Abstract
Age-related hearing impairment (ARHI), or presbycusis, is the most prevalent sensory impairment in the elderly. ARHI is a complex disease caused by an interaction between environmental and genetic factors. Here we describe the results of the first whole genome association study for ARHI. The study was performed using 846 cases and 846 controls selected from 3434 individuals collected by eight centers in six European countries. DNA pools for cases and controls were allelotyped on the Affymetrix 500K GeneChip® for each center separately. The 252 top-ranked single nucleotide polymorphisms (SNPs) identified in a non-Finnish European sample group (1332 samples) and the 177 top-ranked SNPs from a Finnish sample group (360 samples) were confirmed using individual genotyping. Subsequently, the 23 most interesting SNPs were individually genotyped in an independent European replication group (138 samples). This resulted in the identification of a highly significant and replicated SNP located in GRM7, the gene encoding metabotropic glutamate receptor type 7. Also in the Finnish sample group, two GRM7 SNPs were significant, albeit in a different region of the gene. As the Finnish are genetically distinct from the rest of the European population, this may be due to allelic heterogeneity. We performed histochemical studies in human and mouse and showed that mGluR7 is expressed in hair cells and in spiral ganglion cells of the inner ear. Together these data indicate that common alleles of GRM7 contribute to an individual's risk of developing ARHI, possibly through a mechanism of altered susceptibility to glutamate excitotoxicity.
INTRODUCTION
Age-related hearing impairment (ARHI), or presbycusis, is the most common sensory abnormality of the elderly (1,2). In 1999, the World Health Organization estimated that 580 million people worldwide over the age of 65 experienced hearing loss. It is anticipated that by the year 2020 over one billion people over the age of 60 will be affected by ARHI (http://www.who.int/en/). It has been shown that ARHI can lead to social isolation, depression and cognitive impairment (3). The time of onset and the rate of progression of the hearing loss vary greatly between individuals. Similarly, the functions of the ear associated with this decline, such as elevation of pure tone threshold perception and reduction of word recognition, vary among individuals. The etiology of ARHI is multifactorial and includes both genetic and environmental influences, such as noise and ototoxin exposures and chronic medical conditions (4).
The contribution of genetic factors to ARHI has been clearly demonstrated in humans. A heritability of 0.47 for hearing loss after age 64 was shown in a study of twins and ARHI (5), while the use of extended pedigrees from the National Heart, Lung and Blood Institute's Framingham study demonstrated a heritability estimate of approximately 0.5 for medium and low frequency sensory presbycusis (6). Finally, a Danish study identified a heritability of 40% for self reported hearing loss in twins over age 75 (7).
Whole genome analysis of ARHI has been hampered in the past by the limits of genotyping technology and the limitations of phenotypic characterization in well-studied populations. To date, only a few association studies, based upon candidate gene approaches, have been published. In one study, no association was found between ARHI and DFNA5, a gene responsible for one form of autosomal dominant non-syndromic hearing loss (8). Similarly, no association was found between glutathione-related antioxidant enzyme levels and ARHI (9). Lastly, no association was found between ARHI and the 35delG mutation of GJB2, a common cause of recessive hearing loss (10). In contrast, several single nucleotide polymorphisms (SNPs) in a 13 kb region of KCNQ4 were correlated with ARHI in two independent European subject groups (11). An association between a polymorphism in N-acetyltransferase 2 (NAT2*6A) and ARHI was detected in a small group of patients and recently replicated in a larger population, implicating reactive oxygen species (ROS) in ARHI (12,13). Finally, an association study performed on 2418 ARHI samples derived from nine centers from seven European countries resulted in the identification of a highly significant SNP in the GRHL2 gene. Subsequent fine-mapping of this locus demonstrated that the majority of the associated SNPs reside in intron 1 (14).
Mitochondrial DNA (mtDNA) mutations have long been known to be associated with human diseases, including non-syndromic hearing loss (15). A highly significant increase in mitochondrial mutations has been described in aging human auditory tissue (16). Analyses of human temporal bones have shown that a 4977 bp deletion in mtDNA occurs frequently in patients affected by ARHI (17). Finally, a significantly higher prevalence of ARHI in subjects with mtDNA haplogroups U and K has recently been reported (18).
In mice, there is also evidence for a role of genes in ARHI. Clear mouse strain differences in hearing at an advanced age exist and have led to the identification of at least three loci, Ahl1, Ahl2 and Ahl3 (19–21). The characterization of these ARHI strains demonstrates age-dependent degeneration of the organ of Corti and variable loss of cells in the spiral ganglion and the cochlear lateral wall. The Cdh23ahl allele, a hypomorphic synonymous SNP leading to in-frame skipping of exon 7 at the Ahl1 locus, is common to C57BL/6J, BALB/cByJ and 129S6 strains (22).
Genome-wide association study (GWAS) of thousands of well characterized case and control individuals at hundreds of thousands of markers has elucidated the genetic underpinnings of multiple diseases and disorders, including type 2 diabetes, breast cancer and Crohn's disease among others (23–26). Although the concept of utilizing GWAS to better understand human disease is maturing, it remains both a cost and time intensive experimental paradigm. Our team and others have developed methods based on the genotyping of pooled samples to improve both the speed and cost of GWAS with only minimal impact on our ability to identify existing major genomic associations (27). The first report using this approach on the current generation of high density arrays identified a novel gene associated with human episodic memory performance, and this association has since been independently and externally validated (28,29). For orphan and/or under-funded disorders, the pooling approach represents a viable alternative to traditional individual-based GWAS.
Using a pooling-based genomic association study at over 500,000 SNPs, we have identified, and subsequently validated, a significant risk association at the GRM7 locus for ARHI. We show that GRM7 is expressed in key regions of the ear and that its protein product, mGluR7, is also present at these sites. These findings are important for both the management and possible future therapeutic intervention into presbycusis.
RESULTS
Whole genome association study
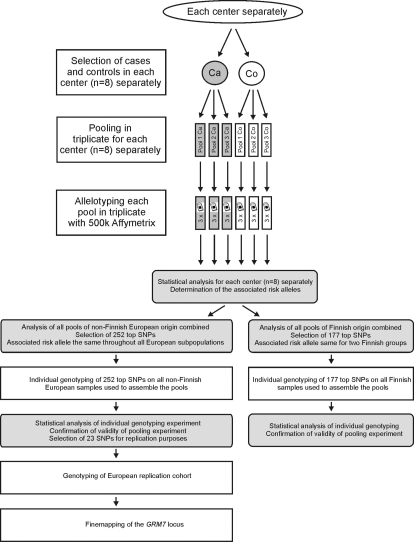
A total of 846 cases and 846 controls out of 3434 individuals collected by eight centers from various European regions, including Finland, were analyzed in this study (Supplementary Material, Table S1). Case and control pools were created in triplicate for each subpopulation (n = 8) separately, and each pool was allelotyped on three replicate Affymetrix Human Mapping 500K arrays containing 506 627 SNPs (Fig. 1).
Figure 1.
Study outline. Laboratory steps are indicated with white rectangles. Statistical analysis steps are indicated with shaded rectangles with rounded corners. Ca, cases; Co, controls.
