generated a human CLN3 construct carrying a stop codon after amino acid 153 and a Drosophila version with a stop codon at the equivalent position based on sequence alignments. There are contrasting views regarding the likelihood that the truncated form of human CLN3 might retain some functionality in vivo (46,47), so first we examined the distribution of the truncated proteins in HEK293 when expressed as N-terminal YFP fusions. We argued that for the truncated forms to retain function, they should localize similarly to the full-length proteins. In contrast to the full-length YFP-CLN3 fusion proteins (Fig. 1), both the human and Drosophila truncated forms localized in a reticular pattern reminiscent of the endoplasmic reticulum (ER) (see Supplementary Material, Fig. S3). These data confirm previous observations for the human protein (22) and suggest the truncated proteins mis-fold. Although neither construct expresses the novel 28 amino acids associated with the C-terminus of the disease-causing CLN3 allele, these are unlikely to restore the localization. Transgenic flies overexpressing the truncated forms either untagged or as YFP-fusions failed to show phenotypes in the eye (Fig. 2L and data not shown) or in the wing (data not shown). Taken together, these data indicate that the pathways and molecules identified in these assays interact with fully functional CLN3.
DISCUSSION
Our understanding of the function of CLN3, the gene disrupted in JNCL, is poor. Few cellular interactions have been identified for CLN3 and very little is known of the signalling pathways that require CLN3 function. Consequently, the cell biology underlying the disease remains poorly understood and options for therapeutic intervention for this fatal disorder are non-existent. To expand the animal models available to study CLN3 biology, we describe the characterization of Drosophila CLN3. We find that Drosophila CLN3 shows considerable similarity to the human form, with significant stretches of homology and an intracellular location that corresponds to that seen for human CLN3. To begin to identify the functional pathways within which CLN3 may function, we have utilized the opportunities provided by Drosophila to develop a gain-of-function genetic system. This screen has identified novel interactions between CLN3 and both Notch and JNK signalling pathways. Furthermore, we have demonstrated interactions between CLN3 and an RNA-binding protein involved in polarity, RNA transport and stability.
Drosophila CLN3 shares many of the properties of human CLN3
The Drosophila CLN3 protein shows considerable homology with the human and mammalian forms and can be fitted to the most current topology predictions. Homology is especially strong in the predicted transmembrane domains, in the large second lumenal loop and in the C-terminal intracellular domain. The Drosophila CLN3 transcript is expressed throughout the animal (FlyAtlas, www.flyatlas.org), in common with that observed for mammalian versions of the gene. When expressed in human HEK293 cells, N-terminally tagged forms of Drosophila CLN3 protein localize partially to late endosomes or lysosomes. On occasion, some CLN3 appeared on the plasma membrane and was also localized within Rab11-positive recycling vesicles. Although Drosophila CLN3 is found in the same intracellular compartments as human CLN3, we consider these results as indicative only because the CLN3 proteins were both tagged and overexpressed. Moreover, fusion of tags to the C-terminus of Drosophila CLN3 led to retention of the protein in the ER (data not shown) as has been reported previously for human CLN3 (22). An antiserum will be required to confirm the localization of the endogenous protein in vivo.
The majority of JNCL patients have a 1 kb deletion within the CLN3 locus that potentially could express a truncated CLN3 protein. This truncated protein may or may not retain some function (46,47). Supporting previous findings (22), we find that similarly truncated forms of either human or Drosophila CLN3 protein fail to localize correctly. The proteins localize into a reticular network, suggesting that they fail to fold correctly and are retained in the ER.
Although JNCL is a recessive trait generally thought to be due to a loss-of-gene function, it is possible to identify functional properties of the CLN3 protein by creating a gain-of-function condition. Overexpresssion of either Drosophila or human CLN3 in the developing Drosophila eye cause neurodegenerative phenotypes, suggesting that both proteins have similar functional properties and activate or inhibit similar downstream pathways. This neurodegenerative activity is specific to the overexpressed gene because the phenotype caused by the increased Drosophila CLN3 is suppressed by co-expression of a Drosophila CLN3 RNAi transgene. In addition, overexpression of the truncated forms of both human and Drosophila CLN3 does not show any neurodegenerative phenotype, suggesting that the observed phenotypes are produced by the functional protein and that the truncated versions have lost this activity. It would seem likely, therefore, that Drosophila CLN3 shares functions with its mammalian counterpart and will prove to be an excellent model for understanding CLN3 cell biology.
