Abstract
Motivation: In this work, we aim to develop a computational approach for predicting DNA-binding sites in proteins from amino acid sequences. To avoid overfitting with this method, all available DNA-binding proteins from the Protein Data Bank (PDB) are used to construct the models. The random forest (RF) algorithm is used because it is fast and has robust performance for different parameter values. A novel hybrid feature is presented which incorporates evolutionary information of the amino acid sequence, secondary structure (SS) information and orthogonal binary vector (OBV) information which reflects the characteristics of 20 kinds of amino acids for two physical–chemical properties (dipoles and volumes of the side chains). The numbers of binding and non-binding residues in proteins are highly unbalanced, so a novel scheme is proposed to deal with the problem of imbalanced datasets by downsizing the majority class.
Results: The results show that the RF model achieves 91.41% overall accuracy with Matthew's correlation coefficient of 0.70 and an area under the receiver operating characteristic curve (AUC) of 0.913. To our knowledge, the RF method using the hybrid feature is currently the computationally optimal approach for predicting DNA-binding sites in proteins from amino acid sequences without using three-dimensional (3D) structural information. We have demonstrated that the prediction results are useful for understanding protein–DNA interactions.
Availability: DBindR web server implementation is freely available at http://www.cbi.seu.edu.cn/DBindR/DBindR.htm.
Contact: xsun@seu.edu.cn
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Protein–DNA interactions control a variety of vital biological processes, such as gene regulation, DNA replication and repair, recombination and other critical steps in cellular development (Luscombe et al., 2000). Mutations of DNA-binding residues, such as those on the tumor repressor protein P53, may be directly involved in human diseases (Bullock and Fersht, 2001). Thus, the ability to identify the amino acid residues that recognize DNA can significantly improve our understanding of these biological processes and affect the potential for guiding site-directed mutagenesis studies for the functional characterization of DNA-binding proteins, and can further contribute to advances in drug discovery, such as aiding the design of artificial transcription factors (Ho et al., 2007; Wang and Brown, 2006a).
The protein–DNA recognition mechanism is complicated and the interactions consist of a variety of atomic contacts involving hydrogen bonds, van der Waals contacts and electrostatic, water-mediated bonds between amino acid residues and nucleotide bases. Such residues that recognize DNA can be identified from the three-dimensional (3D) structure of protein–DNA complexes (Siggers and Honig, 2007). Unfortunately, 3D structures of such complexes are available for <5% of all known DNA-binding proteins (Ofran et al., 2007). Moreover, it is a very expensive and time-consuming process to solve the structure of a protein–DNA complex through experimental methods. A variety of computational methods have been developed to identify DNA-binding interface residues from 3D structures (Jones et al., 2003; Tjong and Zhou, 2007; Tsuchiya et al., 2004). However, these methods have at least two major limitations: (i) One problem with using the structure of an unbound protein to predict sites involved in interactions with DNA is that the actual structure of the DNA-bound protein may differ substantially from the unbound form. (ii) The other problem is that structure-based computational methods still involve the expensive and time-consuming process of experimental determination of protein structure (Kuznetsov et al., 2006; Ofran et al., 2007).
An alternative to structure-based prediction is to predict DNA-binding residues directly from amino acid sequences. Machine learning techniques provide an effective approach for the construction of classifiers for this task, and predicting binding residues can be operated under the assumption that a given protein is bound to the DNA. Recently, neural network models have been constructed to identify protein–DNA interface residues relying on sequence information and residue solvent accessibility (Ahmad et al., 2004). Evolutionary information in terms of position-specific scoring matrices (PSSMs) have contributed to a better prediction performance of DNA-binding sites (Ahmad and Sarai, 2005). Yan et al. (2006) trained a naive Bayes classifier to predict DNA-binding sites based on the identities of sequence and entropy of the target residue. More importantly, a variety of support vector machine (SVM) classifiers have been developed for automated identification of DNA-binding residues with high accuracy (Bhardwaj and Lu, 2007; Ho et al., 2007; Kuznetsov et al., 2006; Ofran et al., 2007; Wang and Brown, 2006a, b).