SNPs from the pooled GWAS were ranked according to a composite score based on their silhouette statistics. For this purpose, each subpopulation was first evaluated separately, which allowed us to determine the associated risk allele for each associated SNP in each subpopulation (Fig. 1). This initial analysis was followed by an analysis of all non-Finnish European samples combined (further called the European group) and an analysis of all Finnish samples combined (further called the Finnish group) (Fig. 1). This was done because the samples collected from Finland (Oulu and Tampere) were considered to be genetically distinct from the remaining European samples. This grouping of samples into two larger sets was based on a previous phylogenetic analysis (13). For the most highly ranked SNPs of this combined analysis, we verified whether the associated risk allele was the same throughout the non-Finnish European subpopulations. If the associated allele was not the same in all subpopulations, that SNP was excluded from further study. The top 252 SNPs identified in the European group and the top 177 SNPs identified in the Finnish group, as ranked by the GenePool software and that fulfilled all criteria, were taken forward for validation by individual genotyping on all samples that were used to assemble the pools. The resulting individual-based χ2 P-values are listed in Supplementary Material, Tables S2 and S3, which also list the pooling ranks. The individual genotyping experiment demonstrated a clear validation of the pooling approach, with more than 35% of the SNPs in the European group and more than 90% of the SNPs in the Finnish group identified in our pooling experiment having P-values less than 0.05 when genotyped individually. DTD1 (D-tyrosyl-tRNA deacylase 1) and PDE9A (phosphodiesterase 9A) were the top-ranked genes for the European group and the Finnish group, respectively. Two genes were significantly associated in both groups: GRM7 (glutamate receptor, metabotropic, 7) and CDH13 (cadherin 13).
Subsequently, a select subset of 23 SNPs (Table 1) was examined in a European replication group consisting of 138 newly recruited cases and controls. After quality control of the genotyping data, only 63 cases and 67 controls were statistically analyzed. SNPs were only included if they passed strict criteria, including (i) being within a gene (with five exceptions), (ii) passing Hardy–Weinberg equilibrium (HWE) measurements at a cut-off of 0.05, (iii) exhibiting an individual call rate >97% and (iv) having a minor allele frequency of a least 5%. In order to correct for multiple testing, we applied Bonferroni correction. Due to linkage disequilibrium (LD), these 23 SNPs represented only 20 true tests, yielding a Bonferroni-corrected significance P-value of 0.0025. One SNP of GRM7 (rs11928865) yielded a P-value of 0.0008 in the European replication group (Table 1). Permutation testing gave a P-value of 0.0011.
Table 1.
Replication genotyping results and statistical analysis
| SNP | Gene | Alleles | Associated allele | HWpval | MAF | Case–control frequencies | Chi-square | P-value | Permuted P-value* |
|---|---|---|---|---|---|---|---|---|---|
| rs11928865 | GRM7 | T:A | T | 0.6287 | 0.32 | 0.793, 0.578 | 12.931 | 8.00E−04 | 0.0011 |
| rs1016730 | ASTN2 | T:A | A | 1 | 0.362 | 0.405, 0.323 | 1.789 | 0.181 | 0.9814 |
| rs6890492 | KIAA0194 | A:G | G | 0.3129 | 0.117 | 0.144, 0.092 | 1.605 | 0.2052 | 0.9897 |
| rs3794261 | METAP2 | G:T | T | 1 | 0.083 | 0.103, 0.063 | 1.272 | 0.2594 | 0.9982 |
| rs2628235 | C:G | G | 0.4231 | 0.045 | 0.061, 0.031 | 1.264 | 0.261 | 0.9983 | |
| rs10193454 | TTC15 | G:A | G | 0.3429 | 0.154 | 0.864, 0.828 | 0.619 | 0.4315 | 1 |
| rs10134299 | RIN3 | T:G | G | 1 | 0.14 | 0.155, 0.127 | 0.397 | 0.5284 | 1 |
| rs2104237 | RIN3 | A:G | G | 1 | 0.138 | 0.153, 0.125 | 0.391 | 0.5318 | 1 |
| rs16834361 | C:T | C | 1 | 0.049 | 0.958, 0.945 | 0.201 | 0.6542 | 1 | |
| rs2295956 | T:G | G | 0.337 | 0.209 | 0.219, 0.200 | 0.137 | 0.7115 | 1 | |
| rs16899799 | FGFR10P | A:G | A | 1 | 0.02 | 0.983, 0.977 | 0.118 | 0.7317 | 1 |
| rs9942 | C:T | C | 0.9165 | 0.143 | 0.864, 0.849 | 0.115 | 0.735 | 1 | |
| rs2280307 | RHOH | G:A | A | 0.5475 | 0.371 | 0.381, 0.362 | 0.104 | 0.747 | 1 |
| rs6035106 | DTD1 | C:T | T | 1 | 0.456 | 0.466, 0.446 | 0.099 | 0.7528 | 1 |
| rs603446 | ZNF259 | C:T | T | 0.4951 | 0.456 | 0.466, 0.446 | 0.099 | 0.7528 | 1 |
| rs2584622 | SMARCD2 | T:C | T | 0.9708 | 0.073 | 0.932, 0.923 | 0.077 | 0.782 | 1 |
| rs1340668 | TINAG | C:T | C | 0.1679 | 0.028 | 0.975, 0.969 | 0.075 | 0.7836 | 1 |
| rs6132107 | DTD1 | A:G | A | 1 | 0.159 | 0.847, 0.836 | 0.061 | 0.8048 | 1 |
| rs8079727 | NLRP1 | A:C | A | 0.4681 | 0.24 | 0.767, 0.754 | 0.06 | 0.806 | 1 |
| rs4813338 | DTD1 | G:A | A | 0.9506 | 0.451 | 0.458, 0.444 | 0.043 | 0.8362 | 1 |
| rs6136423 | DTD1 | G:A | G | 1 | 0.165 | 0.839, 0.831 | 0.03 | 0.8619 | 1 |
| rs10996614 | T:C | C | 1 | 0.024 | 0.025, 0.023 | 0.014 | 0.9044 | 1 | |
| rs9845917 | DLEC1 | T:C | C | 0.9838 | 0.319 | 0.322, 0.315 | 0.013 | 0.9106 | 1 |
HWpval, Hardy–Weinberg P-value; MAF, minor allele frequency.
*The permuted P-value is derived from one million permutations of the data in Haploview.
In both the first European group and the European replication group, the T-allele of rs11928865 (the replicated GRM7 SNP) was associated with ARHI. The frequencies were 76.9% in cases and 71.4% in controls for the first European group, and 79.3% in cases and 57.8% in controls for the European replication group. The odds ratio for this SNP was 2.56 (95% confidence interval of 1.23–5.30) in the replication group. As no Finnish replication subjects could be collected, the SNPs within GRM7, significant in the Finnish group, rs779706 and rs779701, could not be replicated. These latter SNPs could also not be replicated in the European group, nor could the rs11928865 be replicated in the Finnish group. These association signals seem to be independent of each other. The joint analysis P-value for rs11928865 (the replicated GRM7 SNP) in the European, European replication and Finnish group was 9×10E−5. The Cochran–Mantel–Haenzel test yielded a P-value of 0.0001 (Supplementary Material, Table S4).
GRM7 fine mapping
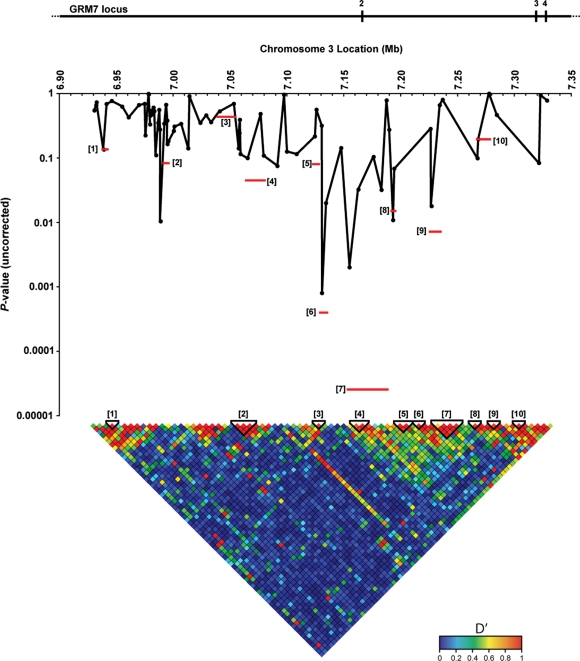
The GRM7 gene measures approximately 880 kb. Fine mapping was restricted to a region of 400 kb surrounding rs11928865. Based on HapMap data, 75 GRM7 tag SNPs were selected and genotyped on all European samples (first European group as well as European replication group). Figure 2 illustrates the fine mapping of the GRM7 locus in the European replication group. rs11928865 remained the most significantly associated individual SNP, while haplotype blocks 7 (consisting of SNPs rs6804466, rs3828472, rs12497688, rs9819783, rs11920109) and 6 (consisting of SNPs rs11928865 and rs9877154) were the most significantly associated haplotype blocks (Table 2). One million permutations of the data resulted in a P-value of 0.0013 and 0.0266 for haplotype blocks 7 and 6, respectively. Because haplotype blocks 6 and 7 were in close proximity and because of the fairly high level of LD between them, we forced both blocks into a single block. This joint 6–7 haplotype had a P-value of 0.0004 which was the most significant of all permuted P-values. We have calculated the odds ratio for the joint 6–7 haplotype in the replication group, resulting in an odds ratio of 12.01 (95% confidence interval of 3.68–20.34). This odds ratio is likely inflated due to the limited size of the replication cohort.