Increase in CLN3 activity modifies Notch and JNK signalling in Drosophila
Drosophila have been used to model a variety of neurodegenerative diseases (reviewed in 16,17). Fly models exists for the infantile and congenital forms of NCL (18,19), whereas Batten disease-like pathologies are also observed in flies bearing mutations in spinster/benchwarmer (48). No mutations exist in Drosophila CLN3 at present and these will be required to fully model JNCL in flies. However we can use Drosophila overexpression as a mechanism to elucidate the types of pathways that are affected by an increase in CLN3 activity. Increased levels of CLN3 cause a neurodegenerative phenotype in the Drosophila eye which is similar to, but more severe than, the phenotype observed when the Drosophila Cln1 gene DmPPT1 is overexpressed in the same tissue (49). In the case of DmPPT1, the loss of photoreceptors is through apoptotic cell death, whereas overexpression of CLN3 seems to cause neurodegeneration via a process that does not require apoptosis. Our preliminary evidence suggests that blocking autophagy also fails to rescue the eye phenotype. The pathways activated to cause this neurodegeneration are under current investigation.
We also identified that when CLN3 is expressed at high levels in all cells in Drosophila, phenotypes reminiscent of a loss of Notch activity are observed. This ability of increased CLN3 to block Notch activity was confirmed by demonstrating that overexpression of CLN3 in the wing inhibits the activation of a Notch target gene, cut. This inhibition of Notch is not rescued by re-expression of full-length Notch but is rescued by Notchintra, the active intracellular fragment of Notch that signals to the nucleus. This suggests that increased levels of CLN3 might affect the regulation of Notch processing or cleavage but probably not expression. Interestingly, we observed Notch inhibition only in certain developmental contexts, rather than a global inhibition throughout the development of the fly, suggesting some degree of cell specificity. Notch mutations underlie several human cognitive disorders and the Notch signalling pathway has been implicated in the regulation of synaptic plasticity and memory (reviewed in 50,51). Better understood is the major role that Notch signalling plays in a wide variety of developmental processes in many organ systems, including neurogenesis and gliogenesis, where Notch regulates decisions of cell fate, differentiation and stem cell renewal. With the NCLs being early onset diseases, they may have a developmental origin that manifests itself as an early failure of brain function. Previous studies have implicated early defects in particular brain regions (7) and it may be of interest to investigate whether defects in Notch signalling during development may compromise the normal function of these regions.
The absence of a global influence of CLN3 expression on all Notch signalling led us to investigate whether increased levels of CLN3 may affect any of the other identified pathways that impinge on Notch signalling. Overexpression of CLN3 in varied cell types gave rise to phenotypes that had been previously associated with alterations in JNK signalling—apoptosis of cells in the developing wing and a failure of dorsal closure. Significantly, crosstalk between the Notch and JNK signalling pathways has been reported during dorsal closure (40). We find that the phenotypes seen when CLN3 is overexpressed are considerably enhanced when the activity of bsk, the Drosophila JNK, is simultaneously increased by the co-overexpression of bsk or of the upstream activating regulatory JNK kinase, hep. In addition, overexpression of CLN3 activates the expression of the JNK pathway downstream target, puckered. These observations are the first indication that CLN3 may play a role in regulating JNK activity and so places the protein within a major signal transduction pathway implicated in neuronal disease. JNK signalling is an important regulator of neuronal apoptosis and differentiation and aberrant signalling and has been implicated in various neurodegenerative disorders including Parkinson’s, Alzheimer’s and Huntington’s diseases (reviewed in 52). Support for the association of JNK signalling pathology with NCL disease also comes from the Drosophila model for infantile NCL (INCL), where a gain-of-function screen for modifiers of a neurodegeneration phenotype induced by DmPPT1 identified misshapen, an upstream kinase in the JNK signalling pathway, and kayak, the Drosophila fos, a component of AP1, a major transcription regulator and target of the JNK pathway (53). The JNK/AP1 pathway has been implicated as a regulator of synaptic activity, dendritic and synapse growth (54–56). Although PPT1 and CLN3 encode very different genes, deficits in their activity lead to INCL and JNCL, respectively, whose pathologies share many common features, making it tempting to speculate that aberrations in JNK signalling may be a common failure leading to synaptic dysfunction. Similarly, loss of spinster/benchwarmer activity in Drosophila results in lipofuscin accumulation and produces synapse growth defects and loss of activity (48,57). It would be interesting to investigate whether JNK signalling is altered in loss-of-function mouse models where alterations in synaptic activity have been speculated to be a possible root cause of Batten’s disease.
