Abstract
Replication and repair of DNA require equilibrated pools of deoxynucleoside triphosphate precursors. This concept has been proven by in vitro studies over many years, but in vivo models are required to demonstrate its relevance to multicellular organisms and to human diseases. Accordingly, we have generated thymidine phosphorylase (TP) and uridine phosphorylase (UP) double knockout (TP−/−UP−/−) mice, which show severe TP deficiency, increased thymidine and deoxyuridine in tissues and elevated mitochondrial deoxythymidine triphosphate. As consequences of the nucleotide pool imbalances, brains of mutant mice developed partial depletion of mtDNA, deficiencies of respiratory chain complexes and encephalopathy. These findings largely account for the pathogenesis of mitochondrial neurogastrointestinal encephalopathy (MNGIE), the first inherited human disorder of nucleoside metabolism associated with somatic DNA instability.
INTRODUCTION
The notion that a balanced pool of deoxyribonucleoside triphosphate (dNTP) precursors is required for DNA replication and repair has been studied in vitro for more than four decades (1,2), but its relevance to human disease was not documented until 1999, when mutations on the TYMP gene were identified as the cause of mitochondrial neurogastrointestinal encephalopathy (MNGIE). This autosomal-recessive disease is characterized by extraocular muscle weakness manifesting as ptosis and ophthalmoplegia, peripheral neuropathy, gastrointestinal dysmotility causing severe cachexia, leukoencephalopathy and mitochondrial abnormalities (3–5). TYMP mutations cause a severe decrease of thymidine phosphorylase (TP) activity; marked elevations of the pyrimidine nucleosides thymidine (Thd) and deoxyuridine (dUrd) in plasma and tissues and somatic multiple deletions, depletion and site-specific point mutations of mtDNA (3,4,6–12). The point mutations are predominantly T-to-C transitions preceded by 5′-AA sequences, suggesting that elevated deoxythymidine triphosphate (dTTP) induces next-nucleotide effects in this disease (9). On the basis of these findings, we hypothesized that elevations of Thd and dUrd in MNGIE cause nucleotide pool imbalance, which, in turn, causes mtDNA instability (4,7). The relationship between increased Thd levels, unbalanced mitochondrial dNTPs and mtDNA alterations has been demonstrated in vitro. HeLa cells and human fibroblasts exposed to 10–50 µm Thd showed unbalanced mitochondrial dNTPs and mtDNA deletions and depletion, which were more prominent in quiescent cells (13–15).
Although the in vitro results support our hypothesis for the pathogenesis of MNGIE, an in vivo model is necessary to validate this mechanism, understand the tissue specificity and test potential therapies. To address these issues, we have generated TP knockout (TP−/−) mice, which have no observable clinical phenotype, but show modest elevations of Thd and dUrd. Because murine uridine phosphorylase (UP) degrades Thd and dUrd, we obtained UP knockout (UP−/−) mice (16) and have generated TP/UP double knockout (TP−/−UP−/−) mice. Similar TP−/−UP−/− mice generated by Haraguchi et al. (17), that show leukoencephalopathy without depletion or deletions of mtDNA, were not available. In the present work, we characterize the biochemical, genetic and histological traits of our TP−/−UP−/− mice, which reproduce features of MNGIE.
RESULTS
Disruption of Tymp gene and generation of TP−/−UP−/− mice
We generated a TP−/− mouse by electroporation of a DNA cassette, which comprised a phosphoglycerate kinase promoter and neomycin resistance gene flanked by LoxP sites (PGK-Neo cassette, LPN) disrupting the TP consensus sequence located in exon 4 of the mouse Tymp gene into embryonic stem (ES) cells (Supplementary Material, Fig. S1). Homologous recombinants were identified by polymerase chain reaction (PCR) screening and confirmed by Southern blot analysis. Mutant ES cells were microinjected into blastocysts and produced four chimeric mutant mice. We successfully obtained germ-line transmission of the mutation and generated homozygous mutant mice (Supplementary Material, Fig. S1).
To avoid remote effects of the PGK-Neo cassette inserted into Tymp gene, we have successfully floxed out the cassette by crossing the TP-knockout mice with CRE-expressing mice (Supplementary Material, Fig. S1). We have backcrossed our mice with C57BL6/J mice for 10 generations to produce TP−/− animals with a homogeneous nuclear DNA background. TP−/− mice have no observable clinical phenotype with increased Thd in lung and dUrd in spleen, but no significant changes of nucleosides in the brain, muscle, small intestine and kidney (Fig. 1).
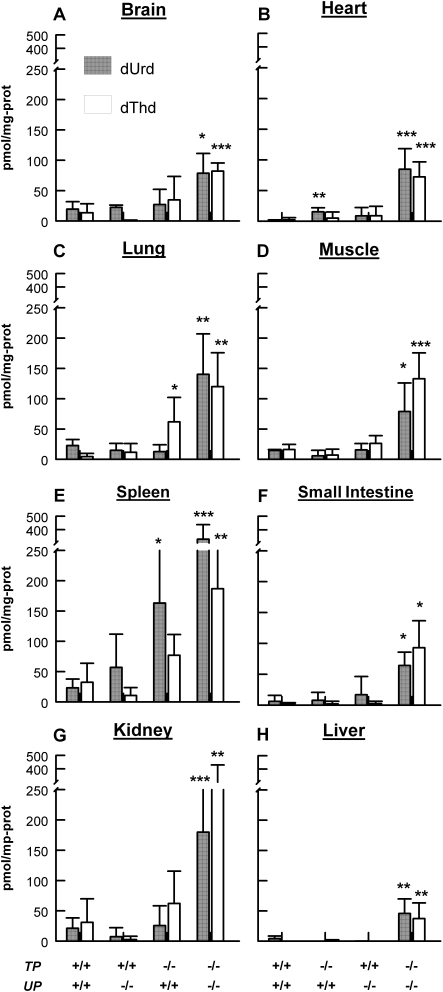
Figure 1.
Thd and dUrd levels were increased in all analyzed tissues of TP−/−UP−/− mice compared with wild-type (TP+/+UP+/+) mice. Bars patterns representing different mice are indicated at the bottom of the figure: TP+/+UP+/+=wild type, TP+/+UP−/−=UP knockout, TP−/−UP+/+=TP knockout, TP−/−UP−/−=TP/UP double knockout. (A) Brain. (B) Heart. (C) Lung. (D) Muscle. (E) Spleen. (F) Small Intestine. (G) Kidney. (H) Liver. All mice were sacrificed between ages 14–18 months. Data are expressed as mean ± standard deviation of five mice per group. *P< 0.05; **P < 0.01; and ***P < 0.001 versus TP+/+UP+/+.