In the present work, we aim to design optimal predictors for DNA-binding sites in proteins directly from amino acid sequences. In designing classifiers, the following points are taken into consideration: (i) Approaches learning from a small training dataset can result in the over-fitting of training data and will then have poor generalization performance for new data. Accordingly, it is necessary that all available datasets of DNA-binding proteins are employed to build a new classifier for recognizing binding residues (Bhardwaj and Lu, 2007; Ofran et al., 2007). (ii) The SVM is a powerful machine-learning algorithm developed from statistical learning theory (Vapnik, 1998) that has been successfully applied in many fields for data classification. However, it is definitely time consuming to find the appropriate kernel function and optimal-free parameters of an SVM for very large training datasets. So it is significant to find a new algorithm that is fast and robust for different parameter values. (iii) Although many methods have been proposed, it is still challenging to predict DNA-binding residues directly from amino acid sequence data. To our knowledge, incorporating more effective features is the most important way of improving the performance of classifiers. (iv) The numbers of binding and non-binding residues in proteins are significantly unbalanced such that the problem of imbalanced datasets should be considered in enhancing the prediction accuracy. (v) The prediction reliability is an important factor that shows more information about the quality of the prediction. (vi) How do we ensure that parameter estimation and model generation are completely independent of the test data? (vii) What about the performance of distinguishing DNA-binding proteins from non-binding proteins?
In this article, we propose a novel method for predicting DNA-binding residues using the random forest (RF) algorithm in conjunction with a hybrid feature by combining evolutionary information of the amino acid sequence, orthogonal binary vector (OBV) information that reflects the characteristics of 20 kinds of amino acids for two physical–chemical properties (dipoles and volumes of the side chains) and secondary structure (SS) information. The results show that the RF-based model achieves 91.41% overall accuracy with Matthew's correlation coefficient of 0.70, and with a sensitivity of 76.57% and a specificity of 94.38%.
2 MATERIALS AND METHODS
2.1 Dataset
DBP-374 is a dataset of 374 structures of representative protein–DNA complexes from the Protein Data Bank (PDB) (Berman et al., 2000). We collected all protein–DNA complexes (released by October 17, 2007) determined by X-ray crystallography with a resolution better than 3.5Å. Redundancy among the amino acid sequences was removed by clustering analysis using the blastclust program in the BLAST package (Altschul et al., 1990) from NCBI (http://www.ncbi.nlm.nih.gov/BLAST/download.shtml) with a threshold of 25% for sequence identity. Thus, the non-redundant DBP-374 dataset listed in the Supplementary Material was created by retaining only the longest sequence in each cluster.
As in the previous studies (Ahmad and Sarai, 2005; Ahmad et al., 2004; Ho et al., 2007; Wang and Brown, 2006a, b), an amino acid residue within a protein sequence is designated as a binding site if it contains at least one atom that falls within the cutoff distance of 3.5Å from any atoms of the DNA molecule in the complex, and all other residues are labeled non-binding sites. The DBP-374 dataset contains 5652 DNA-binding residues and 74 782 non-binding residues.
2.2 Features of data instances
RFs are constructed using residue-wise data instances from the protein sequences of the DBP-374 dataset. Each data instance is a segment of amino acid sequences with length β = 11, where β is the sliding window size. Compared with other window sizes, the RF classifiers constructed with β = 11 present the best performance. From an amino acid sequence with m residues, a total of (m−β+1 − γ) data instances are extracted, where γ is the number of residues that lack information about their atomic coordinates in the PDB entries. A data instance is labeled with ‘P’ (positive) if the central residue is DNA-binding or ‘N’ (negative) if the central residue is non-binding.