Identification of novel CLN3 functional pathways in Drosophila
We have made use of the neurodegeneration phenotype caused by overexpression of CLN3 in the Drosophila eye to perform a non-biased genetic screen to identify novel candidate genes that may form part of the CLN3 functional network. This modifier screen identifies genes that change the phenotypic consequence of CLN3 overexpression when their gene dosage and activity are reduced by half. A number of chromosomal deficiencies were identified that suppress the eye phenotype, suggesting that genes normally required to generate the degenerative phenotype are deleted in these deficiencies. Work is ongoing to identify the individual genes responsible for this suppression. In addition, we identified two deficiencies that enhance the CLN3 phenotype. Testing of individual genes in one of these deficiencies identified that loss of one copy of mago nashi is responsible for this enhancement and suggesting that the mago gene product normally acts antagonistically to the pathways activated by CLN3. mago encodes a conserved component of the EJC that was originally identified for its role in generating polarity in the Drosophila oocyte (43,58). In the absence of mago, there is a failure to transport oskar mRNA to the posterior of the oocyte, suggesting a requirement for Mago to localize RNA (58). Mago requires an interaction with a partner protein Y14/Tsunagi to bind RNA, and a reduction in tsunagi gene dosage also enhances the CLN3 phenotype, suggesting that CLN3 activity influences or is influenced by a pathway-regulating RNA metabolism. Although the Mago/Y14 complex is an essential component of the EJC, it remains unclear whether this splicing role or a role in RNA localization is the most significant function for oogenesis. Transport of mRNA and ribonucleic particles to the synapse is known to be important for synapse function and plasticity because it allows the rapid local translation of crucial components necessary for dendrite/synapse activity. Many of the components necessary for the localization and translational control of these neuronal mRNAs are shared with those that are associated with localized mRNA in the Drosophila oocyte, e.g. Staufen, Barentsz and FMRP (59). In addition, Pumilio, a translational repressor also required for the polarization of the oocyte, has been shown to be associated with neuronal RNPs (59,60) and is necessary for synapse function and dendrite morphogenesis (61–63). We have preliminary evidence that reveals a reduction in pumilio activity suppresses the effect of overexpression of CLN3 in the wing (V.V., R.I.T. and G.T. unpublished data). FMRP, the fragile X mental retardation protein, is also a component of these RNP granules, and evidence for its possible function has come from modelling fragile X in Drosophila where overexpression in the eye also leads to neurodegeneration (64). These results lead to the suggestion that CLN3 may participate in or be regulated by this process of RNA transport. They also demonstrate why this kind of genetic screen will complement existing approaches for identifying the functional networks of CLN3: RNA-binding components of the RNA localization machinery or translation regulators would be more difficult to isolate using, for example, in vitro binding- or yeast-based techniques.
We have demonstrated here that Drosophila CLN3 shares many properties with its human counterpart and that gain-of-function assay provides a valuable tool for studying CLN3 cell biology and for identifying novel interactions. In this study, we have identified a possible involvement for CLN3 in the Notch and JNK signalling pathways, and our dominant modifier screen has identified a potential role for the RNA localization machinery in regulating CLN3 function. These findings are particularly interesting given that misregulation of both JNK signalling and RNA localization affects synapse function and has been associated with neurodegenerative diseases. Further work driving expression of CLN3 in neurons should reveal whether these interactions are important for neuronal function. In addition, further screens in the eye and wing are possible on a large scale and will identify more genetic interactions to direct and instruct future studies on CLN3 cell biology.
MATERIALS AND METHODS
Drosophila stocks and husbandry
Flies were maintained at room temperature on standard cornmeal agar medium. All crosses were maintained at 25°C on standard cornmeal medium or on semi-defined medium (recipes at Bloomington Stock Centre website) except those involving dppGal4, which were maintained at 29°C.
Drosophila stocks detailed in FlyBase (www.flybase.org)
GMRgal4, vgMgal4, enGal4 daGal4 and DlRF were all supplied by the Bloomington Stock Centre. UAS-bsk, UAS-bskDN, UAS-hep, UAS-p35, UAS-CD8::GFP and pucE69 were supplied by J. Ng and pnrMD237 by J. Bateman. The RNAi line to CG5582 was supplied by the Vienna Drosophila RNAi Centre. dpp-Gal4, UAS-Notch full length and UAS-Notchintra were provided by A. Martinez-Arias. UAS-human CLN3 was provided by L. Myllykangas. mago1 was provided by D. St Johnson. UAS-LAMP-HRP and UAS-Hook-RFP were provided by H. Krämer and UAS-Spinster-GFP by S. Sweeney. UAS-CD8::Venus::PARP was provided by D. Williams.
Deficiencies
The dominant modifier screen used predominantly defined deficiencies from the DrosDel collection supplied by the Bloomington and Szeged stock centres. Cytologically defined deficiencies from the Bloomington deficiency kits were used to fill coverage gaps.
Unpublished fly stocks
mago69B and tsuΔ18 were gifts from Drs J.Y. Roignant and J. Treisman (Skirball Institute, NYU Medical Institute) prior to publication.
Generation of CLN3 expression constructs and transgenic flies
Entry clones for full-length and truncated forms of Drosophila CLN3 and human CLN3 were amplified from cDNA RE03692 and IMAGE clone 5764535 respectively by PCR using Platinum Pfx polymerase (Invitrogen) and inserted into pENTR (Invitrogen). Error-free clones were transferred to pDEST53 (Invitrogen) for HEK293 transfection or to pTW and pTVW for transgenic flies (Murphy Gateway collection, supplied by the Drosophila Genetic Resource Center, University of Indiana) using LR Clonase II (Invitrogen). Transgenic flies were generated as a service by BestGene Inc., Chino Hills, CA, USA, using standard procedures and the mini-white marker mapped to chromosomes using standard mating schemes.