Because murine UP degrades Thd and dUrd, we crossed our TP−/− mice with UP−/− mice (knock out of the Upp1 gene encoding UP) (16) to obtain TP+/−UP+/− mice and intercrossed these mice to generate TP−/−UP−/− mice. The cumulative genotype ratios (+/+: +/−: −/−) in the offspring followed the expected Mendelian distribution (1:2:1), indicating that TP−/−UP−/− is not embryonically lethal. The double knockouts are fertile, exhibit normal growth and lifespan, but develop kyphosis at the age of 5 months.
Undetectable TP activity and increased levels of Thd and dUrd in TP−/−UP−/− mice tissues
TP−/−UP−/− mice showed undetectable TP activity in the brain, heart, lung, muscle, spleen, small intestine and kidney, but 17% in liver relative to TP+/+UP+/+ mice (Table 1). As a consequence of the TP deficiency, in all tissues of the double-knockout animals, Thd and dUrd were increased 4–65-fold relative to wild-type littermates (Fig. 1); and the highest levels of both nucleosides were detected in spleen, kidney, lung and muscle and the lowest in liver, small intestine, heart and brain (Fig. 1). In contrast, UP−/− mice showed a significant increase of only dUrd in heart.
Table 1.
TP activity in mice tissues (nmol/h/mg prot)
| TP+/+ UP+/+ | TP−/− UP−/− | |
|---|---|---|
| Brain | 81.6 ± 61.94 | UND |
| Heart | 12.3 ± 2.83 | UND |
| Lung | 51.7 ± 23.19 | UND |
| Muscle | 1.2 ± 1.93 | UND |
| Spleen | 17.1 ± 2.52 | UND |
| Small intestine | 567.8 ± 190.4 | UND |
| Kidney | 40.2 ± 0.63 | UND |
| Liver | 83.9 ± 36.45 | 14.6 ± 5.70 |
Data are mean ± SD, n = 4.
UND, undetectable.
Unbalanced mitochondrial dNTP in liver and brain of TP−/−UP−/− mice
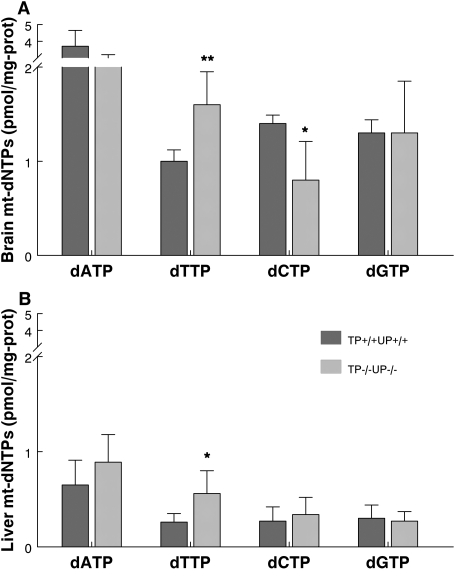
As an unbalance of mitochondrial DNA precursor has been considered the cause of the mtDNA instability in MNGIE, we measured levels of mt-dNTPs in the mouse model. We observed significant increases of dTTP in brain (P < 0.01) and liver mitochondria (P < 0.05) (Fig. 2A and B) and a significant decrease of dCTP in brain mitochondria (P < 0.05) of TP−/−UP−/− mice (Fig. 2A). The ratios of the Tk2-dependent mitochondrial dNTPs, dTTP/dCTP, were higher in mitochondria from brain (2.0) and liver (1.6) of TP−/−UP−/− than in TP+/+UP+/+ brain (0.7) and liver (1.0), indicating that dNTPs imbalance is more severe in brain than in liver.
Figure 2.
TP−/−UP−/− mice show altered mitochondrial dNTPs pools. Levels of dATP, dTTP, dCTP and dGTP in mitochondria from mouse brain (A) and liver (B) are unbalanced in mutant relative to control mice. All mice were sacrificed between ages 14–18 months old. Data are expressed as mean ± SD of 5 mice per group. *P < 0.05 and **P < 0.01 versus TP+/+UP+/+.
To confirm the link between increased Thd and mitochondrial dTTP, we measured the mitochondrial dNTPs in liver 20 min after intraperitoneal injections of Thd (100 mg/kg bw). We noted a 1250% increase of Thd levels in liver over baseline levels in TP−/−UP−/− mice (P < 0.001) (Supplementary Material, Fig. S2A). The elevated Thd provoked a more severe imbalance of mitochondrial dNTPs, with a 2.2-fold increase in the dTTP/dNTP ratio (representing the amount of dTTP relative to total dNTP pool) of Thd-treated versus untreated double-knockout mice (Supplementary Material, Fig. S2B).
MtDNA depletion in the brain of TP−/−UP−/− mice
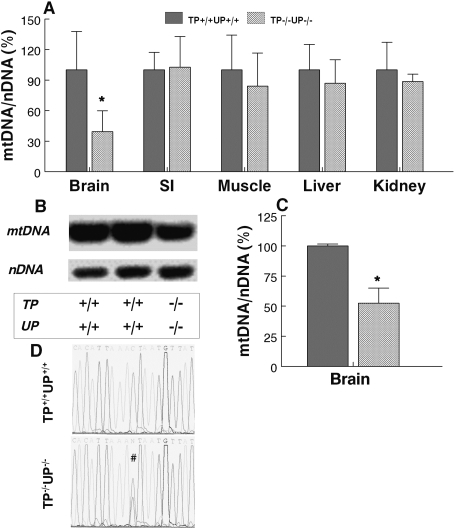
Because MNGIE patients have alterations of mtDNA in affected tissues, we also screened mtDNA for depletion, deletions and point mutations in our mouse model. We observed 27% depletion in the brain of 6-month-old TP−/−UP−/− mice (Supplementary Material, Fig. S3) using quantitative real-time PCR with Taqman® MGB probes for COXI (mtDNA) and GADPH (nDNA). The reduction of mtDNA was more evident in older TP−/−UP−/− mice (14–18 months old), which showed 61% depletion of mtDNA in brain relative to wild-type mice (n = 5 in each group, P < 0.05), but not in other tissues (Fig. 3A). The depletion of mtDNA relative to 18S rRNA (nDNA) in the brain of the oldest mice was confirmed by the Southern blot analysis (Fig. 3B), which demonstrated a 50% reduction of mtDNA relative to nDNA (Fig. 3C) (P < 0.05).
Figure 3.
MtDNA depletion detected in brain of TP−/−UP−/− mice. (A) Quantitative real-time PCR showing mtDNA depletion in brain but not in other tissues. Southern blot image (B) and quantification of band intensities (C) demonstrate mtDNA depletion in brain. (D) Heteroplasmic C-to-T transition at nucleotide 15 588 in the D-loop of a TP−/−UP−/− mouse compared with the TP+/+UP+/+ mouse sequence. All mice sacrificed were between 14 and 18 months old. Data are expressed as mean ± SD of four mice per group. *P < 0.05 versus TP+/+UP+/+.
In contrast, we did not detect deletions of mtDNA in the brain, muscle and small intestine from TP−/−UP−/− mice using long PCR to amplify nearly full-length mouse mtDNA (18) (data not shown).