Electrostatic (including hydrogen bonding) and hydrophobic interactions probably dominate protein–DNA interactions, and can be reflected by the dipoles and volumes of the side chains of amino acids, respectively. In the protein–DNA complex structures, hydrogen bond is a major interaction force and its strength is associated with the dipoles involved hydrogen bond (Coulocheri et al., 2007). Based on the dipoles and volumes of the side chains, the 20 kinds of amino acids can be clustered into seven classes (Shen et al., 2007). The unique amino acid cysteine in the seventh class is called back to the third class in this study because disulfide bonds have no special attribution to protein–DNA interaction. Therefore, the 20 kinds of amino acids are grouped into six classes, namely Class 1:Ala, Gly, Val; Class 2: Ile, Leu, Phe, Pro; Class 3: Tyr, Met, Thr, Ser, Cys; Class 4: His, Asn, Gln, Tpr; Class 5: Arg, Lys; and Class 6: Asp, Glu. Amino acids in each class are encoded as an OBV, for example (1,0,0…) or (0,1,0…) and the vector is 6D. In this article, each data instance is coded with a hybrid feature by combining the OBVs of the amino acids, evolutionary information of the amino acid sequence and SS information of the amino acids.
In this work, evolutionary information of amino acid sequences in terms of their PSSMs are generated for prediction of DNA-binding sites (Bhardwaj and Lu, 2007; Ho et al., 2007; Kuznetsov et al., 2006; Ofran et al., 2007). The PSSM elements are scaled to fall within the range 0–1 by the standard logistic function (Wang et al., 2006):
| (1) |
SS information extracted from the PDB files is encoded as follows: helix (1,0,0), strand (0,1,0) and others (0,0,1). For each data instance, the input vector contains 319 feature values, including 220 (20 × 11) PSSMs, 66 (6 × 11) OBVs and 33 (3 × 11) SS elements. The SS information was, however, predicted by the PREDATOR program (Frishman and Argos, 1997) during the prediction of new proteins.
2.3 Algorithms for classification
RF is a classification algorithm that uses an ensemble of tree-structured classifiers (Breiman, 2001). The RF algorithm is implemented by the randomForest (version 4.5-18) R package (Liaw and Wiener, 2002).
To ensure that parameter estimation and model generation of RFs are completely independent of the test data, a nested cross-validation procedure (Scheffer, 1999) is performed. Nested cross-validation means that there is an outer cross-validation loop for model assessment and an inner loop for model selection. In this study, the original samples are randomly divided into k = 5 parts in the outer loop. Each of these parts is chosen one by one for assessment, and the remaining 4/5 of the samples are for model selection in the inner loop where a type of cross-validation using the so-called out-of-bag (OOB) samples is performed.
2.4 The imbalanced data problem
The problem of imbalanced datasets occurs in many practical classification events where the goal is to detect a rare but important case. All solutions to this problem can roughly be grouped into two main categories (Cohen et al., 2006): the first consists of preprocessing the data to re-establish class balance (either by upsizing the minority class or downsizing the majority class); whereas the second involves modifying the learning algorithm itself to cope with imbalanced data. Kubat and Matwin (1997) presented an algorithm for the one-sided selection of examples by downsizing the majority class. Examples of the majority class can roughly be divided into four groups: class-label noise, borderline, redundant and safe examples, and the algorithm attempts to create a subset retaining the safe examples (Kubat and Matwin, 1997). A similar approach is used in this article to deal with the imbalanced data problem (only 7.25% of the total data instances are positive).
The algorithm for downsizing the majority class can be summarized as follows.
Let Ω be the original training set and ϕ be the set of all negative data instances of Ω.
Let ψ contain all positive data instances and 1% randomly selected negative data instances from ϕ.
Predict ϕ using the model trained by the data instances in ψ based on the RF and then get F+, where F+ is the fraction of the tree votes for the positive class in each data instance.
Repeat steps 2 and 3. 10 times.
Combine the F+ from all 10 iterations, and regard the data instances in ϕ of which 50% of the F+ values are between 1/9 and 2/9 as safe (because the ratio of positive to negative instances in ψ is 5331:681).