Cell culture, transfection and immunofluorescence
HEK293 cells were grown in standard DMEM+10% FBS (Invitrogen) at 37°C, 5% CO2. Cells were subcultured 24 h prior to transfection and seeded at subconfluent density onto acid-washed coverslips in six-well dishes. One microgram plasmid was transfected per well using Fugene-6 according to the manufacturer’s protocol (Roche). Cells were grown for a further 48 h before fixation. For immunofluorescence, coverslips were washed once in cytoskeleton buffer (65) and then fixed for 10 min in 4% EM-grade formaldehyde (Polysciences) in cytoskeleton buffer. After washing in PBS, cells were permeabilized for 10 min in 0.1% NP-40 (Thermo Scientific) and then blocked in 1% BSA in PBS for 30 min. Antibodies were diluted in block and applied for 1 h for primaries or for 45 min for secondaries. After each, coverslips were washed five times for 5 min each in PBS with 0.1% NP-40. Finally, coverslips were mounted in Prolong Gold (Invitrogen). All immunofluorescence steps were performed at room temperature.
Wing disc fixation and immunostaining
Third instar Drosophila were dissected in cold cytoskeleton buffer and fixed for 30 min on ice in 4% EM-grade formaldehyde in the same buffer. After rinsing in PBS, fixed discs were permeabilized in PBS containing 0.1% Triton X-100 (PBST) and then blocked for 30–60 min in 1% BSA in PBST. Antibodies were diluted in blocking solution and applied overnight at 4°C on a nutator. Next morning, discs were washed five to six times in PBST and then secondary antibodies applied for 2 h at room temperature. After washing, discs were transferred to Vectashield (Vector Labs) and allowed to equilibrate overnight at 4°C. Finally, discs were fine-dissected in Vectashield and transferred to bridge slides for viewing.
Quantification of CLN3 overexpression
An indication of the levels of overexpression of CLN3 was established by measuring the relative expression levels of CLN3 driven by enGal4 in the posterior half of wing discs versus basal levels in the anterior half of the disc. Indirect immunofluorescence staining with anti-CLN3 was imaged by confocal microscopy. Nine discs were fixed, processed and imaged identically. Five-micrometre optical slices was captured with gain levels set to ensure that all pixel intensities fell within the 4096 levels of grey detectable. ImageJ software was used subsequently to measure the mean fluorescence in 30 × 30 µm2 areas either side of the compartment boundary. Background fluorescence due to the secondary antibody was calculated from discs stained without primary antibody. The relative overexpression was then calculated as the mean ratio of posterior:anterior expression.
Antibodies
Primary antibodies and dilutions used: rabbit anti-GFP 1:2000 (Invitrogen); mouse anti-LAMP1 H4A3 1:10 (Developmental Studies Hybridoma Bank, University of Iowa); mouse anti-Rab11 (BD Biosciences), 1:500; mouse anti-cut 2B10 1:10 (DSHB); rabbit anti-cleaved PARP 1:500 (AbCam); chicken anti-βgal 1:2000 (AbCam). Rabbit anti-Drosophila CLN3 SK4208 was generated as a service by Eurogentec against two peptides corresponding to amino acids 1–15 and 20–34 of the CLN3 protein sequence. The antiserum was affinity-purified against the N-terminal peptide and used at 1:1000 dilution. Secondary antibodies used were Alexa488 and Alexa555 conjugates (Invitrogen) appropriate to the primaries. Each was used at 1:500 dilution from a stock diluted 1:1 in PBS:glycerol. DNA was visualized by incubating with ToPro3 1:35 000 (Invitrogen) for 10 min.
Histology and microscopy
Adult eyes, thorax and abdomen
Flies were placed in 1.5 ml tubes and snap-frozen in liquid N2. After thawing, the flies were photographed using an Olympus DP71 camera mounted on a Leica MZ16 stereo microscope using ambient light to minimize glare.
Adult wings
Wings were removed at the hinge, transferred to 100% isopropanol and then to 1:1 isopropanol:glycerol. After equilibration, they were mounted in a 1:1 glycerol:lactic acid solution on slides. Images were taken on a SpotRT digital camera mounted on Zeiss AxioPlan2 upright microscope.
Immunofluorescence
Images were captured using a Zeiss LSM510 laser scanning confocal microscope. Sequential scanning was used in all examples to prevent bleed through. HEK293 cells were imaged with a ×63 1.4 NA oil immersion objective, with an optical slice thickness set to 0.7 µm. Serial optical slices of 5 µm through wing discs were imaged with a ×20 0.5 NA objective.
Images were processed post-collection with LSM (Zeiss), Velocity (Improvision) and Photoshop (Adobe) software and figures prepared with Photoshop. No gamma correction or filtering was applied to images.
Scanning electron microscopy
SEM was performed in the Centre for Ultrastructural Imaging at Kings College London by Dr Tony Brain. Samples were fixed and critical point dried as described in Wolff (66) and then sputter-coated in gold and imaged using an FEI Quanta 200F microscope operated at 5 kV in high vacuum mode.