Figure 2.
Finemapping of the GRM7 locus in the European replication group. Haplotype tagging SNPs in the 400 kb region surrounding rs11928865 were identified and assayed. On top of the figure, the positions of the GRM7 exons within the 400 kb region are indicated with numbered vertical bars. The individual significance values for each SNP are indicated with filled black circles on their respective locations within the GRM7 locus. The LD structure is illustrated at the bottom of the figures with the haplotype blocks numbered in brackets. The statistical significances for each haplotype block are shown in the figure as well, with each horizontal red bar representing both the significance level and genomic location.
Table 2.
Associated haplotypes within the GRM7 locus
| Haplotype | Freq. | Case–control ratios | Chi-square | P-value | Permuted P-value* |
|---|---|---|---|---|---|
| Block 6 | |||||
| TC | 0.524 | 71.0:47.0, 59.0:71.0 | 5.421 | 0.0199 | 0.6856 |
| AT | 0.312 | 24.0:94.0, 53.3:76.7 | 12.316 | 4.00E−04 | 0.0266 |
| TT | 0.164 | 23.0:95.0, 17.7:112.3 | 1.561 | 0.2115 | 1 |
| Block 7 | |||||
| CTTCC | 0.361 | 42.0:76.0, 47.5:82.5 | 0.028 | 0.8675 | 1 |
| TGTTT | 0.268 | 23.4:94.6, 43.1:86.9 | 5.599 | 0.018 | 0.6471 |
| CTCTT | 0.145 | 20.9:97.1, 15.0:115.0 | 1.894 | 0.1688 | 0.9999 |
| CTTCT | 0.093 | 20.6:97.4, 2.5:127.5 | 17.729 | 2.55E−05 | 0.0013 |
| CGTTT | 0.071 | 8.0:110.0, 9.6:120.4 | 0.039 | 0.8435 | 1 |
| TTTTT | 0.03 | 0.1:117.9, 7.5:122.5 | 6.68 | 0.0098 | 0.4426 |
| CTTTT | 0.02 | 1.1:116.9, 3.8:126.2 | 1.226 | 0.2682 | 1 |
*The permuted P-value is derived from one million permutations of the data in Haploview.
We have genotyped a large number of closely spaced SNPs across the GRM7 gene locus to further examine the association signal, and this indicated that the putative causative allele is likely positioned within a 150 kb region surrounding GRM7 exon 2. We resequenced exon 2 and the intron–exon boundaries in 100 random ARHI subjects. The only two SNPs that we observed were already known: rs35106713 and rs3749448. Subsequent analysis of these two SNPs on all samples did not result in significant associations (P-value of 0.1 and 0.114, respectively).
mGluR7 expression in the mouse and human inner ear
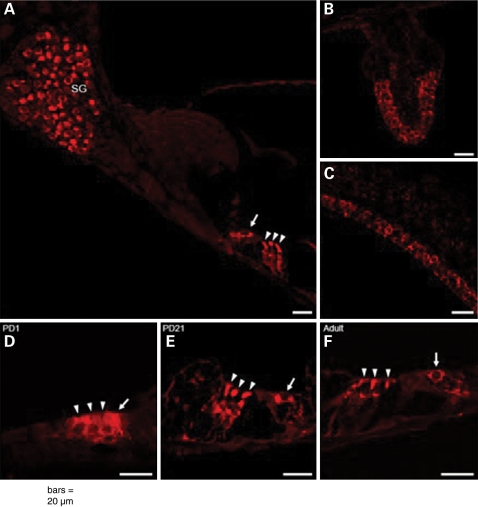
mGluR7 expression was studied by immunohistochemistry in three different stages of development of the mouse inner ear (postnatal day (PD)1, PD21 and adult; Fig. 3). Negative controls without primary antibody and/or without secondary antibody showed no staining (data not shown). In every one of these developmental stages, mGluR7 expression was concentrated in the neural cell bodies of the spiral ganglion (Fig. 3A), in the inner hair cell and outer hair cells of the organ of Corti (Fig. 3A, D, E and F), and the hair cells of the vestibular apparatus: the saccule (Fig. 3C), the utricule (data not shown) and the cristae ampullaris (Fig. 3B). At PD1, while mGluR7 labeling was not as bright as in the older stages, a clear specific signal was detected in the sensory epithelium of the organ of Corti (Fig. 3D), in the hair cells of the vestibular apparatus, and in the spiral ganglion. At PD21 and in adult inner ears, expression of mGluR7 was more abundant. There were no differences observed in staining intensity or staining pattern between basal and apical turns of the cochlea.
Figure 3.
Immunohistochemistry results of mGluR7 in mouse inner ear. (A–C and F) mGluR7 expression in spiral ganglion (SG) neurons (A), IHC (arrow) and OHCs (arrowheads) of the organ of Corti (A and F), hair cells of crista ampullaris (B) and saccule (C) in adult mice—(D–F) comparison of mGluR7 expression in the IHC (arrow) and OHCs (arrowheads) of the organ of Corti at stage PD1 (D), PD21 (E) and in adult (F). Scale bar is 20 µm.
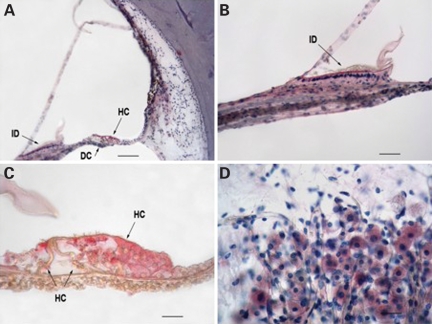
mGluR7 expression was also studied in celloidin embedded adult (83-year-old male with Pure Tone Average of 22 dB) human temporal bone specimens using immunohistochemistry. mGluR7 was detected in the interdental cells of the spiral limbus, the inner and outer hair cells and Hensens’ cells of the organ of Corti and the type II fibrocytes of the spiral ligament (Fig. 4A–C). As in the mouse, mGluR7 was also detected in the spiral ganglion neurons (Fig. 4D).
Figure 4.
Immunohistochemistry results of mGluR7 in human temporal bone of an 83-year-old male with pure tone average of 22 dB. (A) Anterior basal segment of horizontally cut temporal bone labeled with mGluR7 as the primary antibody, biotinylated IgG as the secondary antibody and fast red as the chromagen (MBIC). Hematoxylin (H) was used as a counter stain. Red labeling is visible in the interdental cells of the limbus, the hair and Hensen's cells of the organ of Corti, and fibrocytes in the type II area of the spiral ligament. Moderate distortion is due to difficulty in mounting deceloidinized sections. (B) Higher power showing red labeling of interdental cells of the limbus. (C) Organ of Corti with labeled hair cells and Hensen's cells. (D) Spiral ganglion cells labeled with MBIC & H. Scale bar is 120 µm for (A), 24 µm for (B) and (D), 12 µm for (C). ID, interdental cells; DC, Deiters’ cells; HC, hair and Hensen's cells.