Get the processed dataset ζ containing all positive data instances and the safe negative data instances from ϕ. The processed dataset ζ has 5331 positive data instances and 26 721 negative data instances.
2.5 Measurement of classifier's performance
The overall prediction accuracy (ACC), sensitivity (SE), precision (PR), specificity (SP) (Wang and Brown, 2006a) and Matthew's correlation coefficient (MCC) (Matthews, 1975) are for assessment of the prediction system.
The receiver operating characteristic (ROC) curve is probably the most robust technique for evaluating classifiers and visualizing their performance (Egan, 1975).
2.6 Reliability index
The prediction reliability index (RI) is an important factor in evaluating the quality of prediction. We adopt the RI to show the level of reliability in the prediction of a submitted sequence. RI is defined as:
| (2) |
| (3) |
Here, F+ is the fraction of the tree votes (FV) for the positive class in each sample, and φ is the threshold by which to classify samples according to FV for the positive class and is set to 1/6 in this article, because the ratio of positive to negative samples for training is 1: 5.
3 RESULTS AND DISCUSSION
3.1 Prediction performance of RF method using various features
The prediction results of the RF modules using various features are presented in Table 1. The RF-based classifier with the PSSM feature achieved a prediction accuracy of 88.93% and a 0.883 AUC (area under the ROC curve) value. The classifiers appending either the SS or OBV feature achieved total accuracies of 89.69% and 90.46%, respectively. The combination of all features produced the best performance with a 91.41% total accuracy and a 0.913 AUC value. The results indicate that the combination of all features is capable of capturing more information for discriminating DNA-binding data instances from non-binding ones. Four repeats of the nested cross-validation process were implemented. The mean of the ACC values is 91.54% with a 95% confidence interval (CI) between 91.38% and 91.70%. For a detailed presentation of the prediction of one representative protein–DNA complex can be found in the Supplementary Material.
Table 1.
The prediction performance of the RF model based on various features. The prediction system was evaluated by nested cross-validation and the threshold for sample classification is 1/6
| Features | ACC±SD(%) | SE±SD(%) | PR±SD(%) | SP±SD(%) | MCC |
|---|---|---|---|---|---|
| A | 88.93±0.19 | 76.29±1.40 | 63.39±0.79 | 91.46±0.22 | 0.633 |
| A+B | 89.69±0.41 | 76.58±1.94 | 66.51±1.28 | 92.31±0.35 | 0.652 |
| A+C | 90.46±0.44 | 75.64±0.65 | 69.63±2.36 | 93.41±0.59 | 0.668 |
| A+B+C | 91.41±0.49 | 76.57±1.69 | 73.16±2.78 | 94.38±0.76 | 0.70 |
A: PSSMs; B: SS; C: OBVs of amino acids.
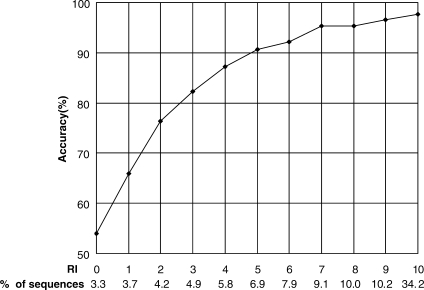
The prediction RI is an important measure in evaluating the quality of prediction. We adopt the RI to indicate the level of certainty in the prediction of a submitted sequence. Figure 1 presents the distribution of the expected prediction accuracy and the fraction of samples with a different RI using the RF approach with nested cross-validation. The higher the RI, the higher the reliability gained by the prediction. When RI≥5, prediction accuracy is ≥90%. About 78.39% of all samples have an RI≥5 and of these samples about 95.77% are correctly predicted by our method.
Fig. 1.
The expected prediction accuracy and the fraction of sequences with each RI by RF. For example, 34.2% of all samples have RI=10 and of these samples 97.63% are predicted correctly.