Brain mtDNA sequence of two TP−/−UP−/− mice and a wild-type brain corresponded precisely to C57BL6/J mtDNA sequence (19), except for a heteroplasmic C-to-T transition at nucleotide (nt) 15 588, which was preceded by three A residues in the D-loop of one mutant mouse (Fig. 3D). This change was also present in the TP−/−UP−/− sibling, but not in non-matrilineal TP−/−UP−/−, TP−/−UP+/+, TP+/+UP−/− or TP+/+UP+/+ mice or SVJ129 ES cells. Published mtDNA sequences from 20 inbred mouse strains (including C57BL6/J) and two mouse cell lines, nt 15 588 contain a cytosine residue; whereas, in three wild-derived inbred strains (CAST/EiJ, MOLF/EiJ and PWD/PhJ) and a divergent common inbred strain (NZB/B1NJ), nt 15 588 harbors a thymine res idue (19,20). These data indicate that the heteroplasmic transition arose in the maternal lineage of the two siblings and is not pathogenic because it is present in four mouse strains, which have the most divergent mtDNA sequences compared with the remaining strains (19). A selection bias against maternal transmission of pathogenic point mutations may be contributing to the absence of coding region mtDNA polymorphisms (21,22).
Sequencing of mtDNA genes ND3, ND6 and Cyt b as well as the D-loop of muscle and small intestine did not reveal point mutations. Because polymorphisms present at <20% heteroplasmy are generally not detected by dideoxy-DNA sequencing, we also performed single-strand conformation polymorphism (SSCP) analysis of PCR-amplified fragments of ND3, ND6, Cyt b and D-Loop of mtDNA from small intestine, but we found no differences between TP−/−UP−/− and TP+/+UP+/+ mice.
Mitochondria respiratory chain deficiency in the brain of TP−/−UP−/− mice
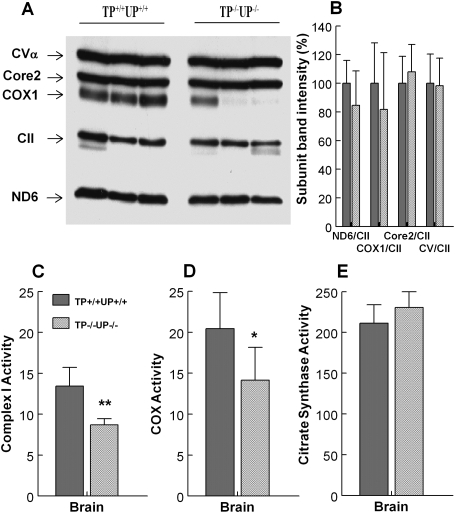
Because brain was the only organ manifesting mtDNA alterations in TP−/−UP−/− mice, we assessed respiratory chain complexes in this tissue biochemically and by western blot. Western blot analysis of periventricular cerebral hemispheres showed a trend toward reduced levels of mtDNA-encoded proteins, ND6 (15% decrease) and COXI (20% decrease), in TP−/−UP−/− mice (n = 7) relative to wild-type mice (n = 5) (Fig. 4A and B). In contrast, levels of nuclear-encoded protein, Core2 and CVα, were similar in TP+/+UP+/+ and TP−/−UP−/− mice. Consistent with the mtDNA depletion in the brain of TP−/−UP−/− mice, this tissue showed ∼30% reduced activities of complexes I (P < 0.005) and IV (P < 0.05) (Fig. 4C and D). Citrate synthase (CS) activity, a marker of mitochondria abundance, was normal in the brain of TP−/−UP−/− mice (Fig. 4E).
Figure 4.
Diminished levels of mtDNA-encoded proteins and decreased mitochondrial respiratory chain function were observed in TP−/−UP−/− mice. (A and B) A trend toward reduced levels of mtDNA-encoded subunits: subunit 6 of NADH dehydrogenase (complex I) and subunit I of cyt c oxidase (complex IV) in periventricular section of the brain were detected by western blot performed with whole-tissue protein extracts (30 µg/lane) from 14- to 18-month-old mice. (A) Representative western blot with three different mice per group. (B) Densitometry of western blot results from TP+/+UP+/+ (n = 5) and TP−/−UP−/− (n = 7) mice. Complexes were detected with Rodent Total OXPHOS Complexes Detection Kit cocktail of antibodies (MitoSciences, Eugene, OR, USA). (C and D) Brain of TP−/−UP−/− mice showed decreased activity of mitochondrial complex I (C) and IV (D) normalized to citrate synthase. (E) Citrate synthase were similar in wild-type and mutant animals. All mice were sacrificed between ages 14 and 18 months. Data are expressed as mean ± SD of five mice per group. **P < 0.01 versus TP+/+UP+/+.
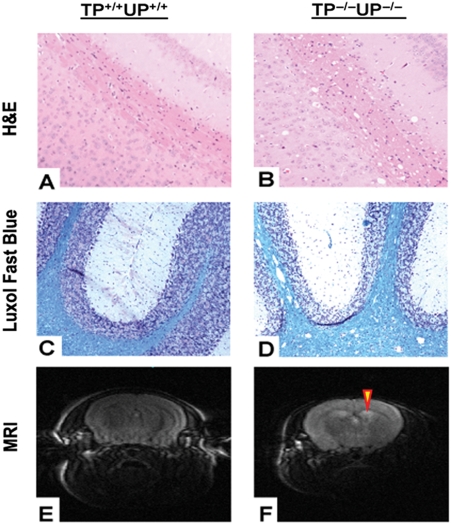
Histology and magnetic resonance imaging
To assess the consequences of the mitochondrial respiratory chain defect in the brain of mutant mice, we performed histological and magnetic resonance imaging (MRI) studies. In young animals (7-month-old TP−/−UP−/− versus 6-month-old TP+/+UP+/+), no histological changes were detected with hematoxylin and eosin (H&E) and Luxol-fast blue (myelin stain) stains (data not shown). In contrast, 16–22-month-old TP−/−UP−/− mice showed white matter abnormalities that were not present in the 11–20-month-old TP+/+UP+/+ mice and consisted of multiple vacuoles in subcortical, periventricular, internal capsule and cerebellar white matter (Fig. 5). The vacuoles were larger and more abundant in the cerebellum than in other brain regions. Basal ganglia and the dentate nucleus in the older double-knockout mice also contained vacuoles, although less prominently than the white matter. Myelin staining showed homogeneous blue intensity throughout the white matter, as well as normal myelinated axons, thus excluding focal demyelination (Fig. 5C and D). MRI showed increased T2 signal in the cerebral white matter of a 22-month-old TP−/−UP−/− mouse (Fig. 5H), compared with an age-matched TP+/+UP+/+ animal (Fig. 5G), an indication of leukoencephalopathy.
Figure 5.