GRM7 mRNA expression in the mouse inner ear
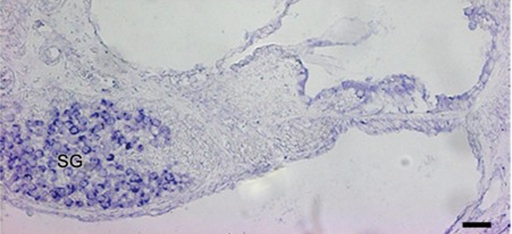
GRM7 mRNA expression was examined by in situ hybridization in the postnatal mouse cochlea (P6). Sense riboprobe showed no staining above background levels (data not shown). Antisense riboprobe demonstrated GRM7 expression in the neurons of the spiral ganglion, but no expression was detected above background in the hair cells or supporting cells of the organ of Corti (Fig. 5). The absence of a signal in these regions, in contrast to the immunohistochemical expression patterns, can likely be explained by low but undetectable mRNA expression in the cochlear epithelium because of the lower sensitivity of in situ hybridization compared to immunohistochemistry.
Figure 5.
In situ hybridization of postnatal (P6) mouse inner ear demonstrating Grm7 mRNA expression in the cells of the spiral ganglion (SG). Scale bar is 20 µm.
DISCUSSION
Presbycusis, or ARHI, was characterized histopathologically by Schuknecht and Gacek (30) in 1993. The two major types, sensory and neural, involve varying degrees of hair cell and cochlear neuronal cell loss and can often be found together. Loss of cochlear neurons is the most consistent finding in the aging ear. Cochlear neuronal counts can be as high as 40 000 in the first decade of life and decrease by more than 50% by the ninth decade in affected individuals (31). The genetic mechanism underlying this age-related loss of approximately 2100 neurons per decade is not understood. The current study provides convincing genetic evidence that variation in a metabotropic glutamate receptor (GRM7) plays a role in ARHI. This includes data obtained in the Finnish population identifying two significant SNPs in GRM7 and significant data obtained in non-Finnish European subjects that replicated in an independent European replication group.
We believe that the GRM7 association is genuine for several reasons. First, we were able to replicate this association in a European replication group. The size of the replication group was small, and it is known that a replication group of smaller size than the original group used for association testing may lead to a lack of power for replication, and subsequent non-replication of results (known as the winner's curse) (32). It is therefore possible that we have missed replication of other genuine results in our replication step. However, the result for GRM7 was significant. A second strong argument is that we have found association for GRM7 in the Finnish population, independently from the (replicated) association found in the non-Finnish European population. It must be noted that the association in the Finnish group was found in a different region of the gene than the association in the European group. GRM7 is a large gene, and the European and the Finnish association signals are approximately 300 kb apart. As the Finnish are known to be genetically distinct from the rest of Europe, possibly due to the documented historical population bottleneck (33), this finding may be the result of allelic heterogeneity at the GRM7 locus. Allelic heterogeneity is frequently mentioned as a factor leading to the inability to replicate the exact SNP discovered and sometimes as the cause of lack of replication at an entire genetic locus. Some recent examples include reports on associations for rheumatoid arthritis (34) and bipolar disorder (35). However, allelic heterogeneity may not be the sole reason for replication of the association signal in a different region of the GRM7 gene. It is possible that the rs11928865 SNP exerts a smaller effect in the Finnish population or, if the effect size is similar, that the disease-associated allele frequency is small in Finland and therefore we may have been underpowered to detect the effect at that specific GRM7 SNP. A similar finding regarding disease allele frequency was discussed in a recent article (36). Population specific differences in allele frequency of putative interacting SNPs could also lead to these findings. If the interacting locus is present at an altered allele frequency, there will be a large decrease in power to detect an association (37). Lastly, we cannot rule out the possibility that our observation is a false positive association.
Individual genotyping of our large sample set was cost-prohibitive at the time this study was initiated; therefore, we based our GWAS on a pooling approach. It is important to recognize the advantages, as well as the disadvantages of such an approach, and we took several measures to overcome some of the limitations of pooled association experiments. First, to control for error, each pool was created de novo in triplicate and allelotyped independently on three separate arrays. Second, we analyzed our data in perhaps an overly strict fashion permitting only those SNPs with alleles associated in the same manner across all populations through to the next stage of analysis. Thirdly, before investigating the top most hits in our replication cohort, we individually genotyped a selected list of SNPs (detailed above) using an entirely different chemistry to provide further evidence that our initial findings were not due to pooling and/or microarray artifacts. This validation step indeed confirmed that the pooling approach led to reliable results as we confirmed the association of more than 35 and 90% of the selected SNPs in the European and Finnish samples, respectively.
The top 252 SNPs identified in the European sample group and the top 177 SNPs from the Finnish sample group did not contain any SNP from the three genes that previously had been implicated in ARHI: NAT2 (12,13), KCNQ4 (11) and GRHL2 (14). One possibility explaining this seeming non-replication is that the previously reported associations may have been spurious. However, this is unlikely as all three reported associations had previously been replicated in some way. In our opinion, the reason for not finding NAT2, KCNQ4 and GRHL2 SNPs among the top SNPs of the pooling experiment is possibly due to the lack of sensitivity of the pooling approach in combination with the large number of SNPs that we screened in the current study (500 K) compared to only a few dozen in candidate gene association studies. As a GWAS will pay a much larger multiple testing cost, less significant associations will always be ignored and only top-ranking SNPs will be considered further.
Metabotropic glutamate receptors (mGluRs) are activated by L-glutamate, the primary excitory amino acid neurotransmitter in the mammalian central nervous system (CNS). mGluRs are G-protein coupled receptors (GPCRs) functioning via secondary messenger pathways to modulate neuronal excitability and synaptic efficacy. The general structure of the metabotropic receptors consists of a glutamate binding site, a cystein-rich region, a seven transmembrane domain and an intracellular C-terminal region. To date, eight subtypes of mGluRs have been identified. GRM7 (mGluR7) is a member of the mGluR III group. mGluR7 is negatively coupled to the enzyme adenylate cyclase, and its activation results in reduced levels of cAMP. Its expression is temporally and spatially regulated in the human fetal brain. It is expressed at much lower levels in the adult brain in the hippocampus, hypothalamus and thalamus (38). In general, the activation of presynaptic mGluRs, such as class III mGluRs, has been found to reduce transmitter release from synapses in many brain regions (39). The mGluRs have diverse roles in the forms of synaptic plasticity that are believed to be involved in learning and memory in vertebrates. As such, mGluRs have attracted attention since their discovery as putative targets for many CNS indications, including anxiety, pain, neuroprotection, epilepsy, Parkinson's disease and cognitive disorders. mGluR7 has become particularly interesting since it was shown that its targeted deletion in mice results in several interesting neurological phenotypes including altered amygdala-dependent conditioned fear and aversion responses and reduced anxiety- and stress-related behaviors (40,41).
To date, little is known about the physiological roles of the metabotropic glutamate receptors in the human inner ear. The mGluRI agonist DHPG produced a pronounced increase in afferent auditory nerve firing in vivo, suggesting a role for metabotropic glutamate receptors in the mammalian inner ear (42). Relatively recently, L-glutamate and ionotropic receptor subunits were detected by immunocytochemistry in human cochlear sections (43), while mRNA of metabotrophic receptors group I (mGluRI) was detected in the mammalian cochlea and spiral ganglion (44–46). Glutamate is known to be the principle neurotransmitter coupling the mechanoelectrical transduction of sound impulses between the inner hair cells and the auditory afferent neurons of the inner ear (47,48). This was further confirmed by the fact that mice lacking Orphan Glutamate Receptor δ1 Subunit (GluRδ1), which is normally expressed robustly in the neurons of the hippocampus, cochlear inner hair cells, outer hair cells and spiral ganglion cells, show high frequency sensorineural hearing loss (49). The current study demonstrates robust mGluR7 expression in inner and outer hair cells and spiral ganglion neurons.