3.2 Performance comparison with other methods
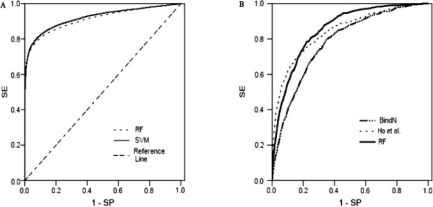
In the past few years, SVMs have successfully been applied to many fields, such as pattern recognition and data mining. Therefore, the SVM method was selected as an alternative algorithm with which to compare the RF classifier in this study. Both classifiers were applied to the same dataset DBP-374 with the same hybrid features and were assessed by nested cross-validation. For the SVM classifier, the model selection in the inner loop of the nested cross-validation procedure is performed by a standard grid parameter search. The ROC analysis shows that the SVM classifier (AUC 0.921) slightly outperforms the RF classifier (AUC 0.913) (Fig. 2A). Thus, we conclude that the hybrid feature contributes most to the excellent prediction performance. Because of time-saving and robust performance for different parameter values, the RF algorithm was chosen in this study.
Fig. 2.
Performance comparisons of ROC graphs with other methods. (A) Both classifiers were preformed on the same dataset DBP-374 with the same features ‘PSSMS+SS+OBVS’. RF: performance from the features (A+B+C) of Table 2; SVM: a SVM-based method evaluated by nested cross-validation. (B) all classifiers were tested on the same testing dataset TS75. The predictors have the following AUC value: BindN 0.782, Ho et al. 0.843 and our RF model 0.855.
BindN (http://bioinfo.ggc.org/bindn/) predicts potential DNA-binding residues with SVMs (Wang and Brown, 2006a). The SVM models used by the BindN web server were constructed with the PDNA-62 dataset (can be found in the Supplementary Material) whose data instances were encoded with three sequence features, including the side chain pKa value, hydrophobicity index and molecular mass of the amino acid. To ensure that model generation is completely independent of the test data, a test dataset TS75 with 75 proteins (can be found in the Supplementary Material) in protein–DNA complexes was randomly selected from the DBP-374 dataset without the proteins of the PDNA-62 dataset. The other 299 proteins were designated as the training dataset TR299 for constructing prediction models. Putative DNA-binding residues in the test dataset TS75 were predicted using BindN, and the default values were used for the optional parameters. Ho et al. (2007) proposed a hybrid method using a SVM in conjunction with evolutionary information of amino acid sequences in terms of their PSSMs for prediction of DNA-binding sites. The SVM models were evaluated by a 6-fold cross-validation and the best performance was achieved with a window size s = 7. We repeated the process of constructing prediction models, based on the training dataset TR299 and which follow the same strategy as Ho's method except for the standard grid parameter search in our process. Putative binding residues in the dataset TS75 were then predicted. We constructed RF models on the TR299 dataset using exactly the same scheme as the original RF models and applied the models to predict putative DNA-binding residues in the test dataset TS75 whose SS information was predicted by the PREDATOR program (Frishman and Argos, 1997). For all three classifiers in this paragraph, 3.5 Å was designated as the cutoff distance in the definition of a binding residue. The AUC were 0.782, 0.843 and 0.855 for the BindN, Ho's method and RF predictor, respectively (Fig. 2B), indicating that our RF model achieves the best performance.
DP-Bind (http://lcg.rit.albany.edu/dp-bind) (Hwang et al., 2007; Kuznetsov et al., 2006) is also a web server that predicts DNA-binding sites in a DNA-binding protein from its amino acid sequence. However, 4.5 Å is regarded as the cutoff distance in the definition of a binding residue in DP-Bind. The models constructed were also based on the PDNA-62 dataset, and predictions were performed using a profile of evolutionary conservation in terms of PSSMs of the input sequence automatically generated by the web server. The model provides three machine learning methods: SVM, KLR (kernel logistic regression) and PLR (penalized logistic regression). Putative DNA-binding residues were predicted by DP-Bind for the test dataset TS75, and the recommended encoding schemes were used and prediction results were received by E-mail (default). Here, we also trained other RF models on the TR299 dataset using exactly the same strategy as the original RF models, except that the training data instances were extracted according to the criterion of a cutoff distance of 4.5 Å in the definition of a binding residue. These models were also used to identify putative.