Histology studies and MRI images. (A and C) Correspond to 14 months TP+/+UP+/+ mice. (B and D) correspond to 18 months TP−/−UP−/− mice. (A and B) H&E staining: subcortical white matter of TP−/−UP−/− mice (B) shows abnormal conspicuous vacuoles compared to the TP+/+UP+/+ mice (A); (C and D) Luxol-fast blue staining for myelin revealed white matter vacuoles in TP−/−UP−/− mice (D), whereas the wild-type (C) white matter is normal. All figures are at approximately ×100 magnification. (E and F) T2-weighted MRI images showing increased signal in cerebral white matter adjacent to the lateral ventricles of 22-month-old TP−/−UP−/−, but not TP+/+UP+/+ mice.
DISCUSSION
TP catalyzes the conversion of the nucleosides, Thd and dUrd, to the bases, thymine and uracil, plus 2-deoxy 1-ribose-phosphate. In MNGIE, TP deficiency caused by mutations in the TYMP gene provokes more than 100-fold elevations of Thd and dUrd in plasma and tissues and pathogenic somatic mtDNA alterations. To understand the pathogenic mechanism of MNGIE, we initially generated TP−/− mice, which showed minor alterations of nucleosides in liver and spleen, but not in other tissues and do not manifest a clinical phenotype. Because rodent UP also catalyzes the conversion of Thd and dUrd to thymine and uracil, we crossed the TP−/− animals with UP−/− mice (16) to obtain TP−/−UP−/− mice, which show no abnormalities of growth, development, sexual maturation or reproductive ability, but develop kyphosis at the age of 5 months. TP activity was undetectable in all studied tissues except liver, which had 17% residual activity that is likely due to UP 2, encoded by Upp2 and expressed in murine liver (23). As a consequence of TP deficiency in mutant mice, Thd and dUrd were significantly increased by 4–65-fold in all tissues tested resulting in 2.8- and 1.7-fold elevations of dTTP:dCTP ratios in brain and liver, respectively. Importantly, brain of TP−/−UP−/− mice showed partial mtDNA depletion at 14–18 months, resulting in reduced activities of mitochondrial respiratory chain complexes I and IV. Neuropathological studies revealed late-onset vacuoles in cerebral and cerebellar white matter without demyelination or axonal loss, whereas MRI demonstrated increased T2 signal in the white matter. Thus, the TP−/−UP−/− mice recapitulated several features of patients with MNGIE including TP deficiency, elevated Thd and dUrd in tissues, mtDNA depletion, respiratory chain defects and white matter changes. Moreover, the mutant mice demonstrated dNTP pool imbalances, which have been hypothesized to exist in MNGIE.
In addition to leukoencephalopathy, patients with MNGIE also manifest extraocular muscle weakness, gastrointestinal dysmotility, cachexia, peripheral neuropathy and myopathy; this tissue-specificity suggests that in addition to TP deficiency, other factors dictate organ involvement (3,5,24). This notion is further supported by the observation that, even among siblings with identical TYPM mutations, age-at-onset, sequence of tissue involvement and progression of the disease can vary considerably (24,25). In contrast to patients with multisystemic MNGIE disease, our TP−/−UP−/− mice with ubiquitous TP deficiency and elevated Thd and dUrd in tissues showed mtDNA depletion, respiratory chain defects and histological alterations only in the brain. The brain-specific phenotype in TP−/−UP−/− mice may be due to multiple factors including: (a) shorter life-span of mice compared with humans; (b) relatively modest increases of nucleosides in mutant mice; and (c) vulnerability of post-mitotic neurons. First, the average age-at-onset of symptoms in MNGIE is 18.7 years (range 5 months to 43 years) (5); therefore, the 2-year lifespan of mice may not be sufficient to allow significant accumulation of somatic mtDNA alterations and consequent respiratory dysfunction in most tissues of TP−/−UP−/− mice. Secondly, in contrast to humans who have undetectable levels of Thd and dUrd in tissues and plasma (<0.05 µm), wild-type mice have significant amounts of these pyrimidine nucleosides in tissues; therefore, TP−/−UP−/− mice tissues show only 4–65-fold increases of Thd and dUrd, whereas MNGIE patients show >100-fold increases (11,12). Consequently, mitochondrial dNTP pool imbalances in MNGIE patients should be more dramatic than in TP−/−UP−/− mice and cause mtDNA instability in vulnerable tissues. Thirdly, quiescent cells in the G0 phase (such as post-mitotic neurons) are more dependent on the TK2-mediated mitochondrial pyrimidine salvage pathway than mitotically active cells that express enzymes of the cytosolic salvage pathway, including TK1, and de novo nucleotide synthesis and hence have expanded dNTP pools, particularly during the S-phase, for DNA synthesis in the nucleus and mitochondria (13). In support of this concept, Rylova et al. (26) have shown low expression of TK1 and high levels of TK2 transcript in the brain of 5–6-week-old CD-1 mice. Moreover, differential expression and activities of others enzymes involved in the pyrimidine metabolism, i.e. mitochondrial 5′-deoxynucleotidase 2, are likely to influence in the balance of the mitochondrial dNTP pools and contribute to the tissue-specific effects of TP deficiency.
In vitro studies performed by two independent groups support the notion that elevations of Thd and dUrd in MNGIE cause nucleotide pool imbalance, which, in turn, causes mtDNA instability. HeLa cells exposed to 50 µm Thd for 4 h showed alterations of mitochondrial pools with increased concentrations of dTTP (260% of controls) and deoxyguanosine triphosphate (dGTP) (220%) and decreased deoxyadenosine triphosphate (dATP) (78%) and deoxycytidine triphosphate (dCTP) (43%) (15) and after 8 months the cells developed multiple deletions of mtDNA. In contrast, cultured quiescent skin and lung fibroblasts maintained in 9–40 μm Thd and 0.1% FCS showed 2–5.5-fold increases in cytosolic dTTP pools and 3.7–8.5-fold expansions of mitochondrial dTTP 50–80% depletion of mtDNA without deletions or point mutations of mtDNA after 14–64 days of treatment (13,14). The more pronounced elevation of mitochondrial dTTP pools in cultured cells relative to tissues of TP−/−UP−/− mice may account for the more rapid depletion and accumulation of deletions in vitro.
In contrast to our findings, Haraguchi et al. (17) described leukoencephalopathy with enlarged myelinated fibers in TP−/−UP−/− mice, but no mtDNA abnormalities, leading them to conclude that ‘encephalopathy in MNGIE patients does not appear to be caused by mtDNA alterations’. These apparent discrepancies may be due to several limitations of the first study: (i) the mtDNA probe used to detect mtDNA depletion by Southern blot was not normalized to a nuclear DNA probe, and partial mtDNA depletion may have been overlooked; (ii) lack of screening mtDNA for point mutations; (iii) while the increase of Thd in the brain of their animals was comparable to that found in our TP−/−UP−/− mice, mitochondrial dNTPs were not measured; (iv) mitochondrial function was not analyzed in the brain of mutant mice; (v) experiments were performed in animals of varying ages; and (vi) the etiology of the white matter changes was not clearly defined.