Sections of adult eyes
Fixation, toluene blue staining and mounting of adult heads were performed according to the protocol of Dr R. Carthew (Northwestern University) available online at www.bioprotocols.info. Images were collected with a 63× objective as before.
Quantification of scutellar macrochaetae
The number of macrochaetae on the scutellum of each adult flies was counted. Control genotypes were enGal4/+ and enGal4;DlRF/+. Flies of genotype enGal4,UAS-CLN3;+/+ were compared with enGal4,UAS-CLN3;DlRF/+ flies by Mann–Whitney test.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Wellcome Trust (WT082004 to G.T.). Funding to Pay the Open Access Charge was provided by The Welcome Trust.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr Jonathon Cooper for help and advice throughout this project and for critical reading of the manuscript. We thank Dr Tony Brain for performing the SEM, Dr. Alfonso Martinez-Arias for helpful discussions and Dr Joe Bateman for help with eye sections. We thank Drs Jean-Yves Roignant and Jessica Treisman for kindly providing alleles of mago nashi and tsunagi prior to their publication and Drs J. Bateman, H. Krämer, L. Myllykangas, A. Martinez-Arias, J. Ng, D. St Johnston, S. Sweeney and D. Williams and the Bloomington and Vienna stock centres for Drosophila stocks.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Wisniewski K.E., Kida E., Golabek A.A., Kaczmarski W., Connell F., Zhong N. Neuronal ceroid lipofuscinoses: classification and diagnosis. Adv. Genet. 2001;45:1–34. doi: 10.1016/s0065-2660(01)45002-4. [DOI] [PubMed] [Google Scholar]
- 2.Cooper J.D. Progress towards understanding the neurobiology of Batten disease or neuronal ceroid lipofuscinosis. Curr. Opin. Neurol. 2003;16:121–128. doi: 10.1097/01.wco.0000063762.15877.9b. [DOI] [PubMed] [Google Scholar]
- 3.Rakheja D., Narayan S.B., Bennett M.J. Juvenile neuronal ceroid-lipofuscinosis (Batten disease): a brief review and update. Curr. Mol. Med. 2007;7:603–608. doi: 10.2174/156652407781695729. [DOI] [PubMed] [Google Scholar]
- 4.Futerman A.H., van Meer G. The cell biology of lysosomal storage disorders. Nat. Rev. Mol. Cell Biol. 2004;5:554–565. doi: 10.1038/nrm1423. [DOI] [PubMed] [Google Scholar]
- 5.Consortium TIBD. Isolation of a novel gene underlying Batten disease, CLN3. Cell. 1995;82:949–957. doi: 10.1016/0092-8674(95)90274-0. [DOI] [PubMed] [Google Scholar]
- 6.Munroe P.B., Mitchison H.M., O’Rawe A.M., Anderson J.W., Boustany R.M., Lerner T.J., Taschner P.E., de Vos N., Breuning M.H., Gardiner R.M., et al. Spectrum of mutations in the Batten disease gene, CLN3. Am. J. Hum. Genet. 1997;61:310–316. doi: 10.1086/514846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pontikis C.C., Cotman S.L., MacDonald M.E., Cooper J.D. Thalamocortical neuron loss and localised astrocytosis in the Cln3Deltaex7/8 knock-in mouse model of Batten disease. Neurobiol. Dis. 2005;20:823–836. doi: 10.1016/j.nbd.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 8.Pontikis C.C., Cella C.V., Parihar N., Lim M.J., Chakrabarti S., Mitchison H.M., Mobley W.C., Rezaie P., Pearce D.A., Cooper J.D. Late onset neurodegeneration in the Cln3−/− mouse model of juvenile neuronal ceroid lipofuscinosis is preceded by low level glial activation. Brain Res. 2004;1023:231–242. doi: 10.1016/j.brainres.2004.07.030. [DOI] [PubMed] [Google Scholar]
- 9.Eliason S.L., Stein C.S., Mao Q., Tecedor L., Ding S.L., Gaines D.M., Davidson B.L. A knock-in reporter model of Batten disease. J. Neurosci. 2007;27:9826–9834. doi: 10.1523/JNEUROSCI.1710-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katz M.L., Johnson G.S., Tullis G.E., Lei B. Phenotypic characterization of a mouse model of juvenile neuronal ceroid lipofuscinosis. Neurobiol. Dis. 2008;29:242–253. doi: 10.1016/j.nbd.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pearce D.A., Nosel S.A., Sherman F. Studies of pH regulation by Btn1p, the yeast homolog of human Cln3p. Mol. Genet. Metab. 1999;66:320–323. doi: 10.1006/mgme.1999.2819. [DOI] [PubMed] [Google Scholar]
- 12.Gachet Y., Codlin S., Hyams J.S., Mole S.E. btn1, the Schizosaccharomyces pombe homologue of the human Batten disease gene CLN3, regulates vacuole homeostasis. J. Cell Sci. 2005;118:5525–5536. doi: 10.1242/jcs.02656. [DOI] [PubMed] [Google Scholar]