DNA-binding residues in the TS75 dataset whose SS information was predicted by the PREDATOR program (Frishman and Argos, 1997). As shown in Table 2, the total accuracy is 75.31%, 78.19% and 76.49% for the SVM, KLR, PLR predictors in DP-Bind, respectively, and 77.98% for the majority consensus prediction of the three predictors. The RF-based method attained 80.47% prediction accuracy with the AUC being 0.834. The results show that our RF model achieves the best performance. Performance comparisons with the DISIS web server (http://cubic.bioc.columbia.edu/services/disis) (Ofran et al., 2007) can be found in the Supplementary Material.
Table 2.
Performance comparisons with the methods in DP-Bind. All classifiers were tested on the same testing dataset TS75, and 4.5 Å was designated as the cutoff distance in the definition of a binding residue
| Classifiersa | ACC(%) | SE(%) | PR(%) | SP(%) | MCC |
|---|---|---|---|---|---|
| SVM | 75.31 | 68.40 | 23.05 | 76.04 | 0.290 |
| KLR | 78.19 | 67.22 | 25.45 | 79.34 | 0.315 |
| PLR | 76.49 | 64.06 | 23.23 | 77.79 | 0.279 |
| MAJ | 77.98 | 67.85 | 25.35 | 79.04 | 0.316 |
| RF | 80.47 | 67.16 | 27.69 | 81.84 | 0.341 |
aSVM, KLR and PLR are the SVM, KLR and PLR predictors in DP-Bind, respectively; MAJ is the majority consensus prediction of the three predictors.
3.3 Comparison of the prediction of DNA binding and non-binding proteins
The use of models for predicting binding residues is based on an inherent assumption that a protein is known to bind DNA. What about the performance of applying the RF model to predict non-binding proteins? For this purpose, we analyzed a set of 100 proteins that do not interact with DNA collected by Wang et al. (Wang and Brown, 2006a), and whose SS information was predicted by the PREDATOR program (Frishman and Argos, 1997). The classifier achieves 80.30% total accuracy which is similar to that of Wang et al. (Wang and Brown, 2006a). For direct comparison of the prediction of DNA binding and non-binding proteins by RF-based models, we examined fractions of the tree votes for the positive class (FVPs) of each data instance from the proteins in the dataset DBP-374 and the dataset NBP-350 including 350 non-binding proteins, namely, 100 xnon-binding proteins from Wang et al. (Wang and Brown, 2006) and a non-redundant set of 250 non-binding proteins obtained from Stawiski et al. (2003). The FVP values of the DBP-374 dataset are sourced from Section 3.1. We computed the average of the FVPs of predicted positive samples (FVPs ≥ 1/6) of each protein in both datasets. The mean of all 374 averages of the DBP-374 dataset is 0.4135 with a 95% CI from 0.3977 to 0.430. However, the mean of the 350 non-binding proteins of the dataset NBP-350 is 0.2765 with a 95% CI from 0.2723 to 0.2807. A two-independent sample t-test was also implemented for all averages of the DBP-374 and those of the dataset NBP-350. The P-value was <0.001, which indicated that the prediction between DNA binding and non-binding proteins was significantly different. The results suggest that the CI values of DNA binding and non-binding proteins would be helpful for predicting whether a given protein is a binder or non-binder.