High concentrations of glutamate are known to be neurotoxic, because of its excitatory properties. Glutamate toxicity has been implicated in several forms of progressive hearing loss, including noise induced hearing loss and ARHI (50). The results of our study implicate that GRM7 variants possibly act on ARHI through excitotoxicity. Because mGluR7 reduces the release of glutamate, we hypothesize that the causative allele of GRM7 alters synaptic autoregulation of glutamate in the synaptic cleft of the sensory cells of the inner hair cells and the auditory neurons, leading to higher levels of glutamate in the cleft over time with subsequent excitotoxic neuronal and/or sensory cell death. Recent support for glutamate toxicity in the ear comes from studies in cultured spiral ganglion explants. In a recent study, cultured spiral ganglion explants were incubated with high concentrations of glutamate (51). The authors demonstrated concentration-dependent glutamate induced apoptosis of these explants that could be selectively blocked by a caspase-3 inhibitor.
GPCRs are the most successful class of drug targets to date. The identification of N,N′-dibenzhydrylethane-1,2-diamine dihydrochloride (AMN082), a highly selective agonist for mGluR7, has provided a valuable tool for studying the role of mGluR7 in stress-related CNS disorders (52). AMN082 is orally active and has been shown to traverse the blood-brain barrier. Studies of hearing in aging mGluR7 deficient mice are currently underway and will provide invaluable initial insights into the molecular pathology of ARHI and a potential model for developing drug therapies.
In conclusion, the current pooling GWAS has provided convincing evidence that variation in GRM7 is associated with ARHI. Fine mapping has positioned the causative allele in the European population in a 150 kb region surrounding GRM7 exon 2. While the causative allele remains to be identified, this finding suggests a possible role for glutamate toxicity in the pathophysiology of the disease.
MATERIALS AND METHODS
Study subjects and presbycusis assessment
The subjects have previously been described (13). Briefly, Caucasian volunteers, 53 to 67 years of age, from eight centers in six European countries, were collected via population registries or via audiological consultations. If the subjects were collected via hearing health services, the subject's spouses were also included. Informed consent was obtained from all volunteers. The study was approved by the ethical committees of the respective recruiting centers.
All subjects underwent an otoscopic investigation. Subjects with ear diseases potentially affecting hearing thresholds were excluded from the study. All subjects completed an extended questionnaire detailing medical history and exposure to environmental factors. In general, subjects with pathologies that could potentially influence their hearing thresholds were excluded according to an extensive exclusion list (11). Air conduction hearing thresholds were measured at 0.125, 0.25, 0.5, 1, 2, 3, 4, 6 and 8 kHz, and bone conduction at 0.5, 1, 2 and 4 kHz for all participating volunteers. Audiological exclusion criteria were an air-bone gap of more than 15 dB averaged over 0.5, 1 and 2 kHz in one or both ears or asymmetrical hearing impairment with a difference in air conduction thresholds exceeding 20 dB in at least two frequencies between 0.5, 1 and 2 kHz.
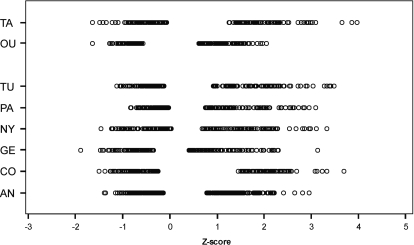
Z-scores were calculated as described previously (11,53). Briefly, frequency-specific thresholds were converted to sex and age independent Z-scores based on the ISO 7029 standard. These Z-scores represent the number of standard deviations the actual hearing threshold differs from the median at a given frequency. Subjects whose hearing is better than the age and sex specific median at a certain frequency have a negative Z-score. As a measure of high frequency hearing impairment, the Z-scores at 2, 4 and 8 kHz were averaged (Zhigh). This was done separately for both ears but only the Z-score for the better hearing ear was used. After excluding phenotypic outliers for Zhigh, we selected the best (controls) and worst (cases) hearing subjects within each center, for males and females separately (Fig. 6).
Figure 6.
Selection of the samples based on Zhigh, a gender- and age-independent measure of high-frequency hearing impairment. The Zhigh-score of each selected individual is represented by a circle. The two Finnish groups, Oulu and Tampere, are separated from the general European groups (AN, Antwerp; CO, Copenhagen; GE, Ghent; NY, Nijmegen; PA, Padova; TU, Tübingen; OU, Oulu; TA, Tampere).
For each of the eight centers separately (each subpopulation), individuals were ranked according to their Zhigh score. The top and the bottom of this ranking were pooled into a control pool and a case pool. At least 84 individuals were included in each pool. The percentile cut-off used at both extremes for making the pools was variable due to cohort size and the requirement to include a minimum number of individuals and ranged from 19 to 33%. Details of the percentile cut-off used for each subpopulation and resulting sample size per pool can be found in Supplementary Material, Table S1.
European replication group
Two centers, Ghent and Antwerp, collected a replication set of subjects using identical exclusion and inclusion criteria as the first set of study subjects. Taking the 33 percentile cut-offs at both extremes of 90 Antwerp and 117 Ghent samples resulted in a total of 138 samples (69 cases and 69 controls) that were used for replication purposes.
SNP genotyping
Genomic DNA concentrations were determined through triplicate measurements with the PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA). Additionally, the quality of the genomic DNA was assessed for each sample by spectrophotometric analysis and gel electrophoresis. Each individual contributed a total of 200 ng of DNA to the pool, and each pool was created de novo a total of three times. Each of these three pools was then genotyped in triplicate on the GeneChip® Human Mapping 500 K array pair from Affymetrix (Santa Clara, CA, USA). Individual-based SNP genotyping was performed by KBiosciences (Hoddesdon, UK) using fluorescence-based competitive allele-specific PCR (KASPar). Resequencing was conducted with standard Sanger sequencing procedures (Applied Biosystems, Foster City, CA, USA) and subsequent rs35106713 and rs3749448 genotyping was performed using Taqman assays (Applied Biosystems) on a real-time PCR machine (Roche, Basel, Switzerland). Individual-based SNP genotype data for the GRM7 locus can be freely downloaded at http://www.tgen.org/neurogenomics/data.
Statistical analysis
SNPs from the pooled GWAS were ranked using the freely available GenePool software developed by our group and reported elsewhere (27). Each subpopulation was evaluated separately (i.e. Antwerp cases were compared to Antwerp controls to generate the SNP scores and rankings). Using k-correction factors, allele frequencies and subsequently the associated allele were determined for each subpopulation separately as previously described.
For the analysis of the European replication group and for the finemapping, tests for deviation from HWE and tests for association (using allele χ2 P-values) were applied for all SNPs using Haploview or SPSS 15.0 software (Chicago, IL, USA; http://www.spss.com). Permutation testing (n = 1 × 106 permutes) was performed using the module from Haploview.