- 13.Kim Y., Ramirez-Montealegre D., Pearce D.A. A role in vacuolar arginine transport for yeast Btn1p and for human CLN3, the protein defective in Batten disease. Proc. Natl Acad. Sci. USA. 2003;100:15458–15462. doi: 10.1073/pnas.2136651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vitiello S.P., Wolfe D.M., Pearce D.A. Absence of Btn1p in the yeast model for juvenile Batten disease may cause arginine to become toxic to yeast cells. Hum. Mol. Genet. 2007;16:1007–1016. doi: 10.1093/hmg/ddm046. [DOI] [PubMed] [Google Scholar]
- 15.Osorio N.S., Carvalho A., Almeida A.J., Padilla-Lopez S., Leao C., Laranjinha J., Ludovico P., Pearce D.A., Rodrigues F. Nitric oxide signaling is disrupted in the yeast model for Batten disease. Mol. Biol. Cell. 2007;18:2755–2767. doi: 10.1091/mbc.E06-11-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bilen J., Bonini N.M. Drosophila as a model for human neurodegenerative disease. Annu. Rev. Genet. 2005;39:153–171. doi: 10.1146/annurev.genet.39.110304.095804. [DOI] [PubMed] [Google Scholar]
- 17.Marsh J.L., Thompson L.M. Drosophila in the study of neurodegenerative disease. Neuron. 2006;52:169–178. doi: 10.1016/j.neuron.2006.09.025. [DOI] [PubMed] [Google Scholar]
- 18.Myllykangas L., Tyynela J., Page-McCaw A., Rubin G.M., Haltia M.J., Feany M.B. Cathepsin D-deficient Drosophila recapitulate the key features of neuronal ceroid lipofuscinoses. Neurobiol. Dis. 2005;19:194–199. doi: 10.1016/j.nbd.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 19.Hickey A.J., Chotkowski H.L., Singh N., Ault J.G., Korey C.A., MacDonald M.E., Glaser R.L. Palmitoyl-protein thioesterase 1 deficiency in Drosophila melanogaster causes accumulation of abnormal storage material and reduced life span. Genetics. 2006;172:2379–2390. doi: 10.1534/genetics.105.053306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nugent T., Mole S.E., Jones D.T. The transmembrane topology of Batten disease protein CLN3 determined by consensus computational prediction constrained by experimental data. FEBS Lett. 2008;582:1019–1024. doi: 10.1016/j.febslet.2008.02.049. [DOI] [PubMed] [Google Scholar]
- 21.Phillips S.N., Benedict J.W., Weimer J.M., Pearce D.A. CLN3, the protein associated with Batten disease: structure, function and localisation. J. Neurosci. Res. 2005;79:573–583. doi: 10.1002/jnr.20367. [DOI] [PubMed] [Google Scholar]
- 22.Jarvela I., Lehtovirta M., Tikkanen R., Kyttala A., Jalanko A. Defective intracellular transport of CLN3 is the molecular basis of Batten disease (JNCL) Hum. Mol. Genet. 1999;8:1091–1098. doi: 10.1093/hmg/8.6.1091. [DOI] [PubMed] [Google Scholar]
- 23.Kyttala A., Yliannala K., Schu P., Jalanko A., Luzio J.P. AP-1 and AP-3 facilitate lysosomal targeting of Batten disease protein CLN3 via its dileucine motif. J. Biol. Chem. 2005;280:10277–10283. doi: 10.1074/jbc.M411862200. [DOI] [PubMed] [Google Scholar]
- 24.Persaud-Sawin D.A., McNamara J.O., II, Rylova S., Vandongen A., Boustany R.M. A galactosylceramide binding domain is involved in trafficking of CLN3 from Golgi to rafts via recycling endosomes. Pediatr. Res. 2004;56:449–463. doi: 10.1203/01.PDR.0000136152.54638.95. [DOI] [PubMed] [Google Scholar]
- 25.Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 26.Stewart B.A. Membrane trafficking in Drosophila wing and eye development. Semin. Cell Dev. Biol. 2002;13:91–97. doi: 10.1016/s1084-9521(02)00013-7. [DOI] [PubMed] [Google Scholar]
- 27.Fischer J.A., Eun S.H., Doolan B.T. Endocytosis, endosome trafficking, and the regulation of Drosophila development. Annu. Rev. Cell Dev. Biol. 2006;22:181–206. doi: 10.1146/annurev.cellbio.22.010605.093205. [DOI] [PubMed] [Google Scholar]
- 28.Bray S. Notch signalling in Drosophila: three ways to use a pathway. Semin. Cell Dev. Biol. 1998;9:591–597. doi: 10.1006/scdb.1998.0262. [DOI] [PubMed] [Google Scholar]
- 29.Hay B.A., Wolff T., Rubin G.M. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- 30.James T., Nichols A.M., Gerry W. Notch Signaling—constantly on the move. Traffic. 2007;8:959–969. doi: 10.1111/j.1600-0854.2007.00592.x. [DOI] [PubMed] [Google Scholar]
- 31.Blair S.S. Wing vein patterning in Drosophila and the analysis of intercellular signaling. Annu. Rev. Cell Dev. Biol. 2007;23:293–319. doi: 10.1146/annurev.cellbio.23.090506.123606. [DOI] [PubMed] [Google Scholar]