Although leukoencephalopathy was identified in the prior TP−/−UP−/− mice study, demyelination was not observed by standard light microscopy but rather enlarged myelin sheaths with normal axons were detected by ultrastructural analyses (17). In contrast, neuropathologic studies in our TP−/−UP−/− mice revealed a late-onset vacuolar leukoencephalopathy not associated with demyelination affecting cerebral and cerebellar white matter. In humans, neuropathologic studies of two MNGIE patients (27) revealed mild cerebral demyelination without cerebellar involvement and prominent accumulations of albumin in neurons and astrocytes, indicating impairment of the blood-brain barrier. White matter involvement has also been observed in other mitochondrial disorders including Kearns–Sayre syndrome (KSS), MELAS, Leigh syndrome, Leber hereditary optic neuropathy and complex I deficiencies (28–30). Importantly, KSS manifests prominent spongiform degeneration of the cerebral and cerebellar white and gray matter with dysfunction of the blood-brain barrier reminiscent of the changes in our TP−/−UP−/− mice (28,29,31). Additionally, in KSS, neuronal loss and degeneration, predominantly in the brainstem and cerebellum, and demyelination in cerebral and cerebellar white matter have been observed. The white matter vacuoles without myelin loss found in our TP−/−UP−/− mice may be due to disruption of the blood-brain barrier causing edema that precedes demyelination. Further neuropathologic studies are necessary to elucidate the nature and evolution of this leukoencephalopathy in this animal model.
In conclusion, this analysis of TP−/−UP−/− mice is the first in vivo study to link increased Thd levels, imbalances of mitochondrial dNTPs, mtDNA depletion and mitochondrial dysfunction. These findings, in large part, account for the pathogenic mechanism of MNGIE, the first inherited human disease of nucleoside metabolism with somatic DNA instability. Moreover, this knockout mouse represents an important model to further investigate the pathophysiology of leukoencephalopathy in mitochondrial diseases and is a useful tool to assess tissue specificity and develop therapeutic strategies for MNGIE.
MATERIALS AND METHODS
Generation of TP−/−UP−/− mice
A summary is described in Results section and further information is listed in Supplementary Material, Methods.
TP activity assay
TP activity was measured using a high-performance liquid chromatography (HPLC) detection method as described previously (12,32).
Thd and dUrd measurements
dThd and dUrd levels were assessed by a gradient-elution HPLC method as described previously (33), with minor modifications. Briefly, deproteinized samples were injected into an Alliance HPLC system (Waters Corporation) with an Alltima C18NUC reversed-phase column (Alltech) at a constant flow rate of 1.5 ml/min (except where indicated) using four buffers: eluent A (20 mm potassium phosphate, pH 5.6), eluent B (20 mm potassium phosphate–60% methanol, pH 5.6), eluent C (water) and eluent D (methanol). Samples were eluted over 60 min with a gradient as follows: 0–5 min, 100% eluent A; 5–25 min, 100–71% eluent A, 29% eluent B; 25–26 min, 0–100% eluent D; 26–30 min, 100% eluent D; 30–31 min, 0–100% eluent C; 31–35 min, 100% eluent C (1.5–2 ml/min); 35–45 min, 100% eluent C (2 ml/min); 45–46 min, 100% eluent C (2-1.5 ml/min); 46–47 min, 0–100% eluent D; 47–50 min, 100% eluent D; 50–51 min, 0–100% eluent A; and 51–60 min, 100% eluent A.
Absorbance of the eluates were monitored at 267 nm and dThd and dUrd peaks were quantified by comparing their peak areas with a calibration curve obtained with aqueous standards. For definitive identification of dThd and dUrd peaks for each sample, we used a second aliquot treated with excess of purified E. coli TP (Sigma) to specifically eliminate Thd and dUrd. The detection limit of this method is 0.05 μmol/l for both deoxynucleosides.
Mitochondria isolation and mt-dNTPs pool determination
We measured mt-dNTPs pool by the DNA polymerase extension assay as described previously (34,35). Briefly, liver and brain homogenates were centrifuged at 1000g for 3 min at 4°C (twice) and supernatants were centrifuged at 9000g (liver) or 15 000g (brain) for 5 min at 4°C (twice). Mitochondrial pellets from brain and liver were re-suspended in 200 µl of cold water and an aliquot of 10 µl was used to measure proteins. Then, the dNTPs were extracted with 60% methanol and after evaporation the dry residue was re-suspended in 60 µl of water (34,35). To measure the dNTPs pool, 2 µm of [3H]-dATP or [3H]-dTTP was used in each reaction. dNTP pools could not be assessed in mitochondria of other tissues due to insufficient yield of isolated organelles.
Intraperitoneal Thd administration
An amount of 100 mg/kg body weight was injected intraperitoneally to each mouse. Because pharmacokinetics of Thd and its analogs show maximal absorption at 20–30 min after intraperitoneal administration (34,36), we sacrificed our mice 20 min after injection. Tissues were collected and processed to determine the Thd levels and mitochondrial dNTPs as described above.
mtDNA quantification
Mouse mtDNA was quantitated by real-time PCR using an ABI PRISM 7000 sequence detection system as described using primers and probes for murine COX I gene (mtDNA) and mouse glyceraldehyde-3-phosphate dehydrogenase (nDNA) (37). The values mtDNA levels were normalized by nDNA, and the data were expressed in terms of percent relative to wild-type mice.
Brain mtDNA was also assessed by Southern blot as described with slight modifications (10). Briefly, 5 μg of total brain DNA was digested with the restriction enzyme NcoI (New England Biolabs, USA) to linearize mtDNA by cutting the circular molecule at unique site 9217–9221. The DNA samples were subjected to electrophoresis through a 0.8% agarose gel and transfer to nylon membrane (BioRad, Hercules, CA, USA). The PCR-amplified templates for the hybridization probes corresponding to whole linearized mtDNA and mouse 18S rRNA (nDNA) (Ambion, USA) were labeled with α-32P-dATP (Random Primed DNA Labeling kit, Roche Diagnostic Corp.). The signals were analyzed in the Molecular Imager System GS-363 (Biorad) to quantitate the mtDNA and nDNA bands.
MtDNA deletions
MtDNA deletions were detected by long PCR as described previously (18). Briefly, mouse mtDNA was amplified from 100 ng of total DNA with the primers at nucleotides 1953–1924 (forward) and 2473–2505 (reverse) of mouse mtDNA and using TaKaRa LA Taq™ (Takara Bio Inc., Japan) with buffer 2 and the following PCR conditions: 98°C for 10 s and 68°C for 15 min, 30 cycles.