3.4 Web server
DBindR is available at http://www.cbi.seu.edu.cn/DBindR/DBindR.htm. All the CGI scripts of models were written in Perl 5.8.4 and the interface was designed using HTML. On the DBindR web page, users can copy/paste amino acid sequences in FASTA format and choose the prediction method (either RF or SVM). The RF and SVM models used for predicting new proteins were constructed from all the data instances in the processed DBP-374 dataset. The RF algorithm is implemented by the randomForest (version 4.5-18) R package (Liaw and Wiener, 2002), and the SVM algorithm was by the e1071 (version 1.5-16) R package (Dimitriadou et al., 2007). An E-mail address is required to receive the results. The program slides a window with length β = 11 along the input sequence (into a segment of amino acid sequences). Each window segment constitutes a sample and each sample will be mapped into a 319-dimension feature space reflecting a hybrid feature by combining evolutionary information of the amino acid sequence, the OBVs of the amino acids and SS information of the amino acids. The web server returns the predicted DNA-binding residues and non-binding residues along the input sequence, and marks them ‘P’ and ‘N’. The prediction RI ranges from the lowest level 0 to the highest level 10 for presentation, and the higher the RI is, the higher reliability the prediction gains.
4 CONCLUSIONS
In this work, we proposed a new RF-based approach combining a hybrid feature for prediction of DNA-binding residues from amino acid sequence data. The results indicate that the RF-reliant models have a prediction accuracy of 91.41% with MCC set at 0.70. To the best of our knowledge, until now, the RF method combined with a hybrid feature has been the most effective method for predicting binding residues in proteins from protein sequences without using 3D structural information, and the prediction results are useful to realize how proteins interact with DNA molecules. The results demonstrate that the hybrid feature contributes most to the excellent prediction performance. The prediction RI would be helpful to biologists for guiding selection of residue candidates in experimental studies of site-directed mutagenesis for functional DNA-binding proteins. A web server, called DBindR, has been developed for efficient online predictions. In the next step, we will attempt to classify protein–DNA complexes by using the conventional motif-based classification and the structural and functional properties-based classification of DNA binding proteins, and then predict their DNA-binding sites in proteins and identify common properties and rules that govern protein–DNA recognition.
Funding: National Natural Science Foundation of China (Project No. 60671018 and No. 60121101). Funding to pay the Open Access publication charges for this article was provided by National Natural Science Foundation of China (60671018).
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Ahmad S, Sarai A. PSSM-based prediction of DNA binding sites in proteins. BMC Bioinformatics. 2005;6:6. doi: 10.1186/1471-2105-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad S, et al. Analysis and prediction of DNA-binding proteins and their binding residues based on composition, sequence and structural information. Bioinformatics. 2004;20:477–486. doi: 10.1093/bioinformatics/btg432. [DOI] [PubMed] [Google Scholar]
- Altschul SF, et al. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj N, Lu H. Residue-level prediction of DNA-binding sites and its application on DNA-binding protein predictions. FEBS Lett. 2007;581:1058–1066. doi: 10.1016/j.febslet.2007.01.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. Random forest. Mach. Learn. 2001;45:5–32. [Google Scholar]
- Bullock AN, Fersht AR. Rescuing the function of mutant p53. Nat. Rev. Cancer. 2001;1:68–76. doi: 10.1038/35094077. [DOI] [PubMed] [Google Scholar]
- Cohen G, et al. Learning from imbalanced data in surveillance of nosocomial infection. Artif. Intell. Med. 2006;37:7–18. doi: 10.1016/j.artmed.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Coulocheri SA, et al. Hydrogen bonds in protein-DNA complexes: where geometry meets plasticity. Biochimie. 2007;89:1291–1303. doi: 10.1016/j.biochi.2007.07.020. [DOI] [PubMed] [Google Scholar]