Mouse immunohistochemistry
Swiss mouse inner ear tissue from PD1, PD21 and adult mice (age ranging from PD42 to PD70) was prepared as described previously (54). Briefly, mice were transcardially perfused, inner ears were removed and postfixed in 4% phosphate-buffered paraformaldehyde. After fixation, P21 and adult inner ears were decalcified in phosphate-buffered saline (PBS) containing 5% ethylenediaminetetraacetic acid. Tissue was paraffin embedded and mounted on uncoated glasses. Sections were blocked using an endogenous avidin/biotin blocking kit (Zymed Laboratories, San Francisco, CA, USA), as described by the manufacturer. To avoid non-specific binding from the primary antibody, sections were blocked with 50 mm glycin/PBS. Then, slides were incubated in blocking buffer containing PBS, 10% normal goat serum, 0.05% thimerosal, 5% bovine serum albumin (BSA) and 0.3% Triton X-100. Subsequently, sections were incubated overnight at 4°C with peptide-affinity purified rabbit polyclonal antibody to mGluR7 (Imgenex, San Diego, CA, USA), at a dilution of 1:150 in PBS with 0.3% Triton X-100 (tx-PBS). This antibody was raised against a synthetic peptide corresponding to the N-terminal extracellular domain of human mGluR7 and was shown to crossreact with mouse (http://www.imgenex.com/antibody_details.php?catalog=IMG-71406). Biotinylated Fab fragments of goat anti-rabbit immunoglobulin (Ig) G (Rockland, Gilbertsville, PA, USA), diluted 1:500 in tx-PBS, were added for 2 h. Sections were washed and incubated in Cy3-conjugated streptavidin (Jackson Immunoresearch Laboratories, West Grove, PA, USA), diluted 1:5000 in PBS. Finally, slides were mounted with Citifluor (Ted Pella, Redding, CA, USA) and studied with fluorescence and confocal microscopy.
Human immunohistochemistry
Human temporal bone sections were embedded in celloidin and stored in 80% ethanol. The sections were washed in ethanol followed by a wash in methanol. A non-heated antigen retrieval reagent [Saturated NaOH (Sigma, St Louis, MO, USA)] and methanol at a concentration of 1:3 were applied to the sections for 5 min. Sections were washed in various concentrations of methanol and rinsed in PBS. Each section was placed in 0.3% Triton-X100 (Sigma, St Louis, MO, USA) and washed in PBS. The sections were incubated in 10% BSA (Sigma) to block non-specific binding. After blocking, sections were incubated with the primary antibody (mGluR7, Imgenex) used at a concentration of 2 µl/ml. Sections were washed in PBS and incubated in 7 µg/ml biotinylated secondary antibody (LSAB2 system, Carpinteria, CA, USA). Sections were then washed in PBS and incubated in a tertiary antibody or labeled antibody–Streptavidin conjugated enzyme (LSAB2 system, Carpinteria, CA, USA). Following this incubation, the sections were washed in PBS and 20 µl of fast red chromogen (Chromogen and Substrate Kit, Biogenex, San Ramon, CA, USA). The sections were washed in PBS to stop the chromogen reaction and placed in distilled water. Each section was then counterstained with Mayer's Hematoxylin (Biogenex), rinsed with tap water and 1% ammonium water. Finally, each section was rinsed in distilled water and mounted onto a glass slide with aqueous mounting media (Biogenex) and cover slipped.
Cloning of GRM7 fragment
Mouse brain RNA was isolated using Trizol (Invitrogen) and reverse-transcribed with the Superscript™ III First-strand synthesis system for RT–PCR (Invitrogen). A 839 bp-fragment of mouse GRM7 [nucleotides 901–1739 (based on numbering by Kosinski) (55)] was amplified using the iProof High-fidelity DNA polymerase kit (BioRad, Hercules, CA, USA) according to the manufacturer's instructions, in the presence of 200 µm dNTP mix (BD Biosciences Clontech, Palo Alto, CA, USA) and 0.5 µm of each primer (5′-AGAGCTGACCAAGTAGGAC-3′ and 5′-GGGATGTTCTGACAGCCAG-3′) (Invitrogen). PCR product was gel-purified using the QIAquick Gel Extraction kit (QIAGEN GmbH, Hilden, Germany) and 3′A-overhangs were generated by adding 1x PCR-buffer, 200 µm dNTP mix (BD Biosciences Clontech) and 0.1 U/µl of Silverstar™ Taq polymerase (Eurogentec, Seraing, Belgium), followed by an incubation at 72°C for 10 min. The fragment was subsequently cloned with the TOPO TA Cloning kit for sequencing (Invitrogen), following the manufacturer's protocol. Inserts were verified by DNA sequencing.
In situ hybridization
In situ hybridization experiments were carried out in wild-type C3HeB/FeJ mouse inner ears. For probe labeling, plasmids were linearized with either NotI or SpeI (Fermentas GmbH, St Leon-Rot, Germany), and digests were purified using the Rapid PCR purification system (Marligen Bioscience Inc., Ijamsville, MD, USA). Subsequently, digoxigenin (DIG)-labeled antisense and sense riboprobes were generated with T3 and T7 polymerase using Riboprobe® in vitro Transcription Systems (Promega, Madison, WI, USA) and DIG-11-UTP (Roche Diagnostics, Brussels, Belgium), as prescribed by the manufacturer. Riboprobes were then hydrolyzed to a fragment length of approximately 150 bp. After ethanol precipitation, pellets were dissolved in diethylpyrocarbonate (DEPC)-treated water. Probe concentrations were determined using the DIG Nonradioactive Nucleic Acid Labeling and Detection System (Roche Diagnostics) according to the instructions of the manufacturer. In situ hybridization on tissue sections was performed with modifications as described (56). Briefly, sections were treated with 2 µg/ml proteinase K. The tissue sections were then delipidated and dehydrated before hybridization. Hybridization with digoxigenin-labeled riboprobes was performed at 60°C overnight in a hybridization buffer solution. After hybridization, sections were incubated in 20% heat-inactivated sheep serum in PBS with 0.1% Triton-X100 (PBT) to block non-specific binding sites. To visualize the hybrids, the sections were incubated with an anti-digoxigenin antibody (conjugated with alkaline phosphatase). The slides were developed after the addition of a chromogenic substrate, BCIP using NBT as a catalyst. Three PD6 mice ears were analyzed using the Grm7 probe.
SUPPLEMENTARY MATERIAL
FUNDING
The first phases of this work were funded by the European Community [5th Framework project QLRT-2001-00331]. Additional funding came from the British Royal National Institute for Deaf and hard of hearing individuals (RNID), the Flemish organization for scientific research [FWO grant G.0131.04], the University of Antwerp [TOP project] and the National Institutes of Health Neuroscience Microarray Consortium [U24NS051872]. The authors would like to acknowledge and sincerely thank the Seaver Institute for their generous gift without which this study would not have been possible.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to express their most sincere gratitude to all the volunteers who have participated in this study and to Vedat Topsakal, Jan-Jaap Hendrickx, Samuli Hannula, Elina Mäki-Torkko, Mona Jensen, Kelly Demeester, Manuela Baur, Amanda Bonaconsa, Manuela Mazzoli, Angeles Espeso, Katia Verbruggen, Joke Huyghe, Sylvia Kunst and Minna Manninen for their efforts during the collection phase of the samples.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Gates G.A., Mills J.H. Presbycusis. Lancet. 2005;366:1111–1120. doi: 10.1016/S0140-6736(05)67423-5. [DOI] [PubMed] [Google Scholar]