MtDNA point mutations
PCR products of the brain mtDNA described above were purified using QIAquick PCR Purification Kit (QIAGEN Inc., USA). The resultant products were used to amplify 28 overlapping segments of 400–650 bp covering 95.6% of the entire mtDNA. The remained 4.4% (819 bp) were amplified directly from brain mtDNA in two overlapping segments that were directly sequenced. Moreover, we determined the mtDNA regions (ND3, ND6, cyt b and D-Loop) with highest frequency of (A)nT sequences, which are commonly mutated in MNGIE patients (9). We sequenced those mtDNA regions in muscle and small intestine. All PCR were performed with 400 pm forward primer (Supplementary Material, Tables S1, S2 and S3) and 400 pm reverse primer (Supplementary Material, Tables S1, S2 and S3). In addition, we performed SSCP analysis of small fragments of ND3, ND6, cyt b and D-Loop in small intestine DNA as described elsewhere (38,39).
Western blot analyses
Periventricular section of the brains were dissected, and the extracts were obtained in 0.5 mm PMSF, 50 mm Tris/HCl (pH 7.6), 2 mm DTT, 5 mm benzamidin, 5% NP-40 and 2.5% Glycerol. Thirty micrograms of periventricular extracts were electrophorezed in an SDS–12% PAGE gel, transferred to Immun-Blot™ PVDF membranes (Biorad) and probed with Rodent Total OXPHOS Complexes Detection Kit cocktail of antibodies (MitoSciences, Eugene, OR, USA). Protein–antibody interactions were detected with peroxidase-conjugated mouse anti-mouse IgG antibody (Sigma-Aldrich, St. Louis, MO, USA), using SuperSignal® chemi-luminiscence detection kit (Thermo Fisher, Waltham, MA, USA). Quantification of proteins was carried out using NIH ImageJ 1.37V software.
Mitochondrial respiratory chain complexes activities
To measure the complex activities, 40–70 mg tissue was homogenized in 500 µl of CPT medium, sonicated during 10 s and centrifuged at 1000g for 10 min at 4°C. The supernatant was used for protein determination and enzymatic assays. Complex I (CI) activity was measured following the oxidation of NADH at 340 nm (40) and subtracting residual activity in the presence of rotenone (10 µg/ml) from total activity. Complex IV (COX) activity was measured following the reduction of cyt c at 550 nm (41). Citrate synthase activity was measured following the reduction of 100 mm 5,5′-dithiobis(2-nitrobenzoic acid) at 412 nm (30°C) in the presence of 60 µg of sample protein (41). Results of CI and COX activities were normalized to CS activity.
Histological studies
Six mice, three TP+/+UP+/+ (sacrificed at ages 7, 11 and 14 months) and three TP−/−UP−/− (sacrificed at 6, 17 and 18 months), were analyzed. Brains were removed, fixed in formalin, and embedded on paraffin. Serial sections of brains were cut in a cryostat at 6 µm thickness and were deparaffinized with xylene. To evaluate neuropathologic alterations, sections were stained with hematoxylin and eosin (H&E) and Luxol-fast blue (myelin), according to the standard methods.
Immunostains were applied to sections of the cerebral cortex, white matter, hippocampus, basal ganglia, choroid plexus and cerebellum. The intensity of the immunostain was analyzed using a computer-assisted image analysis system (42). Sections were examined at ×100–400 magnifications with an Olympus BX51 microscope, and the images were scanned under equal light conditions with the Qcapture computer program.
MRI analysis
MRI studies were conducted on a General Electric 3.0 T whole-body MRI scanner with a high-performance, actively shielded gradient coil system capable of magnetic field gradients of up to 50 mT/m and slew rates of up to 200 mT/m/s. Radiofrequency transmission and signal detection were performed with a 20 × 14 mm elliptical surface coil that was placed directly over the mouse head. Scans were carried out in a custom-designed Plexiglas chamber that allowed continuous delivery of isoflurane gas to keep the animals anesthetized during the measurements. To eliminate motion artifacts, the RF coil and the mouse were secured in a tightly fitting styrofoam former inside the Plexiglas anesthesia chamber, which was in turn secured to the scanner table using Velcro strips. A fast-spin echo (FSE) sequence was used to acquire interleaved multislice T2-weighted, high-resolution mouse brain images in approximately 40 min with: TE/TR = 3000/102 ms, field-of-view = 12 × 20 mm, raw data matrix size = 192 × 320, slice thickness = 1.0 mm, inter-slice gap = 0.5 mm, and 48 excitations per phase-encoding step. The data were zero-filled to yield a final image size of 512 × 512 pixels, with a 60 μm in-plane resolution.
Statistical analysis
Data are expressed as the mean ± SD of five experiments per group. Two-tailed Student’s t-test was used to compare the mean between groups. A P-value of <0.05 was considered to be statistically significant.
Accession codes
The published complete mtDNA sequences of the different mouse strains are available in GenBank: (i) common inbred strains: C57BL6/J (NC_005089), C3H/He (AB049457), AKR/J (AB042432), Balb/cJ (AJ512208), NOD/LtJ (AY533107), SAMR1 (AB042524), SAMP1 (AB042524), SAMP8 (AB042809), BTBRT+ tfJ (EF108334), ALR/Lt (AY533105), ALS/Lt (AY533106), VM (DQ106412), NON/Lt (AY533108), KK/HlJ (EF108339), NZW/LacJ (EF108341), FVB/NJ (EF108338), DBA/2J (EF108337), C3H/HeJ (EF108335), BALB/cByJ (EF108333), 129S1/SvImJ (EF108330), A/J (EF108331); (ii) wild-derived inbred strain: CAST/EiJ (EF108342), PWD/PhJ (EF108343), WSB/EiJ (EF108344), MOLF/EiJ (EF108345); (iii) divergent common inbred strain: NZB/B1NJ (L07095); MilP (L07096); (iv) cell lines: LA91981 (J01442), LA92002 (AY339599), L929 (AJ489607).