- 32.Ludlow C., Choy R., Blochlinger K. Functional analysis of Drosophila and mammalian cut proteins in files. Dev. Biol. 1996;178:149–159. doi: 10.1006/dbio.1996.0205. [DOI] [PubMed] [Google Scholar]
- 33.de Celis J.F., Garcia-Bellido A., Bray S.J. Activation and function of Notch at the dorsal–ventral boundary of the wing imaginal disc. Development. 1996;122:359–369. doi: 10.1242/dev.122.1.359. [DOI] [PubMed] [Google Scholar]
- 34.Seugnet L., Simpson P., Haenlin M. Requirement for dynamin during Notch signaling in Drosophila neurogenesis. Dev. Biol. 1997;192:585–598. doi: 10.1006/dbio.1997.8723. [DOI] [PubMed] [Google Scholar]
- 35.Goto S., Taniguchi M., Muraoka M., Toyoda H., Sado Y., Kawakita M., Hayashi S. UDP-sugar transporter implicated in glycosylation and processing of Notch. Nat. Cell Biol. 2001;3:816–822. doi: 10.1038/ncb0901-816. [DOI] [PubMed] [Google Scholar]
- 36.Ye Y., Lukinova N., Fortini M.E. Neurogenic phenotypes and altered Notch processing in Drosophila presenilin mutants. Nature. 1999;398:525–529. doi: 10.1038/19096. [DOI] [PubMed] [Google Scholar]
- 37.Polaski S., Whitney L., Barker B.W., Stronach B. Genetic analysis of slipper/mixed lineage kinase reveals requirements in multiple Jun-N-terminal kinase-dependent morphogenetic events during Drosophila development. Genetics. 2006;174:719–733. doi: 10.1534/genetics.106.056564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin-Blanco E., Pastor-Pareja J.C., Garcia-Bellido A. JNK and decapentaplegic signaling control adhesiveness and cytoskeleton dynamics during thorax closure in Drosophila. Proc. Natl Acad. Sci. USA. 2000;97:7888–7893. doi: 10.1073/pnas.97.14.7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin-Blanco E., Gampel A., Ring J., Virdee K., Kirov N., Tolkovsky A.M., Martinez-Arias A. puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev. 1998;12:557–570. doi: 10.1101/gad.12.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zecchini V., Brennan K., Martinez-Arias A. An activity of Notch regulates JNK signalling and affects dorsal closure in Drosophila. Curr. Biol. 1999;9:460–469. doi: 10.1016/s0960-9822(99)80211-5. [DOI] [PubMed] [Google Scholar]
- 41.Kuranaga E., Miura M. Molecular genetic control of caspases and JNK-mediated neural cell death. Arch. Histol. Cytol. 2002;65:291–300. doi: 10.1679/aohc.65.291. [DOI] [PubMed] [Google Scholar]
- 42.Williams D.W., Kondo S., Krzyzanowska A., Hiromi Y., Truman J.W. Local caspase activity directs engulfment of dendrites during pruning. Nat. Neurosci. 2006;9:1234–1236. doi: 10.1038/nn1774. [DOI] [PubMed] [Google Scholar]
- 43.Kataoka N., Diem M.D., Kim V.N., Yong J., Dreyfuss G. Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon–exon junction complex. EMBO J. 2001;20:6424–6433. doi: 10.1093/emboj/20.22.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao X.F., Nowak N.J., Shows T.B., Aplan P.D. MAGOH interacts with a novel RNA-binding protein. Genomics. 2000;63:145–148. doi: 10.1006/geno.1999.6064. [DOI] [PubMed] [Google Scholar]
- 45.Hachet O., Ephrussi A. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr. Biol. 2001;11:1666–1674. doi: 10.1016/s0960-9822(01)00508-5. [DOI] [PubMed] [Google Scholar]
- 46.Kitzmuller C., Haines R.L., Codlin S., Cutler D.F., Mole S.E. A function retained by the common mutant CLN3 protein is responsible for the late onset of juvenile neuronal ceroid lipofuscinosis. Hum. Mol. Genet. 2008;17:303–312. doi: 10.1093/hmg/ddm306. [DOI] [PubMed] [Google Scholar]
- 47.Chan C.H., Mitchison H.M., Pearce D.A. Transcript and in silico analysis of CLN3 in juvenile neuronal ceroid lipofuscinoses and associated mouse models. Hum. Mol. Genet. 2008;17:3332–3339. doi: 10.1093/hmg/ddn228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dermaut B., Norga K.K., Kania A., Verstreken P., Pan H., Zhou Y., Callaerts P., Bellen H.J. Aberrant lysosomal carbohydrate storage accompanies endocytic defects and neurodegeneration in Drosophila benchwarmer. J. Cell Biol. 2005;170:127–139. doi: 10.1083/jcb.200412001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Korey C.A., MacDonald M.E. An over-expression system for characterizing Ppt1 function in Drosophila. BMC Neurosci. 2003;4:30. doi: 10.1186/1471-2202-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Costa R.M., Drew C., Silva A.J. Notch to remember. Trends Neurosci. 2005;28:429–435. doi: 10.1016/j.tins.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 51.Yoon K., Gaiano N. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat. Neurosci. 2005;8:709–715. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]