- 2.Heine C., Browning C.J. Communication and psychosocial consequences of sensory loss in older adults: overview and rehabilitation directions. Disabil. Rehabil. 2002;24:763–773. doi: 10.1080/09638280210129162. [DOI] [PubMed] [Google Scholar]
- 3.Dalton D.S., Cruickshanks K.J., Klein B.E., Klein R., Wiley T.L., Nondahl D.M. The impact of hearing loss on quality of life in older adults. Gerontologist. 2003;43:661–668. doi: 10.1093/geront/43.5.661. [DOI] [PubMed] [Google Scholar]
- 4.Van Eyken E., Van Camp G., Van Laer L. The complexity of age-related hearing impairment: contributing environmental and genetic factors. Audiol. Neurootol. 2007;12:345–358. doi: 10.1159/000106478. [DOI] [PubMed] [Google Scholar]
- 5.Karlsson K.K., Harris J.R., Svartengren M. Description and primary results from an audiometric study of male twins. Ear Hear. 1997;18:114–120. doi: 10.1097/00003446-199704000-00003. [DOI] [PubMed] [Google Scholar]
- 6.Gates G.A., Couropmitree N.N., Myers R.H. Genetic associations in age-related hearing thresholds. Arch Otolaryngol. Head Neck Surg. 1999;125:654–659. doi: 10.1001/archotol.125.6.654. [DOI] [PubMed] [Google Scholar]
- 7.Christensen K., Frederiksen H., Hoffman H.J. Genetic and environmental influences on self-reported reduced hearing in the old and oldest old. J. Am. Geriatr. Soc. 2001;49:1512–1517. doi: 10.1046/j.1532-5415.2001.4911245.x. [DOI] [PubMed] [Google Scholar]
- 8.Van Laer L., DeStefano A.L., Myers R.H., Flothmann K., Thys S., Fransen E., Gates G.A., Van Camp G., Baldwin C.T. Is DFNA5 a susceptibility gene for age-related hearing impairment? Eur. J. Hum. Genet. 2002;10:883–886. doi: 10.1038/sj.ejhg.5200878. [DOI] [PubMed] [Google Scholar]
- 9.Ates N.A., Unal M., Tamer L., Derici E., Karakas S., Ercan B., Pata Y.S., Akbas Y., Vayisoglu Y., Camdeviren H. Glutathione S-transferase gene polymorphisms in presbycusis. Otol. Neurotol. 2005;26:392–397. doi: 10.1097/01.mao.0000169774.23668.f1. [DOI] [PubMed] [Google Scholar]
- 10.Van Eyken E., Van Laer L., Fransen E., Topsakal V., Hendrickx J.J., Demeester K., Van de Heyning P., Maki-Torkko E., Hannula S., Sorri M., et al. The contribution of GJB2 (Connexin 26) 35delG to age-related hearing impairment and noise-induced hearing loss. Otol. Neurotol. 2007;28:970–975. doi: 10.197/MAO.0b013e3180dca1b9. [DOI] [PubMed] [Google Scholar]
- 11.Van Eyken E., Van Laer L., Fransen E., Topsakal V., Lemkens N., Laureys W., Nelissen N., Vandevelde A., Wienker T., Van De Heyning P., et al. KCNQ4: a gene for age-related hearing impairment? Hum. Mutat. 2006;27:1007–1016. doi: 10.1002/humu.20375. [DOI] [PubMed] [Google Scholar]
- 12.Unal M., Tamer L., Dogruer Z.N., Yildirim H., Vayisoglu Y., Camdeviren H. N-acetyltransferase 2 gene polymorphism and presbycusis. Laryngoscope. 2005;115:2238–2241. doi: 10.1097/01.mlg.0000183694.10583.12. [DOI] [PubMed] [Google Scholar]
- 13.Van Eyken E., Van Camp G., Fransen E., Topsakal V., Hendrickx J.J., Demeester K., Van de Heyning P., Maki-Torkko E., Hannula S., Sorri M., et al. Contribution of the N-acetyltransferase 2 polymorphism NAT2*6A to age-related hearing impairment. J. Med. Genet. 2007;44:570–578. doi: 10.1136/jmg.2007.049205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Laer L., Van Eyken E., Fransen E., Huyghe J.R., Topsakal V., Hendrickx J.J., Hannula S., Maki-Torkko E., Jensen M., Demeester K., et al. The grainyhead like 2 gene (GRHL2), alias TFCP2L3, is associated with age-related hearing impairment. Hum. Mol. Genet. 2008;17:159–169. doi: 10.1093/hmg/ddm292. [DOI] [PubMed] [Google Scholar]
- 15.Wallace D.C. Mitochondrial DNA in aging and disease. Sci. Am. 1997;277:40–47. doi: 10.1038/scientificamerican0897-40. [DOI] [PubMed] [Google Scholar]
- 16.Fischel-Ghodsian N., Bykhovskaya Y., Taylor K., Kahen T., Cantor R., Ehrenman K., Smith R., Keithley E. Temporal bone analysis of patients with presbycusis reveals high frequency of mitochondrial mutations. Hear. Res. 1997;110:147–154. doi: 10.1016/s0378-5955(97)00077-4. [DOI] [PubMed] [Google Scholar]
- 17.Bai U., Seidman M.D., Hinojosa R., Quirk W.S. Mitochondrial DNA deletions associated with aging and possibly presbycusis: a human archival temporal bone study. Am. J. Otol. 1997;18:449–453. [PubMed] [Google Scholar]
- 18.Manwaring N., Jones M.M., Wang J.J., Rochtchina E., Howard C., Newall P., Mitchell P., Sue C.M. Mitochondrial DNA haplogroups and age-related hearing loss. Arch Otolaryngol. Head Neck Surg. 2007;133:929–933. doi: 10.1001/archotol.133.9.929. [DOI] [PubMed] [Google Scholar]
- 19.Johnson K.R., Erway L.C., Cook S.A., Willott J.F., Zheng Q.Y. A major gene affecting age-related hearing loss in C57BL/6J mice. Hear. Res. 1997;114:83–92. doi: 10.1016/s0378-5955(97)00155-x. [DOI] [PubMed] [Google Scholar]
- 20.Johnson K.R., Zheng Q.Y. Ahl2, a second locus affecting age-related hearing loss in mice. Genomics. 2002;80:461–464. [PMC free article] [PubMed] [Google Scholar]
- 21.Nemoto M., Morita Y., Mishima Y., Takahashi S., Nomura T., Ushiki T., Shiroishi T., Kikkawa Y., Yonekawa H., Kominami R. Ahl3, a third locus on mouse chromosome 17 affecting age-related hearing loss. Biochem. Biophys. Res. Commun. 2004;324:1283–1288. doi: 10.1016/j.bbrc.2004.09.186. [DOI] [PubMed] [Google Scholar]
- 22.Noben-Trauth K., Zheng Q.Y., Johnson K.R. Association of cadherin 23 with polygenic inheritance and genetic modification of sensorineural hearing loss. Nat. Genet. 2003;35:21–23. doi: 10.1038/ng1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Easton D.F., Pooley K.A., Dunning A.M., Pharoah P.D., Thompson D., Ballinger D.G., Struewing J.P., Morrison J., Field H., Luben R., et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunter D.J., Kraft P., Jacobs K.B., Cox D.G., Yeager M., Hankinson S.E., Wacholder S., Wang Z., Welch R., Hutchinson A., et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rioux J.D., Xavier R.J., Taylor K.D., Silverberg M.S., Goyette P., Huett A., Green T., Kuballa P., Barmada M.M., Datta L.W., et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scott L.J., Mohlke K.L., Bonnycastle L.L., Willer C.J., Li Y., Duren W.L., Erdos M.R., Stringham H.M., Chines P.S., Jackson A.U., et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pearson J.V., Huentelman M.J., Halperin R.F., Tembe W.D., Melquist S., Homer N., Brun M., Szelinger S., Coon K.D., Zismann V.L., et al. Identification of the genetic basis for complex disorders by use of pooling-based genomewide single-nucleotide-polymorphism association studies. Am. J. Hum. Genet. 2007;80:126–139. doi: 10.1086/510686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papassotiropoulos A., Stephan D.A., Huentelman M.J., Hoerndli F.J., Craig D.W., Pearson J.V., Huynh K.D., Brunner F., Corneveaux J., Osborne D., et al. Common Kibra alleles are associated with human memory performance. Science. 2006;314:475–478. doi: 10.1126/science.1129837. [DOI] [PubMed] [Google Scholar]