SUPPLEMENTARY MATERIAL
FUNDING
The work was supported by grants from the National Institutes of Health (grant number P01NS11766) and Muscular Dystrophy Association USA (grant number 4174). The authors are supported by grants from the Muscular Dystrophy Association, National Institutes of Health, Marriott Mitochondrial Disorder Clinical Research Fund (MMDCRF) and FUNDISMUN Foundation. L.C.L. and A.G.C. were supported by postdoctoral fellowships from the Ministerio de Educación y Ciencia, Spain.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Jorida Coku, Dr. Ali B. Naini and Dr. Catarina M. Quinzii for technical support and Drs. Eric Schon and Salvatore DiMauro for comments on the manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Bridges B.A., Law J., Munson R.J. Mutagenesis in Escherichia coli, II. Evidence for a common pathway for mutagenesis by ultraviolet light, ionizing radiation and thymine deprivation. Mol. Gen. Genet. 1968;103:266–273. doi: 10.1007/BF00273698. [DOI] [PubMed] [Google Scholar]
- 2.Kunz B.A., Kohalmi S.E., Kunkel T.A., Mathews C.K., McIntosh E.M., Reidy J.A. International Commission for protection against environmental mutagens and carcinogens. Deoxyribonucleoside triphosphate levels: a critical factor in the maintenance of genetic stability. Mutat. Res. 1994;318:1–64. doi: 10.1016/0165-1110(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 3.Hirano M., Silvestri G., Blake D.M., Lombes A., Minetti C., Bonilla E., Hays A.P., Lovelace R.E., Butler I., Bertorini T.E., et al. Mitochondrial neurogastrointestinal encephalomyopathy (MNGIE): clinical, biochemical, and genetic features of an autosomal recessive mitochondrial disorder. Neurology. 1994;44:721–727. doi: 10.1212/wnl.44.4.721. [DOI] [PubMed] [Google Scholar]
- 4.Nishino I., Spinazzola A., Hirano M. Thymidine phosphorylase gene mutations in MNGIE, a human mitochondrial disorder. Science. 1999;283:689–692. doi: 10.1126/science.283.5402.689. [DOI] [PubMed] [Google Scholar]
- 5.Hirano M., Nishigaki Y., Martí R. MNGIE: a disease of two genomes. Neurologist. 2004;10:8–17. doi: 10.1097/01.nrl.0000106919.06469.04. [DOI] [PubMed] [Google Scholar]
- 6.Papadimitriou A., Comi G.P., Hadjigeorgiou G.M., Bordoni A., Sciacco M., Napoli L., Prelle A., Moggio M., Fagiolari G., Bresolin N., et al. Partial depletion and multiple deletions of muscle mtDNA in familial MNGIE syndrome. Neurology. 1998;51:1086–1092. doi: 10.1212/wnl.51.4.1086. [DOI] [PubMed] [Google Scholar]
- 7.Spinazzola A., Marti R., Nishino I., Andreu A., Naini A., Tadesse S., Pela I., Zammarchi E., Donati M.A., Oliver J.A., et al. Altered thymidine metabolism due to defects of thymidine phosphorylase. J. Biol. Chem. 2002;277:4128–4133. doi: 10.1074/jbc.M111028200. [DOI] [PubMed] [Google Scholar]
- 8.Martí R., Nishigaki Y., Hirano M. Elevated plasma deoxyuridine in patients with thymidine phosphorylase deficiency. Biochem. Biophys. Res. Commun. 2003;303:14–18. doi: 10.1016/s0006-291x(03)00294-8. [DOI] [PubMed] [Google Scholar]
- 9.Nishigaki Y., Marti R., Copeland W.C., Hirano M. Site-specific somatic mitochondrial DNA point mutations in patients with thymidine phosphorylase deficiency. J. Clin. Invest. 2003;111:1913–1921. doi: 10.1172/JCI17828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishigaki Y., Martí R., Hirano M. ND5 is a hot-spot for multiple atypical mitochondrial DNA deletions in mitochondrial neurogastrointestinal encephalomyopathy. Hum. Mol. Genet. 2004;13:91–101. doi: 10.1093/hmg/ddh010. [DOI] [PubMed] [Google Scholar]
- 11.Blazquez A., Martin M.A., Lara M.C., Marti R., Campos Y., Cabello A., Garesse R., Bautista J., Andreu A.L., Arenas J. Increased muscle nucleoside levels associated with a novel frameshift mutation in the thymidine phosphorylase gene in a Spanish patient with MNGIE. Neuromuscul. Disord. 2005;15:775–778. doi: 10.1016/j.nmd.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 12.Valentino M.L., Marti R., Tadesse S., Lopez L.C., Manes J.L., Lyzak J., Hahn A., Carelli V., Hirano M. Thymidine and deoxyuridine accumulate in tissues of patients with mitochondrial neurogastrointestinal encephalomyopathy (MNGIE) FEBS Lett. 2007;581:3410–3414. doi: 10.1016/j.febslet.2007.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferraro P., Pontarin G., Crocco L., Fabris S., Reichard P., Bianchi V. Mitochondrial deoxynucleotide pools in quiescent fibroblasts: a possible model for mitochondrial neurogastrointestinal encephalomyopathy (MNGIE) J. Biol. Chem. 2005;280:24472–24480. doi: 10.1074/jbc.M502869200. [DOI] [PubMed] [Google Scholar]
- 14.Pontarin G., Ferraro P., Valentino M.L., Hirano M., Reichard P., Bianchi V. Mitochondrial DNA depletion and thymidine phosphate pool dynamics in a cellular model of mitochondrial neurogastrointestinal encephalomyopathy (MNGIE) J. Biol. Chem. 2006;281:22720–22728. doi: 10.1074/jbc.M604498200. [DOI] [PubMed] [Google Scholar]
- 15.Song S., Wheeler L.J., Mathews C.K. Deoxyribonucleotide pool imbalance stimulates deletions in HeLa cell mitochondrial DNA. J. Biol. Chem. 2003;278:43893–43896. doi: 10.1074/jbc.C300401200. [DOI] [PubMed] [Google Scholar]
- 16.Cao D., Leffert J.J., McCabe J., Kim B., Pizzorno G. Abnormalities in uridine homeostatic regulation and pyrimidine nucleotide metabolism as a consequence of the deletion of the uridine phosphorylase gene. J. Biol. Chem. 2005;280:21169–21175. doi: 10.1074/jbc.M412343200. [DOI] [PubMed] [Google Scholar]
- 17.Haraguchi M., Tsujimoto H., Fukushima M., Higuchi I., Kuribayashi H., Utsumi H., Nakayama A., Hashizume Y., Hirato J., Yoshida H., et al. Targeted deletion of both thymidine phosphorylase and uridine phosphorylase and consequent disorders in mice. Mol. Cell. Biol. 2002;22:5212–5221. doi: 10.1128/MCB.22.14.5212-5221.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tyynismaa H., Sembongi H., Bokori-Brown M., Granycome C., Ashley N., Poulton J., Jalanko A., Spelbrink J.N., Holt I.J., Suomalainen A. Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Hum. Mol. Genet. 2004;13:3219–3227. doi: 10.1093/hmg/ddh342. [DOI] [PubMed] [Google Scholar]