- 52.Johnson G.L., Nakamura K. The c-jun kinase/stress-activated pathway: regulation, function and role in human disease. Biochim. Biophys. Acta. 2007;1773:1341–1348. doi: 10.1016/j.bbamcr.2006.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buff H., Smith A.C., Korey C.A. Genetic modifiers of Drosophila palmitoyl-protein thioesterase 1-induced degeneration. Genetics. 2007;176:209–220. doi: 10.1534/genetics.106.067983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanyal S., Sandstrom D.J., Hoeffer C.A., Ramaswami M. AP-1 functions upstream of CREB to control synaptic plasticity in Drosophila. Nature. 2002;416:870–874. doi: 10.1038/416870a. [DOI] [PubMed] [Google Scholar]
- 55.Etter P.D., Narayanan R., Navratilova Z., Patel C., Bohmann D., Jasper H., Ramaswami M. Synaptic and genomic responses to JNK and AP-1 signaling in Drosophila neurons. BMC Neurosci. 2005;6:39. doi: 10.1186/1471-2202-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hartwig C.L., Worrell J., Levine R.B., Ramaswami M., Sanyal S. Normal dendrite growth in Drosophila motor neurons requires the AP-1 transcription factor. Dev. Neurobiol. 2008;68:1225–1242. doi: 10.1002/dneu.20655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sweeney S.T., Davis G.W. Unrestricted synaptic growth in spinster—a late endosomal protein implicated in TGF-beta-mediated synaptic growth regulation. Neuron. 2002;36:403–416. doi: 10.1016/s0896-6273(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 58.Micklem D.R., Dasgupta R., Elliott H., Gergely F., Davidson C., Brand A., Gonzalez-Reyes A., St Johnston D. The mago nashi gene is required for the polarisation of the oocyte and the formation of perpendicular axes in Drosophila. Curr. Biol. 1997;7:468–478. doi: 10.1016/s0960-9822(06)00218-1. [DOI] [PubMed] [Google Scholar]
- 59.Barbee S.A., Estes P.S., Cziko A.M., Hillebrand J., Luedeman R.A., Coller J.M., Johnson N., Howlett I.C., Geng C., Ueda R., et al. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006;52:997–1009. doi: 10.1016/j.neuron.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vessey J.P., Vaccani A., Xie Y., Dahm R., Karra D., Kiebler M.A., Macchi P. Dendritic localisation of the translational repressor Pumilio 2 and its contribution to dendritic stress granules. J. Neurosci. 2006;26:6496–6508. doi: 10.1523/JNEUROSCI.0649-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ye B., Petritsch C., Clark I.E., Gavis E.R., Jan L.Y., Jan Y.N. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr. Biol. 2004;14:314–321. doi: 10.1016/j.cub.2004.01.052. [DOI] [PubMed] [Google Scholar]
- 62.Menon K.P., Sanyal S., Habara Y., Sanchez R., Wharton R.P., Ramaswami M., Zinn K. The translational repressor Pumilio regulates presynaptic morphology and controls postsynaptic accumulation of translation factor eIF-4E. Neuron. 2004;44:663–676. doi: 10.1016/j.neuron.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 63.Muraro N.I., Weston A.J., Gerber A.P., Luschnig S., Moffat K.G., Baines R.A. Pumilio binds para mRNA and requires Nanos and Brat to regulate sodium current in Drosophila motoneurons. J. Neurosci. 2008;28:2099–2109. doi: 10.1523/JNEUROSCI.5092-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wan L., Dockendorff T.C., Jongens T.A., Dreyfuss G. Characterization of dFMR1, a Drosophila melanogaster homolog of the fragile X mental retardation protein. Mol. Cell Biol. 2000;20:8536–8547. doi: 10.1128/mcb.20.22.8536-8547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Biyasheva A., Svitkina T., Kunda P., Baum B., Borisy G. Cascade pathway of filopodia formation downstream of SCAR. J. Cell Sci. 2004;117:837–848. doi: 10.1242/jcs.00921. [DOI] [PubMed] [Google Scholar]
- 66.Wolff T. In: Drosophila Protocols. Sullivan W., Ashburner M., Hawley R.S., editors. Cold Spring Harbor Laboratory Press; 2000. pp. 229–243. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.