- 29.Schaper K., Kolsch H., Popp J., Wagner M., Jessen F. KIBRA gene variants are associated with episodic memory in healthy elderly. Neurobiol. Aging. 2008;29:1123–1125. doi: 10.1016/j.neurobiolaging.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 30.Schuknecht H.F., Gacek M.R. Cochlear pathology in presbycusis. Ann. Otol. Rhinol. Laryngol. 1993;102:1–16. doi: 10.1177/00034894931020S101. [DOI] [PubMed] [Google Scholar]
- 31.Otte J., Schunknecht H.F., Kerr A.G. Ganglion cell populations in normal and pathological human cochleae. Implications for cochlear implantation. Laryngoscope. 1978;88:1231–1246. doi: 10.1288/00005537-197808000-00004. [DOI] [PubMed] [Google Scholar]
- 32.Lohmueller K.E., Pearce C.L., Pike M., Lander E.S., Hirschhorn J.N. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat. Genet. 2003;33:177–182. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 33.Sajantila A., Salem A.H., Savolainen P., Bauer K., Gierig C., Paabo S. Paternal and maternal DNA lineages reveal a bottleneck in the founding of the Finnish population. Proc. Natl Acad. Sci. USA. 1996;93:12035–12039. doi: 10.1073/pnas.93.21.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plenge R.M., Cotsapas C., Davies L., Price A.L., de Bakker P.I., Maller J., Pe'er I., Burtt N.P., Blumenstiel B., DeFelice M., et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat. Genet. 2007;39:1477–1482. doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palo O.M., Antila M., Silander K., Hennah W., Kilpinen H., Soronen P., Tuulio-Henriksson A., Kieseppa T., Partonen T., Lonnqvist J., et al. Association of distinct allelic haplotypes of DISC1 with psychotic and bipolar spectrum disorders and with underlying cognitive impairments. Hum. Mol. Genet. 2007;16:2517–2528. doi: 10.1093/hmg/ddm207. [DOI] [PubMed] [Google Scholar]
- 36.McCarthy M.I. Casting a wider net for diabetes susceptibility genes. Nat. Genet. 2008;40:1039–1040. doi: 10.1038/ng0908-1039. [DOI] [PubMed] [Google Scholar]
- 37.Marchini J., Donnelly P., Cardon L.R. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat. Genet. 2005;37:413–417. doi: 10.1038/ng1537. [DOI] [PubMed] [Google Scholar]
- 38.Makoff A., Pilling C., Harrington K., Emson P. Human metabotropic glutamate receptor type 7: molecular cloning and mRNA distribution in the CNS. Brain Res. Mol. Brain Res. 1996;40:165–170. doi: 10.1016/0169-328x(96)00110-6. [DOI] [PubMed] [Google Scholar]
- 39.Shigemoto R., Mizuno N., Ottersen O.P., Mathisen J.S. Handbook of Chemical Neuroanatomy. Amsterdam: Elsevier; 2000. [Google Scholar]
- 40.Cryan J.F., Kelly P.H., Neijt H.C., Sansig G., Flor P.J., van Der Putten H. Antidepressant and anxiolytic-like effects in mice lacking the group III metabotropic glutamate receptor mGluR7. Eur. J. Neurosci. 2003;17:2409–2417. doi: 10.1046/j.1460-9568.2003.02667.x. [DOI] [PubMed] [Google Scholar]
- 41.Masugi M., Yokoi M., Shigemoto R., Muguruma K., Watanabe Y., Sansig G., van der Putten H., Nakanishi S. Metabotropic glutamate receptor subtype 7 ablation causes deficit in fear response and conditioned taste aversion. J. Neurosci. 1999;19:955–963. doi: 10.1523/JNEUROSCI.19-03-00955.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kleinlogel S., Oestreicher E., Arnold T., Ehrenberger K., Felix D. Metabotropic glutamate receptors group I are involved in cochlear neurotransmission. Neuroreport. 1999;10:1879–1882. doi: 10.1097/00001756-199906230-00015. [DOI] [PubMed] [Google Scholar]
- 43.Nordang L., Cestreicher E., Arnold W., Anniko M. Glutamate is the afferent neurotransmitter in the human cochlea. Acta Otolaryngol. 2000;120:359–362. doi: 10.1080/000164800750000568. [DOI] [PubMed] [Google Scholar]
- 44.Bilak S.R., Morest D.K. Differential expression of the metabotropic glutamate receptor mGluR1alpha by neurons and axons in the cochlear nucleus: in situ hybridization and immunohistochemistry. Synapse. 1998;28:251–270. doi: 10.1002/(SICI)1098-2396(199804)28:4<251::AID-SYN1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 45.Niedzielski A.S., Safieddine S., Wenthold R.J. Molecular analysis of excitatory amino acid receptor expression in the cochlea. Audiol. Neurootol. 1997;2:79–91. doi: 10.1159/000259232. [DOI] [PubMed] [Google Scholar]
- 46.Safieddine S., Eybalin M. Expression of mGluR1 alpha mRNA receptor in rat and guinea pig cochlear neurons. Neuroreport. 1995;7:193–196. [PubMed] [Google Scholar]
- 47.Eybalin M., Norenberg M.D., Renard N. Glutamine synthetase and glutamate metabolism in the guinea pig cochlea. Hear. Res. 1996;101:93–101. doi: 10.1016/s0378-5955(96)00136-0. [DOI] [PubMed] [Google Scholar]
- 48.Fex J., Kachar B., Rubio J.A., Parakkal M.H., Altschuler R.A. Glutaminase-like immunoreactivity in the organ of Corti of guinea pig. Hear. Res. 1985;17:101–113. doi: 10.1016/0378-5955(85)90014-0. [DOI] [PubMed] [Google Scholar]
- 49.Gao J., Maison S.F., Wu X., Hirose K., Jones S.M., Bayazitov I., Tian Y., Mittleman G., Matthews D.B., Zakharenko S.S., et al. Orphan glutamate receptor delta1 subunit required for high-frequency hearing. Mol. Cell Biol. 2007;27:4500–4512. doi: 10.1128/MCB.02051-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pujol R., Puel J.L., Gervais d'Aldin C., Eybalin M. Pathophysiology of the glutamatergic synapses in the cochlea. Acta Otolaryngol. 1993;113:330–334. doi: 10.3109/00016489309135819. [DOI] [PubMed] [Google Scholar]
- 51.Steinbach S., Lutz J. Glutamate induces apoptosis in cultured spiral ganglion explants. Biochem. Biophys. Res. Commun. 2007;357:14–19. doi: 10.1016/j.bbrc.2007.03.098. [DOI] [PubMed] [Google Scholar]
- 52.Mitsukawa K., Yamamoto R., Ofner S., Nozulak J., Pescott O., Lukic S., Stoehr N., Mombereau C., Kuhn R., McAllister K.H., et al. A selective metabotropic glutamate receptor 7 agonist: activation of receptor signaling via an allosteric site modulates stress parameters in vivo. Proc. Natl Acad. Sci. USA. 2005;102:18712–18717. doi: 10.1073/pnas.0508063102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fransen E., Van Laer L., Lemkens N., Caethoven G., Flothmann K., Govaerts P., Van de Heyning P., Van Camp G. A novel Z-score-based method to analyze candidate genes for age-related hearing impairment. Ear Hear. 2004;25:133–141. doi: 10.1097/01.aud.0000120362.69077.0b. [DOI] [PubMed] [Google Scholar]
- 54.Cryns K., Thys S., Van Laer L., Oka Y., Pfister M., Van Nassauw L., Smith R.J., Timmermans J.P., Van Camp G. The WFS1 gene, responsible for low frequency sensorineural hearing loss and Wolfram syndrome, is expressed in a variety of inner ear cells. Histochem. Cell Biol. 2003;119:247–256. doi: 10.1007/s00418-003-0495-6. [DOI] [PubMed] [Google Scholar]
- 55.Kosinski C.M., Risso Bradley S., Conn P.J., Levey A.I., Landwehrmeyer G.B., Penney J.B., Jr, Young A.B., Standaert D.G. Localization of metabotropic glutamate receptor 7 mRNA and mGluR7a protein in the rat basal ganglia. J. Comp. Neurol. 1999;415:266–284. [PubMed] [Google Scholar]
- 56.Groves A.K., Bronner-Fraser M. Competence, specification and commitment in otic placode induction. Development. 2000;127:3489–3499. doi: 10.1242/dev.127.16.3489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.