- 19.Goios A., Pereira L., Bogue M., Macaulay V., Amorim A. mtDNA phylogeny and evolution of laboratory mouse strains. Genome Res. 2007;17:293–298. doi: 10.1101/gr.5941007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bayona-Bafaluy M.P., Manfredi G., Moraes C.T. A chemical enucleation method for the transfer of mitochondrial DNA to rho(o) cells. Nucleic Acids Res. 2003;31:e98. doi: 10.1093/nar/gng100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan W., Waymire K.G., Narula N., Li P., Rocher C., Coskun P.E., Vannan M.A., Narula J., Macgregor G.R., Wallace D.C. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science. 2008;319:958–962. doi: 10.1126/science.1147786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stewart J.B., Freyer C., Elson J.L., Wredenberg A., Cansu Z., Trifunovic A., Larsson N.G. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol. 2008;6:e10. doi: 10.1371/journal.pbio.0060010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansson M. Identification of a novel human uridine phosphorylase. Biochem. Biophys. Res. Commun. 2003;307:41–46. doi: 10.1016/s0006-291x(03)01062-3. [DOI] [PubMed] [Google Scholar]
- 24.Nishino I., Spinazzola A., Papadimitriou A., Hammans S., Steiner I., Hahn C.D., Connolly A.M., Verloes A., Guimarães J., Maillard I., et al. MNGIE: an autosomal recessive disorder due to thymidine phosphorylase mutations. Ann. Neurol. 2000;47:792–800. [PubMed] [Google Scholar]
- 25.Gamez J., Ferreiro C., Accarino M.L., Guarner L., Tadesse S., Marti R.A., Andreu A.L., Raguer N., Cervera C., Hirano M. Phenotypic variability in a Spanish family with MNGIE. Neurology. 2002;59:455–457. doi: 10.1212/wnl.59.3.455. [DOI] [PubMed] [Google Scholar]
- 26.Rylova S.N., Mirzaee S., Albertioni F., Eriksson S. Expression of deoxynucleoside kinases and 5′-nucleotidases in mouse tissues: implications for mitochondrial toxicity. Biochem. Pharmacol. 2007;74:169–175. doi: 10.1016/j.bcp.2007.03.029. [DOI] [PubMed] [Google Scholar]
- 27.Szigeti K., Sule N., Adesina A.M., Armstrong D.L., Saifi G.M., Bonilla E., Hirano M., Lupski J.R. Increased blood–brain barrier permeability with thymidine phosphorylase deficiency. Ann. Neurol. 2004;56:881–886. doi: 10.1002/ana.20302. [DOI] [PubMed] [Google Scholar]
- 28.Oldfors A., Fyhr I.M., Holme E., Larsson N.G., Tulinius M. Neuropathology in Kearns–Sayre syndrome. Acta Neuropathol. 1990;80:541–546. doi: 10.1007/BF00294616. [DOI] [PubMed] [Google Scholar]
- 29.Tanji K., Kunimatsu T., Vu T.H., Bonilla E. Neuropathological features of mitochondrial disorders. Semin. Cell Dev. Biol. 2001;12:429–439. doi: 10.1006/scdb.2001.0280. [DOI] [PubMed] [Google Scholar]
- 30.Lerman-Sagie T., Leshinsky-Silver E., Watemberg N., Luckman Y., Lev D. White matter involvement in mitochondrial diseases. Mol. Genet. Metab. 2005;84:127–136. doi: 10.1016/j.ymgme.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 31.Tanji K., Schon E.A., DiMauro S., Bonilla E. Kearns–Sayre syndrome: oncocytic transformation of choroid plexus epithelium. J. Neurol. Sci. 2000;178:29–36. doi: 10.1016/s0022-510x(00)00354-3. [DOI] [PubMed] [Google Scholar]
- 32.Marti R., Verschuuren J.J., Buchman A., Hirano I., Tadesse S., van Kuilenburg A.B., van Gennip A.H., Poorthuis B.J., Hirano M. Late-onset MNGIE due to partial loss of thymidine phosphorylase activity. Ann. Neurol. 2005;58:649–652. doi: 10.1002/ana.20615. [DOI] [PubMed] [Google Scholar]
- 33.Marti R., Spinazzola A., Tadesse S., Nishino I., Nishigaki Y., Hirano M. Definitive diagnosis of mitochondrial neurogastrointestinal encephalomyopathy by biochemical assays. Clin. Chem. 2004;50:120–124. doi: 10.1373/clinchem.2003.026179. [DOI] [PubMed] [Google Scholar]
- 34.Ferraro P., Nicolosi L., Bernardi P., Reichard P., Bianchi V. Mitochondrial deoxynucleotide pool sizes in mouse liver and evidence for a transport mechanism for thymidine monophosphate. Proc. Natl Acad. Sci. USA. 2006;103:18586–18591. doi: 10.1073/pnas.0609020103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song S., Pursell Z.F., Copeland W.C., Longley M.J., Kunkel T.A., Mathews C.K. DNA precursor asymmetries in mammalian tissue mitochondria and possible contribution to mutagenesis through reduced replication fidelity. Proc. Natl Acad. Sci. USA. 2005;102:4990–4995. doi: 10.1073/pnas.0500253102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noy R., Ben-Zvi Z., Elezra M., Candotti F., Ford H., Jr, Morris J.C., Marquez V.E., Johns D.G., Agbaria R. Pharmacokinetics and organ distribution of N-methanocarbathymidine, a novel thymidine analog, in mice bearing tumors transduced with the herpes simplex thymidine kinase gene. Cancer. Chemother. Pharmacol. 2002;50:360–366. doi: 10.1007/s00280-002-0505-8. [DOI] [PubMed] [Google Scholar]
- 37.Spinazzola A., Viscomi C., Fernandez-Vizarra E., Carrara F., D’Adamo P., Calvo S., Marsano R.M., Donnini C., Weiher H., Strisciuglio P., et al. MPV17 encodes an inner mitochondrial membrane protein and is mutated in infantile hepatic mitochondrial DNA depletion. Nat. Genet. 2006;38:570–575. doi: 10.1038/ng1765. [DOI] [PubMed] [Google Scholar]
- 38.Hamblet N.S., Ragland B., Ali M., Conyers B., Castora F.J. Mutations in mitochondrial-encoded cytochrome c oxidase subunits I, II, and III genes detected in Alzheimer’s disease using single-strand conformation polymorphism. Electrophoresis. 2006;27:398–408. doi: 10.1002/elps.200500420. [DOI] [PubMed] [Google Scholar]
- 39.Sheffield V.C., Beck J.S., Kwitek A.E., Sandstrom D.W., Stone E.M. The sensitivity of single-strand conformation polymorphism analysis for the detection of single base substitutions. Genomics. 1993;16:325–332. doi: 10.1006/geno.1993.1193. [DOI] [PubMed] [Google Scholar]
- 40.Kirby D.M., Thorburn D.R., Turnbull D.M., Taylor R.W. Biochemical assays of respiratory chain complex activity. Methods Cell Biol. 2007;80:93–119. doi: 10.1016/S0091-679X(06)80004-X. [DOI] [PubMed] [Google Scholar]
- 41.Barrientos A. In vivo and in organello assessment of OXPHOS activities. Methods. 2002;26:307–316. doi: 10.1016/S1046-2023(02)00036-1. [DOI] [PubMed] [Google Scholar]
- 42.Tanji K., DiMauro S., Bonilla E. Disconnection of cerebellar Purkinje cells in Kearns–Sayre syndrome. J. Neurol. Sci. 1999;166:64–70. doi: 10.1016/s0022-510x(99)00114-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.