- Dimitriadou E, et al. e1071: Misc Functions of the Department of Statistics (e1071), TU Wien. R package, Version 1.5-16. 2007. Available at http://cran.r-project.org/ [Google Scholar]
- Egan JP. Signal Detection Theory and ROC-Analysis. New York: Academic Press; 1975. [Google Scholar]
- Frishman D, Argos P. Seventy-five percent accuracy in protein secondary structure prediction. Proteins. 1997;27:329–335. doi: 10.1002/(sici)1097-0134(199703)27:3<329::aid-prot1>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Ho SY, et al. Design of accurate predictors for DNA-binding sites in proteins using hybrid SVM-PSSM method. Biosystems. 2007;90:234–241. doi: 10.1016/j.biosystems.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Hwang S, et al. DP-Bind: a web server for sequence-based prediction of DNA-binding residues in DNA-binding proteins. Bioinformatics. 2007;23:634–636. doi: 10.1093/bioinformatics/btl672. [DOI] [PubMed] [Google Scholar]
- Jones S, et al. Using electrostatic potentials to predict DNA-binding sites on DNA-binding proteins. Nucleic Acids Res. 2003;31:7189–7198. doi: 10.1093/nar/gkg922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubat M, Matwin S. Addressing the curse of imbalanced training sets: one-sided selection. In: Fisher D, editor. Machine Learning: Proceedings of the Fourteenth International Conference (ICML'97). San Francisco, CA: Morgan Kaufmann Publishers; 1997. pp. 179–186. [Google Scholar]
- Kuznetsov IB, et al. Using evolutionary and structural information to predict DNA-binding sites on DNA-binding proteins. Proteins. 2006;64:19–27. doi: 10.1002/prot.20977. [DOI] [PubMed] [Google Scholar]
- Liaw A, Wiener M. Classification and Regression by randomForest. R News. 2002;2:18–22. [Google Scholar]
- Luscombe NM, et al. An overview of the structures of protein-DNA complexes. Genome Biol. 2000;1:REVIEWS001. doi: 10.1186/gb-2000-1-1-reviews001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BW. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta. 1975;405:442–451. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- Ofran Y, et al. Prediction of DNA-binding residues from sequence. Bioinformatics. 2007;23:i347–i353. doi: 10.1093/bioinformatics/btm174. [DOI] [PubMed] [Google Scholar]
- Scheffer T. School of Computer Science. Berlin: Technischen University; 1999. Error Estimation and Model Selection. [Google Scholar]
- Shen J, et al. Predicting protein-protein interactions based only on sequences information. Proc. Natl Acad. Sci. USA. 2007;104:4337–4341. doi: 10.1073/pnas.0607879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggers TW, Honig B. Structure-based prediction of C2H2 zinc-finger binding specificity: sensitivity to docking geometry. Nucleic Acids Res. 2007;35:1085–1097. doi: 10.1093/nar/gkl1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stawiski EW, et al. Annotating nucleic acid-binding function based on protein structure. J. Mol. Biol. 2003;326:1065–1079. doi: 10.1016/s0022-2836(03)00031-7. [DOI] [PubMed] [Google Scholar]
- Tjong H, Zhou HX. DISPLAR: an accurate method for predicting DNA-binding sites on protein surfaces. Nucleic Acids Res. 2007;35:1465–1477. doi: 10.1093/nar/gkm008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya Y, et al. Structure-based prediction of DNA-binding sites on proteins using the empirical preference of electrostatic potential and the shape of molecular surfaces. Proteins. 2004;55:885–894. doi: 10.1002/prot.20111. [DOI] [PubMed] [Google Scholar]
- Vapnik VN. Statistical Learning Theory. New York: Wiley; 1998. [Google Scholar]
- Wang L, Brown SJ. BindN: a web-based tool for efficient prediction of DNA and RNA binding sites in amino acid sequences. Nucleic Acids Res. 2006a;34:W243–W248. doi: 10.1093/nar/gkl298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Brown SJ. Prediction of DNA-binding residues from sequence features. J. Bioinform. Comput. Biol. 2006b;4:1141–1158. doi: 10.1142/s0219720006002387. [DOI] [PubMed] [Google Scholar]
- Wang Y, et al. Better prediction of the location of alpha-turns in proteins with support vector machine. Proteins. 2006;65:49–54. doi: 10.1002/prot.21062. [DOI] [PubMed] [Google Scholar]
- Yan C, et al. Predicting DNA-binding sites of proteins from amino acid sequence. BMC Bioinformatics. 2006;7:262. doi: 10.1186/1471-2105-7-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.